Introduction
Pancreatic cancer is the fourth most common cause of
cancer-related mortality in the United States and the eighth
worldwide (1). This disease has an
extremely poor prognosis with a median survival of 4–12 months and
a dismal 5-year survival rate of 5% (2,3). The
lethal nature of pancreatic adenocarcinoma is mainly due to the
high frequency of local or regional spread and distant metastasis
(4). Although various molecular
changes have been revealed in this malignancy, mechanisms
underlying the aggressiveness of this neoplasm remain largely
unclear.
CD9, a member of the tetraspanin family, is widely
expressed on the plasma membrane of various cell types, including
hematopoietic cells, endothelial cells, epidermal cells, and many
tumor cell lines (5–8). Like other members of the tetraspanins,
CD9 interacts with a number of transmembrane proteins, and forms
functional complexes, which facilitate cell adhesion, motility and
signaling (5,9–14). In
pancreatic cancer, CD9 expression is reduced in pancreatic cancer
cells but is elevated in the stroma (15) and reduced CD9 expression levels are
associated with high tumor grade and lymph node metastasis
(16), although contradictory
findings have also been reported (17). However, mechanisms underlying the
involvement of CD9 in pancreatic cancer metastasis remain
incompletely understood.
In the present study, using the two closely
associated pancreatic cancer cell lines, PaTu-8988t and PaTu-8988s,
which have been shown to be non-metastatic and metastatic in
vivo, respectively (18), we
demonstrated an inverse correlation between CD9 expression and cell
proliferation and motility. PaTu-8988s cells expressed a lower
level of CD9 but had higher proliferation and migration rates than
the PaTu-8898t cells. We further demonstrated an inverse
correlation between CD9 level and the cell surface expression of
epidermal growth factor receptor (EGFR). CD9 overexpression reduced
the cell surface expression of EGFR, associated with increased
expression of dynamin-2, whereas CD9 knockdown increased the cell
surface expression of EGFR, associated with decreased expression of
dynamin-2. While forced expression of EGFR did not reverse CD9
overexpression-mediated inhibition of cell proliferation, CD9
knockdown-promoted pancreatic cancer cell proliferation and
migration were blocked by EGFR RNAi both in vitro and in
vivo. Thus, targeting CD9 may offer therapeutic benefits to
patients with pancreatic cancer.
Materials and methods
Animals
Female severe combined immunodeficient (SCID) mice
(4–6 weeks of age) were purchased from jackson Laboratory.
Temperature (20–21°C) and humidity (50–60%) were controlled. Daily
light cycle consisted of 12 h of light and 12 h of dark. Animals
were manipulated under sterile conditions. All animal experiments
were carried out after approval by the Institutional Review board
of Shanghai Tenth People’s Hospital and were performed in
accordance with the Guidelines for the Care and Use of Laboratory
Animals published by the National Institutes of Health.
Cell lines
PaTu-8988s and PaTu-8988t cells were obtained from
the Cancer Research UK Central Cell Service (Clare Hall
Laboratories, Potters bar, UK). Cells were routinely maintained in
Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal
bovine serum (FBS) in a humidified incubator at 37°C with an
atmosphere of 5% CO2.
Immunostaining and confocal
microscopy
Cells growing on coverslips were fixed with 4%
paraformaldehyde (PFA) for 15 min at room temperature. They were
later blocked with 5% normal donkey serum containing 0.1% Triton
X-100 in phosphate-buffered saline (PBS) for 1 h at room
temperature. Cells were incubated with an antibody mixture
containing a mouse monoclonal anti-CD9 (Abcam) and a rabbit
polyclonal anti-EGFR (R&D Systems) antibody at 4°C overnight.
After washing 3 times with PBS containing 0.1% Triton X-100, the
cells were incubated with a mixture of Alexa Fluor 488-conjugated
donkey anti-mouse IgG (Life Technologies) and Alexa Fluor
594-conjugated donkey anti-rabbit IgG (Life Technologies) for 2 h
at room temperature, and washed 3 times with PBS containing 0.1%
Triton X-100 and incubated with 1 µg/ml
4′,6-diamidino-2-phenylindole (DAPI; Sigma Aldrich) for 10 min.
Confocal microscopy was performed on an LSM-710 laser scanning
microscope (Zeiss, Oberkochen, Germany) with a 40×1.3 numerical
aperture oil immersion lens. All digital images were captured at
the same settings. Final images were processed using Adobe
PhotoShop software.
Western blot analysis
Tissues or cells were lysed in 2X Laemmli buffer
containing a cocktail of protease inhibitors (Bio-Rad, Hercules,
CA, USA). The proteins were extracted, boiled for 10 min at 100°C,
and then resolved by SDS-PAGE on a 12% gel. The protein signals
were detected by luminal detection reagent (Santa Cruz
biotechnologies, Santa Cruz, CA, USA). The following primary
antibodies were used: anti-CD9 antibody (1:1,000), anti-β-actin
antibody (1:2,000), anti-EGFR (1:1,000), anti-dynamin-2 antibody
(1:1,000) and anti-Na-K-ATPase (1:1,000) (all from Abcam).
Construction of recombinant lentiviral
vectors, preparation of lentiviral particles and cell
infection
To construct lentiviral CD9 shRNAs, three
oligonucleotides sequences: 5′-GCTGTT CGGATTTAACTTCAT-3′ (shCD9-1),
5′-TTCTACACAGGA GTCTATATT-3′ (shCD9-2), 5′-CACAAGGATGAGGTGATT
AAG-3′ (shCD9-3), which specifically target CD9 mRNA (Genbank
accession no. NM_001769.3) were synthesized. A scrambled sequence
of the CD9 target sequence was used as a negative control. The
lentiviral shRNA constructs were prepared at Genomeditech
biotechnology Co. Ltd. (www.genomeditech.com) using the vector
U6-MCS-CMV-EGFP. The lentiviral EGFR-shRNA (shEGFR) was constructed
similarly using the specific oligonucleotide sequence:
5′-GCAAAGUGUGUA ACGGAAUAG-3′) which targets a sequence starting at
nucleotide 1247 and lying at the junction of exon 8 and 9 (19). To construct the lentiviral CD9 and
EGFR constructs, the CD9 cDNA and EGFR cDNA were amplified by PCR
using Pfu polymerase (Promega, USA), and were inserted into the
BamHI and XhoI digestion sites in the lentiviral
vector CMV-MCS-IRES-EGFP. Lentiviral particles were prepared at
Genomeditech biotechnology Co. Ltd. Cells were transfected with the
lentiviral vectors using Lipofectamine 2000 (Invitrogen Corp, Long
Island, NY, USA) according to the manufacturer’s instructions.
Cell proliferation assay
PaTu-8898s and PaTu-8898t cells were plated in a
96-well plate (2,000 cells/well), which was incubated for 1–7 days
at 37°C in an atmosphere of 5% CO2 in air. Thereafter,
20 µl of CCK-8 solution was added to each well of the plate
and incubated for an additional 4 h under the same conditions.
Then, the absorbance at 450 nm was measured using a microplate
reader, wherein the absorbance value indicated the proliferative
capacity.
Scratch-wound assay
The scratch-wound assay was performed as previously
described (20). Cells were plated
onto 6-well plates at a concentration of 2×105
cells/well. Cell monolayers were carefully wounded by scratching
with a sterile plastic pipette tip. The cells were washed twice
with PBS and incubated for a further 48 h. Photographs of each
wound were captured in the same fields at different times up to 48
h. The distance between the wound edges was analyzed using Image J
version 1.42 (National Institutes of Health, Bethesda, MD, USA).
The percentage of wound occupied was calculated by dividing the
non-recovered area at 24 h by the initial wound area at 0 h and
subtracting this value as a percentage from 100%.
