Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignant disease worldwide and is the second leading cause
of cancer-related deaths (~700,000 annually) (1–3).
Although some patients will benefit from surgery (4), the long-term survival of most is
unsatisfactory due to the high incidence of postoperative
recurrence and metastasis (5).
Unfortunately, effective measures to predict or prevent recurrence
and metastasis of HCC are unavailable, mainly since the detailed
mechanisms of postoperative metastasis and relapse remain to be
determined.
Due to the absence of widely accepted nucleic acid
and protein markers for predicting the prognosis of HCC, we focused
our research on microRNAs (miRNAs), which are highly conserved
RNAs, comprised of ~19–22 nucleotides. iRNAs inhibit gene
expression at the posttranscriptional level by binding to the 3′
untranslated region (3′UTR) of target mRNAs (6). Different miRNAs are involved in the
regulation of diverse physiological processes or contribute to
pathological processes. Among these miRNAs, more than 50% are
encoded by cancer-associated genomic regions or fragile sites
(7), indicating that dysregulation
of miRNA expression plays important roles in oncogenesis.
The epithelial-mesenchymal transition (EMT), which
is implicated in pathologies such as organ fibrosis, tumor
progression and metastasis (8,9), is an
important step in HCC metastasis (10). Among numerous miRNAs, miR-99b, which
is encoded by chromosome 19, has attracted our research interest,
since it promotes the EMT of normal murine mammary gland cells and
increases their ability to migrate (11). Moreover, the expression of miR-99b
is upregulated significantly in tumors such as multiple myeloma
(12,13) and esophageal cancer (14) and is associated with lymph node
metastasis (15). These findings
led us to propose the hypothesis that miR-99b functions as an
oncogene and contributes to the metastatic phenotype of HCC,
although we are unaware of any studies indicating a role of miR-99b
in HCC.
To test this hypothesis, we conducted experiments to
detect the expression of miR-99b in HCC samples. We uncovered
compelling evidence that miR-99b may serve as a prognostic marker
for overall survival (OS) and disease-free survival (DFS) of
patients with HCC. We analyzed the role of miR-99b in promoting the
migration and metastasis of HCC cells. Furthermore, using a
dual-luciferase assay, we showed in the present study that the mRNA
encoding claudin 11 (CLDN11), which is an integral membrane protein
and a component of tight junction strands, was a direct target of
miR-99b and we further demonstrated that the expression of
CLDN11 was regulated by miR-99b in HCC cell lines and in HCC
tissues. These results indicated that overexpression of miR-99b
promotes metastatic growth and the migration of HCC cells through
inhibition of CLDN11 expression, thus providing a better
understanding of the underlying mechanism of the role of miR-99b in
the pathogenesis of HCC.
Materials and methods
Human tumor samples
A total of 104 pairs of HCC and adjacent non-tumor
liver tissue (ANLT) samples were obtained from patients who
underwent hepatectomy between January 2007 and December 2011 at
Xiangya Hospital, Central South University in China. Sixteen cases
of normal liver tissues were obtained from tissues distant from the
liver hemangioma tissues. Tumor or normal liver tissues were
snap-frozen immediately after surgery and stored in liquid nitrogen
at −80°C. Histopathology was evaluated by pathologists at the
Pathology Department of Xiangya Hospital. Patient informed consents
were obtained from every patient before surgery. The study protocol
was approved by the Ethics Committee of Central South
University.
Cell lines
HCC cell lines (HepG2, HUH7, HCCLM3 and SMMC7721)
and a normal liver cell line (L02) were obtained from the Central
Experimental Laboratory of Xiangya Medical School in Central South
University and cultured in Dulbecco′s modified Eagle′s medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) (both from
Sigma-Aldrich, St. Louis, MO, USA), 1% penicillin/streptomycin (50
U/ml) and 1% L-glutamine (2 mmol/l) (both from Invitrogen, Basel,
Switzerland). All cell lines were cultured at 37°C in humidified
air with 5% CO2.
Real-time PCR
Total RNA of HCC tissues and cultured cells was
extracted using TRIzol (Beyotime Institute of Biotechnology,
Beijin, China) according to the manufacturer′s instructions.
Reverse transcription was performed using the First Strand cDNA
Synthesis ReverTra Ace kit (Toyobo, Tokyo, Japan). Real-time PCR
was performed with the SYBR-Green PCR Master Mix (Roche Applied
Science, Indianapolis, IN, USA) according to the manufacturer′s
instructions using the ABI 7300 Real-Time PCR System. U6 snRNA was
measured as an internal control for miRNA and GAPDH was used for
normalization for mRNA. The primers for CLDN11 were: forward primer
5′-ACCATCGTGAGCTTTGGCTA-3′ and reverse primer
5′-GCTAGAGCCCGCAGTGTAGT-3′.
Transfection and luciferase assay
The 293T cell line was seeded in 24-well plates and
infected with miR-99b overexpression lentivirus. pGL3 vector with
mutant (Mut) CLDN11-3′UTR or wild-type (Wt) CLDN11-3′UTR as well as
2 ng/well of plasmid pRL-TK were cotransfected using Lipofectamine
LTX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer′s
instructions. Three days later, the cell lines were extracted and
the relative luciferase activity assay was performed using a
Dual-Luciferase Reporter Assay system (Promega, Madison, WI,
USA).
