Introduction
Mitochondria are the intracellular organelles
responsible for ATP production to meet the energy demands (1). Generally, there are several hundred to
1,000 mitochondria in a human cell and 2–10 copies of mitochondrial
DNA (mtDNA) in a human mitochondrion to compose the mitochondrial
network (2). The amount of mtDNA
copies in a human cell is highly variable, depending on the cell
type and the surrounding pathophysiological conditions, and usually
is positively correlated to the energy demands (3). Different from the heterozygotic nature
of nuclear DNA (nDNA), the mtDNA is exclusively transmitted through
maternal lineage with a single origin and the majority of mtDNA
copies in the post-mitotic tissues are assumed identical after
birth. This specific feature is termed as homoplasmy (4). Due to the lack of introns, naked DNA
exposure without adequate histone protection, impaired DNA repair
system and an environment with high reactive oxygen species (ROS)
concentration in the inner membrane of mitochondria, human mtDNA is
far more susceptible to oxidative damages or mutations than nDNA
(5–7). When the damaged or the mutated mtDNA
variants coexist with the wild-type inborn mtDNA molecules, the
homoplasmy is disrupted and shifted to a condition termed as
heteroplasmy (8).
Human mtDNA, containing a coding and a non-coding
region, is a circular and double-stranded DNA structure 16.6-kb in
size. The inner strand is called the light strand (L) and the outer
strand is called the heavy strand (H). Based on the L strand, the
entire mtDNA has been sequenced completely as the revised Cambridge
Reference Sequence (rCRS) (9). The
coding region of mtDNA codes for 13 polypeptides that are required
for the composition of respiratory chain complexes, and 2 rRNAs
plus a set of 22 tRNAs necessary for protein synthesis in the
mitochondria. All the other ~90 polypeptides constituting the
respiratory chain complexes are encoded in nDNA. The non-coding
region 1.1-kb in size, also called the displacement loop (D-loop),
is the regulatory region for specific protein binding to trigger
mtDNA replication and transcription (10,11).
Although damages can occur anywhere throughout the
entire mtDNA, they are frequently found in the D-loop, particularly
in the D310 region (12). Between
nucleotide position (np) 303 and np 316 of the D-loop, there is a
poly-cytidine (C) tract with a thymidine (T) interrupted at np 310
(−C303CCCCCCT310 CCCCCC316 − =
C7TC6). The C number remains constant as 6
after T, however, it is highly variable before T with a range from
6 to 12 (C6, C7, C8,
C9, C10, C11 and C12),
and 7 (C7, wild-type) being the most common one. These
variations with a T shifting over np 310 of the D-loop in mtDNA are
termed as D310 polymorphism or D310 sequence variations (9).
Breast invasive ductal carcinoma (BIDC) is the most
commonly diagnosed malignancy worldwide among women (13). Due to the multidisciplinary
treatment modalities, including surgical resection and combinations
of perioperative chemotherapy, radiotherapy, hormone-ablation or
targeted therapies, and the advance in the understanding of its
underlying molecular pathogenesis, the associated morbidity and
mortality associated with BIDC have been reduced gradually during
the past decades (14). In order to
achieve an optimal therapeutic result, surgical-pathological T-, N-
and M-status and the cancer stage, and the immunological expression
levels of estrogen receptor (ER), progesterone receptor (PR), human
epidermal growth factor receptor-2 (HER-2/neu), protein p53 and
protein Ki-67 are routinely assessed in BIDC patients, at present
(15–18). However, the roles of mtDNA
alterations in BIDC remain speculative. In this retrospective
study, we aimed to appraise the associations among mtDNA
alterations, including the change in mtDNA copy number and the
shifting of mtDNA D310 sequence variations (D310 mutation), the
pathological status, and the immunological ER, PR, HER-2/neu, p53
and Ki-67 expression levels in BICDs, respectively.
Materials and methods
Patient selection, tissue preparation and
DNA extraction
A total of 51 women diagnosed with BIDC, without
obvious distant organ metastasis on preoperative assessments, who
underwent curative modified radical mastectomy plus axillary lymph
node dissection as the primary treatment modality between January
2009 and June 2011 were enrolled. None of the patients received
preoperative neoadjuvant chemotherapy, radiotherapy or both. Their
pathologic status (TNM and cancer stage according to the American
Joint Committee on Cancer; AJCC, 7th edition) and ER, PR,
HER-2/neu, p53 and Ki-67 expression levels, and clinical data were
recorded in detail for systemic analysis. Approval from the
Institutional Review Board was obtained to conduct the present
study.
As reviewed by an experienced pathologist,
representative areas harboring BIDC and paired non-cancerous breast
tissue on pathologic slides were located, and thin sections
(~5-μm) from matched formalin-fixed and paraffin-embedded
tissue blocks were prepared for DNA extraction. These tissue
samples were stored in 1.5-ml Eppendorf vials and mixed with 500
μl xylene (Merck KGaA, Darmstadt, Germany) at room
temperature for 16 h. After centrifuging at 10,000 × g for 10 min
at room temperature and discarding the supernatants, these tissue
samples were re-hydrated with 100, 80, 60 and 40% alcohol aqueous
solution and then pure distilled water for 5 min in steps. Finally,
the hydrated tissue samples were mixed with 200 μl
QuickExtract DNA extraction solution (Epicenter, Madison, WI, USA)
plus 3 μl of 5% butylated hydroxytoluene in methanol to
extract total cellular DNA at 65°C for 3 h as previously described
(19,20). The DNA sample was kept at −20°C
until use.
Standard curves for mtDNA and nDNA
quantifications
Quantitative real-time polymerase chain reaction
(q-PCR) using LightCycler® FastStart DNA Master
SYBR-Green I (Roche Applied Science, Mannheim, Germany) to detect
threshold cycle (Ct) was applied for mtDNA and nDNA standard curve
establishment and subsequent quantification. Briefly, genomic DNA
of 143B osteosarcoma cells were 4-fold diluted from 20 to 0.0048828
ng/μl and then subjected to q-PCR for Ct value
determination. The sequences of the primers used for mtDNA
amplification (near the ND1 region, mainly coding for tRNA leucine
1) were: mtF3212, 5′-CACCCAAGAACAGGGTTTGT-3′; and mtR3319,
5′-TGGCCATGGGATTGTTGTTAA-3′. The sequences of primers used for nDNA
amplification (18S rRNA region) were: 18SF1546,
5′-TAGAGGGACAAGTGGCGTTC-3′; and 18SR1650, 5′-CGCTGAGCCAGTCAGTGT-3′
(21). The equations of standard
curves set for mtDNA and nDNA quantification were set as previously
described (19,20,22).
Then the mtDNA and nDNA copies of the clinical samples relative to
mtDNA and nDNA copies of the 143B osteosarcoma cells were
calculated.
