Introduction
Epithelial-mesenchymal transition (EMT) is an
essential process for driving plasticity during development, and an
unintentional behavior of cells during the progression of malignant
tumor (1–3). The EMT-associated reprogramming of
cancer cells suggests that fundamental changes occur to several
regulatory networks and that an intimate interplay exists between
them. Disturbance of a controlled epithelial balance is triggered
by altering several layers of regulation, including the
transcriptional and translational machinery, expression of
non-coding RNAs, alternative splicing and protein stability
(4–6).
ARK5, known as KIAA0537/Novel (nua) kinase family 1
(NUAK1) has been identified as the fifth member of the adenosine
monophosphate-activated protein kinase (AMPK)-related kinase (ARK)
family (7). Akt phosphorylates ARK5
at Ser600, a C-terminal site outside the catalytic domain, which
leads to activation of the 74-kDa kinase. During glucose
deprivation or response to adenosine monophosphate, ARK5 supports
cell survival in an Akt-dependent manner (7). It suppresses cell death induced by
nutrient starvation and activation of death receptors through
inhibition of caspase 8, and the negative regulation of procaspase
6 (8,9). ARK5 is also strongly associated with
tumor invasion and metastasis, and is a tumor survival and tumor
progression factor (10–18).
MicroRNAs (miRNAs) are small non-coding RNA
molecules that suppress gene expression by interacting with the 3′
untranslated regions (3′UTRs) of target mRNAs (19–21).
The aberrant expression of miRNAs are reported in various human
types of cancer and are known to have an oncogenic or
tumor-suppressor role and have been shown to play key roles in cell
survival, proliferation, apoptosis, migration, invasion and various
other characteristic features that are altered in human cancer
types (22,23). Over 50% of the known miRNAs have
been shown to participate in human tumorigenesis and/or metastasis
by directly targeting oncogenes or tumor-suppressor genes (24,25).
miR-1181 inhibits stem cell-like phenotypes and suppresses SOX2 and
STAT3 in human pancreatic cancer, although its role has not been
reported in ovarian cancer (26).
In addition, EMT is associated with acquisition of malignant and
stem-cell characteristics (27).
In the present study, we examined the role of ARK5
in in ovarian cancer tissues, compared with normal matched tissues.
The results showed that ARK5 was upregulated in the cancer tissues
versus the healthy tissues. Moreover, ARK5 promoted EMT and
inhibited miR-1181 expression in ovarian cancer cells. Subsequent
investigations showed that miR-1181 promoted MET in ovarian cancer
cells. We also searched downstream target genes of miR-1181, and
found that miR-1181 degraded HOXA10 by targeting its 3′UTR in
ovarian cancer cells. Additionally, we confirmed that HOXA10
promoted EMT in ovarian cancer cells. Thus, activation of the
ARK5/miR-1181/HOXA10 axis may be positively associated with EMT in
ovarian cancer.
Materials and methods
Ovarian cancer tissues
Six ovarian cancer patients diagnosed with ovarian
cancer were recruited from the Department of Obstetrics and
Gynaecology, Linyi People's Hospital and Hubei Cancer Center. Human
tissue samples were utilized according to internationally
recognized guidelines as well as local and national regulations.
Studies carried out on human subjects were conducted according to
international and national regulations. The Medical Ethics
Committee approved the experiments undertaken. Informed consent was
obtained from each participant.
Cell lines, plasmids,
pre-miR-1181/control miR, anti-miR-1181/scramble and
transfection
Human EG, OVCAR8, OVCAR3, OCC1, HEY and SKOV3
ovarian cancer cell lines were obtained from the MD Anderson Cancer
Center (Houston, TX, USA). Briefly, the cells were maintained in
RPMI-1640 medium supplemented with 5% fetal bovine serum (FBS)
(Gibco, Grand Island, NY, USA) and penicillin/streptomycin at 37°C
in a humidified atmosphere with 5% CO2. ARK5- and
HOXA10-expressing plasmids/empty vectors (pcDNA3.1) were purchased
from the National RNAi Core Facility in Academic Sinica, Taipei,
Taiwan. The expressing plasmids or empty vector (pcDNA3.1) used for
each transfection was 10 µg. Pre-miR-1181/control miR and
anti-miR-1181/scramble were purchased from Ambion, Inc. (Austin,
TX, USA). Transfection was performed with Lipofectamine 2000
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions.