In vitro invasion assay
Cells were detached and resuspended in serum-free
medium. Cells (4×104 cells/well) were then plated into
Matrigel-coated invasion chambers (Becton-Dickinson) and allowed to
invade for 24 h. The remaining cells in the chambers were removed
by Q-tips and the invading cells on the lower surface of the
chambers were stained with 0.2% crystal violet solution. The number
of invading cells was calculated by counting three different fields
under a phase-contrast microscope and were plotted as the
percentage of invading cells of the total number of plated
cells.
In vivo tumor growth and metastasis
experiments
PaTu-8988t (1×107) cells infected with
the lentiviral scramble shRNA or CD9-specific shRNA (shRNA3) were
subcutaneously implanted into one flank of the SCID mice (n=12,
respectively). The presence of tumors and their size (length ×
width2 × π/6) were monitored twice weekly. When the
animals died or were sacrificed due to tumor burden, the tumors,
draining lymph nodes, liver and lungs were harvested and then
subjected to histopathological examination. PaTu-8988s
(1×106) cells infected with the lentiviral CD9 construct
or the lentiviral vector were intravenously injected into the tail
vein of SCID mice (n=12, respectively). At 120 days after
injection, the mice were sacrificed and lungs and livers were
collected for H&E staining. Three cross-sections of each lung
at different levels (every 100 mm) were investigated for assessment
of micrometastasis. The percentage of mice with lung metastases and
the total number of micrometastatic nodules from the three sections
are presented.
Statistical analysis
For the in vitro experiments, the values are
expressed as the mean ± SEM. Comparison between two groups was
carried out using the Student’s t-test. All experiments were
repeated three times. For the in vivo studies, tumor volumes
were calculated as the mean ± SEM. Wilcoxon rank sum test and
Kruskal-Wallis test were used to compare treatment differences.
Results
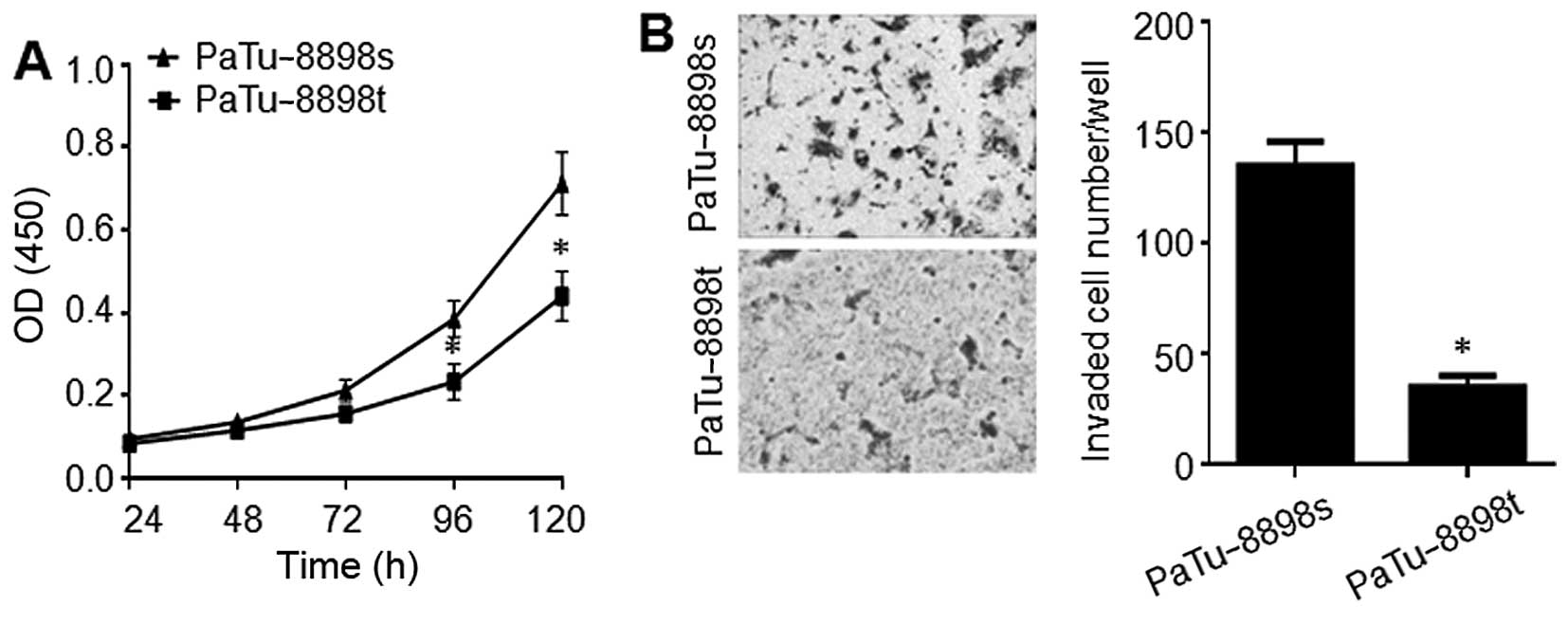
Different proliferation and migration
capabilities between the PaTu-8898s and PaTu-8898t cells
Although PaTu-8898s and PaTu-8898t, two cell lines
originating from liver metastases of the same human pancreatic
adenocarcinoma, have been shown to be metastatic and non-metastatic
in vivo, respectively (18),
whether they behave differentially in vitro has rarely been
investigated. The proliferation and invasion of both cell lines
were investigated by conducting in vitro proliferation and
Matrigel invasion assays, respectively. As shown in Fig. 1, PaTu-8898s cells underwent a
significantly higher rate of proliferation and invasion compared to
the Patu-8898t cells.
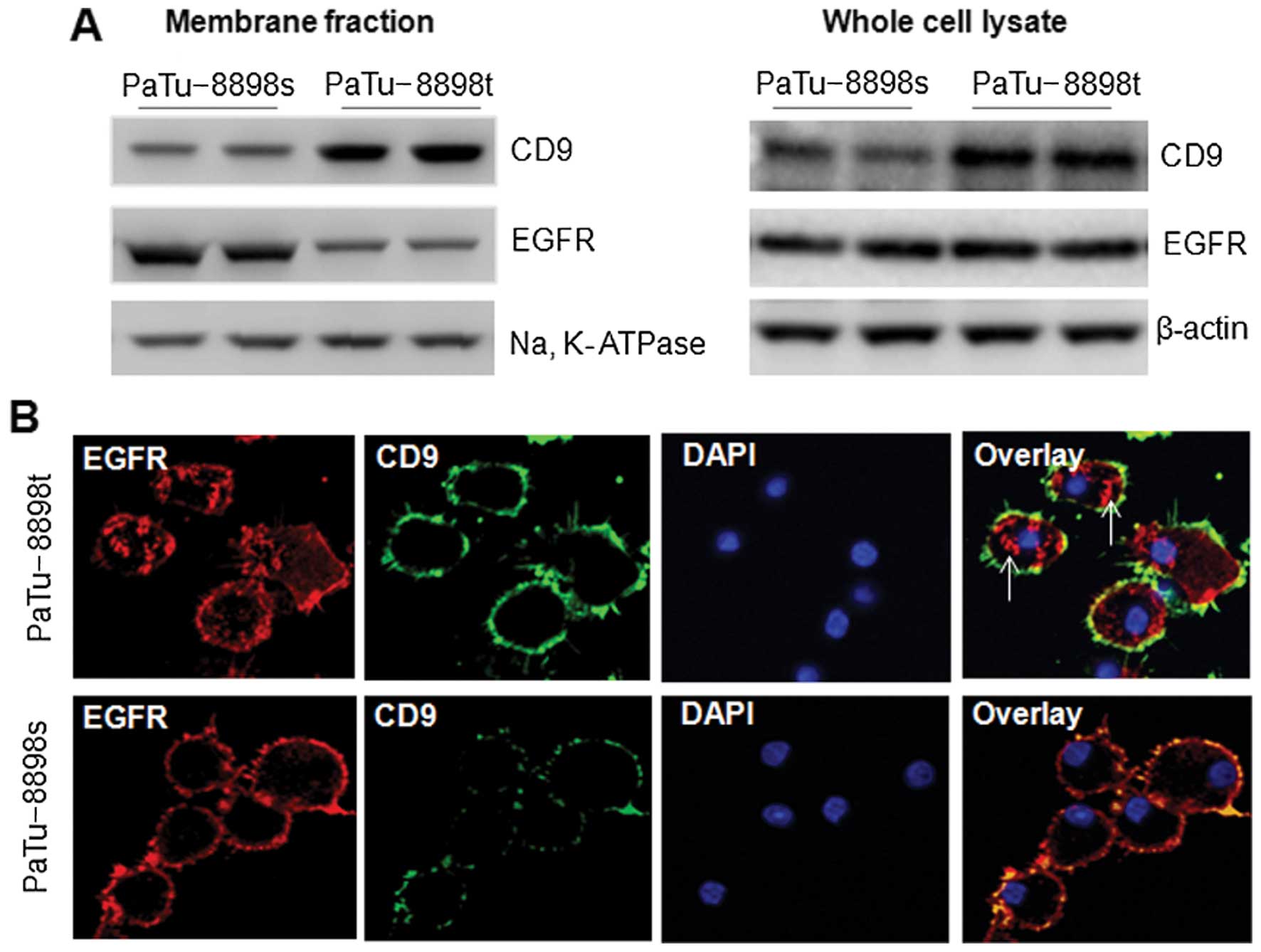
Inverse correlation between CD9
expression and cell surface expression of EGFR in the PaTu-8898s
and PaTu-8898t cells
To examine the expression of CD9 in both cell lines,
the membrane fraction and whole cell lysate were subjected to
western blot analysis. As shown in Fig.