Determination of mtDNA copy number and
mtDNA copy ratio
The mtDNA copy number was defined as the total mtDNA
copies divided by the total nDNA copies for each clinical sample.
q-PCR was applied for sample mtDNA and nDNA quantification. For
each reaction, 1 μl (10 ng/μl) of sample DNA was
amplified in a 10 μl mixture that containing 0.25 μl
of each primer (20 μM, mtF3212 and mtR3319 for mtDNA
quantification; 18SF1546 and 18SR1650 for nDNA quantification), 1.2
μl of 3 mM MgCl2, 1 μl of
LightCycler® FastStart DNA Master SYBR-Green I and 6.3
μl of PCR grade H2O. Simultaneously, 1 μl
of DNA from 143B cells (1 ng/μl) and PCR grade
H2O were included as positive and negative controls,
respectively. The PCR conditions were set as: hot start at 95°C for
10 min followed by 40 cycles of 95°C for 20 sec, 62°C for 20 sec
and 72°C for 20 sec. Fluorescence intensity for Ct value detection
was measured at the end of every extension phase at 79°C. Using the
equations of the established standard curves, the mtDNA copies and
nDNA copies of sample DNA relative to those of 143B cells (1
ng/μl) were determined. The mtDNA copy number (total mtDNA
copies/total nDNA copies; i.e. relative mtDNA copies to
143B/relative nDNA copies to 143B) of each clinical sample was
determined after adjusting the mtDNA copy number of the 143B cells
to 1.000. Each reaction was carried out in duplicate and the mean
value was used for data presentation. To evaluate the change in
mtDNA copy number between the BIDC and paired non-cancerous breast
tissues, we defined mtDNA copy ratio as the mtDNA copy number of
the BIDC divided by mtDNA copy number of the paired non-cancerous
breast tissue.
Sequencing of the D310 region
The D310 region of mtDNA was amplified by PCR and
then subjected to direct sequencing as previously described
(20,23,24).
Each 50 μl PCR reaction contained 25 μl of RBC
SensiZyme® Hotstart Taq Premix (RBC Bioscience,
New Taipei City, Taiwan), 22 μl of PCR-grade H2O,
1 μl of each primer (H76-1, 5′-CACGCGATAGCATTGCGA-3′; and
L335, 5′-TAAGTGCTGTGGCCAGAAGC-3′), and 1 μl of sample DNA
(10 ng/μl). The PCR procedures included a hot start at 95°C
for 10 min, 40 cycles of 95°C for 15 sec, 58°C for 15 sec and 72°C
for 30 sec, and a final extension at 72°C for 7 min. After
confirmation by 3% agarose gel electrophoresis, the PCR products
were subjected to direct sequencing (MB Mission Biotech, Taipei,
Taiwan). The D310 sequence variations, including patterns of
homoplasmy or heteroplasmy, number of detected D310 variants, and
the predominant D310 variant were determined as previously
described (20,23,24).
Compared to the D310 sequences of the paired non-cancerous breast
tissue, any sequence alterations detected in the BIDC were defined
as D310 mutations (20).
Immunohistochemical staining
The expression levels of ER, PR, HER-2/neu, p53 and
Ki-67 were determined by immunohistochemical (IHC) staining, and
these parameters are routinely determined in the pathological
examinations in our hospital. Thin sections (~4-μm) from the
formalin-fixed and paraffin-embedded tissues blocks were cut for
IHC staining. Characteristics of the primary antibodies used for
IHC and their criteria for positive/high expression are illustrated
in Table I. Briefly, the sections
were de-paraffinized with xylene (Merck KGaA) and then rehydrated
with 100% alcohol, 95% alcohol solution and pure distilled running
water in steps. After reacting with target retrieval solution
(EnVision™ FLEX, pH 9.0, 50X, K8004; pH 6.0, 10X, S2369; both from
Dako, Carpinteria, CA, USA) at 99°C for 20 min, they were cooled at
room temperature for 20 min and incubated in TBS buffer (EnVision™
FLEX Wash Buffer, 20X, K8007; Dako) for 5 min. Endogenous
peroxidase was blocked with peroxidase-blocking reagent (DM821;
Dako) for 5 min. After washing with TBS, the primary antibodies
were applied at room temperature for 20 min (HER-2/neu, 30 min).
After washing with TBS, the sections were incubated with
horseradish peroxidase (HRP; DM822; Dako) at room temperature for
20 min, and the sections were again washed in TBS. The color was
developed by 10 min of incubation with 3,3-diaminobenzidine
tetrahydrochloride solution (DAB; + chromogen, DM827; diluted in
substrate buffer, SM803; both from Dako). The sections were weakly
counterstained with hematoxylin (Merck).
 | Table IDetails of the antibodies, dilutions,
antigen retrieval methods and expression criteria used in this
study. |
Table I
Details of the antibodies, dilutions,
antigen retrieval methods and expression criteria used in this
study.
| Primary
antibody | Dilution/incubation
time | Antigen
retrival | Specificity | Source | Product code | High expression
criteria |
|---|
| ER | Ready to use/20
min | pH 9.0/heat at
99°C | Monoclonal rabbit
anti-human ERα | Dakoa | IS084 | >10% cancer
cells with strong nuclear staining |
| PR | Ready to use/20
min | pH 9.0/heat at
99°C | Monoclonal mouse
anti-human PR | Dakoa | IS068 | >10% cancer
cells with strong nuclear staining |
| HER-2/neu | 1:800/30 min | pH 6.0/heat at
99°C | Polyclonal rabbit
anti-human c-erbB-2 | Dakoa | A0485 | HercepTest™
criteria |
| Ki-67 | Ready to use/20
min | pH 6.0/heat at
99°C | Monoclonal mouse
anti-Ki-67 | Dakoa | IS626 | >20% cancer
cells with strong nuclear staining |
| p53 | Ready to use/20
min | pH 9.0/heat at
99°C | Monoclonal mouse
anti-p53 | Dakoa | IS616 | >50% cancer
cells with strong nuclear staining |
Statistical analyses
All the statistical analyses were performed using
the Statistical Package for the Social Sciences (SPSS), version
15.0, software (SPSS, Inc., Chicago, IL, USA). The continuous
variables were compared using the Student’s t-test/Mann-Whitney u
test between two groups or ANOVA/Kruskal-Wallis H test among three
or more groups when appropriate. Categorical variables between
groups were compared using the Chi-square/Fisher’s exact tests or
Chi-square test for trend when appropriate. The difference between
groups was considered to indicate a statistically significant
result when the p-value was <0.05.
Results
mtDNA D310 sequence variations and mtDNA
copy numbers of examined tissues and their alterations
The results of the mtDNA D310 sequence variations
and mtDNA copy numbers of the non-cancerous breast tissues and
paired BIDCs and their alterations are listed in Table II and classified in Fig. 1. For non-cancerous breast tissues,
the mean detected D310 variants and mean mtDNA copy numbers were
2.0 and 0.317, respectively, and 29 (56.9%) harbored a
heteroplasmic D310 pattern. For BIDCs, their mean detected D310
variants and mtDNA copy numbers were 2.1 and 0.295, respectively,
and 27 (52.9%) harbored a heteroplasmic D310 pattern (Table III). When compared to the paired
non-cancerous breast tissues, 29 (56.9%) BIDCs harbored D310
mutations, and 24 (47.1%) had an increased mtDNA copy number with
mtDNA copy ratio >1.000 (Table
IV).
