Introduction
Colorectal carcinoma (CRC) is the third most
frequently diagnosed malignancy and the third leading cause of
mortality among cancer patients in the USA (1). Despite current therapeutic strategies
combing adjuvant chemotherapy, surgery, radiotherapy and sometimes
immunotherapy, the prognosis of CRC remains poor. The main reason
for the current situation is that most CRC patients have distant
metastasis at diagnosis or develop recurrent metastatic CRC
following surgical treatment. Furthermore, lack of an accurate
prognosis biomarker makes it difficult to predict the patients
survival time after surgery. Although recent developments in
molecular biology have provided insight into the molecular
mechanisms of CRC, the fundamental molecular mechanisms underlying
progression and metastasis in CRC have not been fully elucidated.
Thus, identification of CRC metastasis-associated molecules may be
beneficial for a further prediction of clinical outcomes and may
ultimately be used to identify subsets of patients that may benefit
from specific targeted therapies.
MicroRNAs (miRNAs) are small non-coding RNAs (~22
nucleotides in length), transcribed from non-protein coding genes
or introns, which regulate gene expression through repressing
translation and cleaving their mRNAs by binding to complementary
sites in their 3′-untranslated region (3′-UTR) (2). Currently, several aberrant expressed
miRNAs have proven to be associated with carcinogenesis, tumor
progression and metastasis in CRC and are taken as prognostic
indicators for CRC including miR-150 (3), miR-28-5p/-3p (4), miR-200 (5), miR-494 (6) and many others. miR-326 was found
downregulated and functioned as a tumor suppressor in multiple
types of solid tumor and hematologic neoplasms, such as glioma
(7,8), medulloblastoma (9), cholangiocarcinoma (10) and chronic lymphocytic leukemia
(11). Yet, the clarification of
the function and clinical significance of miR-326 in CRC are still
at an early stage.
The yeast nin one binding protein (NOB1) is an
evolutionarily conserved protein (12) and is required for the biogenesis and
function of 26s proteasome (13).
It has been observed that genetic depletion of NOB1 suppresses the
process of 20S pre-rRNA to mature 18S rRNA, producing markedly high
levels of the 20S pre-RNA with novel degradation intermediates
(14). These studies showed that
the NOB1 played a crucial role in protease function and RNA
metabolism. NOB1 expression was found upregulated and it functioned
as an oncogenic molecule in various types of solid tumors, such as
breast (15), ovarian (16) and non-small cell lung cancer
(17), hepatocellular carcinoma
(18) and glioma (7). Importantly, NOB1 was also found
upregulated in CRC tissues (19).
Yet, up to date, the physiological and pathological function of
NOB1 in CRC remains unclear and its relationship with miR-326 in
CRC has not been examined.
The aim of the present study was to investigate the
relationship between miR-326 and NOB1 in CRC, and, furthermore, to
identify the effect of miR-326 and NOB1 on cell proliferation,
migration and invasion of the CRC cell lines. We also discuss the
clinical significance of miR-326 and NOB1 in CRC. We found that
miR-326 inhibited cell proliferation, migration and cell invasion,
and induced cell apoptosis and cell cycle arrest of CRC cells by
directly targeting NOB1. Importantly, we found that the CRC
patients with high expression of miR-326 or low expression of NOB1
tend to obtain better prognosis.
Materials and methods
Patients and tissue samples
The present study was approved by the Research
Ethics Committee of Xi’an Jiaotong University. A written informed
consent was obtained from all the participating patients. All the
specimens were handled and made anonymous according to the ethical
and legal standards. A total of 114 patients were enrolled in the
present study. Patients received curative resection for CRC at the
First Affiliated Hospital, Xi’an Jiaotong University (Xi’an,
China). None of the patients that enrolled in the present study
received blood transfusion or chemotherapy prior to surgery. The
follow-up information of all the participants was updated every 3
months by telephone. The overall survival was defined as the time
elapsed from surgery to death. Information regarding the death of
the patients was ascertained by their family.
Quantitative reverse transcriptase PCR
(qRT-PCR) assay
The expression of miR-326 in CRC and the
corresponding adjacent tissues were detected by qRT-PCR assay.
Briefly, the total RNA was extracted from the tissues using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer protocol. Then, miRNA expression levels were
quantitated using a TaqMan miRNA Real-Time RT-PCR kit (Applied
Biosystems) according to the manufacturer protocol. Data were
analyzed with 7500 software v.2.0.1 (Applied Biosystems), with the
automatic Ct setting for adapting baseline and threshold for Ct
determination. The universal small nuclear RNA U6 (RNU6B) was used
as an endogenous control for miRNAs. Each sample was examined in
triplicate and the amounts of PCR products produced were
non-neoplastic to RNU6B.
MTT assays
Cell proliferation was analyzed in vitro with
the tetrazolium salt
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
reagent. Briefly, 2,000 cells from each group were plated in each
well of five 96-well plates in 200 μl of medium. To analyze
cell proliferation, 20 μl of MTT substrate at a
concentration of 2.5 mg/ml in phosphate-buffered saline (PBS) was
added to each well. The plates were then returned to a standard
tissue incubator for an additional 4 h. The medium was then removed
and the cells were solubilized in 150 μl of dimethyl
sulfoxide for the colorimetric analysis (wavelength, 490 nm). One
plate was analyzed immediately after the cells adhered (~4 h after
plating). Then, one plate per day was examined for the next 4
consecutive days.
Cell apoptosis and cell cycle assays
CRC cells were transfected with NOB1 siRNA, miR-326
and their control for 48 h, and the cells were then suspended in an
incubation buffer at a density of 1×106 cells/ml. The
cells were incubated with Annexin V-FITC and propidium iodide (PI)
(BD Biosciences, San Jose, CA, USA) for 15 min in the dark at room
temperature. Cell apoptosis was then analyzed using FACSCalibur (BD
Biosciences, San Diego, CA, USA). For cell cycle distribution, the
cells of each group were stained with PI and analyzed by flow
cytometry using FACSCalibur. For each group, 10,000 events were
acquired. The percentage of the cells in G1, S and G2 phases of the
cell cycle was calculated.
Cell migration and invasion assays
Cell migration and invasion capacity were measured
using Transwell migration assays (Millipore, Billerica, MA, USA)
in vitro. The CRC cells were transfected with miR-326 and
miR-control for 48 h, and then the cells were suspended in
RPMI-1640 with 10 g/l BSA at a density of 1×106
cells/ml. Then, the cell suspensions (150 μl) were seeded in
the upper chamber with a porous membrane coated with (for the
Transwell invasion assay) or without (for the migration assay)
Matrigel (BD Biosciences). To attract the cells, 500 μl of
RPMI-1640 with 10% serum was added to the lower chamber. After
allowing the cells to migrate for 24 h or to invade for 48 h, the
penetrated cells on the filters were fixed in dried methanol and
stained with 4 g/l crystal violet. The numbers of the migrated or
invasive cells were determined from five random fields using a
microscope (Olympus) at a magnification of ×10.