2A, the PaTu-8898s cells expressed a markedly lower level of
CD9 in both the membrane fraction and the whole cell lysate than
did the PaTu-8898t cells. We also examined the expression of EGFR,
whose aberrant expression is linked to the etiology of several
human epidermal cancers, including pancreatic cancer (21). Intriguingly, a markedly higher level
of EGFR was observed in the membrane fraction of the PaTu-8898s
than that in the PaTu-8898t cells, while the EGFR level in the
whole cell lysate did not differ between the two cell lines
(Fig. 2A). The inverse correlation
between CD9 and EGFR cell surface expression was confirmed with
confocal microscopy. In the PaTu-8898t cells in which CD9 was
highly expressed on the cell surface, a large proportion of EGFR
was localized in the cytosol (Fig.
2b, arrows), whereas in the PaTu-8898s cells in which a low
level of CD9 was observed on the cell surface, EGFR was
predominantly localized on the cell surface (Fig. 2b).
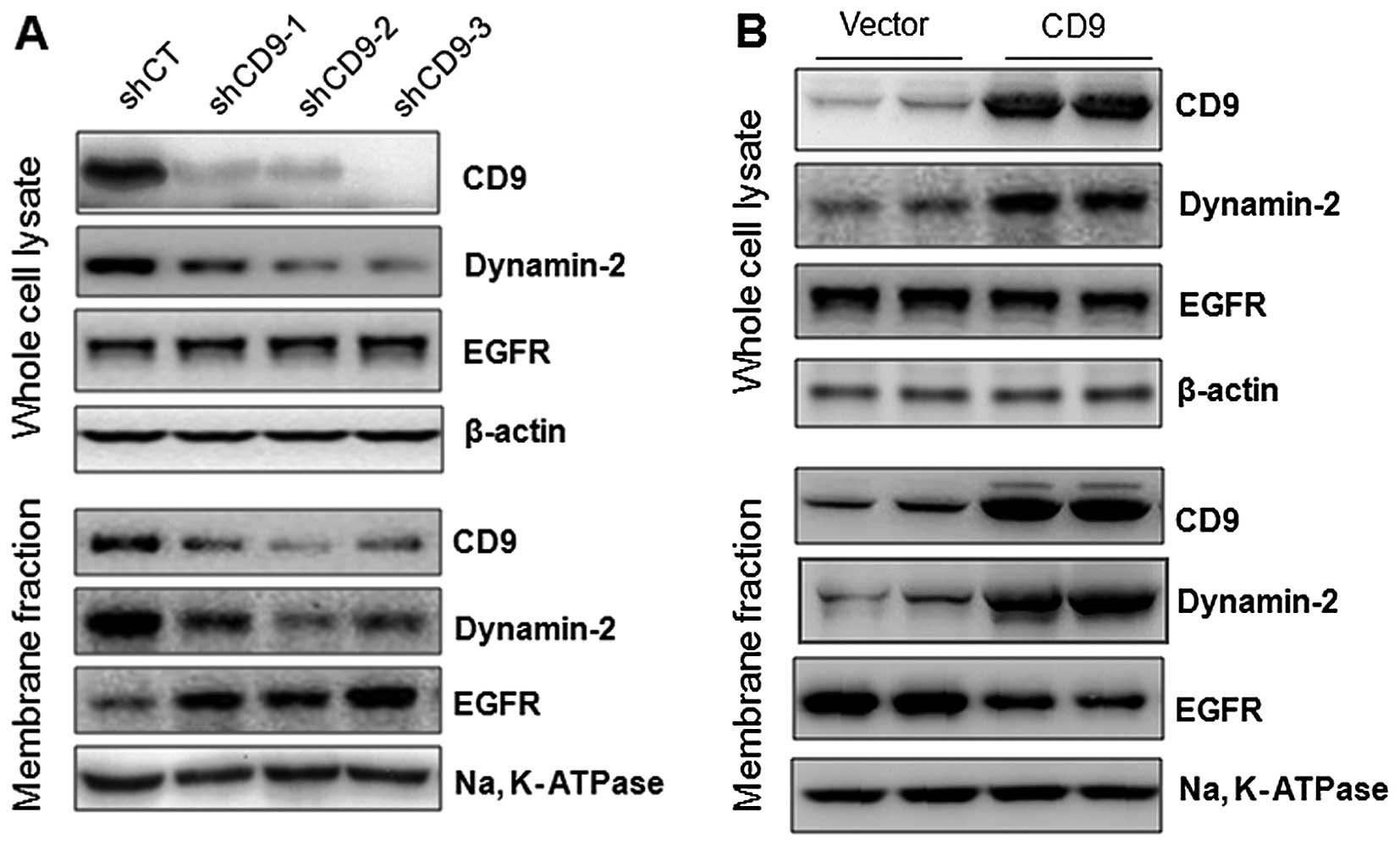
CD9 plays a role in EGFR endocytosis
To examine whether CD9 plays a role in the cell
surface expression of EGFR, we first examined the effect of CD9
knockdown on the cell surface expression of EGFR. PaTu-8898t cells
were infected with lentiviral scramble shRNA (shCT) or CD9 shRNA1-3
(shCD1-3), and the whole cell lysate and membrane fraction were
subjected to western blot analysis. Compared to the lentiviral
scramble shRNA (shCT)-infected cells, the cells infected with
either of the lentiviral shCD9s resulted in a robust downregulation
of CD9. Intriguingly, CD9 knockdown did not change the total EGFR
expression level in the whole cell lysate but robustly increased
the expression of EGFR in the membrane fraction. This was
associated with a marked decrease in the expression of dynamin-2,
which is known to regulate EGFR endocytosis (22,23),
in both the whole cell lysate and membrane fraction (Fig. 3A). Next, we examined the effect of
CD9 overexpression on the cell surface expression of EGFR in the
PaTu-8898s cells. As shown in Fig.