Western blot analysis
Western blot analysis was performed as previously
described (28). Briefly, after
incubation with primary antibody anti-ARK5 (1:500), anti-E-cadherin
(1:500), anti-α-catenin (1:500), anti-N-cadherin (1:500),
anti-vimentin (1:500), anti-α-SMA (1:500), anti-β-catenin (1:500),
anti-fibronectin (1:500), anti-SNAI1 (1:500), anti-TWIST (1:500),
anti-TGFB1 (1:500), anti-ZEB1 (1:500), anti-TGFB2 (1:500),
anti-HOXA10 (1:500) and anti-β-actin (1:500) (all from Abcam,
Cambridge, MA, USA) overnight at 4°C, IRDye™ 800-conjugated
anti-rabbit secondary antibodies (LI-COR Biosciences, Lincoln, NE,
USA) were used for 30 min at room temperature. The specific
proteins were visualized by the Odyssey™ Infrared Imaging System
(Gene Company, Lincoln, NE, USA).
Migration and invasion assays
For the Transwell migration assays, 3×104
cells were plated in the top chamber with the non-coated membrane
(24-well insert; pore size, 8-mm; BD Biosciences, San Jose, CA,
USA). For the invasion assays, 1.25×105 cells were
plated in the top chamber with Matrigel-coated membrane (24-well
insert; pore size, 8 mm; BD Biosciences). In the two assays, the
cells were plated in medium without serum or growth factors, and
medium supplemented with serum was used as a chemoattractant in the
lower chamber. The cells were incubated for 36 h and any cells that
did not migrate or invade through the pores were removed using a
cotton swab. Cells on the lower surface of the membrane were
stained with the Diff-Quik Staining Set (Dade Behring, Newark, DE,
USA) and counted.
Wound-healing assay
Cells (5×105) were seeded in each 35-mm
glass bottom dish (MatTek Corporation, Ashland, MA, USA) and
cultured at 37°C with 5% CO2 for 24 h. The confluent
monolayer of cells was wounded. Cell monolayers were wounded with
yellow pipette tips. After washing with warm phosphate-buffered
saline (PBS), the cells were incubated in fresh culture medium. The
wounded areas were photographed at the beginning (0 h, top panels)
and the end (10 h, bottom panels) of the assay using a Nikon
inverted microscope (Eclipse TE-2000U) equipped with a video camera
(DS-U1) (both from Nikon, Japan).
Immunocytochemistry
Cells transfected as indicated and grown in chamber
slides (Laboratory-Tek; Nalge Nunc International, Rochester, NY,
USA) were washed 3–4 times with PBS and fixed with 4% formaldehyde
for 15 min at room temperature. After 3–4 PBS washes, the cells
were permeabilized with 0.25% Triton X-100 in PBS. The cells were
incubated in PBS containing 2% BSA, followed by overnight
incubation at 4°C with anti-E-cadherin or anti-vimentin antibodies.
The secondary antibody was incubated for 2 h at room
temperature.
miRNA microarray
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit (Ambion, Inc.). cRNA for each sample was synthesized
using a 3′ IVT Express kit (Affymetrix, Santa Clara, CA, USA)
according to the manufacturer's instructions. The purified cRNA was
fragmented by incubation in fragmentation buffer (provided in the
3′IVT Express kit) at 95°C for 35 min and chilled on ice. The
fragmented labeled cRNA was applied to MicroRNA2.0 array and
hybridized in a GeneChip Hybridization Oven 640 (both from
Affymetrix) at 45°C for 20 h. After washing and staining in a
GeneChip fluidics station 450, the arrays were scanned using a
GeneChip Scanner 3000 (both from Affymetrix). The gene expression
levels of samples were normalized and compared using Partek GS 6.5
(Partek Inc., St. Louis, MO, USA). Average-linkage hierarchical
clustering of the data was applied using the cluster and the
results were shown using Treeview (both from Stanford, Stanford
University, CA, USA; http://rana.lbl.gov).
Reverse transcription-qPCR for miRNA
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit. Detection of the mature form of miRNAs was performed
using the mirvana RT-qPCR miRNA Detection kit, according to the
manufacturer's instructions (Ambion, Inc.). The U6 small nuclear
RNA was used as an internal control.