Luciferase reporter assay
CRC cells were seeded in 96-well plates at 60%
confluence. After 24 h, the cells were transfected with 120 ng of
WT or MT 3′-UTR of PTEN mRNA expression vector pgL3-target-3′UTR
and 30 ng of miR-326 mimics using Lipofectamine 2000. The cells
were collected 48 h after the transfection and the luciferase
activity was measured using a Dual-Luciferase Reporter Assay System
according to the manufacturer protocol (Promega).
Xenograft model in nude mice
For the tumorigenesis assays, we engineered HCT116
cells to stably express high-miR-326 or low-NOB1. Xenograft tumors
were generated by subcutaneous injection of HCT116 cells
(2×106) into the hind limbs of 4–6 weeks old BALB/c
athymic nude mice (nu/nu; Animal Center of Xi’an Jiaotong
University, Xi’an, China; n=5 for each group). All the mice were
housed and maintained under specific pathogen-free conditions and
all the experiments were approved by the Animal Care and Use
Committee of Xi’an Jiaotong University and were performed in
accordance with the institutional guidelines. The tumor size was
measured using a slide caliper and the tumor volume was determined
by the formula: 0.44 × A × B2, where A represents the diameter of
the base of the tumor and B represents the corresponding
perpendicular value.
Statistical analysis
Statistical analysis was performed using IBM SPSS
statistical software (version 21.0). Survival curves were estimated
using the Kaplan-Meier method and distributions were evaluated by
the long-rank test. Cox proportional hazard models of the factors
related to survival were used to calculate RRs and identify the
factors that affect survival. The differences in the
characteristics between the 2 groups were examined by the
χ2 test. All the P-values were determined from the
two-sided tests and statistical significance was based on a P-value
of <0.05.
Results
NOB1 is the direct target of miR-326 in
CRC cells
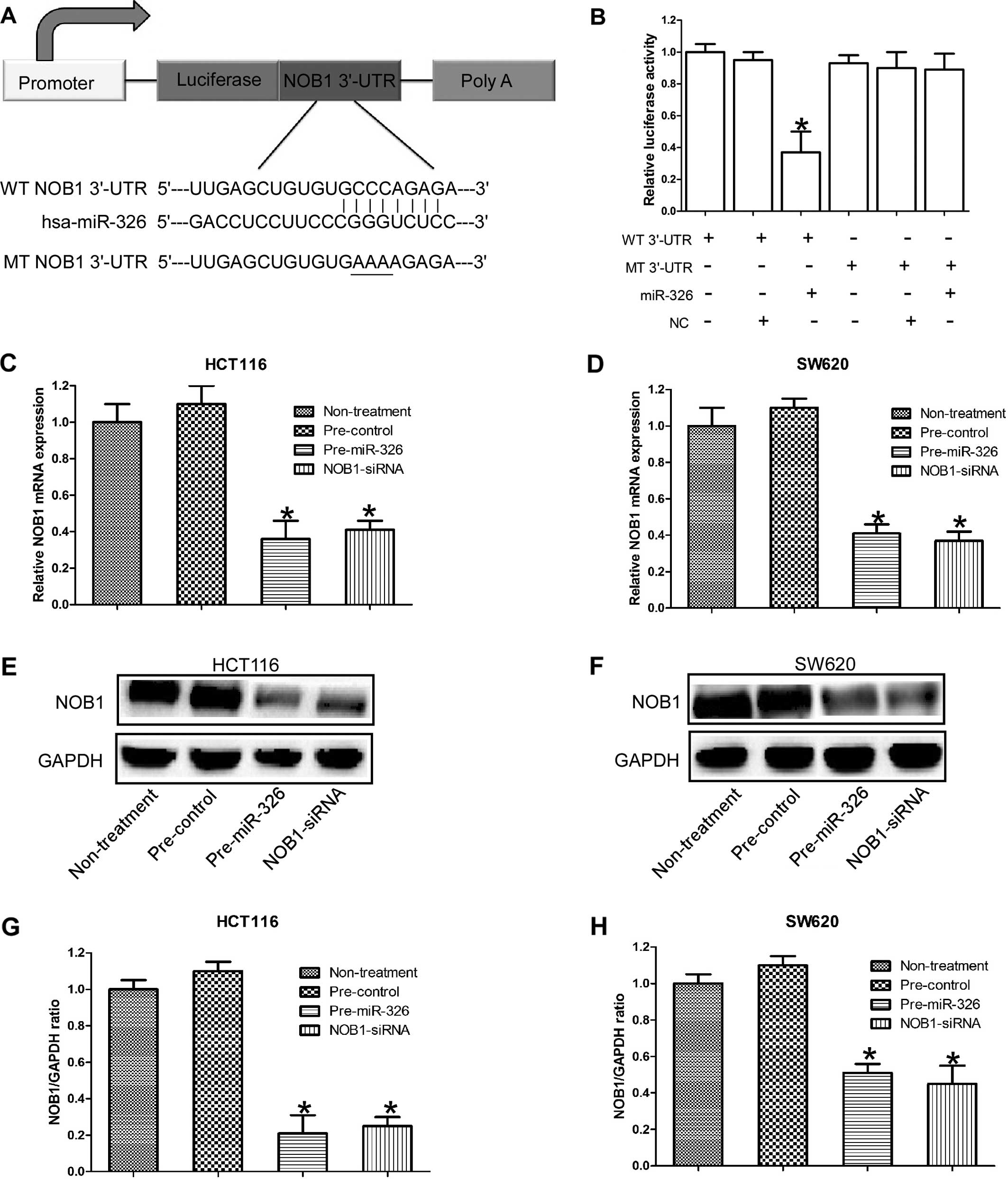
It has been proven that NOB1 was the direct target
of miR-326 in glioma. Considering the developmental stage- and
tissue-specific manner of miRNAs, we decided to investigate the
relationship between NOB1 and miR-326 in CRC. Using TargetScan,
miRanda and PicTar software, NOB1 was identified as a likely target
of miR-326, since it contains a putative miR-326 target site in the
3′-UTR. Then, we performed a luciferase reporter assay to further
verify whether miR-326 directly targeted the 3′-UTR of NOB1 in CRC.
The target sequence of wild-type PTEN 3′-UTR (WT 3′-UTR) or mutant
PTEN 3′-UTR (MT 3′-UTR) was cloned into a luciferase reporter
vector (Fig. 1A). Pre-hsa-miR-326
or non-functional control miR-NC were co-transfected with the
reporter vectors into the HEK 293T cells. The miR-326 target
sequences and the wild-type NOB1 3′-UTR reduced the relative
luciferase activity only when miR-326 was present, but not when the
corresponding mutant was introduced with the miR-326 (Fig. 1B). The luciferase reporter data
indicated that NOB1 is a specific target of miR-326.