3b, CD9 overexpression did not change the total EGFR expression
level in the whole cell lysate but robustly decreased the
expression of EGFR in the membrane fraction, associated with a
marked increase in the expression of dynamin-2 in both the whole
cell lysate and membrane fraction. The effect of CD9 on EGFR cell
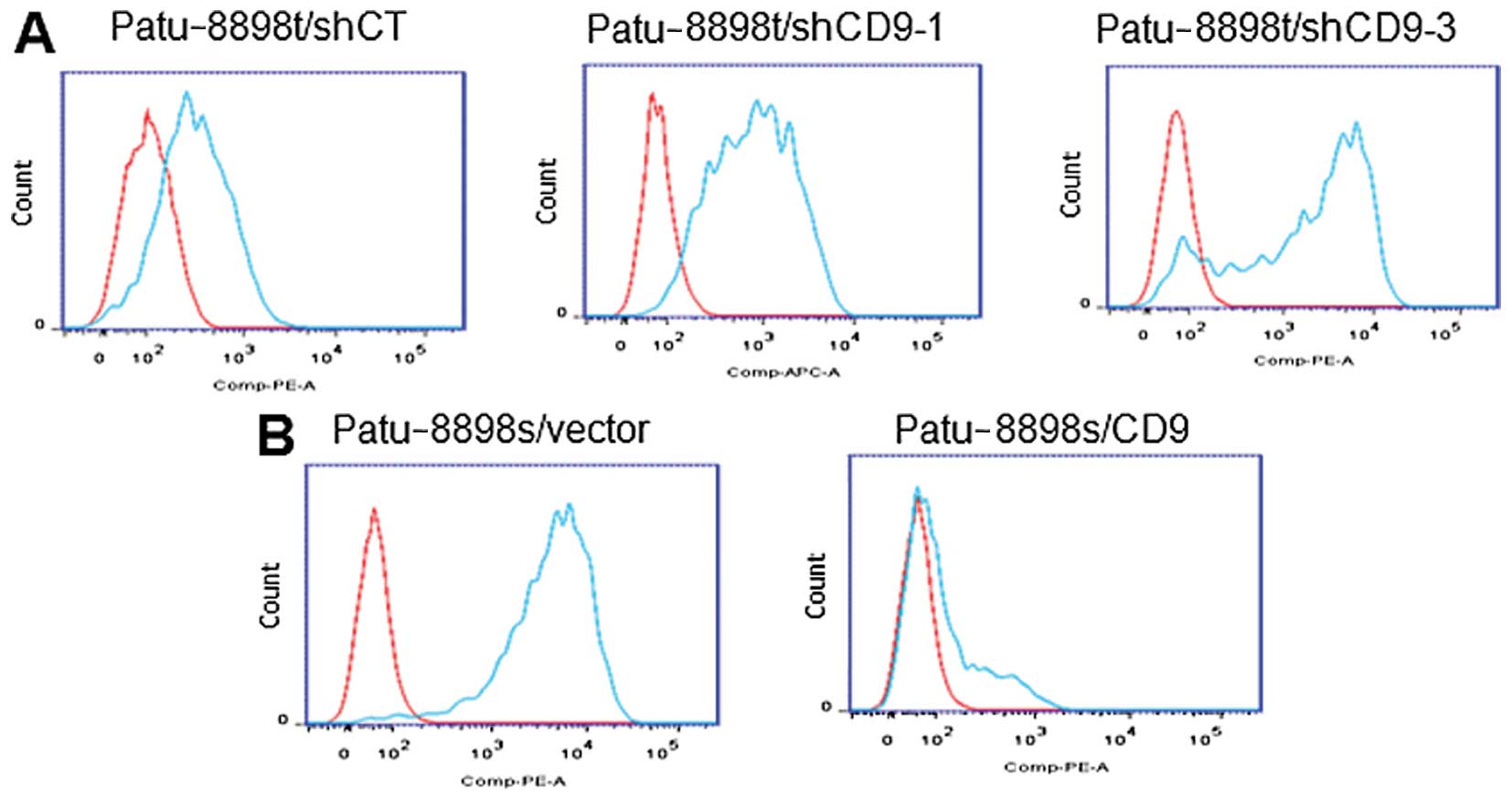
surface expression was further visualized with FACS analysis. As
shown in Fig. 4, CD9 knockdown in
the PaTu-8898t cells resulted in a marked increase in the
expression of EGFR on the cell surface. In contrast, CD9
overexpression decreased the cell surface expression of EGFR.
CD9 overexpression in the PaTu-8898s
cells attenuates cell proliferation, migration and invasion, which
is not reversed by EGFR overexpression
To further address the role of CD9 in pancreatic
cell proliferation, the PaTu-8988s cells infected with the
lentiviral vector, CD9 construct, EGFR construct, or a combination
of CD9 and EGFR constructs (1:1 ratio) were subjected to
proliferation assay. As shown in Fig.
5A, CD9 overexpression significantly reduced the cell
proliferation rate, whereas EGFR overexpression enhanced the cell
proliferation. Intriguingly, the CD9 overexpression-mediated
inhibition of cell proliferation was not reversed by
co-overexpression of EGFR. To examine the effect of CD9 and/or EGFR
overexpression on cell migration, PaTu-8988s cells infected with
the lentiviral vector, CD9 construct, EGFR construct, or a
combination of CD9 and EGFR constructs (1:1 ratio) were subjected
to scratch-wound and invasion assays. While EGFR overexpression
enhanced cell migration and invasion, CD9 overexpression
significantly reduced the cell migration and invasion, which were
not reversed by co-overexpression of EGFR. Consistently, western
blot analysis showed that while CD9 overexpression reduced the cell
surface expression of EGFR, co-overexpression of CD9 and EGFR did
not result in any significant increase in EGFR expression on the
cell surface (Fig. 5F).