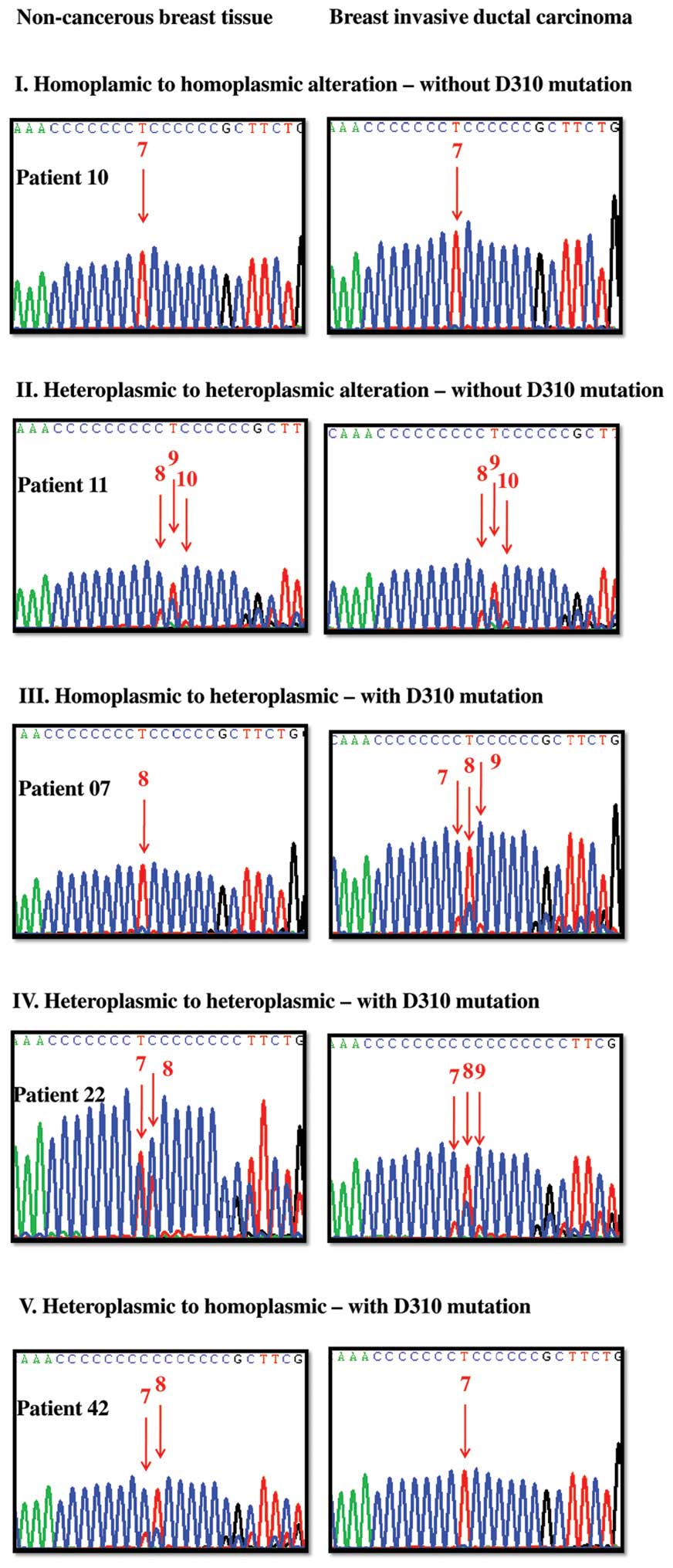 | Figure 1Representative cases to demonstrate
the mitochondrial DNA D310 sequence variations and their shifting
between non-cancerous breast tissue (left) and paired breast IDC
(right). T (thymidine) is shown in red, A (adenine) in green, C
(cytidine) in blue and G (guanine) in black during sequencing. The
Arabic number above the red arrow denotes the C number before the
indicated T peak and the T peak height represents the relative
quantity of the D310 variant. Type I, homoplasmic to homoplasmic
(first row) without D310 mutation; patient 10 as an example. Her
non-cancerous breast tissue harbors one type of D310 variant, the
C7TC6 and is classified as homoplasmic D310
with C7TC6 as the major one. Her BIDC also
harbors one type of D310 variant, the C7TC6,
and is classified as homoplasmic D310 with
C7TC6 as the major one. As a result, patient
10 was defined as type I homoplasmic to homoplasmic alteration
without D310 mutation. Type II, heteroplasmic to heteroplasmic
(second row) without D310 mutation; patient 11 as an example. Her
non-cancerous breast tissue harbors 3 types of D310 variants, the
C9TC6, C8TC6 and
C10TC6 in order, and is classified as
heteroplasmic D310 with C9TC6 as the major
one. Her BIDC also harbors 3 types of D310 variants, the
C9TC6, C8TC6 and
C10TC6 in order, and is classified as
heteroplasmic D310 with C9TC6 as the major
one. No significant shifting of the D310 sequence variations was
noted. As a result, patient 11 was defined as heteroplasmic to
heteroplasmic alteration without D310 mutation. Type III,
homoplasmic to heteroplasmic (third row) with D310 mutation;
patient 07 as an example. Her non-cancerous breast tissue harbors
one type of D310 variants, the C8TC6, and is
classified as homoplasmic D310 with C8TC6 as
the major one. Her BIDC harbors 3 types of D310 variants, the
C8TC6, C7TC6 and
C9TC6 in order, and is classified as
heteroplasmic D310 with C8TC6 as the major
one. Significant shifting of the D310 sequence variations was
noted. As a result, patient 07 was defined as homoplasmic to
heteroplasmic alteration with D310 mutation. Type IV, heteroplasmic
to heteroplasmic (fourth row) with D310 mutation; patient 22 as an
example. Her non-cancerous breast tissue harbors 2 types of D310
variants, the C7TC6 and
C8TC6 in order, and is classified as
heteroplasmic D310 with C7TC6 as the major
one. Her BIDC harbors 3 types of D310 variants, the
C8TC6, C7TC6 and
C9TC6 in order, and is classified as
heteroplasmic D310 with C8TC6 as the major
one. Significant shifting of the D310 sequence variations were
noted. As a result, patient 22 was defined as heteroplasmic to
heteroplasmic alteration with D310 mutation. Type V, heteroplasmic
to homoplasmic (fifth row) with D310 mutation; patient 42 as an
example. Her non-cancerous breast tissue harbors 2 types of D310
variants, the C8TC6 and
C7TC6 in order, and is classified as
heteroplasmic D310 with C8TC6 as the major
one. Her breast IDC harbors one type of D310 variant, the
C7TC6, and is classified as homoplasmic D310
with C7TC6 as the major one. Significant
shifting of the D310 sequence variations were noted. As a result,
patient 42 was defined as heteroplasmic to homoplasmic alteration
with D310 mutation. |
 | Table IIDetails of the mitochondrial DNA D310
sequence variations and mitochondrial DNA copy number of the
non-cancerous breast tissue and paired breast invasive ductal
carcinoma and their alterations in the 51 women. |
Table II
Details of the mitochondrial DNA D310
sequence variations and mitochondrial DNA copy number of the
non-cancerous breast tissue and paired breast invasive ductal
carcinoma and their alterations in the 51 women.
| D310 mutation/type
of alteration | Mitochondrial DNA
D310 region
| Mitochondrial DNA
copy no.