In order to further confirm that NOB1 is the target
gene for miR-326 in CRC, qRT-PCR and western blotting were used to
detect the expression of NOB1 in CRC cell lines HCT116 and SW620.
As expected, the expression of NOB1 at both mRNA and protein level
was significantly downregulated in CRC cells transfected with
pre-miR-326, the intervention effects of which were similar to NOB1
RNA interference (Fig. 1C–H).
Collectively, these results suggest that NOB1 is the direct target
gene of miR-326 in CRC cells.
miR-326 inhibits the proliferation of CRC
cells by directly targeting NOB1
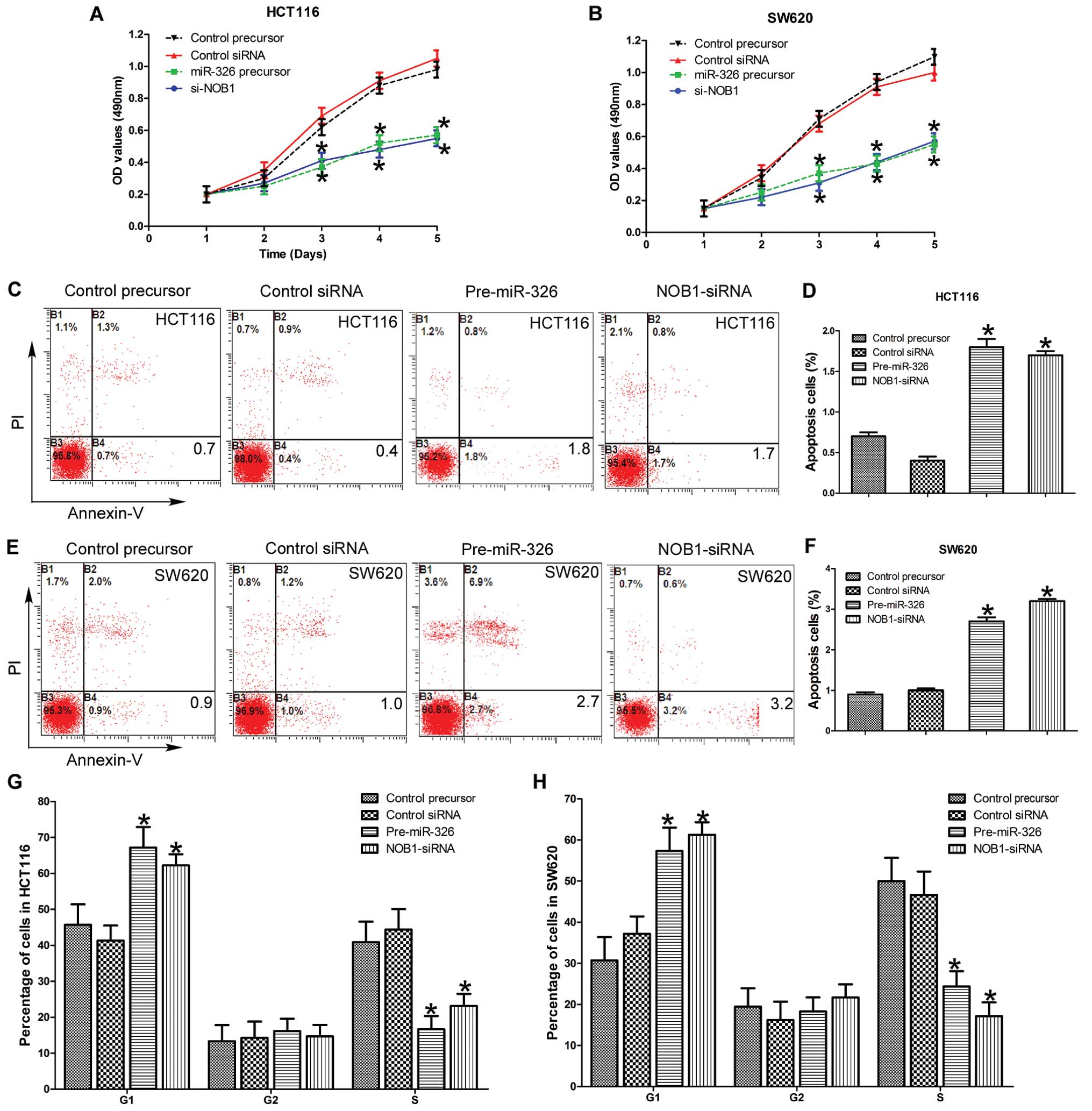
To analyze the effects of miR-326 on CRC cell
proliferation, we transfected miR-326, miR-NC, NOB1- or
control-siRNA into HCT116 and SW620 cells. As shown in Fig. 2A and B, ectopic expression of
miR-326 inhibited the growth of HCT116 and SW620 cell lines
compared to the negative control 5 days after transfection
(P<0.05), and no statistically significant differences in the
growth rate between the miR-326 overexpressing and the
NOB1-siRNA-infected cells was observed. There were no significant
differences in the cell growth detected between the miR-NC and the
control siRNA. Overall, these data suggest that miR-326 and NOB1
play an important role in regulating the proliferation of CRC
cells.
miR-326 induces cell apoptosis and cell
cycle arrest in CRC cell lines by directly targeting NOB1
We then examined the effect of miR-326 on apoptosis,
HCT116 and SW620 cells were stained with Annexin V and PI after
transfection with miR-326, miR-NC, NOB1 siRNA or control siRNA for
72 h. As shown in Fig. 2C–F, the
early apoptosis rate in miR-326 and NOB1 siRNA groups was
significantly higher than miR-NC and control siRNA groups
(P<0.05). We found that miR-326 induced early apoptosis in CRC
cells by targeting NOB1.
To evaluate the effect of miR-326 on the cell cycle
of the CRC cells, we examined the cell cycle distribution by FACS
after transfection. Compared with miR- and siRNA-control cells,
miR-326 overexpressing and NOB1 silencing cells showed a
substantial decrease in the proportion of the cells in the S phase
and an increase in the number of the cells in the G1 phase in the
HCT116 and SW620 cell lines (Fig.