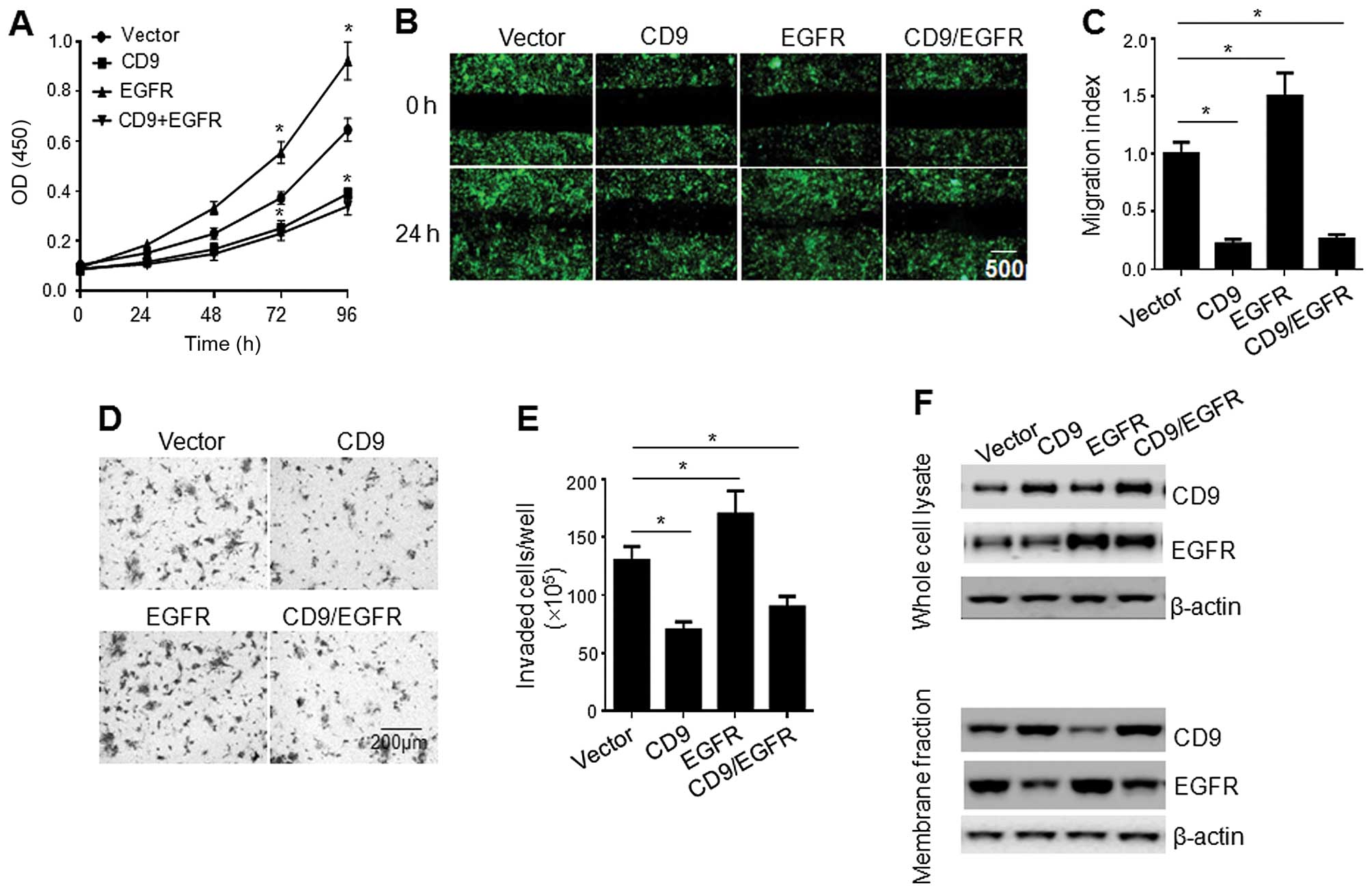 | Figure 5Effect of CD9 overexpression on the
proliferation and migration of the PaTu-8898s cells. (A)
Proliferation of the PaTu-8898s cells infected with the lentiviral
vector, CD9 construct, EGFR construct, or a combination of CD9 and
EGFR constructs (1:1 ratio). Data are the mean ± SEM from three
independent experiments. *p<0.05 compared with the
PaTu-8898s cells infected with the lentiviral vector. (B)
Scratch-wound assay showing the migration of the PaTu-8898s cells
infected with the lentiviral vector, CD9 construct, EGFR construct,
or a combination of CD9 and EGFR constructs (1:1 ratio). Shown are
representatives of three independent experiments with similar
results. (C) Quantification of the migration of the PaTu-8898s
cells infected with the lentiviral vector, CD9 construct, EGFR
construct, or a combination of CD9 and EGFR constructs (1:1 ratio).
Data are the mean ± SEM from three independent experiments.
*p<0.05 compared with the PaTu-8898s cells infected
with the lentiviral vector. (D) Invasion of the PaTu-8898s cells
infected with the lentiviral vector, CD9 construct, EGFR construct
or a combination of CD9 and EGFR constructs (1:1 ratio) in
Matrigel-coated invasion chambers. Shown are representatives of
three independent experiments with similar results. (E)
Quantification of the invasion of the PaTu-8898s cells infected
with the lentiviral vector, CD9 construct, EGFR construct, or a
combination of CD9 and EGFR constructs (1:1 ratio). Data are the
mean ± SEM from three independent experiments.
*p<0.05 compared with the PaTu-8898s cells infected
with the lentiviral vector. (F) Western blot analysis of CD9 and
EGFR in the whole cell lysate and membrane fraction of the
PaTu-8898s cells infected with the lentiviral vector, CD9
construct, EGFR construct, or a combination of CD9 and EGFR
constructs (1:1 ratio). Shown are representatives of three
independent experiments with similar results. |
CD9 knockdown promotes pancreatic cancer
cell proliferation, migration and invasion, which are reversed by
EGFR RNAi
We also examined the effect of CD9 knockdown on the
proliferation, migration and invasion of PaTu-8898t cells.
PaTu-8988t cells infected with the lentiviral shCD9-1 or shCD9-3
exhibited significantly increased proliferation (Fig. 6A), migration (Fig. 6b and C) and invasion (Fig. 6D and E) compared to the cells
infected with the lentiviral shCT, whereas the PaTu-8988t cells
infected with the lentiviral shEGFR exhibited significantly
decreased proliferation (Fig. 6A),
migration (Fig. 6b and C) and
invasion (Fig. 6D and E) compared
to the cells infected with the lentiviral shCT. Intriguingly, the
co-infection of the cells with the lentiviral EGFR with the
lentiviral shCD9-1 or shCD9-3 reversed the increases in cell
proliferation, migration and invasion compared to the cells
infected with the lentiviral CD9-shRNA1 or lentiviral CD9-shRNA3
alone (Fig. 6A–E). Western blot
analysis indicated that co-infection of the cells with lentiviral
EGFR shRNA with either lentiviral CD9-shRNA1 or lentiviral
CD9-shRNA3 resulted in a marked decrease in EGFR expression on the
cell surface compared to the cells infected with the lentiviral
CD9-shRNA1 or lentiviral CD9-shRNA3 alone (Fig. 6F).