Lentivirus
The miR-99b overexpression lentivirus and the
miR-99b inhibition lentivirus as well as their control lentiviruses
were purchased from GenePharma Biotechnology Corp. (Shanghai,
China). The conservation and usage of the lentiviruses were
performed according to the manufacturer′s instructions. GFP in the
HCC cells was used to verify the infection efficiency of the
cells.
Matrigel invasion assays
Cell motility and migratory abilities were assessed
using a Transwell plate with Matrigel (Corning Life Sciences,
Bedford, MA, USA). Matrigel invasion assays were performed as
previously described. Cells (2×104) were suspended in
serum-free culture medium and seeded into the upper chamber.
Culture medium supplemented with 10% FBS was added to the lower
chamber. Cells were incubated in a CO2 incubator at 37°C
for 24 h. Cells in the upper chamber and Matrigel were removed.
Cells that migrated to the underside of the membrane were fixed and
stained with 0.1% crystal violet. Cells that migrated through the
membrane pores to the lower surface of the membrane were counted in
10 microscopic fields. Each experiment was performed in replicate
inserts.
Western blot analysis
Total cellular lysates were obtained by lysing cells
in RIPA buffer containing proteinase and phosphatase inhibitors.
Proteins were extracted from the total cellular lysates. Extracts
(5–30 µg) were separated on 12% polyacrylamide gels and
transferred onto PVDF membranes. Western blot analyses were
performed as previously described (16). Anti-CLDN11 antibody (Santa Cruz) and
β-actin antibody (Sigma) were used in western blot analysis
according to the manufacturer′s instructions.
Immunohistochemistry (IHC) analysis
The paraffin-embedded tissue samples from
postoperative patients were cut into 5-µm sections. Then the
samples were deparaffinized in xylene and rehydrated using a series
of graded alcohols. Slides were blocked with 10% goat serum before
incubating with anti-CLDN11 antibody (dilution 1:50) (Santa Cruz).
The samples were incubated overnight with a primary antibody, and
subsequent secondary antibody followed by 3,3′-diaminobenzidine
(DAB).
Cell migration assay
Cell motility and migratory abilities were assessed
by Transwell assay (Corning Life Sciences). Cell migration assays
were performed as previously described. Cells (2×104)
were suspended in serum-free culture medium and seeded into the
upper chamber. Culture medium supplemented with 10% FBS was added
to the lower chamber. Cells were incubated in a CO2
incubator at 37°C for 24 h. Cells that migrated to the underside of
the membrane were fixed and stained with 0.1% crystal violet. Cells
that migrated through the membrane pores to the lower surface of
the membrane were counted in 10 microscopic fields. Each experiment
was performed in replicate inserts.
Statistical analysis
Statistical analysis was performed using SPSS
version 16.0 (SPSS, Inc., Chicago, IL, USA). Values are expressed
as the mean ± SD. The difference between groups was analyzed using
an independent Student's t-test when comparing only two groups or
one-way ANOVA analysis of variance when comparing more than two
groups. Fisher's exact test was adopted for statistical analysis of
categorical data. Spearman correlation test was used for analyzing
the correlations between the miR-99b expression level and the
clinical and pathological variables. Kaplan-Meier's method was used
to construct survival curves. Equivalences of the survival curves
were tested by log-rank statistics. The independent risk factors
for OS and DFS were identified by Cox proportional hazard
regression model. A two-tailed P<0.05 was considered to a
statistically significant result.
Results
miR-99b is expressed at elevated levels