Bioinformatics analysis
An analysis of potential miRNA target sites was
performed using prediction algorithms by miRanda (http://www.microrna.org/microrna/home.do).
Immunofluorescence analysis
For the immunofluorescence analysis, the cells were
plated on glass coverslips in 6-well plates and transfected as
indicated. At 48 h after transfection, the coverslips were stained
with the abovementioned antiHOXA10 antibodies. Alexa Fluor 488 goat
anti-rabbit IgG antibody was used as the secondary antibody
(Invitrogen). The coverslips were counterstained with DAPI
(Invitrogen Molecular Probes, Eugene, Oregon, USA) for
visualization of the nuclei. Microscopic analysis was performed
with a confocal laser-scanning microscope (Leica Microsystems,
Bensheim, Germany). Fluorescence intensities were measured in a few
viewing areas for 200–300 cells/coverslip and analyzed using ImageJ
1.37v software (http://rsb.info.nih.gov/ij/index.html).
Reverse-transcription polymerase chain
reaction (RT-PCR) and RT-qPCR for HOXA10
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen). First-strand cDNA was synthesized from the
total RNA using M-MLV reverse transcriptase (Promega, Madison, WI,
USA) and random hexamer primers (Sangon, Shanghai, China). The
thermal cycle profile used was: denaturation for 30 sec at 95°C,
annealing for 45 sec at 53–58°C depending on the primers used, and
extension for 45 sec at 72°C. PCR products were visualized on 2%
agarose gels stained with ethidium bromide under UV
transillumination. RT-qPCR was carried out with a Power SYBR-Green
PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) according to
the manufacturer's instructions. The primer sequences used for
HOXA10 were: forward, 5′-GCCCTTCCGAGAGCAGCAAAG-3′ and reverse,
5′-AGGTGGACGCTGCGGCTAATCTCTA-3′.
Luciferase reporter assay
The 3′-UTR of human HOXA10 mRNA was cloned in pRL-TK
(Promega) using a PCR-generated fragment. Site-directed mutagenesis
of the miR-1181 target-site in the HOXA10-3′-UTR was carried out
using a QuikChange mutagenesis kit (Stratagene, Heidelberg,
Germany), with HOXA10-WT-luc as a template. For reporter assays,
cells was transiently transfected with WT or mutant reporter
plasmids and miRNA or anti-miRNA (as shown in Fig. 5H and I) using Lipofectamine 2000
(Invitrogen). Reporter assays were performed 36 h post-transfection
using the Dual-Luciferase Reporter Assay System (Promega),
normalized for transfection efficiency by co-transfected
Renilla luciferase.
 | Figure 5miR-1181 degrades HOXA10 by targeting
its 3′UTR in ovarian cancer cells. (A) vin Diagram showing the
predicted miR-1181 targeting 3′UTR of HOXA10 mRNA from the database
of miRanda. (B) Western blotting for HOXA10 protein in EG and HEY