2G and H). Overall, these results indicate the miR-326 inhibits
the proliferation and promotes cell apoptosis of the CRC cells by
inducing cell cycle arrest.
miR-326 inhibits the migration and
invasion of CRC cells by directly targeting NOB1
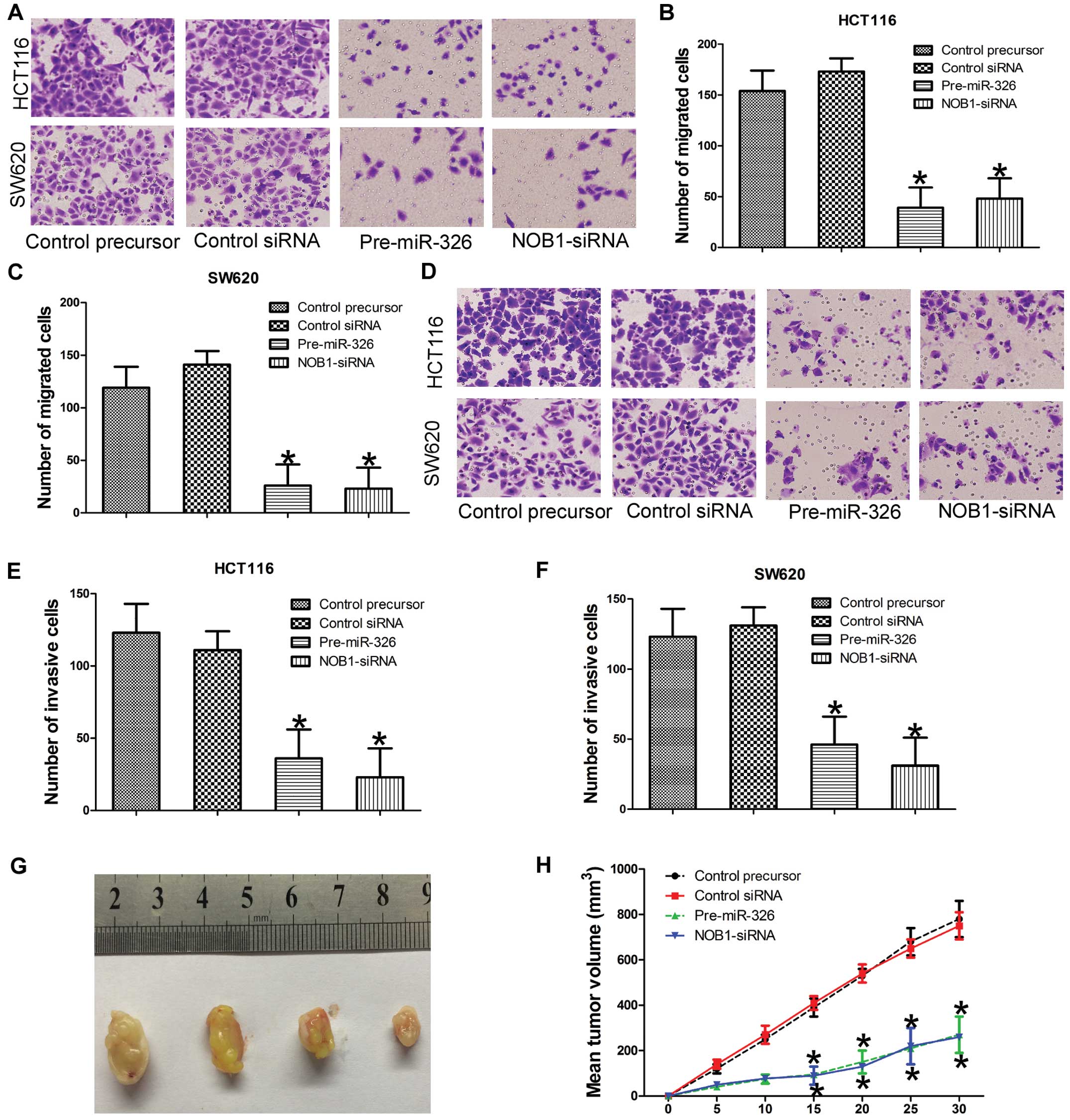
We then determined whether miR-326 was associated
with cell migration and cell invasion of the CRC cells. To test the
effect of miR-326 on cell invasion and migration, we used standard
Matrigel-coated or uncoated Transwell chamber assays. We found,
compared with miR- and siRNA-control cells, that the migration and
invasion ability was significantly reduced in HCT116 and SW620
cells that were transfected with miR-326 or NOB1 siRNA (Fig. 3A–F). Collectively, these data
suggest that silencing of NOB1 mimics the phenotype induced by
overexpression of miR-326 in CRC cells, which further indicates
that NOB1 serves as a downstream functional target of miR-326.
miR-326 inhibits tumor formation and
tumor growth of CRC cells in vivo
In order to further verify the effect of miR-326 on
tumor formation and tumor growth of CRC cells in vivo, we
subcutaneously injected the genetically modified (overexpressed
miR-326 or NOB1 siRNA) HCT116 cells, as well as their control
cells, into the hind limbs of nude mice. Tumor sizes were measured
every 5 days; after 30 days, the mice were sacrificed and the
tumors were collected. Analysis of the tumor growth curves
confirmed that the tumors in the miR-326 or NOB1 siRNA group grew
significantly more slowly than those in the miR-NC and siRNA
control group (Fig. 3G and H).
Collectively, these results further indicate that miR-326
significantly inhibited the tumor formation and tumor growth in
human CRC cells, suggesting that miR-326 played a vital function in
the tumorigenicity of CRC cells in vitro and in
vivo.