in HCC cell lines and in tissues of patients with HCC
We collected 104 pairs of HCC tissues and adult
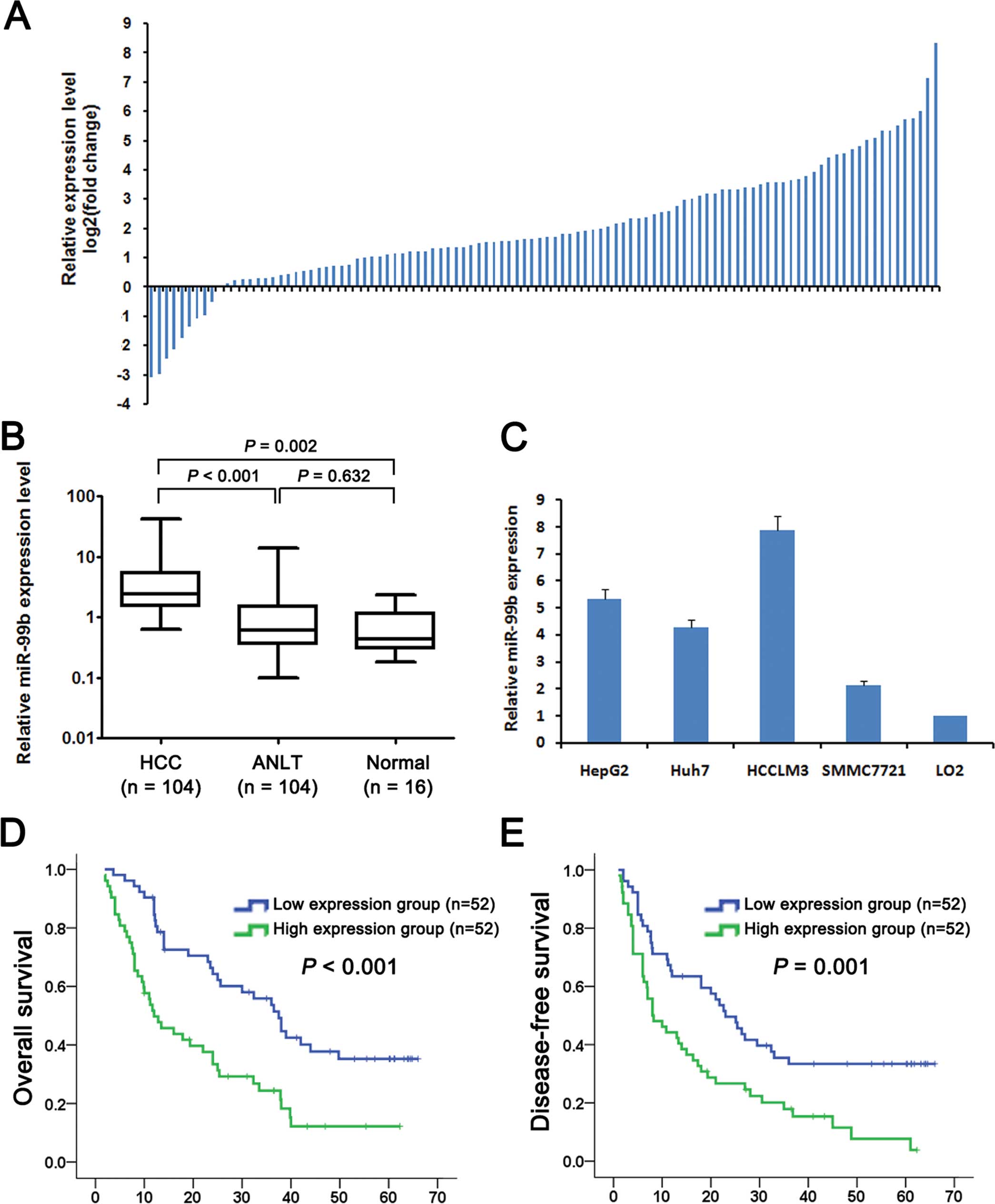
normal liver tissue (ANLT) to assess miR-99b expression. miR-99b
was expressed at >2-fold higher levels in 75 (72.1%) HCC tissues
compared with the level in the ANLT samples (Fig. 1A). The median level of miR-99b
expression in the HCC tissues was significantly higher (3.8-fold)
compared with that of the ANLT samples (5.53±0.80 vs. 1.44±0.24,
P<0.001) or ~8.0-fold higher compared with that in the normal
liver tissues (5.53±0.80 vs. 0.69±0.15, P=0.002) (Fig. 1B). However, there was no significant
difference between miR-99b levels in the ANLT and normal liver
tissues (1.44±0.24 vs. 0.69±0.15, P=0.632). We determined miR-99b
expression levels in four HCC cell lines (HepG2, Huh7, HCCLM3 and
SMMC7721) and a normal liver cell line (LO2) that served as a
control. miR-99b was expressed in the four HCC lines at levels
significantly higher compared with the LO2 cells (5.34-,4.28-,
7.88- and 2.15-fold, respectively) (Fig. 1C).
High levels of miR-99b expression predict
poor prognosis of patients with HCC
We determined the levels of miR-99b expression in
tissues acquired from 104 patients with HCC and classified the
patients into a high expression (greater than the median) and a low
expression (less than the median) group, and analyzed the group
clinicopathological characteristics and clinical courses. We
acquired data for gender, age, tumor size, tumor number,
α-fetoprotein (AFP), hepatitis B virus (HBV) infection, liver
cirrhosis, capsule formation, Edmondson stage, microvascular
invasion and TNM stage, and found a significant association with
capsule formation (P=0.030) and microvascular invasion (P=0.017)
(Table I).
 | Table ICorrelation between miR-99b expression
and clinico-pathological features of the HCC patients. |
Table I
Correlation between miR-99b expression
and clinico-pathological features of the HCC patients.
| Parameters | Cases | miR-99b expression
| χ2 | P-value |
|---|
| Low | High |
|---|
| Gender | | | | | |
| Female | 13 | 7 | 6 | | |
| Male | 91 | 45 | 46 | 0.088 | 0.767 |
| Age (years) | | | | | |
| <60 | 84 | 43 | 41 | | |
| >60 | 20 | 9 | 11 | 0.248 | 0.619 |
| Tumor size
(cm) | | | | | |
| <5 | 18 | 10 | 8 | | |
| ≥5 | 86 | 42 | 44 | 0.269 | 0.604 |
| Tumor no. | | | | | |
| Single | 56 | 30 | 26 | | |
| Multiple | 48 | 22 | 26 | 0.619 | 0.431 |
| AFP (ng/ml) | | | | | |
| <20 | 39 | 18 | 21 | | |
| ≥20 | 65 | 34 | 31 | 0.369 | 0.543 |
| HBV infection | | | | | |
| Negative | 10 | 6 | 4 | | |
| Positive | 92 | 46 | 46 | 1.390 | 0.499 |
| Liver
cirrhosis | | | | | |
| Absence | 22 | 14 | 8 | | |
| Presence | 82 | 38 | 44 | 2.075 | 0.150 |
| Capsular
formation | | | | | |
| Presence | 47 | 29 | 18 | | |
| Absence | 57 | 23 | 34 | 4.697 | 0.030 |
| Edmondson
stage | | | | | |
| I–II | 43 | 20 | 23 | | |
| III–IV | 61 | 32 | 29 | 0.357 | 0.691 |
| Microvascular
invasion | | | | | |
| No | 59 | 36 | 23 | | |
| Yes | 45 | 16 | 29 | 6.620 | 0.017 |
| TNM stage | | | | | |
| I | 38 | 22 | 16 | | |
| II | 25 | 12 | 13 | | |
| III | 41 | 18 | 23 | 1.597 | 0.450 |
To assess the feasibility of miR-99b as a marker for
postoperative prognosis, we performed univariate and multivariate
analyses of OS (Table II) and DFS
(Table III). Univariate analysis
revealed that the tumor size (P=0.006), liver cirrhosis (P=0.010),
microvascular invasion (P=0.001), TNM stage (P=0.002) and the
miR-99b expression (P<0.001) were significantly associated with
the OS of patients with HCC and that liver cirrhosis (P=0.014),
microvascular invasion (P=0.004), TNM stage (P=0.003) and miR-99b
expression (P=0.002) were significantly associated with DFS.