cells. β-actin was a loading control, n=3. (C) Immunofluorescence
analyses for EG and HEY cells. Top panel shows microscopic images
of immunofluorescence staining of one representative experiment
(magnification, ×100). Bottom panel shows graphic presentation of
mean fluorescence intensities, n=3. (D) Western blotting for HOXA10
protein in HEY cells infected as indicated. Mock groups were
transfected with control miR. β-actin served as a loading control,
n=3. (E) Immunofluorescence analyses for HEY cells transfected with
pre-miR-1181 or control miR (mock). Left panel shows microscopic
images of immunofluorescence staining of one representative
experiment magnification, ×100). Top panel shows graphic
presentation of mean fluorescence intensities, n=3. (F) RT-PCR for
HOXA10 mRNA in HEY cells infected as indicated. GAPDH served as a
loading control, n=3. (G) Diagram of HOXA10-3′UTR-containing
reporter constructs. MUT, contains 4-base-mutation at the
miR-1181-target region, abolishing its binding. (H) Reporter assay
for HEY cells, with co-transfection of 500 ng WT-or MUT-reporter
and 50 nM control-miR (mock) or pre-miR-1181 as indicated, n=3. (I)
Reporter assay for EG cells, with co-transfection of 500 ng WT-or
MUT-reporter and 50 nM scramble or anti-miR-1181 as indicated,
n=3. |
Results
ARK5 is upregulated in ovarian cancer
tissues
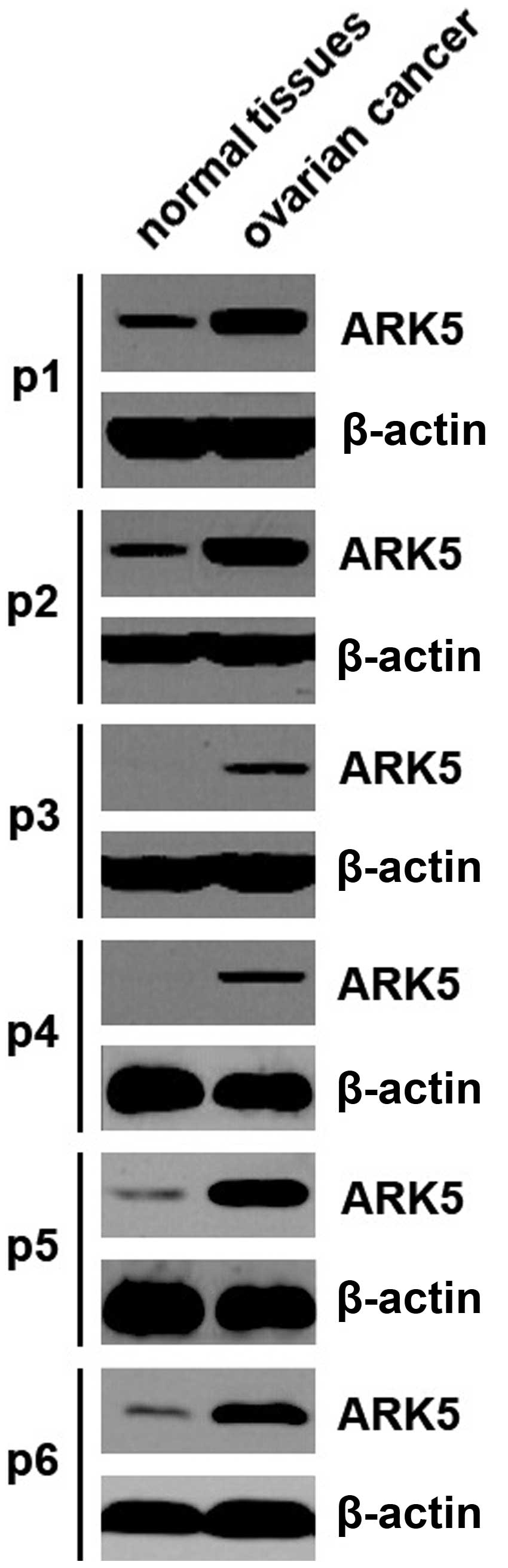
To identify ARK5 protein expression in ovarian
cancer tissues, we performed western blot analysis to detect ARK5
protein between ovarian cancer and adjacent normal tissues. We
found that ARK5 was increased in the cancer tissues of 6 patients,
compared with the adjacent normal tissues (Fig. 1). The results suggested that ARK5
may be an oncogene in ovarian cancer.
ARK5 promotes EMT in ovarian cancer
cells
Western blotting was performed to identify the ARK5
protein expression in the EG, OVCAR8, OVCAR3, OCC1, HEY and SKOV3
ovarian cancer cell lines. Protein isolated from the six cell lines
was detected by western blotting and the results showed that the
expression of ARK5 was lowest in EG cells, whereas its expression
was highest in HEY cells among the six ovarian cancer cell lines
(Fig. 2A). To identify whether ARK5
was associated with EMT in ovarian cancer, we performed western
blotting to detect epithelial and mesenchymal markers in EG cells
(ARK5-negative) and HEY cells (ARK5-positive). The results showed
that the expression of E-cadherin and α-catenin were significantly
elevated in EG cells (ARK5-negative cells), compared with HEY cells
(ARK5-positive cells) (Fig. 2B). By
contrast, the expression of N-cadherin, vimentin and α-SMA protein
was upregulated in HEY cells (ARK5-positive cells) (Fig. 2B). Thus, ARK5 may be associated with
EMT in ovarian cancer.