| Copy ratio | Change |
|---|
Non-cancerous
breast tissue
| Breast invasive
ductal carcinoma
| Non- cancerous
breast tissue | Breast invasive
ductal carcinoma |
|---|
| Pts. | Pattern | Variants | No. | Major | Pattern | Variants | No. | Major |
|---|
| No (n=20) |
| I. Homoplasmic to
homoplasmic | 02 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.322 | 0.223 | 0.692 | Decrease |
| 09 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.313 | 0.280 | 0.895 | Decrease |
| 10 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.152 | 0.214 | 1.413 | Increase |
| 14 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.173 | 0.214 | 1.234 | Increase |
| 18 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.295 | 0.239 | 0.809 | Decrease |
| 19 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.276 | 0.210 | 0.763 | Decrease |
| 21 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.343 | 0.281 | 0.819 | Decrease |
| 23 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.396 | 0.259 | 0.654 | Decrease |
| 25 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.303 | 0.163 | 0.538 | Decrease |
| 26 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.290 | 0.235 | 0.811 | Decrease |
| 27 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.310 | 0.291 | 0.939 | Decrease |
| 32 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.230 | 0.268 | 1.168 | Increase |
| 33 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.252 | 0.361 | 1.435 | Increase |
| 50 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 1.250 | 0.329 | 0.263 | Decrease |
| 51 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.471 | 0.229 | 0.485 | Decrease |
| 52 | Homoplasmy | 7 | 1 | 7 | Homoplasmy | 7 | 1 | 7 | 0.401 | 0.233 | 0.582 | Decrease |
| 04 | Homoplasmy | 8 | 1 | 8 | Homoplasmy | 8 | 1 | 8 | 0.262 | 0.324 | 1.237 | Increase |
| 20 | Homoplasmy | 8 | 1 | 8 | Homoplasmy | 8 | 1 | 8 | 0.324 | 0.224 | 0.692 | Decrease |
| 29 | Homoplasmy | 8 | 1 | 8 | Homoplasmy | 8 | 1 | 8 | 0.435 | 0.815 | 1.872 | Increase |
| 47 | Homoplasmy | 8 | 1 | 8 | Homoplasmy | 8 | 1 | 8 | 0.232 | 0.363 | 1.565 | Increase |
| No (n=2) |
| II. Heteroplasmic
to heteroplasmic | 38 | Heteroplasmy | 8,7,9 | 3 | 8 | Heteroplasmy | 8,7,9 | 3 | 8 | 0.169 | 0.320 | 1.899 | Increase |
| 11 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.397 | 0.281 | 0.710 | Decrease |
| Yes (n=2) |
| III. Homoplasmic
to heteroplasmic | 05 | Homoplasmy | 1 | 1 | 7 | Heteroplasmy | 7,10,9 | 3 | 7 | 0.069 | 0.097 | 1.421 | Increase |
| 07 | Homoplasmy | 8 | 1 | 8 | Heteroplasmy | 8,7.9 | 3 | 8 | 0.260 | 0.311 | 1.194 | Increase |
| Yes (n=23) |
| Heteroplasmic to
heteroplasmic | 22 | Heteroplasmy | 7,8 | 2 | 7 | Heteroplasmy | 8,7,9 | 3 | 8 | 0.126 | 0.262 | 2.088 | Increase |
| 06 | Heteroplasmy | 8,7 | 2 | 8 | Heteroplasmy | 8,7,9 | 3 | 8 | 0.170 | 0.238 | 1.407 | Increase |
| 08 | Heteroplasmy | 8,7,9 | 3 | 8 | Heteroplasmy | 7,8 | 2 | 7 | 0.252 | 0.332 | 1.320 | Increase |
| 15 | Heteroplasmy | 8,9 | 2 | 8 | Heteroplasmy | 8,9,7,10 | 4 | 8 | 0.827 | 0.560 | 0.676 | Decrease |
| 37 | Heteroplasmy | 8,9, | 4 | 8 | Heteroplasmy | 16,17,15, | 6 | 16 | 0.131 | 0.294 | 2.257 | Increase |
| | 7,10 | | | | 18,19,14 | | | | | | |
| 39 | Heteroplasmy | 8,9 | 2 | 8 | Heteroplasmy | 7,8,9,10 | 4 | 7 | 0.193 | 0.117 | 0.609 | Decrease |
| 44 | Heteroplasmy | 8,7 | 2 | 8 | Heteroplasmy | 8.9 | 2 | 8 | 0.326 | 0.151 | 0.463 | Decrease |
| 49 | Heteroplasmy | 8,7 | 2 | 8 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.407 | 0.452 | 1.109 | Increase |
| 53 | Heteroplasmy | 8,7,9 | 3 | 8 | Heteroplasmy | 8,9 | 2 | 8 | 0.263 | 0.312 | 1.186 | Decrease |
| 56 | Heteroplasmy | 8,9,7 | 3 | 8 | Heteroplasmy | 9,8,7,10 | 4 | 9 | 0.290 | 0.241 | 0.833 | Decrease |
| 61 | Heteroplasmy | 8,9 | 2 | 8 | Heteroplasmy | 8,9,10 | 3 | 8 | 0.254 | 0.228 | 0.896 | Decrease |
| 62 | Heteroplasmy | 8,9,7 | 3 | 8 | Heteroplasmy | 8,9 | 2 | 8 | 0.214 | 0.275 | 1.285 | Increase |
| 01 | Heteroplasmy | 9,8 | 2 | 9 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.514 | 0.430 | 0.837 | Decrease |
| 03 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.192 | 0.211 | 1.102 | Increase |
| 13 | Heteroplasmy | 9,8 | 2 | 9 | Heteroplasmy | 8,9 | 2 | 8 | 0.250 | 0.351 | 1.404 | Increase |
| 17 | Heteroplasmy | 9,8 | 2 | 9 | Heteroplasmy | 9,10,8,11 | 4 | 9 | 0.216 | 0.116 | 0.535 | Decrease |
| 28 | Heteroplasmy | 9,8 | 2 | 9 | Heteroplasmy | 9,10,8 | 3 | 9 | 0.334 | 0.358 | 1.075 | Increase |
| 40 | Heteroplasmy | 9,8,10,7 | 4 | 9 | Heteroplasmy | 8,9 | 2 | 8 | 0.511 | 0.235 | 0.460 | Decrease |
| 41 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 8,9,10 | 3 | 8 | 0.339 | 0.383 | 1.128 | Increase |
| 45 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.363 | 0.485 | 1.336 | Increase |
| 57 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 8,9 | 2 | 8 | 0.437 | 0.619 | 1.417 | Decrease |
| 60 | Heteroplasmy | 9,8,10 | 3 | 9 | Heteroplasmy | 9,8,10 | 3 | 9 | 0.271 | 0.253 | 0.932 | Decrease |
| 36 | Heteroplasmy | 16,17,15,
18,19,14 | 6 | 16 | Heteroplasmy | 16,15,17,14,
18,13,20 | 7 | 16 | 0.245 | 0.147 | 0.603 | Decrease |
| Yes (n=4) |
| Heteroplasmic to
homoplasmic | 58 | Heteroplasmy | 7,9,8 | 3 | 7 | Homoplasmy | 7 | 1 | 7 | 0.489 | 0.413 | 0.845 | Increase |
| 42 | Heteroplasmy | 8,7 | 2 | 8 | Homoplasmy | 7 | 1 | 7 | 0.234 | 0.406 | 1.741 | Increase |
| 43 | Heteroplasmy | 8,7 | 2 | 8 | Homoplasmy | 8 | 1 | 8 | 0.268 | 0.268 | 1.000 | Decrease |
| 12 | Heteroplasmy | 9,8,7 | 3 | 9 | Homoplasmy | 8 | 1 | 8 | 0.124 | 0.125 | 1.008 | Increase |
 | Table IIIDistribution of mitochondrial DNA
D310 and mitochondrial DNA copy number of the non-cancerous breast
tissue and paired breast invasive ductal carcinoma in the 51 BIDC
women. |
Table III
Distribution of mitochondrial DNA
D310 and mitochondrial DNA copy number of the non-cancerous breast
tissue and paired breast invasive ductal carcinoma in the 51 BIDC
women.