 | Table IIUnivariate and multivariate analyses
of overall survival by a Cox proportional hazards regression
model. |
Table II
Univariate and multivariate analyses
of overall survival by a Cox proportional hazards regression
model.
| Parameters | Case | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender | | | | | |
| Female | 13 | 1 | | | |
| Male | 91 | 1.079
(0.537–2.169) | 0.831 | | NA |
| Age (years) | | | | | |
| <60 | 84 | 1 | | | |
| >60 | 20 | 1.023
(0.571–1.832) | 0.940 | | NA |
| Tumor size
(cm) | | | | | |
| <5 | 18 | 1 | | 1 | |
| ≥5 | 86 | 3.201
(1.385–7.397) | 0.006 | 2.550
(1.140–5.704) | 0.023 |
| Tumor no. | | | | | |
| Single | 56 | 1 | | | |
| Multiple | 48 | 1.563
(0.986–2.476) | 0.057 | | NA |
| AFP (ng/ml) | | | | | |
| <20 | 39 | 1 | | | |
| ≥20 | 65 | 1.264
(0.779–2.052) | 0.343 | | NA |
| HBV infection | | | | | |
| Negative | 10 | 1 | | | |
| Positive | 92 | 1.714
(0.736–3.990) | 0.211 | | NA |
| Liver
cirrhosis | | | | | |
| Absence | 22 | 1 | | 1 | |
| Presence | 82 | 2.297
(1.225–4.307) | 0.010 | 2.793
(1.442–5.413) | 0.002 |
| Capsular
formation | | | | | |
| Presence | 47 | 1 | | | |
| Absence | 57 | 1.433
(0.896–2.292) | 0.133 | | NA |
| Edmondson
stage | | | | | |
| I–II | 43 | 1 | | | |
| III–IV | 61 | 0.773
(0.478–1.226) | 0.274 | | NA |
| Microvascular
invasion | | | | | |
| No | 59 | 1 | | 1 | |
| Yes | 45 | 2.138
(1.346–3.394) | 0.001 | 1.899
(1.106–3.258) | 0.020 |
| TNM stage | | | | | |
| I–II | 63 | 1 | | 1 | |
| III | 41 | 1.526
(1.167–1.994) | 0.002 | 1.244
(0.907–1.706) | 0.175 |
| miR-99b
expression | | | | | |
| Low | 52 | 1 | | 1 | |
| High | 52 | 2.408
(1.501–3.864) |
<0.001 | 2.447
(1.506–3.976) |
<0.001 |
 | Table IIIUnivariate and multivariate analyses
of disease-free survival by a Cox proportional hazards regression
model. |
Table III
Univariate and multivariate analyses
of disease-free survival by a Cox proportional hazards regression
model.
| Parameters | Case | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Gender | | | | | |
| Female | 13 | 1 | | | |
| Male | 91 | 0.969
(0.484–1.941) | 0.930 | | NA |
| Age (years) | | | | | |
| <60 | 84 | 1 | | | |
| >60 | 20 | 1.002
(0.984–1.020) | 0.816 | | NA |
| Tumor size
(cm) | | | | | |
| <5 | 18 | 1 | | | |
| ≥5 | 86 | 1.836
(0.970–3.478) | 0.062 | | NA |
| Tumor no. | | | | | |
| Single | 56 | 1 | | | |
| Multiple | 48 | 1.538
(0.990–2.390) | 0.056 | | NA |
| AFP (ng/ml) | | | | | |
| <20 | 39 | 1 | | | |
| ≥20 | 65 | 1.247
(0.787–1.976) | 0.348 | | NA |
| HBV infection | | | | | |
| Negative | 10 | 1 | | | |
| Positive | 92 | 1.680
(0.756–3.773) | 0.203 | | NA |
| Liver
cirrhosis | | | | | |
| Absence | 22 | 1 | | 1 | |
| Presence | 82 | 2.120
(1.164–3.858) | 0.014 | 2.091
(1.133–3.859) | 0.018 |
| Capsular
formation | | | | | |
| Presence | 47 | 1 | | | |
| Absence | 57 | 1.307
(0.837–2.041) | 0.239 | | NA |
| Edmondson
stage | | | | | |
| I–II | 43 | 1 | | | |
| III–IV | 61 | 0.712
(0.456–1.114) | 0.137 | | NA |
| Microvascular
invasion | | | | | |
| No | 59 | 1 | | 1 | |
| Yes | 45 | 1.919
(1.233–2.986) | 0.004 | 1.522
(0.923–2.509) | 0.099 |
| TNM stage | | | | | |
| I–II | 63 | 1 | | 1 | |
| III–IV | 41 | 1.469
(1.135–1.902) | 0.003 | 1.326
(0.980–1.794) | 0.067 |
| miR-99b
expression | | | | | |
| Low | 52 | 1 | | 1 | |
| High | 52 | 2.049
(1.308–3.208) | 0.002 | 1.694
(1.068–2.685) | 0.025 |
Multivariate analysis revealed that tumor size
(P=0.023), liver cirrhosis (P=0.002), microvascular invasion
(P=0.020) and miR-99b expression (P<0.001) were independent
prognostic indicators of OS and that miR-99b expression (P=0.025)
and liver cirrhosis (P=0.018) were independent prognostic
indicators of DFS. When we performed Kaplan-Meier analysis, we
found that OS and DFS were shorter for patients with HCC who
expressed high levels of miR-99b (Fig.