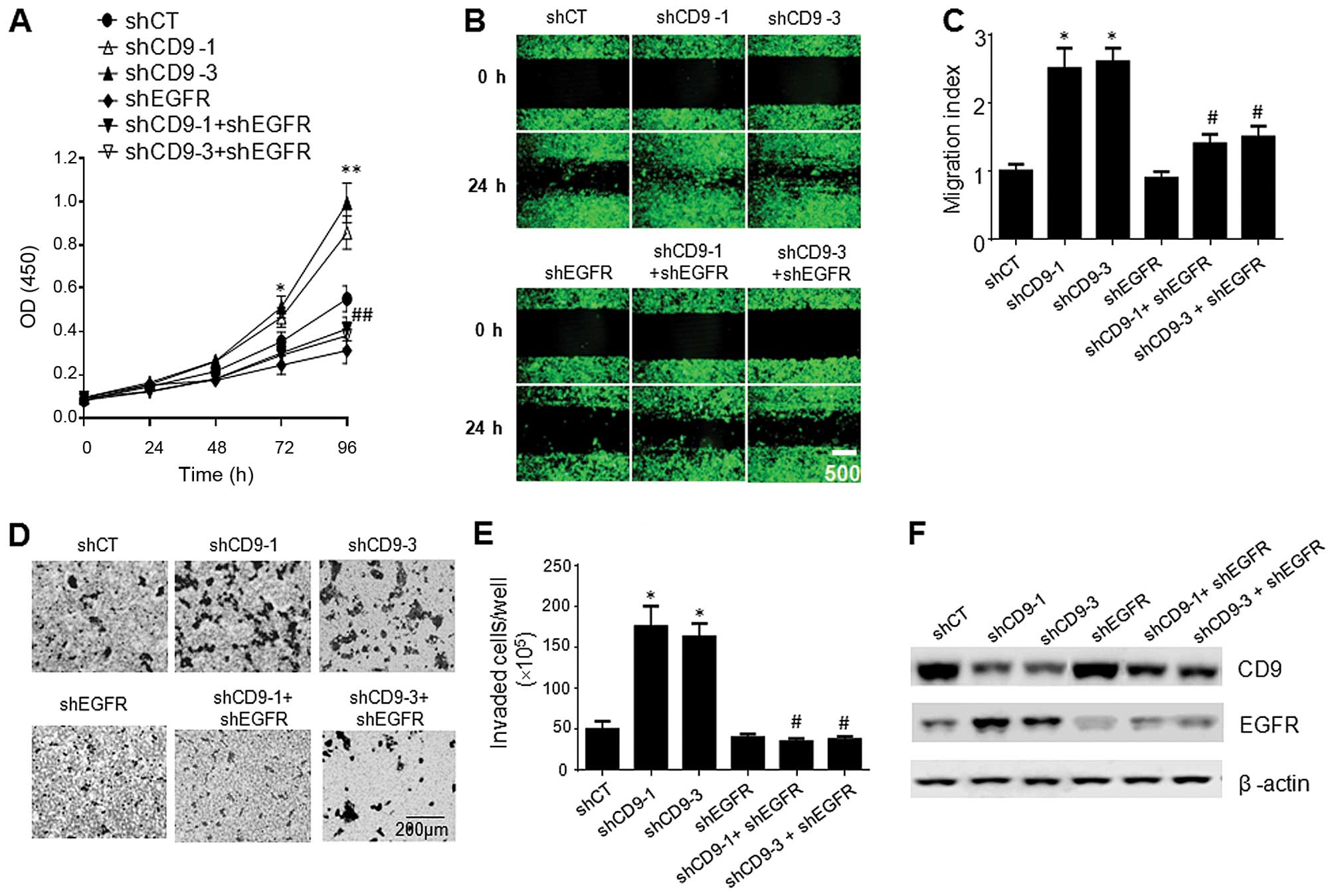 | Figure 6Effect of CD9 knockdown on the
proliferation and migration of the PaTu-8898t cells. (A)
Proliferation of the PaTu-8898t cells infected with the lentiviral
shCT, shCD9-1, shCD9-3, shEGFR, shCD9-1 + shEGFR (1:1 ratio), or
shCD9-3 + shEGFR (1:1 ratio). Data are the mean ± SEM from three
independent experiments. **p<0.01 compared with the
PaTu-8898s cells infected with the lentiviral shCT;
##p<0.01 compared with the PaTu-8898s cells infected
with the lentiviral shCD9-1 or shCD9-3 alone. (B) Scratch-wound
assay showing the migration of the PaTu-8898t cells infected with
the lentiviral shCT, shCD9-1, shCD9-3, shEGFR, shCD9-1 + shEGFR
(1:1 ratio), or shCD9-3 + shEGFR (1:1 ratio). Shown are
representatives of three independent experiments with similar
results. (C) Quantification of the migration of the PaTu-8898t
cells infected with the lentiviral shCT, shCD9-1, shCD9-3, shEGFR,
shCD9-1 + shEGFR (1:1 ratio), or shCD9-3 + shEGFR (1:1 ratio). Data
are the mean ± SEM from three independent experiments.
*p<0.05 compared with the PaTu-8898s cells infected
with the lentiviral shCT; #p<0.05 compared with the
PaTu-8898s cells infected with the lentiviral shCD9-1 or shCD9-3
alone. (D) Invasion of the PaTu-8898t cells infected with the
lentiviral shCT, shCD9-1, shCD9-3, shEGFR, shCD9-1 + shEGFR (1:1
ratio), or shCD9-3 + shEGFR (1:1 ratio) in the Matrigel-coated
invasion chambers. Shown are representatives of three independent
experiments with similar results. (E) Quantification of the
invasion of the PaTu-8898s cells infected with the lentiviral shCT,
shCD9-1, shCD9-3, shEGFR, shCD9-1 + shEGFR (1:1 ratio), or shCD9-3
+ shEGFR (1:1 ratio). Data are the mean ± SEM from three
independent experiments. *p<0.05 compared with the
PaTu-8898s cells infected with the lentiviral shCT;
#p<0.05 compared with the PaTu-8898s cells infected
with the lentiviral shCD9-1 or shCD9-3 alone. (F) Western blots
analysis of CD9 and EGFR in the whole cell lysate and membrane
fraction of the PaTu-8898s cells. p<0.05 compared with the
PaTu-8898s cells infected with the lentiviral shCT;
#p<0.05 compared with the PaTu-8898s cells infected
with the lentiviral shCD9-1 or shCD9-3 alone. Shown are
representatives of three independent experiments with similar
results. |
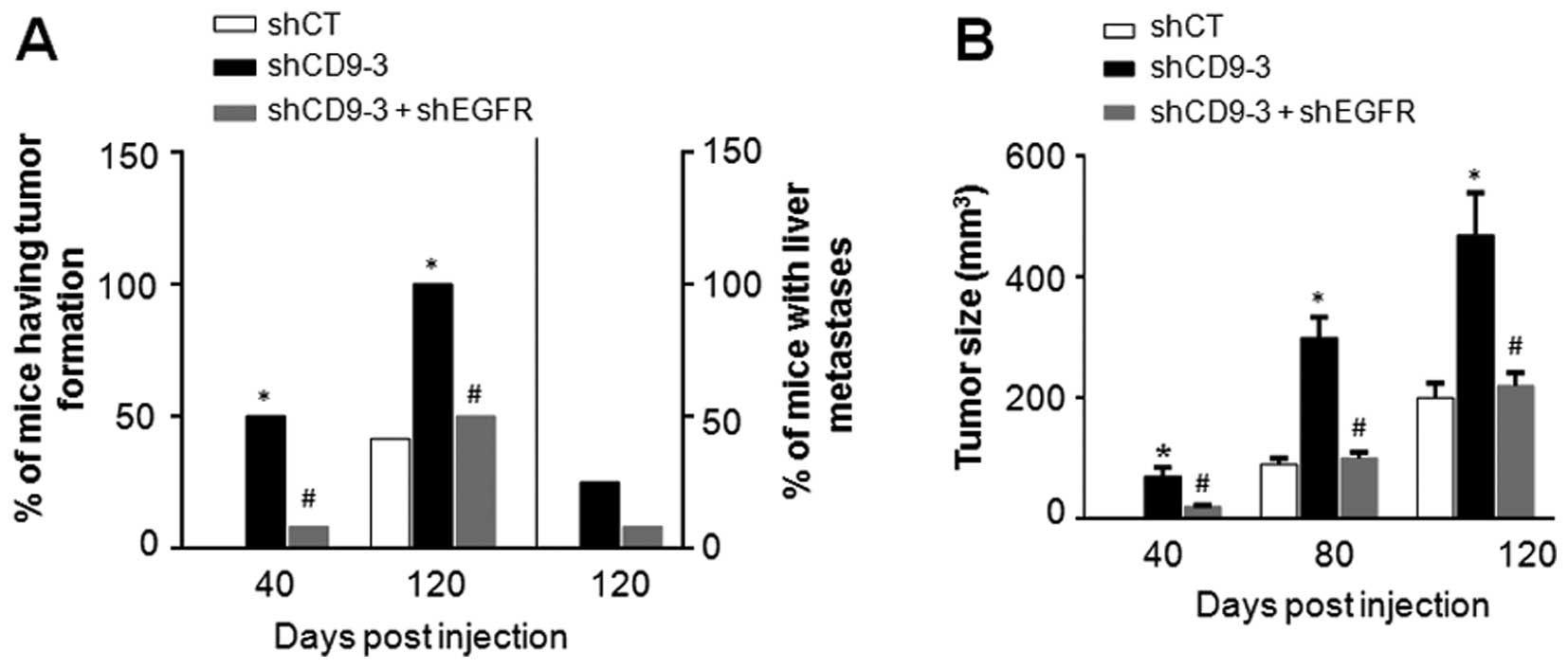
CD9 knockdown promotes tumor growth and
metastasis in vivo
To examine whether CD9 knockdown affects pancreatic
cancer growth and metastasis in vivo, PaTu-8988t cells
infected with the lentiviral shCT, shCD9-3, or a combination of
lentiviral shCD9-3 and shEGFR (1:1 ratio) were subcutaneously
implanted into the right flank of SCID mice. Among the animals
injected with the PaTu-8988t cells infected with the lentiviral
shCD9-3, 50% (6/12) of the mice had tumor formation on day 40, and
100% (12/12) of mice had tumor formation on day 120. However, among
the animals injected with the PaTu-8988t cells infected with a
combination of the lentiviral shCD9-3 and shEGFR, only 8% (1/12) of
mice had tumor formation on day 40, and 50% (6/12) of mice had
tumor formation on day 120. In contrast, among the animals injected
with the PaTu-8988t cells infected with the lentiviral shCT, no
mice had tumor formation on day 40, and only 41.6% (5/12) of mice
had tumor formation on day 120 (Fig.