| Mitochondrial
DNA | Non-cancerous
breast tissue | Breast invasive
ductal carcinoma |
|---|
| D310 pattern (n,
%) |
| Homoplasmic | 22 (43.1) | 24 (47.1) |
| Heteroplasmic | 29 (56.9) | 27 (52.9) |
| No. of D310
variants |
| Mean ± SD | 2.0±1.1 | 2.1±1.4 |
| (95% CI of
mean) | (1.7-2.3) | (1.8-2.6) |
| Copy no. |
| Mean ± SD | 0.317±0.184 | 0.295±0.130 |
| (95% CI of
mean) | (0.269–0.385) | (0.250–0.360) |
 | Table IVDemographic data concerning clinical,
pathological and mitochondrial DNA features of the 51 BIDC
women. |
Table IV
Demographic data concerning clinical,
pathological and mitochondrial DNA features of the 51 BIDC
women.
| Demographic
data | Data |
|---|
| Age (years) mean ±
SD (95% CI of mean) | 53.6±11.4
(50.5–56.8) |
| Tumor side, n
(%) |
| Left | 30 (58.8) |
| Right | 21 (41.2) |
| Tumor location, n
(%) |
| Outer-upper | 28 (54.9) |
| Outer-lower | 9 (17.6) |
| Inner-upper | 9 (17.6) |
| Inner-lower | 3 (5.9) |
| Sub-areolar | 2 (3.9) |
|
Surgical-pathological findings mean ± SD
(95% CI of mean) |
| Tumor diameter
(cm) | 2.5±1.6
(2.2–3.0) |
| Total dissected
lymph nodes | 17.0±10.0
(14.5–19.0) |
| Positive-dissected
lymph nodes | 1.8±4.2
(0.8–3.3) |
| T-status, n
(%) |
| T1 | 24 (47.1) |
| T2 | 21(41.2) |
| T3 | 6 (11.8) |
| N-status, n
(%) |
| N0 | 30 (58.8) |
| N1 | 13 (25.5) |
| N2 | 6 (11.8) |
| N3 | 2 (3.9) |
| Cancer stage, n
(%) |
| I | 17 (33.3) |
| II | 26 (51.0) |
| III | 8 (15.7) |
| Histological grade,
n (%) |
| I | 4 (7.8) |
| II | 39 (76.5) |
| III | 8 (15.7) |
| Estrogen receptor,
n (%) |
| Negative | 14 (27.5) |
| Positive | 37 (72.5) |
| Progesterone
receptor |
| Negative | 18 (35.3) |
| Positive | 33 (64.7) |
| HER-2/neu |
| Negative | 37 (72.5) |
| Positive | 14 (27.5) |
| p53 |
| Negative | 37 (72.5) |
| Positive | 14 (27.5) |
| Ki-67 |
| Low | 42 (82.4) |
| High | 9 (17.6) |
| Mitochondrial DNA
alteration, n (%) D310 mutation |
| Yes | 29 (56.9) |
| No | 22 (43.1) |
| Mitochondrial DNA
copy ratio | 1.052±0.441 |
| mtDNA copy number,
n (%) |
| Increase (ratio
>1.000) | 24 (47.1) |
| Decrease (ratio
≤1.000) | 27 (52.9) |
Clinicopathological demographic data
The demographic data concerning the clinical,
pathological and biochemical analyses of the 51 BIDC women are
listed in Table IV. The mean age
was 53.6 and the mean tumor diameter was 2.5 cm. After pathological
examinations, the mean total dissected axillary lymph nodes was
17.0 with a mean number of positive nodes of 1.8. Concerning the
pathological T- and N-status and cancer stage, 24 (47.1%), 21
(41.2%) and 6 (11.8%) belonged to T1, T2 and T3; 30 (58.8%), 13
(25.5), 6 (11.8%) and 2 (3.9%) to N0, N1, N2 and N3; and 17
(33.3%), 26 (51.%) and 8 (15.7%) to stages I–III, respectively. For
the histological grade, 4 (7.8%) belonged to grade I, 39 (76.5%) to
II and 8 (15.7%) to III, respectively.
Concerning the expression of therapeutic biomarkers,
37 (72.5%), 33 (64.7%) and 14 (27.5%) were positive for ER, PR and
HER-2/neu, respectively. For the biomarkers refecting tumor
aggressiveness and proliferation, 14 (27.5%) were positive for p53
expression and 9 (17.6%) had a high Ki-67 expression.
Factors affecting mtDNA copy ratio
The possible factors that affect mtDNA copy ratio
are listed in Table V. Advanced
T-status (p=0.056), negative-ER expression (p=0.005), negative-PR
expression (p=0.007), positive-p53 (p=0.050) and Ki-67 with high
expression (p=0.004) were related to a higher mtDNA copy ratio in
the BIDCs, respectively.
 | Table VPossible factors affecting
mitochondrial DNA copy number ratio. |
Table V
Possible factors affecting
mitochondrial DNA copy number ratio.
| Factors (n, %) | Mitochondrial DNA
copy no. ratio | P-value |
|---|
| Age (years) | | 0.832 |
| ≤50 (n=24,
47.1) | 1.066±0.502 | |
| >50 (n=27,
52.9) | 1.039±0.388 | |
| Pathological
findings |
| T-status | | 0.056 |
| T1 (n=24,
47.1) | 1.144±0.460 | |
| T2 (n=21,
41.2) | 0.878±0.341 | |
| T3 (n=6,
11.8) | 1.293±0.520 | |
| N-status | | 0.131 |
| N0 (n=30,
58.8) | 0.969±0.399 | |
| N1 (n=13,
25.5) | 1.218±0.470 | |
| N2 (n=6,
11.8) | 0.912±0.369 | |
| N3 (n=2,
3.9) | 1.637±0.637 | |
| Cancer stage | | 0.975 |
| I (n=17,
33.3) | 1.053±0.412 | |
| II (n=26,
51.0) | 1.038±0.452 | |
| III (n=8,
15.7) | 1.093±0.518 | |
| Histological
grade | | 0.215 |
| I (n=4, 7.8) | 0.834±0.488 | |
| II (n=39,
76.5) | 1.042±0.465 | |
| III (n=8,
15.7) | 1.206±0.245 | |
| Estrogen
receptor | | 0.005 |
| Negative (n=14,
27.5) | 1.291±0.357 | |
| Positive (n=37,
72.5) | 0.961±0.440 | |
| Progesterone
receptor | | 0.007 |
| Negative (n=18,
35.3) | 1.233±0.372 | |
| Positive (n=33,
64.7) | 0.953±0.449 | |
| HER-2/neu | | 0.301 |
| Negative (n=37,
72.5) | 1.012±0.433 | |
| Positive (n=14,
27.5) | 1.158±0.459 | |
| p53 | | 0.050 |
| Negative (n=37,
72.5) | 0.983±0.426 | |
| Positive (n=14,
27.5) | 1.233±0.444 | |
| Ki-67 | | 0.004 |
| Low (n=42,
82.4) | 0.977±0.420 | |
| High (n=9,
17.6) | 1.399±0.382 | |
Factors related to the mtDNA D310
mutation
As shown in Table
VI, advanced T-status (p= 0.019) and negative-HER-2/neu
expression (p=0.061) were associated with a higher rate of D310
mutations in the human BIDCs.