1D and E). The 1-, 3- and 5-year OS values of patients in the
high miR-99b expression group were 51.5, 24.2 and 12.1%,
respectively, and significantly lower compared with those of
patients in the low miR-99b expression group (90.3, 55.5 and 34.7%,
respectively, P<0.001). Furthermore, the values for the 1-, 3-
and 5-year DFS of the high miR-99b group were 44.2, 18.0 and 8.3%,
respectively, and significantly lower compared with those of the
low miR-99b group (65.4, 35.5 and 33.4%, respectively;
P=0.001).
miR-99b promotes the metastatic phenotype
of HCC cell lines
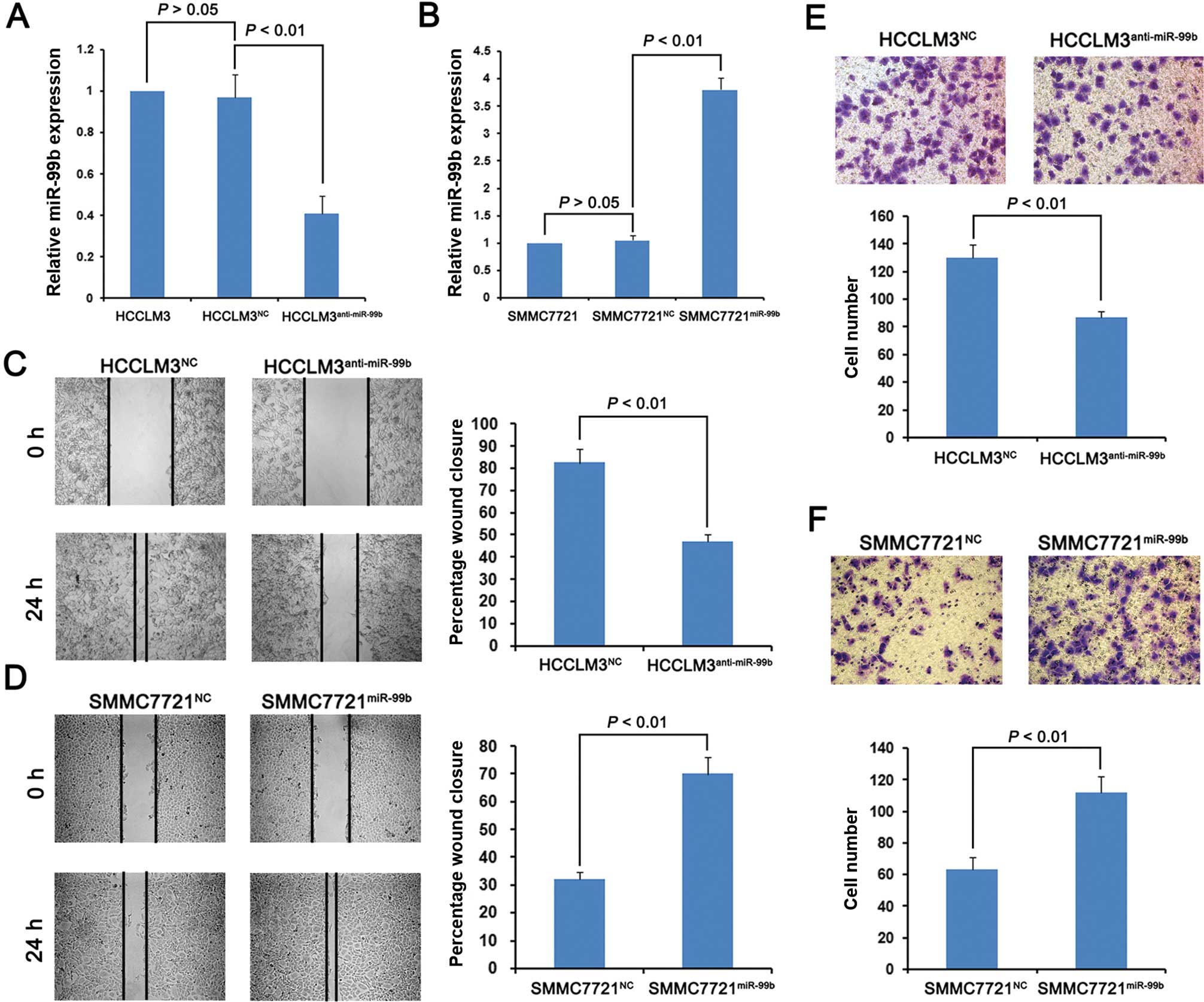
The HCC HCCLM3 and SMMC7721 cell lines that
expressed the highest and lowest levels of miR-99b, respectively,
among the four cell lines tested, were infected with lentiviruses
that expressed the antisense version of miR-99b or overexpressed
miR-99b as well as with the respective cognate negative control
(NC). We isolated stably transduced cell lines of each infectant,
designated HCCLM3anti-miR-99b,
SMMC7721miR-99b, HCCLM3NC and
SMMC7721NC. Real-time PCR analysis showed that miR-99b
HCCLM3anti-miR-99b cells expressed miR-99b at a level
40% lower compared with that of the HCCLM3NC cells
(Fig. 2A, P<0.01). Furthermore,
SMMC7721miR-99b cells expressed miR-99b at a level
3-fold higher compared with the SMMC7721NC cells
(Fig. 2B, P<0.01).