7A). On day 120, three mice bearing PaTu-8988t cells infected
with the lentiviral shCD9-3 had liver metastasis, one mouse bearing
PaTu-8988t cells infected with a mixture of lentiviral shCD9-3 and
lentiviral shEGFR had liver metastasis, whereas none of the mice
bearing PaTu-8988t cells infected with the lentiviral shCT had
metastasis (Fig. 7A). At all of the
time points, a significantly increased tumor volume was observed in
animals bearing the PaTu-8988t cells infected with the lentiviral
shCD9-3 compared to those bearing the PaTu-8988t cells infected
with the lentiviral shCT (Fig. 7b).
However, a marked decrease in tumor volume was observed in the
animals bearing the PaTu-8988t cells infected with a mixture of
lentiviral shCD9-3 and lentiviral shEGFR compared to those bearing
the PaTu-8988t cells infected with the lentiviral shCD9-3 alone
(Fig. 7b).
Discussion
Echoing the previous finding that reduced CD9
expression in pancreatic cancer is associated with high tumor grade
and lymph node metastasis (16), we
demonstrated an inverse correlation between the CD9 expression
level and pancreatic cancer cell proliferation and migration using
the two closely related pancreatic cancer cell lines, PaTu-8898s
and PaTu-8898t. We further demonstrated an inverse correlation
between the CD9 level and the cell surface expression of EGFR. CD9
overexpression reduced the cell surface expression of EGFR,
associated with increased expression of dynamin-2, whereas CD9
knockdown increased the cell surface expression of EGFR, associated
with decreased expression of dynamin-2. While forced expression of
EGFR did not reverse CD9-overexpression-mediated inhibition of cell
proliferation, CD9 knockdown-promoted pancreatic cancer cell
proliferation and migration were blocked by EGFR RNAi both in
vitro and in vivo.
Mounting evidence indicates that CD9 negatively
regulates cell motility in various cancer cell types (24–26),
and CD9 knockout has been shown to increase spontaneous metastasis
(27). However, direct evidence for
a negative role of CD9 in pancreatic cancer motility has been
lacking until the present study. Here, we demonstrated that CD9
overexpression reduced cell motility, whereas its knockdown
promoted pancreatic cancer cell motility in vitro and
enhanced pancreatic cancer metastasis in vivo. In contrast
to the well-established role of CD9 in metastasis, very little is
known concerning the involvement of this tetraspanin in the process
of cancer cell proliferation. Here, we showed that CD9 knockdown
promoted pancreatic cancer cell proliferation, whereas its
overexpression inhibited cell proliferation. Our data are in line
with the previous finding in other cancer cells, such as colon
carcinoma cells (28) and small
cell lung cancer cells (29).
However, controversial results have also been reported. For
instance, in lymphoma cell lines, introduction of CD9 expression
resulted in significantly increased cell proliferation (30). This discrepancy is not surprising,
as tetraspanins have recently been postulated as both cancer
suppressors and promoters. This late appreciation is probably due
to their capacity to associate with various molecules, which they
recruit into special membrane microdomains, and their abundant
presence in tumor-derived small vesicles that aid intercellular
communication (31).
It is likely that CD9 plays a negative role in
pancreatic cancer proliferation and migration through, at least in
part, modulation of cell surface expression of EGFR. This notion is
based on the findings that i) there was an inverse correlation
noted between the CD9 level and cell surface expression of EGFR in
pancreatic cancer cells; ii) forced expression of CD9 reduced the
cell surface expression of EGFR, whereas knockdown of CD9 promoted
EGFR expression on the cell surface; and iii) the CD9
knockdown-mediated increase in pancreatic cancer cell proliferation
and migration were blocked by EGFR RNAi. EGFR activation via ligand
binding results in signaling through various pathways ultimately
resulting in cellular proliferation, survival, angiogenesis,
invasion and metastasis. EGFR overexpression is detected in up to
90% of pancreatic tumors (32), and
aberrant expression or activity of EGFR has been strongly linked to
the etiology of many other cancers including but not limited to
head and neck squamous cell carcinoma (HNSCC), non-small cell lung
cancer (NSCLC), colorectal cancer (CRC), breast cancer, pancreatic
cancer and brain cancer (21). In
line with our findings, previous studies have shown that a decrease
inCD9 and an increase in EGFR predict malignant progression of
cancer (33), that CD9 associates
with EGFR in hepatocellular carcinoma cells and Chinese hamster
ovary cancer cells transfected with EGFR/CD9, and that expression
of CD9 specifically attenuated EGFR signaling through
downregulation of surface expression of EGFR (34).
Based on our findings that CD9 knockdown did not
alter EGFR expression in total but rather increased its cell
surface expression, we propose that CD9 may play a role in EGFR
endocytosis. In support of this hypothesis, we showed that CD9
knockdown resulted in marked decrease in dynamin-2 expression,
whereas CD9 overexpression increased dynamin-2 expression. Dynamin,
a large GTPase that deforms lipid bilayers, is well known to be
involved in the endocytosis of EGFR (22). A recent study has shown that
dynamin-2 depletion leads to a strong inhibition of EGFR
endocytosis, robust enhancement of EGFR autophosphorylation and
ubiquitination, and slower kinetics of EGFR degradation, and that
dynamin-mediated endocytosis leads to attenuation of EGFR
activation and downstream signaling (23). Thus, CD9 may play a role in EGFR
endocytosis through modulation of dynamin expression. However, the
mechanisms by which CD9 modulates dynamin-2 expression are largely
unknown and should be investigated in future studies.
In summary, we demonstrated that CD9 downregulation,
which is evident in many cases of pancreatic cancer, promoted
pancreatic cancer cell proliferation and migration through at least
in part, enhancing the cell surface expression of EGFR, a
well-established target for pancreatic cancer therapy. CD9 played a
role in EGFR endocytosis likely through regulating the expression
of dynamin, which is known to regulate EGFR endocytosis, although
the underlying mechanisms remain to be investigated. These findings
lead to the hypothesis that targeting CD9 may offer therapeutic
benefits to pancreatic cancer patients.
Acknowledgments
This study was supported by grants from the Shanghai
Science and Technology Commission (no. 11JC1410000), Fund of the
Shanghai Health bureau (no. 20114315) and Training Plan of
Excellent Academic Researcher of Shanghai Tenth People’s Hospital
(nos. 12XSGG105 and 04.01.13037). We appreciate Dr Rui Li from
Genomeditech Co. Ltd. (www.genomeditech.com) for the generation of the
lentiviral particles.