miR-326 is downregulated and NOB1 is
upregulated in CRC tissues
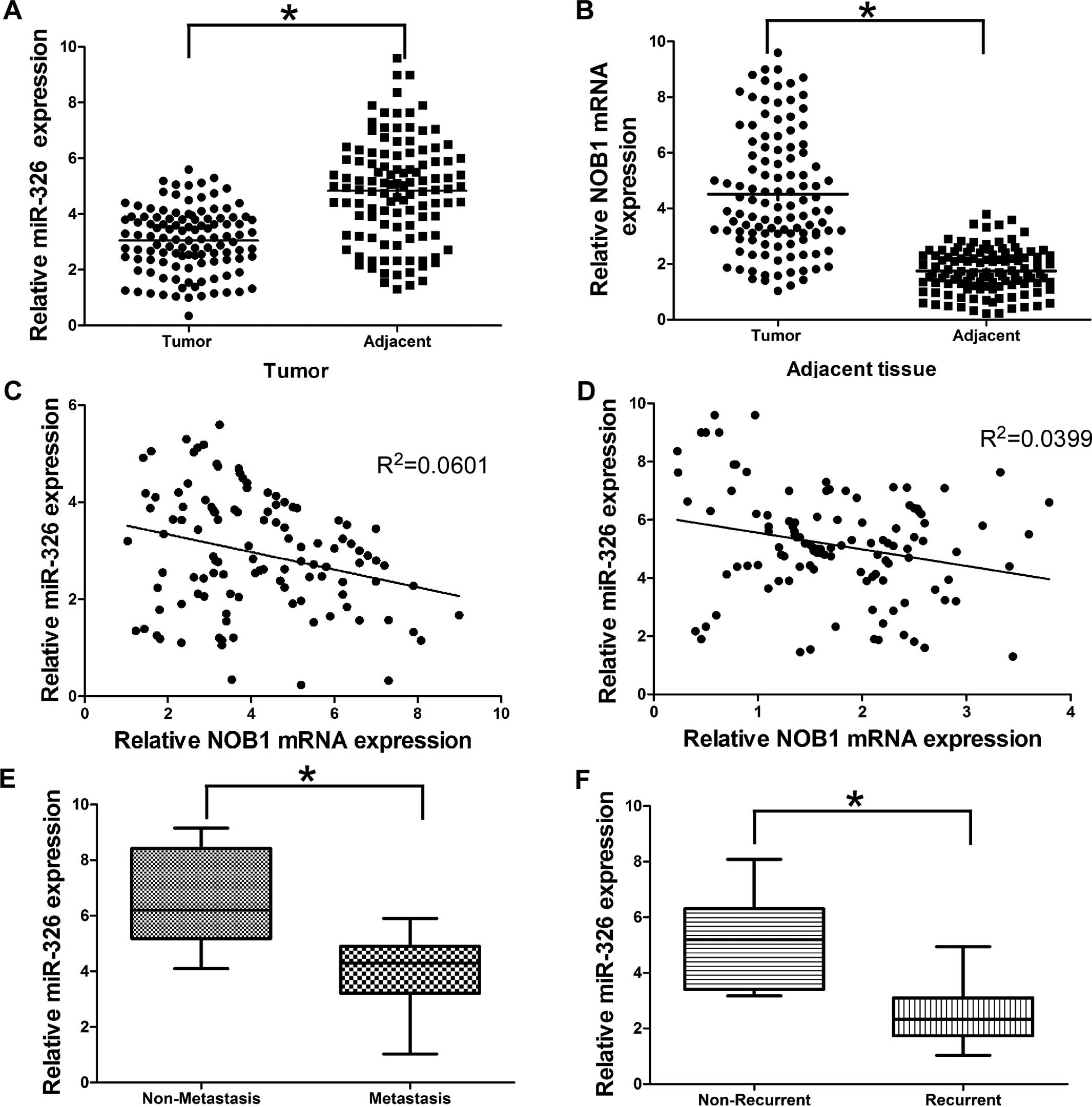
To further evaluate the clinical significance of
miR-326 expression in CRC, we conducted qRT-PCR to measure the
miR-326 expression in tumor tissues and their corresponding
adjacent tissues of CRC patients. We found that miR-326 expression
was significantly downregulated in 114 CRC tumor tissues compared
with their corresponding adjacent tissues (Fig. 4A). In contrast, NOB1 expression was
significantly upregulated in CRC tumor tissues compared with their
corresponding adjacent tissues (Fig.
4B), which was consistent with previous literature. More
importantly, statistically significant inverse correlations were
revealed by Spearman’s correlation analysis between mRNA levels of
miR-326 and NOB1 in CRC tumor and adjacent specimens (Fig. 4C and D; r=−0.2452; P=0.0086 and
r=-0.1998; P=0.0331). Collectively, these results suggest that
miR-326 plays a tumor suppressor role and NOB1 has an oncogenic
role in CRC.
miR-326 is associated with tumor
metastasis and recurrence in CRC patients
We then determined whether miR-326 was associated
with tumor metastasis and recurrence. Tumor specimens were divided
into two groups according to their metastatic or recurrent status.
Our data showed that miR-326 expression decreased in the metastatic
group (Fig. 4E). Moreover, miR-326
levels were also significantly decreased in the specimens obtained
from the patients who suffered CRC recurrence (Fig. 4F). Collectively, our findings showed
that downregulation of miR-326 may increase the risk of metastasis
and recurrence in CRC patients.
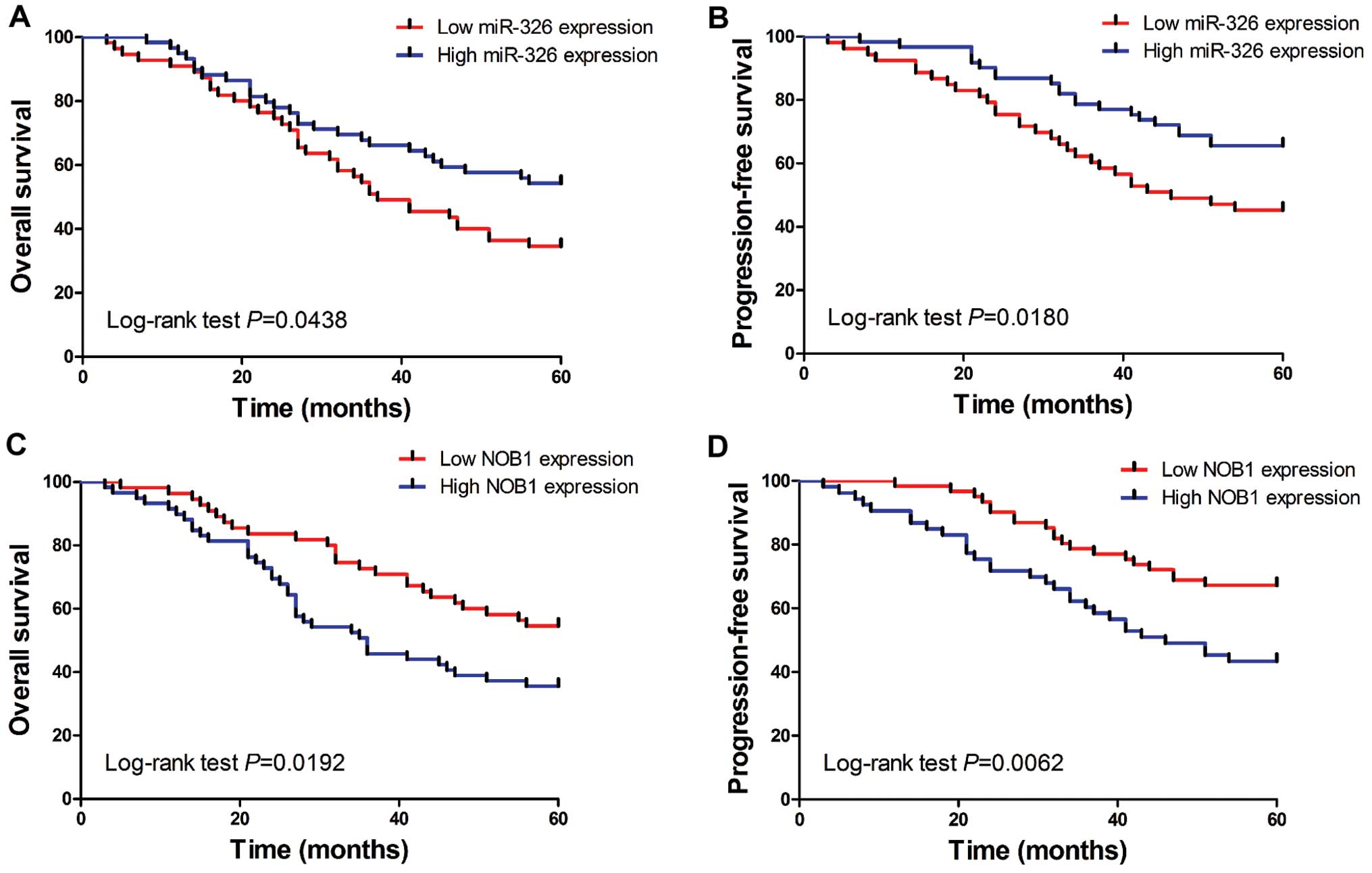
miR-326 is associated with survival and
prognosis in CRC patients
In order to further determine the clinical
significance of miR-326 in CRC, we split the 114 patients into two
groups based on miR-326 or NOB1 expression levels (low vs. high)
with their median expression levels as a cut-off point. The
Kaplan-Meier analysis revealed that low miR-326 expression was
significantly correlated with reduced overall and progression-free
survival in 114 CRC patients (Fig.