Using wound healing and Matrigel invasion assays, we
evaluated whether the levels of miR-99b expression in these cell
lines imparted a metastatic phenotype.
HCCLM3anti-miR-99b cells promoted wound healing that was
slower compared with the HCCLM3NC cells. Similarly, the
SMMC7721miR-99b cells promoted more rapid wound healing
compared with the SMMC7721NC cells (Fig. 2D). The Matrigel assays revealed that
the HCCLM3anti-miR-99b cells migrated more slowly
compared with the HCCLM3NC cells (Fig. 2E), while the
SMMC7721miR-99b migrated faster compared with the
SMMC7721NC cells (Fig.
2F). These data indicate that cells with elevated levels of
miR-99b exhibited phenotypic characteristics of metastatic
cells.
miR-99b directly inhibits CLDN11
expression
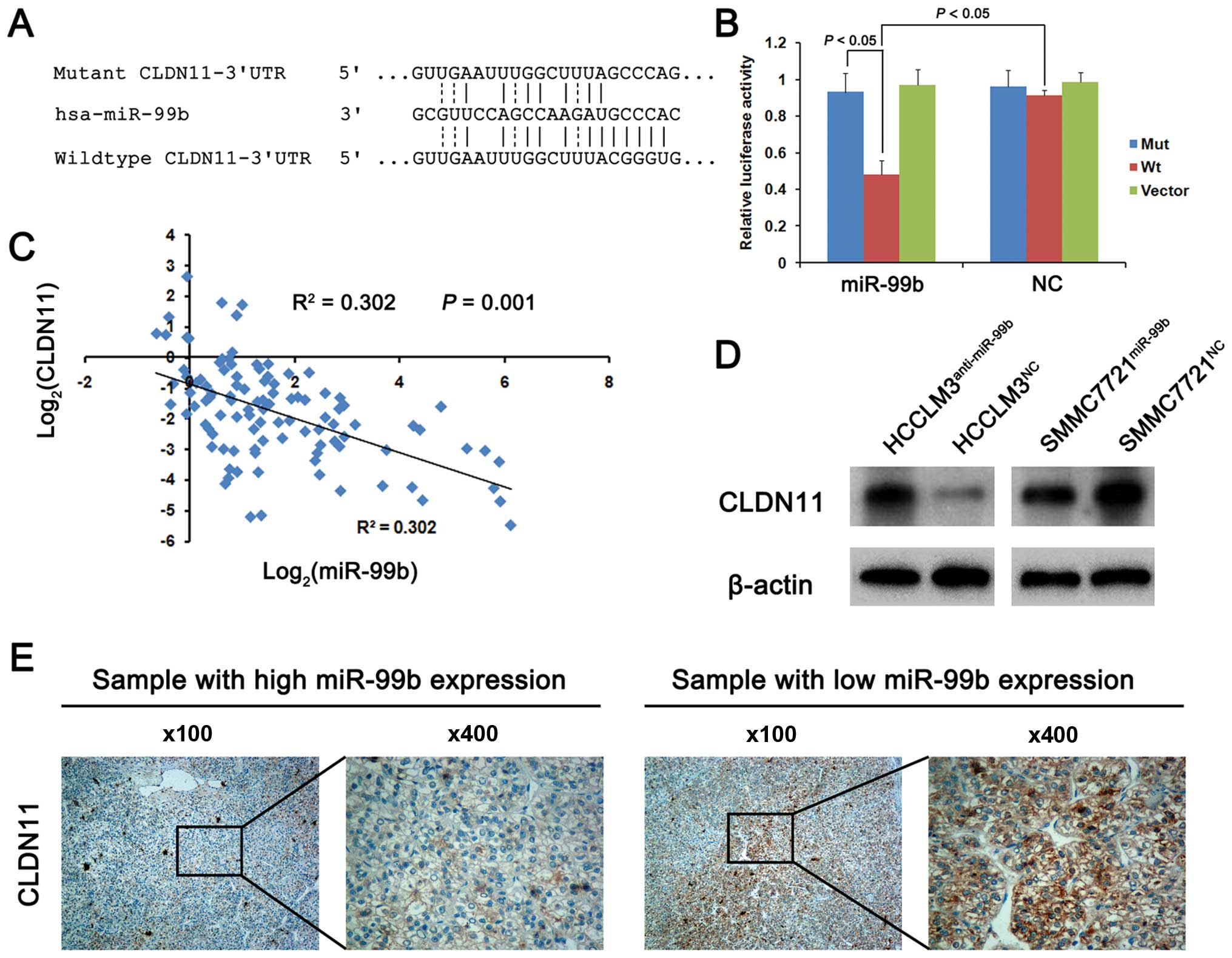
To determine whether miR-99b expression contributes
to the metastatic phenotype of HCC cells, we searched the
TargetScan (www.targetscan.org), PicTar
(www.pictar.org) and miRanda (www.microrna.org) databases for genes that are
potential targets of miR-99b. We focused on CLDN11, since it
was predicted as a target by our analysis of each database. To
determine whether the 3′UTR of CLDN11 mRNA binds miR-99b, we
mutated five bases of the CLDN11-3′UTR (Fig. 3A). The Wt and Mut sequences were
each cloned into the luciferase reporter vector pGL3 and each was
used with the control vector pRL-TK to cotransfect 293T cells. The
result of the dual-luciferase assay showed that miR-99b
overexpression significantly inhibited the luciferase activity of
Wt 3′UTR of CLDN11 mRNA but not that of Mut 3′-UTR of
CLDN11 mRNA (Fig. 3B).