 | Table VIPossible factors related to
mitochondrial DNA D310 mutation. |
Table VI
Possible factors related to
mitochondrial DNA D310 mutation.
| Factors (n, %) | Mitochondrial DNA
D310 mutation
| P-value |
|---|
No
n (%) | Yes
n (%) |
|---|
| Total | 22 (100) | 29 (100) | |
| Age (years) | | | 0.714 |
| ≤50 (n=24,
100.0) | 11 (45.8) | 13 (54.2) | |
| >50 (n=27,
100.0) | 11 (40.7) | 16 (59.3) | |
| Pathological
findings |
| T-status | | | 0.019 |
| T1 (n=24,
100.0) | 9 (37.5) | 15 (62.5) | |
| T2 (n=21,
100.0) | 13 (61.9) | 8 (38.1) 6
(100.0) | |
| T3 (n=6,
100.0) | 0 (0.0) | | |
| N-status | | | 0.593 |
| N0 (n=30,
100.0) | 14 (46.7) | 16 (53.3) | |
| N1 (n=13,
100.0) | 5 (38.5) | 8 (61.5) | |
| N2 (n=6,
100.0) | 3 (50.0) | 3 (50.0) 2
(100.0) | |
| N3 (n=2,
100.0) | 0 (0.0) | | |
| Cancer stage | | | 0.598 |
| I (n=17,
100.0) | 6 (35.3) | 11 (64.7) | |
| II (n=26,
100.0) | 13 (50.0) | 13 (50.0) | |
| III (n=8,
100.0) | 3 (37.5) | 5 (62.5) | |
| Histological
grade | | | 0.707 |
| I (n=4,
100.0) | 1 (25.0) | 3 (75.0) | |
| II (n=39,
100.0) | 17 (43.6) | 22 (56.4) | |
| III (n=8,
100.0) | 4 (50.0) | 4 (50.0) | |
| Estrogen
receptor | | | |
| Negative (n=14,
100.0) | 6 (42.9) | 8 (57.1) | |
| Positive (n=37,
100.0) | 16 (43.2) | 21 (56.8) | |
| Progesterone
receptor | | | 0.889 |
| Negative (n=18,
100.0) | 8 (44.4) | 10 (55.6) | |
| Positive (n=33,
100.0) | 14 (42.4) | 19 (57.6) | |
| HER-2/neu | | | 0.061 |
| Negative (n=37,
100.0) | 13 (35.1) | 24 (64.9) | |
| Positive (n=14,
100.0) | 9 (64.3) | 5 (35.7) | |
| P53 | | | 0.214 |
| Negative (n=37,
100.0) | 14 (37.8) | 23 (62.2) | |
| Positive (n=14,
100.0) | 8 (57.1) | 6 (42.9) | |
| Ki-67 | | | 0.163 |
| Low (n=42,
100.0) | 20 (47.6) | 22 (52.4) | |
| High (n=9,
100.0) | 2 (22.2) | 7 (77.8) | |
Discussion
Surgical-pathological T-, N- and M-status and cancer
stage of AJCC remain as the gold standard to predict the prognosis
of BIDC women. With the advance in cancer research, the oncogenic
process and the proliferative aggressiveness of BIDCs were found to
be correlated to positive-p53 (tumor suppressor) and high Ki-67
(cell proliferation) expression, respectively. Furthermore, due to
the breakthrough of modern molecular biology in the evaluation of
specific cellular receptors, hormone ablation or targeted therapy
have been advocated for BIDC women harboring positive-ER/PR or
positive-HER-2/neu expression to improve their outcomes, in
addition to routine adjuvant chemotherapy (15,18,25–29).
In this retrospective study, we demonstrated that: i) advanced
T-status, negative-ER, negative-PR, positive-p53 and high Ki-67
expression are related to higher mtDNA copy ratios in BIDCs; and
ii) advanced T-status and negative-HER-2/neu are related to higher
rates of mtDNA D310 mutations in BIDCs. Since mitochondria are the
cellular powerhouse for energy production, whether these mtDNA
alterations in human BIDCs are related to a metabolic shift
warrants further appraisal and discussion. Metabolic shift in human
cancer was first described by Dr Warburg 7 decades ago (Warburg
effect), and he contended that human cancer tissues exhibited
increased glycolysis yet decreased mitochondrial respiration to
generate ATP (30–32). It also became the basic theory of
positron emission tomography (PET) scan in cancer evaluation.
The changes in mtDNA copy numbers have been analyzed
in several human type of cancers. Compared to the paired
non-cancerous counterparts, a decrease in mtDNA copy number was
reported in lung cancer (33),
hepatocellular carcinoma (34) and
gastric cancer (35). In lung
cancer tissues after neoadjuvant chemotherapy, a progressive
decrease in mtDNA copy number was correlated with disease
progression (22). These decreases
were thought to be decreases in mitochondrial function. On the
contrary, an increase in mtDNA copy number was noted in the
carcinogenesis of head and neck cancers (36), and the progression of esophageal
squamous cell carcinomas (20),
particularly in patients who smoked cigarettes. These increases
were regarded as a compensation process to overcome the oxidative
damages from cigarette smoking and to maintain proper mitochondrial
function (19). Concerning BIDCs, a
decrease in mtDNA copy number was reported in some research
articles (37–39). Nevertheless, we demonstrated a
significantly higher mtDNA copy ratio in BIDCs that harbored
negative-ER, negative-PR, positive-p53 or high Ki-67 expression.
Thus, we focused on the role of ER, PR, p53 and Ki-67 and their
relationships to mtDNA in BIDCs.
ER and PR both belong to the nuclear receptors,
which can interact with the hormone response element (HRE) in nDNA,
after the binding of estrogen or progesterone. With regards to ER,
it not only can activate estrogen HRE in nDNA to control cell
proliferation and apoptosis in several tissues yet also can be
imported into the mitochondria to interact with the mtDNA to
increase the expression of mtDNA-encoded peptides, and
mitochondrial biogenesis due to mtDNA also harbors sequences
similar to the HRE of nDNA (HRE-like motif) (40). Furthermore, an in vitro study
denoted that the interactions among the estrogen, ER and HRE-like
motif in mtDNA were crucial to maintain mitochondrial function,
inhibit cell apoptosis, and overcome oxidative damage in breast
cancer cell line, MCF-7 (41).
Concerning PR, similar effects were also observed in benign breast
epithelial cells (42). As a
result, we proposed that an increase in mtDNA copy numbers in BIDCs
that harbor negative-ER or negative-PR expression are thought to be
a compensation process to enhance the interaction between ER/PR and
HRE motifs on mtDNA to maintain mitochondrial function.
Concerning the associations among p53, mitochondria
and mtDNA, pilot studies have shown that functional p53 may
participate in the regulation of cellular mitochondrial biogenesis
and respiration (43–45), maintenance of mtDNA integrity
(46), maintaining mtDNA copy
number abundance (46) and the
homeostasis of reactive oxygen species (47). In a normal cell, p53 is inactivated
by its negative regulator, mdm2, as a result, the protein p53 is
usually not detectable (48). Under
stress or pathological situations, various pathways lead to the
dissociation of the p53 and mdm2 complex, which leads to an
activated, accumulated and detectable p53. The activated p53
induces cell cycle arrest to allow either repair and survival of
the cell or apoptosis to discard the damaged cells. Although the
primary antibody that we used in the present study cannot
distinguish wild-type or mutant p53, the positively detected p53
did suggest an elevated stress in BIDCs. It is postulated that the
elevated mtDNA copy ratio in BIDCs may indicate an enhanced
interaction between p53 and mtDNA toward a possible metabolic shift
or survival benefit.