5A and B; log-rank test, P=0.0438 and P=0.0180, respectively).
Whereas, the patients with low NOB1 expression tended to obtain
better overall and progression-free survival time, than patients
with high NOB1 expression (Fig. 5C
and D; log-rank test, P=0.0192 and P=0.0062, respectively).
Next, the univariate analysis demonstrated that the
overall and progression-free survival of CRC patients was
associated with tumor-node-metastasis (TNM) stage, NOB1 and miR-326
expression (Tables I and II). To test whether the prognostic value
of miR-326 and NOB1 was independent of other clinicopathological
parameters for poor overall and progression-free survival in CRC
patients, a multivariate analysis was performed using a Cox
proportional hazard model. Multivariate analysis including miR-326
expression, NOB1 expression, age, gender, tumor location, TNM stage
and differentiation status, demonstrated that miR-494 or NOB1
expression was an independent prognostic biomarker for poor overall
and progression-free survival in CRC patients (Tables I and II). These results suggest that patients
with high-miR-326 expression tend to obtain a better prognosis than
patients with low-miR-326 expression, yet the patients with high
NOB1 expression tend to obtain a worse prognosis than the patients
with low-NOB1 expression. Statistically significant results were
also obtained for the TNM stage and lymph node metastasis, where
all the other parameters were not independent prognostic biomarkers
for overall and progression-free survival in CRC patients.
Collectively, these results indicate that miR-326 or its target
NOB1 is an independent biomarker for survival and prognosis of CRC
patients.
 | Table ICox regression analysis of prognostic
factors for overall survival in CRC patients (n=114). |
Table I
Cox regression analysis of prognostic
factors for overall survival in CRC patients (n=114).
| Univariate
| Multivariate
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-326
expression | 0.679 | 0.329–0.875 | 0.018a | 0.582 | 0.282–0.798 | 0.011a |
| NOB1
expression | 3.271 | 1.478–4.182 | 0.031a | 2.894 | 1.391–3.829 | 0.037a |
| Age (years) | 1.723 | 0.674–2.147 | 0.421 | 2.007 | 0.771–2.317 | 0.328 |
| Gender | 1.682 | 0.819–2.781 | 0.391 | 1.713 | 0.671–2.178 | 0.482 |
| Tumor location | 2.398 | 0.519–3.298 | 0.127 | 2.128 | 0.661–2.891 | 0.228 |
| TNM stage | 3.751 | 1.874–5.118 | 0.008a | 4.217 | 2.114–5.981 | 0.011a |
|
Differentiation | 2.511 | 0.754–3.187 | 0.073 | 1.927 | 0.678–3.722 | 0.067 |
 | Table IICox regression analysis of prognostic
factors progression-free survival in CRC patients (n=114). |
Table II
Cox regression analysis of prognostic
factors progression-free survival in CRC patients (n=114).
| Univariate
| Multivariate
|
|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| miR-326
expression | 0.718 | 0.327–0.911 | 0.021a | 0.811 | 0.418–0.951 | 0.017a |
| NOB1
expression | 3.518 | 1.587–4.291 | 0.027a | 3.891 | 1.981–5.155 | 0.032a |
| Age (years) | 1.784 | 0.781–2.871 | 0.325 | 2.214 | 0.881–2.959 | 0.411 |
| Gender | 2.187 | 0.891–3.128 | 0.237 | 1.895 | 0.711–2.623 | 0.229 |
| Tumor location | 1.671 | 0.687–2.887 | 0.229 | 2.181 | 0.667–3.193 | 0.14 |
| TNM stage | 4.789 | 1.871–5.918 | 0.014a | 3.726 | 1.771–4.918 | 0.007a |
|
Differentiation | 2.198 | 0.518–3.115 | 0.074 | 2.481 | 0.784–3.214 | 0.051 |
Discussion
MicroRNAs (miRNAs) are unique in their ability to
regulate many protein-coding genes. Several miRNA have been
identified as candidate components of oncogene and tumor suppressor
networks in CRC, these miRNA and their targets play an important
role in the occurrence and development of CRC. The changes in miRNA
profiling are associated with almost all the aspects of CRC cell
biology, such as cell proliferation (20), angiogenesis (21), apoptosis (22), cell cycle (23), migration and invasion (6). In the present study, we focused on the
role of miR-326 in CRC, which has been proven to be a tumor
suppressor miRNA in multiple types of cancer. We found that miR-326
inhibited cell proliferation, migration and invasion of CRC cells,
yet it induced cell apoptosis and cell cycle arrested of CRC cells.
These results indicate that miR-326 also functions as a tumor
suppressor in CRC. Furthermore, the upregulation of miR-326 in the
CRC cells was revealed to be associated with a feedback loop
involving downregulation NOB1 which mimics the phenotype induced by
miR-326. We identified miR-326 downregulation or NOB1 upregulation,
which was usually associated with poor survival and prognosis of
CRC patients.