Furthermore, there was a significant inverse association between
the expression of miR-99b and CLDN11 mRNA in the 104 matched
pairs of HCC specimens (R2=0.302, P=0.001; Fig. 3C). To determine the functional
significance of CLDN11 mRNA expression, we performed western
blot analyses of the levels of CLDN11 in the HCCLM3 and SMMC7721
cell lines infected with the lentivirus expression vectors as
previously described, and found that knockdown of miR-99b
expression significantly increased CLDN11 levels in the HCCLM3
cells. In contrast, CLDN11 levels were decreased in the infected
cells that expressed miR-99b (Fig.
3D). Thus, these data indicate that miR-99b negatively
regulates the expression of CLDN11 mRNA by directly
targeting the 3′-UTR.
To determine whether miR-99b influences CLDN11
expression in human HCC tissues, we used immunohistochemistry to
analyze three samples each with high or low miR-99b expression and
found that CLDN11 was expressed at significantly lower levels in
the tissues with high miR-99b expression, consistent with the cell
line data (Fig. 3E).
Discussion
In the present study, we showed that the median
level of miR-99b expression in HCC tissues was significantly higher
when compared with that of ANLT or normal liver tissues.
Furthermore, miR-99b was expressed at significantly higher levels
compared with the controls in 72.1% (75/104) of HCC tissues and in
four HCC cell lines. Moreover, increased miR-99b expression was
significantly associated with capsule formation and microvascular
invasion, which indicated poor prognosis. We further showed that
miR-99b expression levels were inversely related to the OS and DFS
of patients, and served as an independent risk factor for OS and
DFS, indicating that miR-99b is a potential biomarker for HCC
recurrence.
CLDN11 is a crucial component of tight junction
strands (17,18). The tight junction is often altered
in human carcinomas, and the loss of the tight junction leads to
cancer progression (19,20). Moreover, the tight junction plays a
number of diverse roles in tumorigenesis, including cell signaling,
cytoskeletal regulation, cell growth, cell motility,
differentiation and maintenance of cell polarity (21–23).
Agarwal et al reported that silencing of CLDN11
expression contributes to gastric carcinogenesis by increasing cell
motility and invasiveness (24).
Moreover, the loss of CLDN11 is a putative indicator of recurrence
and more aggressive behavior of meningiomas (25). Consistent with these findings,
enforced expression of CLDN11 decreased the invasiveness of
bladder cancer cell lines (26),
indicating its role in preventing cancer progression and its
potential as a therapeutic target for inhibiting metastatic
growth.
miRNAs contribute to homeostasis and pathologies via
inhibiting target genes at the posttranscriptional level (27). In the present study, we investigated
the mechanism by which miR-99b promotes the progression of HCC.
Using bioinformatics, we predicted that CLDN11 mRNA is an
miR-99b target. The results of dual-luciferase reporter assays
support this conclusion. Moreover, overexpression of miR-99b
significantly inhibited CLDN11 expression and enhanced the
metastatic phenotype of the HCC cell lines, and knockdown of
miR-99b expression increased that of CLDN11 and attenuated the
metastatic phenotype of the HCC cell lines. In agreement with a
study of bladder cancer cells (26), CLDN11 mRNA expression was
downregulated in 90.4% (94/104) of HCC tissues studied in the
present study, suggesting the contribution to HCC pathogenesis.
Furthermore, we showed in the present study that CLDN11 mRNA
was expressed at low levels in HCC tissues with relatively high
levels of miR-99b expression. Conversely, CLDN11 mRNA was
expressed at high levels in HCC tissues with low levels of miR-99b
expression. Collectively, these data indicate that miR-99b inhibits
CLDN11 expression at the post-transcriptional level and
contributes to HCC metastasis.
Epithelial cells adhere through intercellular
adhesion complexes such as the tight junction. Mesenchymal cells
are non-polarized, lack intercellular junctions and migrate
individually throughout the extracellular matrix (28). The tight junction maintains cell
polarity and influences cell motility (29). Tight junction-associated proteins
promote the EMT through the regulation of tight junctions in human
breast cancer cell lines (30). In
the present study, we showed that miR-99b promoted the metastatic
phenotype of HCC cell lines through the tight junction-associated
protein CLDN11. Therefore, we suggested that miR-99b inhibits
CLDN11 expression and reduces the abundance of tight junctions to
promote the EMT of HCC cells, leading to increased metastasis of
HCC. Further studies are required to confirm these
speculations.