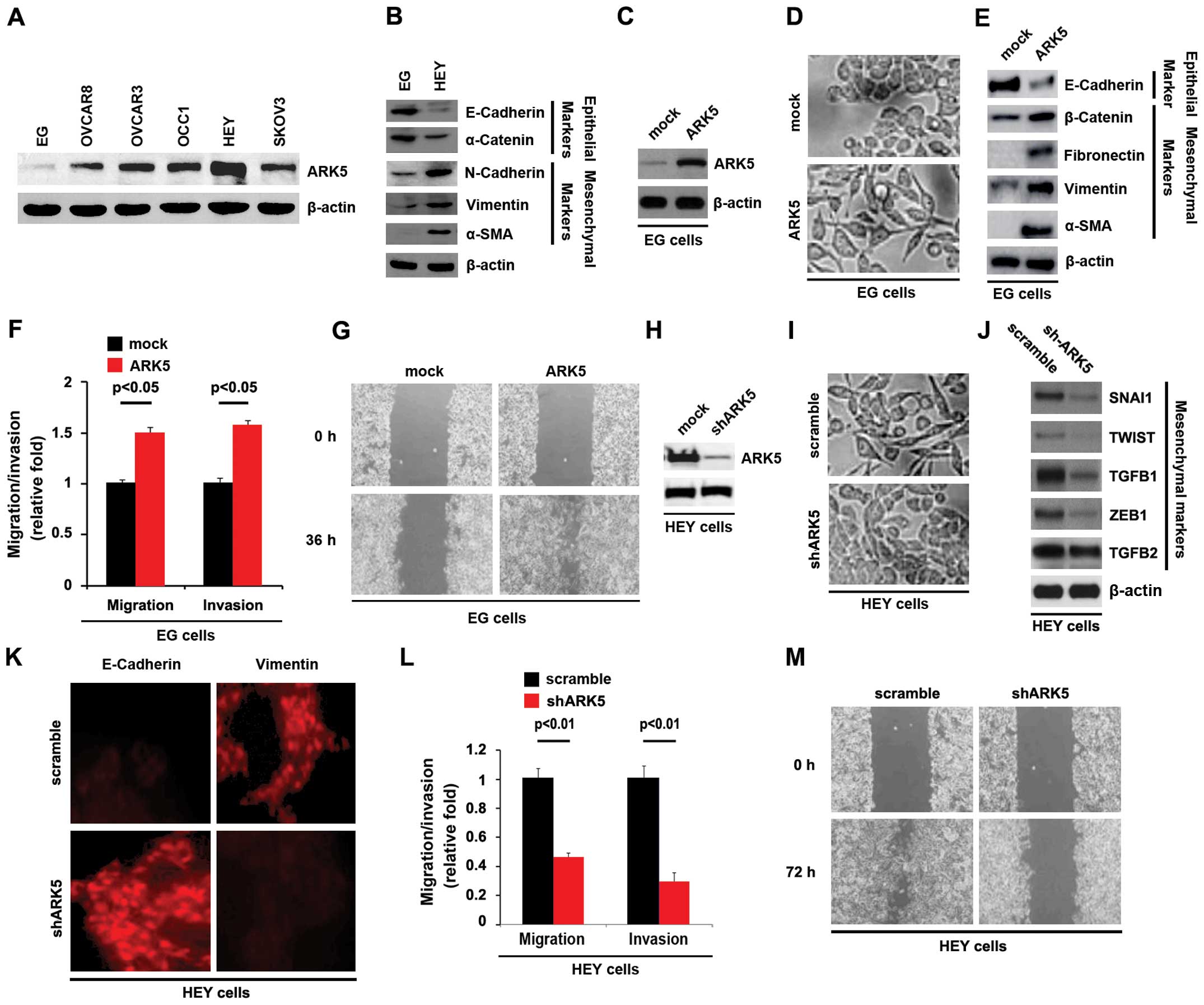 | Figure 2(A) ARK5 promotes EMT in ovarian
cancer cells. Western blotting for ARK5 in EG, OVCAR8, OVCAR3,
OCC1, HEY and SKOV3 ovarian cancer cell lines. β-actin served as a
loading control, n=3. (B) Western blotting for E-cadherin,
α-catenin, N-cadherin, vimentin and α-SMA in ovarian cancer EG and
HEY cells. β-actin served as a loading control, n=3. (C) Western
blotting for ARK5 in EG cells transfected with ARK5-expressing
plasmids. Mock groups were transfected with empty vectors. β-actin
served as a loading control, n=3. (D) EG cells were transfected as
indicated. Cells were then photographed, n=3. (E) Western blotting
for E-cadherin, β-catenin, fibronectin, vimentin and α-SMA in
ovarian cancer cells transfected with ARK5-expressing plasmids.