A previous study confirmed that NOB1, an essential
protein, was associated with 26S proteasome and hence played an
essential role in the universal biological process. In the present
study, we showed that miR-326 inhibited carcinogenesis and
progression by blocking a novel target, NOB1, both in vitro
and in vivo. When the cell cycle of CRC cells was assessed
by FACS, we observed that upregulated miR-326 expression or
downregulated NOB1 expression induced a significant decrease in the
S phase and an increase in the G1 phase population in the CRC cells
with significant cell proliferation arrest. The growth inhibitory
effect was further confirmed by MTT assays and nude mouse xenograft
assays, indicating that miR-326 and NOB1 are crucial for
proliferation and carcinogenesis of human CRC. In addition,
upregulated miR-326 expression or downregulated NOB1 expression
inhibited cell migration and invasion of CRC cells. Metastasis is
the major reason for poor prognosis of CRC patients. Thus, our
findings indicated that miR-326 and/or NOB1 may be a potential
therapeutic target for CRC. Thus, we investigated the clinical
significance of miR-326 and NOB1. As a previous study confirmed
NOB1 expression was upregulated in tissues of CRC and promoted
tumor growth and carcinogenesis of CRC (24,25),
thus we focused on whether miR-326 and/or NOB1 could be the
prognostic indicators for CRC. We found that miR-326 downregulation
or NOB1 upregulation was usually associated with poor survival.
Univariate and multivariate results showed that a high miR-326
expression or a low NOB1 expression was associated with poor
prognosis of CRC patients. Each was an independent prognostic
biomarker for CRC.
Since the identification of NOB1 protein, increased
NOB1 expression has been reported in ovarian (16), colorectal (24), breast (15), thyroid (26) and hepatocellular carcinoma (18). As the genetic depletion of NOB1
strongly suppressed the processing of 20S pre-rRNA to mature 18S
rRNA, producing markedly high levels of 20S pre-RNA with novel
degradation intermediates (14) the
effect corrects the RNA synthesis and affects the cell cycle
regulation (27), since it is
necessary for synthesis of rRNA for cell cycle process (28,29).
Moreover, NOB1 also plays an important role in proteasomes by
forming a complex between 19S regulatory particle of 26S
proteasome, where the latter catalyzes the protein degradation
through the ubiquitin proteasome pathway for cell cycle progression
(30,31). All these studies indicate that NOB1
expression has an essential role in cell cycle regulation. Our
study supported the pivotal role of NOB1 in cell cycle regulation
of CRC cells. Our results also indicated that NOB1 affected cell
proliferation and tumor growth of CRC in vivo and in
vitro by regulating the cell cycle.
Recently, several profiling analyses reported that
NOB1 was involved in carcinogenesis and tumor metastasis. Notably,
NOB1 was found to play a critical role in tumor metastasis of clear
cell renal cell carcinoma (32).
Consistent with a previous study, we also found that NOB1 was
associated with cell migration and invasion of the CRC cell. Yet,
the molecular mechanisms underlying this phenomenon are still
largely unknown and need further specific investigation. Several
miRNAs have also been proven to be associated with carcinogenesis,
progression and metastasis of CRC. Recently, miR-494 was found to
be associated with tumor metastasis and tumor aggressiveness of CRC
(6). Moreover, miR-139-5p
suppressed CRC cell invasion and metastasis by targeting AMFR and
NOTCH1 (33). Our study also
indicates that miR-326 plays an important role in cell metastasis
by directly targeting NOB1. We identified that miR-326 and NOB1
were the metastasis-associated molecules and may be considered as
potential therapeutic targets for CRC. These results shed new light
on the understanding of the molecular mechanism underlying the
metastasis of CRC.
Though miR-326 was found downregulated and as it
functions as a tumor suppressor in multiple types of solid tumor
and hematologic neoplasms, the biological effects and clinical
significance of miR-326 in CRC are still not fully understood.
Consistent with the above studies, our results identified that
miR-326 functioned as a tumor suppressor in CRC. miR-326 inhibited
cell proliferation, cell migration and cell invasion of CRC cells.
Importantly, miR-326 was also an independent prognostic biomarker
for CRC patients. However, a recent report exists with
contradictory conclusion (34). In
the present study, plasma miR-326 was found upregulated in CRC, and
high expression of plasma miR-326 was associated with a decreased
overall and progression-free survival, indicating miR-326 may play
an oncogenic role in CRC. The discrepancies between the studies may
reflect the different cohorts selected for each study. We focused
on the role that miR-326 played in CRC cells and the tissues of the
CRC patients. Yet other authors put their emphasis on whether
plasma miRNAs before treatment may serve as non-invasive markers
predicting the outcome in the metastatic CRC patients treated with
5-FU and oxaliplatin-based chemotherapy. Circulating expression of
miRNAs has been described in many solid cancers including CRC, but
it is still difficult to differentiate whether circulating miRNAs
expression is specifically associated with CRC itself or if this is
a common phenomenon that manifests during progression of any cancer
as a result of perturbations in host immune response (35). Thus, when investigating the role
that circulating miRNA played in CRC, we must make clear whether a
direct correlation exists between tissue and serum levels of miRNA
and their specificity in CRC. Considering the contradicting
conclusion, retrospective or prospective studies with a sufficient
number of samples should be conducted to make clear the role that
miR-326 plays in CRC.
In summary, we showed that miR-326 was downregulated
and its target NOB1 was upregulated in CRC tissues. miR-326
decreased carcinogenesis and metastasis of CRC cells through
directly targeting NOB1. Clinically, miR-326 or NOB1 was
independently prognostic biomarker for CRC patients. Our findings
suggest that exogenous overexpression of miR-326 may be considered
as a promising strategy for targeted therapies in CRC.
Acknowledgments
The present study was supported in part by the
Health Research Projects 2014B1 of Shaanxi Province Health and the
Family Planning Commission. The authors would like to thank the
local doctors and the patients who participated in the present
study.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
miRNAs
|
microRNAs
|
|
qRT-PCR
|
quantitative real-time polymerase
chain reaction
|
|
NOB1
|
nin one binding protein
|
|
3′-UTR
|
3′-untranslated region
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
MT
|
mutant
|
|
WT
|
wild-type
|
|
PI
|
propidium iodide
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar
|
|
4
|
Almeida MI, Nicoloso MS, Zeng L, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896.e9. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pichler M, Ress AL, Winter E, Stiegelbauer
V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn SW,
Kiesslich T, et al: MiR-200a regulates epithelial to mesenchymal
transition-related gene expression and determines prognosis in
colorectal cancer patients. Br J Cancer. 110:1614–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun HB, Chen X, Ji H, Wu T, Lu HW, Zhang
Y, Li H and Li YM: miR-494 is an independent prognostic factor and
promotes cell migration and invasion in colorectal cancer by
directly targeting PTEN. Int J Oncol. 45:2486–2494. 2014.PubMed/NCBI
|
|
7
|
Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang
X, Huang Y, Wang Y, Lu Y, Fu D, et al: MicroRNA-326 functions as a
tumor suppressor in glioma by targeting the Nin one binding protein
(NOB1). PLoS One. 8:e684692013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du W, Liu X, Chen L, et al: Targeting the
SMO oncogene by miR-326 inhibits glioma biological behaviors and
stemness. Neuro Oncol. 17:243–253. 2015. View Article : Google Scholar
|
|
9
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Babashah S, Sadeghizadeh M, Hajifathali A,
Tavirani MR, Zomorod MS, Ghadiani M and Soleimani M: Targeting of
the signal transducer Smo links microRNA-326 to the oncogenic
Hedgehog pathway in CD34+ CML stem/progenitor cells. Int
J Cancer. 133:579–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Makarova KS, Aravind L, Galperin MY,
Grishin NV, Tatusov RL, Wolf YI and Koonin EV: Comparative genomics
of the Archaea (Euryarchaeota): Evolution of conserved protein
families, the stable core, and the variable shell. Genome Res.