In conclusion, we demonstrated that miR-99b is
expressed at high levels in HCC tissues compared with ANLT.
Upregulation of miR-99b expression was positively correlated with
poor clinicopathological characteristics of the patients with HCC
and was associated with poor OS and DFS of postoperative patients.
Our data indicated that miR-99b functions as an oncogene that
promotes the metastasis of HCC via inhibiting CLDN11
expression. Therefore, miR-99b serves as a diagnostic marker as
well as a therapeutic target for HCC.
References
|
1
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: A global and regional perspective. Oncologist. 15(Suppl
4): S5–S13. 2010. View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhong JH, Ke Y, Gong WF, Xiang BD, Ma L,
Ye XP, Peng T, Xie GS and Li LQ: Hepatic resection associated with
good survival for selected patients with intermediate and
advanced-stage hepatocellular carcinoma. Ann Surg. 260:329–340.
2014. View Article : Google Scholar
|
|
5
|
Pang RW, Joh JW, Johnson PJ, Monden M,
Pawlik TM and Poon RT: Biology of hepatocellular carcinoma. Ann
Surg Oncol. 15:962–971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Turcatel G, Rubin N, El-Hashash A and
Warburton D: miR-99a and miR-99b modulate TGF-β induced epithelial
to mesenchymal plasticity in normal murine mammary gland cells.
PLoS One. 7:e310322012. View Article : Google Scholar
|
|
12
|
Huang JJ, Yu J, Li JY, Liu YT and Zhong
RQ: Circulating microRNA expression is associated with genetic
subtype and survival of multiple myeloma. Med Oncol. 29:2402–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lionetti M, Biasiolo M, Agnelli L,
Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S,
Lombardi L, et al: Identification of microRNA expression patterns
and definition of a microRNA/mRNA regulatory network in distinct
molecular groups of multiple myeloma. Blood. 114:e20–e26. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013.PubMed/NCBI
|
|
15
|
Feber A, Xi L, Pennathur A, Gooding WE,
Bandla S, Wu M, Luketich JD, Godfrey TE and Litle VR: MicroRNA
prognostic signature for nodal metastases and survival in
esophageal adenocarcinoma. Ann Thorac Surg. 91:1523–1530. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et
al: VHL-regulated miR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Furuse M, Fujita K, Hiiragi T, Fujimoto K
and Tsukita S: Claudin-1 and -2: Novel integral membrane proteins
localizing at tight junctions with no sequence similarity to
occludin. J Cell Biol. 141:1539–1550. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morita K, Sasaki H, Fujimoto K, Furuse M
and Tsukita S: Claudin-11/OSP-based tight junctions of myelin
sheaths in brain and Sertoli cells in testis. J Cell Biol.
145:579–588. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou W, Fong MY, Min Y, Somlo G, Liu L,
Palomares MR, Yu Y, Chow A, O'Connor ST, Chin AR, et al:
Cancer-secreted miR-105 destroys vascular endothelial barriers to
promote metastasis. Cancer Cell. 25:501–515. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao Y, Gu X, Liu H, Wu G, Yuan D, Yang X
and Song Y: Metadherin regulates proliferation and metastasis via
actin cytoskeletal remodelling in non-small cell lung cancer. Br J
Cancer. 111:355–364. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alshbool FZ and Mohan S: Emerging
multifunctional roles of Claudin tight junction proteins in bone.
Endocrinology. 155:2363–2376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garrido-Urbani S, Bradfield PF and Imhof
BA: Tight junction dynamics: The role of junctional adhesion
molecules (JAMs). Cell Tissue Res. 355:701–715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shang X, Lin X, Alvarez E, Manorek G and
Howell SB: Tight junction proteins claudin-3 and claudin-4 control
tumor growth and metastases. Neoplasia. 14:974–985. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Agarwal R, Mori Y, Cheng Y, Jin Z, Olaru
AV, Hamilton JP, David S, Selaru FM, Yang J, Abraham JM, et al:
Silencing of claudin-11 is associated with increased invasiveness
of gastric cancer cells. PLoS One. 4:e80022009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Soini Y, Rauramaa T, Alafuzoff I, Sandell
PJ and Kärjä V: Claudins 1, 11 and twist in meningiomas.
Histopathology. 56:821–824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Awsare NS, Martin TA, Haynes MD, Matthews
PN and Jiang WG: Claudin-11 decreases the invasiveness of bladder
cancer cells. Oncol Rep. 25:1503–1509. 2011.PubMed/NCBI
|
|
27
|
Giordano S and Columbano A: MicroRNAs: New
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar
|
|
28
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
The importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Artus C, Glacial F, Ganeshamoorthy K,
Ziegler N, Godet M, Guilbert T, Liebner S and Couraud PO: The
Wnt/planar cell polarity signaling pathway contributes to the
integrity of tight junctions in brain endothelial cells. J Cereb
Blood Flow Metab. 34:433–440. 2014. View Article : Google Scholar :
|
|
30
|
Zhao JL, Liang SQ, Fu W, Zhu BK, Li SZ,
Han H and Qin HY: The LIM domain protein FHL1C interacts with tight
junction protein ZO-1 contributing to the epithelial-mesenchymal
transition (EMT) of a breast adenocarcinoma cell line. Gene.
542:182–189. 2014. View Article : Google Scholar : PubMed/NCBI
|