High protein Ki-67 expression denotes a high
proliferative status (49). Ki-67
is detectable during all active phases of the cell cycle
(G1, S, G2 and mitosis) except the resting
status (G0). High Ki-67 expression in BIDCs means a
highly proliferation situation of the cancer cells and is
associated with a poor outcome (15,16).
It is reasonable to hypothesize that the elevated mtDNA copy ratio
in BIDCs with high Ki-67 expression is regarded as increased energy
expenditure for the rapid growth of cancer cells.
In addition to quantitative change of mtDNA copy
number, several studies also evaluated the qualitative mtDNA D310
mutations in human types of cancers, including prostate (50,51),
head and neck (52) and lung cancer
(53,54), and esophageal squamous cell
carcinoma (20), and some showed
clinical significance. In breast cancer, a D-loop mutation,
including D310 mutation, was identified as a poor prognostic factor
(39). Intriguingly, our results
showed that negative-HER-2/neu expression was related to a higher
rate of D310 mutation in BIDCs. Due to the fact that the mtDNA D310
is located near the binding side of mitochondrial transcriptional
factor to control mitochondrial biogenesis (55), whether negative-HER-2/neu expression
and D310 mutation are related to the functional alterations in BIDC
mitochondria deserve further evaluation. Although the detailed
interaction between HER-2/neu and mtDNA remains speculative, a
recent novel study revealed that HER-2/neu can translocate into
mitochondria to negatively regulate mitochondrial respiratory
functions (56). Thus, it is
assumed that an altered mitochondrial function may exist in BIDCs
with negative HER-2/neu expression and mtDNA D310 mutation.
In addition to the above mentioned biomarkers of ER,
PR, HER-2/neu, p53 and Ki-67, TNM status and cancer stages of AJCC
remain the gold standard criteria to predict the prognosis of BIDC
women. In our preliminary result, advanced T status was associated
with a higher incidence of D310 mutation and an elevated mtDNA copy
ratio. It is reasonable to assume that the increase in mtDNA copy
number compensated for the mtDNA with D310 mutations when the BIDCs
underwent progression, which was proposed in head and neck cancer
and esophageal cancer (19,20,36).
To verify whether these increases in mtDNA copy number and mtDNA
D310 mutations confer enhanced aggressiveness to BIDCs, further
in vitro study is warrant.
Based on our preliminary results, we conclude that
elevated mtDNA copy ratios and D310 mutations, suggesting an
altered mitochondrial function, may be relevant biomarkers
correlated with pathological T status and ER, PR, HER-2/neu, p53
and Ki-67 expression in BIDCs.
Acknowledgments
We would like to express our sincere appreciations
to Hsin-Mei Kao for her excellent technique in the preparations of
patholocial sections. This study was supported by grants from the
Department of Health (DOH), Executive Yuan, Taiwan (nos.
Taipei-100-03 and Taipei-101-02) to DOH Affiliated Taipei Hospital
(DOH was named as the Ministry of Health and Welfare as of July 23,
2013).
References
|
1
|
Lee HC and Wei YH: Mitochondrial role in
life and death of the cell. J Biomed Sci. 7:2–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chan DC: Mitochondria: Dynamic organelles
in disease, aging, and development. Cell. 125:1241–1252. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee HC and Wei YH: Mitochondrial
biogenesis and mitochondrial DNA maintenance of mammalian cells
under oxidative stress. Int J Biochem Cell Biol. 37:822–834. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: Heredity,
heteroplasmy and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar
|
|
5
|
Khrapko K, Coller HA, André PC, Li XC,
Hanekamp JS and Thilly WG: Mitochondrial mutational spectra in
human cells and tissues. Proc Natl Acad Sci USA. 94:13798–13803.
1997. View Article : Google Scholar
|
|
6
|
Yakes FM and Van Houten B: Mitochondrial
DNA damage is more extensive and persists longer than nuclear DNA
damage in human cells following oxidative stress. Proc Natl Acad
Sci USA. 94:514–519. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croteau DL, Stierum RH and Bohr VA:
Mitochondrial DNA repair pathways. Mutat Res. 434:137–148. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chinnery PF, Thorburn DR, Samuels DC,
White SL, Dahl HM, Turnbull DM, Lightowlers RN and Howell N: The
inheritance of mitochondrial DNA heteroplasmy: Random drift,
selection or both? Trends Genet. 16:500–505. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Andrews RM, Kubacka I, Chinnery PF,
Lightowlers RN, Turnbull DM and Howell N: Reanalysis and revision
of the Cambridge reference sequence for human mitochondrial DNA.
Nat Genet. 23:1471999. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Asin-Cayuela J and Gustafsson CM:
Mitochondrial transcription and its regulation in mammalian cells.
Trends Biochem Sci. 32:111–117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moraes CT: What regulates mitochondrial
DNA copy number in animal cells? Trends Genet. 17:199–205. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mambo E, Gao X, Cohen Y, Guo Z, Talalay P
and Sidransky D: Electrophile and oxidant damage of mitochondrial
DNA leading to rapid evolution of homoplasmic mutations. Proc Natl
Acad Sci USA. 100:1838–1843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Berry DA, Inoue L, Shen Y, Venier J, Cohen
D, Bondy M, Theriault R and Munsell MF: Modeling the impact of
treatment and screening on U.S. breast cancer mortality: A Bayesian
approach. J Natl Cancer Inst Monogr. 2006:30–36. 2006. View Article : Google Scholar
|
|
15
|
Mylonas I, Makovitzky J, Jeschke U, Briese
V, Friese K and Gerber B: Expression of Her2/neu, steroid receptors
(ER and PR), Ki67 and p53 in invasive mammary ductal carcinoma
associated with ductal carcinoma In Situ (DCIS) Versus invasive
breast cancer alone. Anticancer Res. 25:1719–1723. 2005.PubMed/NCBI
|
|
16
|
Cheang MC, Chia SK, Voduc D, Gao D, Leung
S, Snider J, Watson M, Davies S, Bernard PS, Parker JS, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hugh J, Hanson J, Cheang MC, Nielsen TO,
Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, et
al: Breast cancer subtypes and response to docetaxel in
node-positive breast cancer: Use of an immunohistochemical
definition in the BCIRG 001 trial. J Clin Oncol. 27:1168–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dawood S: Triple-negative breast cancer:
Epidemiology and management options. Drugs. 70:2247–2258. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin CS, Wang LS, Chou TY, Hsu WH, Lin HC,
Lee SY, Lee MH, Chang SC and Wei YH: Cigarette smoking and hOGG1
Ser326Cys polymorphism are associated with 8-OHdG accumulation on
mitochondrial DNA in thoracic esophageal squamous cell carcinoma.
Ann Surg Oncol. 20(Suppl 3): S379–S388. 2013. View Article : Google Scholar
|
|
20
|
Lin CS, Chang SC, Wang LS, Chou TY, Hsu
WH, Wu YC and Wei YH: The role of mitochondrial DNA alterations in
esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg.