References
|
1
|
Hezel AF, Kimmelman AC, Stanger BZ,
Bardeesy N and Depinho RA: Genetics and biology of pancreatic
ductal adenocarcinoma. Genes Dev. 20:1218–1249. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wray CJ, Ahmad SA, Matthews JB and Lowy
AM: Surgery for pancreatic cancer: Recent controversies and current
practice. Gastroenterology. 128:1626–1641. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Breslin TM, Hess KR, Harbison DB, Jean ME,
Cleary KR, Dackiw AP, Wolff RA, Abbruzzese JL, Janjan NA, Crane CH,
et al: Neoadjuvant chemoradiotherapy for adenocarcinoma of the
pancreas: Treatment variables and survival duration. Ann Surg
Oncol. 8:123–132. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
6
|
Berditchevski F: Complexes of tetraspanins
with integrins: More than meets the eye. J Cell Sci. 114:4143–4151.
2001.PubMed/NCBI
|
|
7
|
Boucheix C and Rubinstein E: Tetraspanins.
Cell Mol Life Sci. 58:1189–1205. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hemler ME: Tetraspanin proteins mediate
cellular penetration, invasion, and fusion events and define a
novel type of membrane microdomain. Annu Rev Cell Dev Biol.
19:397–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shaw AR, Domanska A, Mak A, Gilchrist A,
Dobler K, Visser L, Poppema S, Fliegel L, Letarte M and Willett BJ:
Ectopic expression of human and feline CD9 in a human B cell line
confers beta 1 integrin-dependent motility on fibronectin and
laminin substrates and enhanced tyrosine phosphorylation. J Biol
Chem. 270:24092–24099. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones PH, Bishop LA and Watt FM:
Functional significance of CD9 association with beta 1 integrins in
human epidermal keratinocytes. Cell Adhes Commun. 4:297–305. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Inui S, Higashiyama S, Hashimoto K,
Higashiyama M, Yoshikawa K and Taniguchi N: Possible role of
coexpression of CD9 with membrane-anchored heparin-binding EGF-like
growth factor and amphiregulin in cultured human keratinocyte
growth. J Cell Physiol. 171:291–298. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berditchevski F and Odintsova E:
Characterization of integrintetraspanin adhesion complexes: Role of
tetraspanins in integrin signaling. J Cell Biol. 146:477–492. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baudoux B, Castanares-Zapatero D,
Leclercq-Smekens M, Berna N and Poumay Y: The tetraspanin CD9
associates with the integrin alpha6beta4 in cultured human
epidermal keratinocytes and is involved in cell motility. Eur J
Cell Biol. 79:41–51. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi W, Fan H, Shum L and Derynck R: The
tetraspanin CD9 associates with transmembrane TGF-alpha and
regulates TGF-alpha-induced EGF receptor activation and cell
proliferation. J Cell Biol. 148:591–602. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Crnogorac-Jurcevic T, Efthimiou E, Capelli
P, Blaveri E, baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa
A, et al: Gene expression profiles of pancreatic cancer and stromal
desmoplasia. Oncogene. 20:7437–7446. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sho M, Adachi M, Taki T, Hashida H,
Konishi T, Huang CL, Ikeda N, Nakajima Y, Kanehiro H, Hisanaga M,
et al: Transmembrane 4 superfamily as a prognostic factor in
pancreatic cancer. Int J Cancer. 79:509–516. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grønborg M, Kristiansen TZ, Iwahori A,
Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins
MG, et al: Biomarker discovery from pancreatic cancer secretome
using a differential proteomic approach. Mol Cell Proteomics.
5:157–171. 2006. View Article : Google Scholar
|
|
18
|
Elsässer HP, Lehr U, Agricola B and Kern
HF: Establishment and characterisation of two cell lines with
different grade of differentiation derived from one primary human
pancreatic adenocarcinoma. Virchows Arch B Cell Pathol Incl Mol
Pathol. 61:295–306. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen G, Kronenberger P, Teugels E, Umelo
IA and De Grève J: Targeting the epidermal growth factor receptor
in non-small cell lung cancer cells: The effect of combining RNA
interference with tyrosine kinase inhibitors or cetuximab. BMC Med.
10:282012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang CC, Park AY and Guan JL: In vitro
scratch assay: A convenient and inexpensive method for analysis of
cell migration in vitro. Nat Protoc. 2:329–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brand TM, Iida M, Li C and Wheeler DL: The
nuclear epidermal growth factor receptor signaling network and its
role in cancer. Discov Med. 12:419–432. 2011.PubMed/NCBI
|
|
22
|
Sousa LP, Lax I, Shen H, Ferguson SM, De
Camilli P and Schlessinger J: Suppression of EGFR endocytosis by
dynamin depletion reveals that EGFR signaling occurs primarily at
the plasma membrane. Proc Natl Acad Sci USA. 109:4419–4424. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schroeder B, Weller SG, Chen J, Billadeau
D and McNiven MA: A Dyn2-CIN85 complex mediates degradative traffic
of the EGFR by regulation of late endosomal budding. EMBO J.
29:3039–3053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen S, Sun Y, Jin Z and Jing X:
Functional and biochemical studies of CD9 in fibrosarcoma cell
line. Mol Cell Biochem. 350:89–99. 2011. View Article : Google Scholar
|
|
25
|
Fan J, Zhu GZ and Niles RM: Expression and
function of CD9 in melanoma cells. Mol Carcinog. 49:85–93.
2010.
|
|
26
|
Ono M, Handa K, Withers DA and Hakomori S:
Motility inhibition and apoptosis are induced by
metastasis-suppressing gene product CD82 and its analogue CD9, with
concurrent glycosylation. Cancer Res. 59:2335–2339. 1999.PubMed/NCBI
|
|
27
|
Copeland BT, Bowman MJ, Boucheix C and
Ashman LK: Knockout of the tetraspanin Cd9 in the TRAMP model of de
novo prostate cancer increases spontaneous metastases in an
organ-specific manner. Int J Cancer. 133:1803–1812. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ovalle S, Gutiérrez-López MD, Olmo N,
Turnay J, Lizarbe MA, Majano P, Molina-Jiménez F, López-Cabrera M,
Yáñez-Mó M, Sánchez-Madrid F, et al: The tetraspanin CD9 inhibits
the proliferation and tumorigenicity of human colon carcinoma
cells. Int J Cancer. 121:2140–2152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng R, Yano S, Zhang H, Nakataki E,
Tachibana I, Kawase I, Hayashi S and Sone S: CD9 overexpression
suppressed the liver metastasis and malignant ascites via
inhibition of proliferation and motility of small-cell lung cancer
cells in NK cell-depleted SCID mice. Oncol Res. 15:365–372.
2005.
|
|
30
|
Herr MJ, Longhurst CM, Baker B, Homayouni
R, Speich HE, Kotha J and Jennings LK: Tetraspanin CD9 modulates
human lymphoma cellular proliferation via histone deacetylase
activity. Biochem Biophys Res Commun. 447:616–620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zöller M: Tetraspanins: Push and pull in
suppressing and promoting metastasis. Nat Rev Cancer. 9:40–55.
2009. View
Article : Google Scholar
|
|
32
|
Troiani T, Martinelli E, Capasso A,
Morgillo F, Orditura M, De Vita F and Ciardiello F: Targeting EGFR
in pancreatic cancer treatment. Curr Drug Targets. 13:802–810.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nankivell P, Williams H, McConkey C,
Webster K, High A, MacLennan K, Senguven B, Rabbitts P and Mehanna
H: Tetraspanins CD9 and CD151, epidermal growth factor receptor and
cyclooxygenase-2 expression predict malignant progression in oral
epithelial dysplasia. Br J Cancer. 109:2864–2874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Murayama Y, Shinomura Y, Oritani K,
Miyagawa J, Yoshida H, Nishida M, Katsube F, Shiraga M, Miyazaki T,
Nakamoto T, et al: The tetraspanin CD9 modulates epidermal growth
factor receptor signaling in cancer cells. J Cell Physiol.
216:135–143. 2008. View Article : Google Scholar : PubMed/NCBI
|