9:608–628. 1999.PubMed/NCBI
|
|
13
|
Zhang Y, Ni J, Zhou G, Yuan J, Ren W, Shan
Y, Tang W, Yu L and Zhao S: Cloning, expression and
characterization of the human NOB1 gene. Mol Biol Rep. 32:185–189.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fatica A, Oeffinger M, Dlakić M and
Tollervey D: Nob1p is required for cleavage of the 3′ end of 18S
rRNA. Mol Cell Biol. 23:1798–1807. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang WY, Chen DH, Ning L and Wang LW:
siRNA mediated silencing of NIN1/RPN12 binding protein 1 homolog
inhibits proliferation and growth of breast cancer cells. Asian Pac
J Cancer Prev. 13:1823–1827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin Y, Peng S, Yu H, Teng H and Cui M:
RNAi-mediated downregulation of NOB1 suppresses the growth and
colony-formation ability of human ovarian cancer cells. Med Oncol.
29:311–317. 2012. View Article : Google Scholar
|
|
17
|
Li Y, Ma C, Qian M, Wen Z, Jing H and Qian
D: Downregulation of NOB1 suppresses the proliferation and tumor
growth of non-small cell lung cancer in vitro and in vivo. Oncol
Rep. 31:1271–1276. 2014.PubMed/NCBI
|
|
18
|
Lu Z, Guo Q, Shi A, Xie F and Lu Q:
Downregulation of NIN/RPN12 binding protein inhibit the growth of
human hepatocellular carcinoma cells. Mol Biol Rep. 39:501–507.
2012. View Article : Google Scholar
|
|
19
|
He XW, Tao F, Luo SS and Yu XK: Effect of
lentivirus-mediated NOB1 gene silencing by RNA interference on
proliferation and apoptosis of human colon cancer cells. Zhonghua
Wei Chang Wai Ke Za Zhi. 15:1182–1186. 2012.In Chinese. PubMed/NCBI
|
|
20
|
Gao F and Wang W: MicroRNA-96 promotes the
proliferation of colorectal cancer cells and targets tumor protein
p53 inducible nuclear protein S1, forkhead box protein O1 (FOXO1)
and FOXO3a. Mol Med Rep. 11:1200–1206. 2015.
|
|
21
|
Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT,
Mei Q and Sun SH: c-Myc-mediated repression of miR-15-16 in hypoxia
is induced by increased HIF-2α and promotes tumor angiogenesis and
metastasis by upregulating FGF2. Oncogene. Apr 7–2014.Epub ahead of
print. View Article : Google Scholar
|
|
22
|
Fernandez S, Risolino M, Mandia N, et al:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 0:2014.PubMed/NCBI
|
|
23
|
Li T, Yang J, Lv X, Liu K, Gao C, Xing Y
and Xi T: miR-155 regulates the proliferation and cell cycle of
colorectal carcinoma cells by targeting E2F2. Biotechnol Lett.
36:1743–1752. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu DP and He XW: Expression of NOB1 and
its significance in colorectal cancer. Nan Fang Yi Ke Da Xue Xue
Bao. 32:420–422. 2012.In Chinese. PubMed/NCBI
|
|
25
|
Liu Y, Huang H, Yuan B, Zhuang LY, Luo TP
and Zhang Q: Lentivirus-mediated knockdown of NOB1 suppresses the
proliferation of colon cancer cells. Z Gastroenterol. 52:429–435.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng W, Wang PS, Liu J, Xue S, Wang GM,
Meng XY and Chen G: Adenovirus-mediated siRNA targeting NOB1
inhibits tumor growth and enhances radiosensitivity of human
papillary thyroid carcinoma in vitro and in vivo. Oncol Rep.
32:2411–2420. 2014.PubMed/NCBI
|
|
27
|
Granneman S, Nandineni MR and Baserga SJ:
The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA
processing through a direct interaction with Rps14. Mol Cell Biol.
25:10352–10364. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raychaudhuri S, Fontanes V, Barat B and
Dasgupta A: Activation of ribosomal RNA transcription by hepatitis
C virus involves upstream binding factor phosphorylation via
induction of cyclin D1. Cancer Res. 69:2057–2064. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meraner J, Lechner M, Loidl A,
Goralik-Schramel M, Voit R, Grummt I and Loidl P: Acetylation of
UBF changes during the cell cycle and regulates the interaction of
UBF with RNA polymerase I. Nucleic Acids Res. 34:1798–1806. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fasanaro P, Capogrossi MC and Martelli F:
Regulation of the endothelial cell cycle by the
ubiquitin-proteasome system. Cardiovasc Res. 85:272–280. 2010.
View Article : Google Scholar
|
|
31
|
Xu G, Bernaudo S, Fu G, Lee DY, Yang BB
and Peng C: Cyclin G2 is degraded through the ubiquitin-proteasome
pathway and mediates the antiproliferative effect of activin
receptor-like kinase 7. Mol Biol Cell. 19:4968–4979. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li W, Liu M, Feng Y, Xu YF, Huang YF, Che
JP, Wang GC, Yao XD and Zheng JH: Downregulated miR-646 in clear
cell renal carcinoma correlated with tumour metastasis by targeting
the nin one binding protein (NOB1). Br J Cancer. 111:1188–1200.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Song M, Yin Y, Zhang J, Zhang B, Bian Z,
Quan C, Zhou L, Hu Y, Wang Q, Ni S, et al: MiR-139-5p inhibits
migration and invasion of colorectal cancer by downregulating AMFR
and NOTCH1. Protein Cell. 5:851–861. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kjersem JB, Ikdahl T, Lingjaerde OC, Guren
T, Tveit KM and Kure EH: Plasma microRNAs predicting clinical
outcome in metastatic colorectal cancer patients receiving
first-line oxaliplatin-based treatment. Mol Oncol. 8:59–67. 2014.
View Article : Google Scholar
|
|
35
|
No authors listed. MicroRNA miR-133
represses HERG K+ channel expression contributing to QT
prolongation in diabetic hearts. J Biol Chem. 286:286562011.
|