139:189.e4–197.e4. 2010. View Article : Google Scholar
|
|
21
|
Bai RK, Perng CL, Hsu CH and Wong LJ:
Quantitative PCR analysis of mitochondrial DNA content in patients
with mitochondrial disease. Ann NY Acad Sci. 1011:304–309. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin CS, Wang LS, Tsai CM and Wei YH: Low
copy number and low oxidative damage of mitochondrial DNA are
associated with tumor progression in lung cancer tissues after
neoadjuvant chemotherapy. Interact Cardiovasc Thorac Surg.
7:954–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee HT, Lin CS, Chen WS, Liao HT, Tsai CY
and Wei YH: Leukocyte mitochondrial DNA alteration in systemic
lupus erythematosus and its relevance to the susceptibility to
lupus nephritis. Int J Mol Sci. 13:8853–8868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang PN, Lee HC, Wang CH, Ping YH, Liu TY,
Chi CW, Lin KN and Liu HC: Heteroplasmy of mitochondrial D310
mononucleotide repeat region in the blood of patients with
Alzheimer’s disease. J Alzheimers Dis. 18:345–353. 2009.
|
|
25
|
Nishimura R, Osako T, Okumura Y, Tashima
R, Toyozumi Y and Arima N: Changes in the ER, PgR, HER2, p53 and
Ki-67 biological markers between primary and recurrent breast
cancer: Discordance rates and prognosis. World J Surg Oncol.
9:1312011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taneja P, Maglic D, Kai F, Zhu S, Kendig
RD, Fry EA and Inoue K: Classical and novel prognostic markers for
breast cancer and their clinical significance. Clin Med Insights
Oncol. 4:15–34. 2010.PubMed/NCBI
|
|
27
|
Carter CL, Allen C and Henson DE: Relation
of tumor size, lymph node status, and survival in 24,740 breast
cancer cases. Cancer. 63:181–187. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cianfrocca M and Goldstein LJ: Prognostic
and predictive factors in early-stage breast cancer. Oncologist.
9:606–616. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cianfrocca M and Gradishar W: New
molecular classifications of breast cancer. CA Cancer J Clin.
59:303–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
31
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warburg O: Origin of cancer cells.
Oncologia. 9:75–83. 1956.In German. View Article : Google Scholar
|
|
33
|
Lee HC, Yin PH, Lin JC, Wu CC, Chen CY, Wu
CW, Chi CW, Tam TN and Wei YH: Mitochondrial genome instability and
mtDNA depletion in human cancers. Ann NY Acad Sci. 1042:109–122.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui
WY, Wei YH, Liu TY and Chi CW: Alteration of the copy number and
deletion of mitochondrial DNA in human hepatocellular carcinoma. Br
J Cancer. 90:2390–2396. 2004.PubMed/NCBI
|
|
35
|
Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi
CW, Wei YH and Lee HC: Mitochondrial DNA mutations and
mitochondrial DNA depletion in gastric cancer. Genes Chromosomes
Cancer. 44:19–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim MM, Clinger JD, Masayesva BG, Ha PK,
Zahurak ML, Westra WH and Califano JA: Mitochondrial DNA quantity
increases with histopathologic grade in premalignant and malignant
head and neck lesions. Clin Cancer Res. 10:8512–8515. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan AX, Radpour R, Haghighi MM, Kohler C,
Xia P, Hahn S, Holzgreve W and Zhong XY: Mitochondrial DNA content
in paired normal and cancerous breast tissue samples from patients
with breast cancer. J Cancer Res Clin Oncol. 135:983–989. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bai RK, Chang J, Yeh KT, Lou MA, Lu JF,
Tan DJ, Liu H and Wong LJ: Mitochondrial DNA content varies with
pathological characteristics of breast cancer. J Oncol.
2011:4961892011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW,
Lee LM, Wei YH and Lee HC: Mitochondrial DNA mutations and
mitochondrial DNA depletion in breast cancer. Genes Chromosomes
Cancer. 45:629–638. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen JQ, Yager JD and Russo J: Regulation
of mitochondrial respiratory chain structure and function by
estrogens/estrogen receptors and potential
physiological/pathophysiological implications. Biochim Biophys
Acta. 1746:1–17. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pedram A, Razandi M, Wallace DC and Levin
ER: Functional estrogen receptors in the mitochondria of breast
cancer cells. Mol Biol Cell. 17:2125–2137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Behera MA, Dai Q, Garde R, Saner C,
Jungheim E and Price TM: Progesterone stimulates mitochondrial
activity with subsequent inhibition of apoptosis in MCF-10A benign
breast epithelial cells. Am J Physiol Endocrinol Metab.
297:E1089–E1096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bensaad K and Vousden KH: p53: New roles
in metabolism. Trends Cell Biol. 17:286–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Matoba S, Kang JG, Patino WD, Wragg A,
Boehm M, Gavrilova O, Hurley PJ, Bunz F and Hwang PM: p53 regulates
mitochondrial respiration. Science. 312:1650–1653. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Saleem A, Adhihetty PJ and Hood DA: Role
of p53 in mitochondrial biogenesis and apoptosis in skeletal
muscle. Physiol Genomics. 37:58–66. 2009. View Article : Google Scholar
|
|
46
|
Kulawiec M, Ayyasamy V and Singh KK: p53
regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog.
8:82009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lebedeva MA, Eaton JS and Shadel GS: Loss
of p53 causes mitochondrial DNA depletion and altered mitochondrial
reactive oxygen species homeostasis. Biochim Biophys Acta.
1787:328–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Moll UM and Petrenko O: The MDM2-p53
interaction. Mol Cancer Res. 1:1001–1008. 2003.
|
|
49
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen JZ, Gokden N, Greene GF, Mukunyadzi P
and Kadlubar FF: Extensive somatic mitochondrial mutations in
primary prostate cancer using laser capture microdissection. Cancer
Res. 62:6470–6474. 2002.PubMed/NCBI
|
|
51
|
Chen JZ and Kadlubar FF: Mitochondrial
mutagenesis and oxidative stress in human prostate cancer. J
Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 22:1–12.
2004. View Article : Google Scholar
|
|
52
|
Lièvre A, Blons H, Houllier AM,
Laccourreye O, Brasnu D, Beaune P and Laurent-Puig P:
Clinicopathological significance of mitochondrial D-Loop mutations
in head and neck carcinoma. Br J Cancer. 94:692–697.
2006.PubMed/NCBI
|
|
53
|
Suzuki M, Toyooka S, Miyajima K, Iizasa T,
Fujisawa T, Bekele NB and Gazdar AF: Alterations in the
mitochondrial displacement loop in lung cancers. Clin Cancer Res.
9:5636–5641. 2003.PubMed/NCBI
|
|
54
|
Onishi M, Saito M, Sokuza Y, Mori C,
Nishikawa T, Shimizu K, Sugata E and Tsujiuchi T: Numerical changes
in the mitochondrial DNA displacement loop in lung lesions induced
by N-nitrosobis(2-hydroxypropyl)amine in rats. Mutat Res.
638:133–138. 2008. View Article : Google Scholar
|
|
55
|
Fisher RP, Topper JN and Clayton DA:
Promoter selection in human mitochondria involves binding of a
transcription factor to orientation-independent upstream regulatory
elements. Cell. 50:247–258. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ding Y, Liu Z, Desai S, Zhao Y, Liu H,
Pannell LK, Yi H, Wright ER, Owen LB, Dean-Colomb W, et al:
Receptor tyrosine kinase ErbB2 translocates into mitochondria and
regulates cellular metabolism. Nat Commun. 3:12712012. View Article : Google Scholar : PubMed/NCBI
|