Mock groups were transfected with empty vectors. β-actin served as
a loading control, n=3. (F) Invasion and migration assays for EG
cells transfected as indicated, n=3. (G) Wound-healing assays for
EG cells transfected with ARK5-expressing plasmids and empty vector
(mock). The cell layer was photographed, n=3. (H) Western blotting
for ARK5 in ovarian cancer HEY cells transfected as indicated.
β-actin served as a loading control, n=3. (I) HEY cells were
transfected as indicated. Cells were then photographed, n=3. (J)
Western blotting for SNAI1, TWIST, TGFB1, ZEB1 and TGFB2 in HEY
cells transfected as indicated. β-actin served as a loading
control, n=3. (K) Immunocytochemistry for HEY cells transfected as
indicated, n=3. (L) Invasion and migration assays for HEY cells
transfected as indicated, n=3. (M) Wound-healing assays for HEY
cells transfected as indicated. The cell layer was photographed,
n=3. EMT, epithelial-mesenchymal transition. |
In order to assess the role of ARK5 in ovarian
cancer, we transfected EG cells with ARK5-expressing plasmids,
followed by western blotting. ARK5 protein was significantly
increased in the cells transfected with ARK5-expressing plasmids
(Fig. 2C) and its overexpression
caused significant changes in EG cell morphology (EMT) (Fig. 2D). To verify that the changes in
cell morphology were caused by EMT, the expression levels of
epithelial and mesenchymal markers were compared in EG cells
transfected with ARK5-expressing plasmids with EG cells transfected
with empty vectors. The results revealed that the epithelial
markers (E-cadherin) were consistently repressed, whereas the
mesenchymal markers (β-catenin, fibronectin, vimentin and α-SMA)
were induced by ARK5 overexpression in EG cells (Fig. 2E). EMT resulted in increased cell
invasion and migration (29–31).
Thus, ARK5 also affected invasion and migration in EG cells. To
determine the reason for this result, we performed invasion,
migration, and wound-healing assays. We found that ARK5 resulted in
enhanced invasion (Fig. 2F) and
migration (Fig. 2F and G) in the
cells.
As shown above ARK5 overexpression promoted EMT in
EG cells. To provide further evidence that ARK5 was involved in EMT
of ovarian cancer, we studied the effects of an inhibitor of ARK5,
shARK5. After stable transfection, ARK5 expression was detected by
western blotting. The results showed that shARK5 significantly
downregulated ARK5 protein expression in HEY cells (Fig. 2H). After transfection, we observed
that silencing ARK5 caused significant changes in HEY cell
morphology (MET) (Fig. 2I). To
confirm that silencing ARK5 was associated with MET in ovarian
cancer, we performed western blotting to detect the expression of
mesenchymal markers (SNAI1, TWIST, TGFB1, ZEB1 and TGFB2). The
results demonstrated that the expression of SNAI1, TWIST, TGFB1 and
ZEB1 was evidently attenuated by silencing ARK5 (Fig. 2J). We also performed
immunocytochemistry to detect the expression of E-cadherin
(epithelial markers) and vimentin (mesenchymal markers). The
results showed that silencing ARK5 significantly upregulated
E-cadherin expression and downregulated vimentin expression
(Fig. 2K). Given that ARK5
overexpression promoted migration and invasion in EG cells
(ARK5-negative), we hypothesized that silencing ARK5 impaired the
ability of migration and invasion in HEY cells (ARK5-positive).
Thus, we performed invasion, migration, and wound-healing assays to
observe the effect of silencing ARK5 on invasion and migration. The
results confirmed that silencing ARK5 inhibited invasion (Fig. 2L) and migration (Fig. 2L and M) in HEY cells. Thus, ARK5
promoted EMT in ovarian cancer cells.
ARK5 suppresses miR-1181 expression in
ovarian cancer cells
miRNAs, the small non-coding RNA molecules that
suppress gene expression by interacting with the 3′UTRs of target
mRNAs, have also been associated with EMT and cancer (4,32–34).
Moreover, since oncogenes exert their functions by regulating miRNA
expression in tumor (35), we
investigated whether ARK5 affected miRNA expression in ovarian
cancer cells.
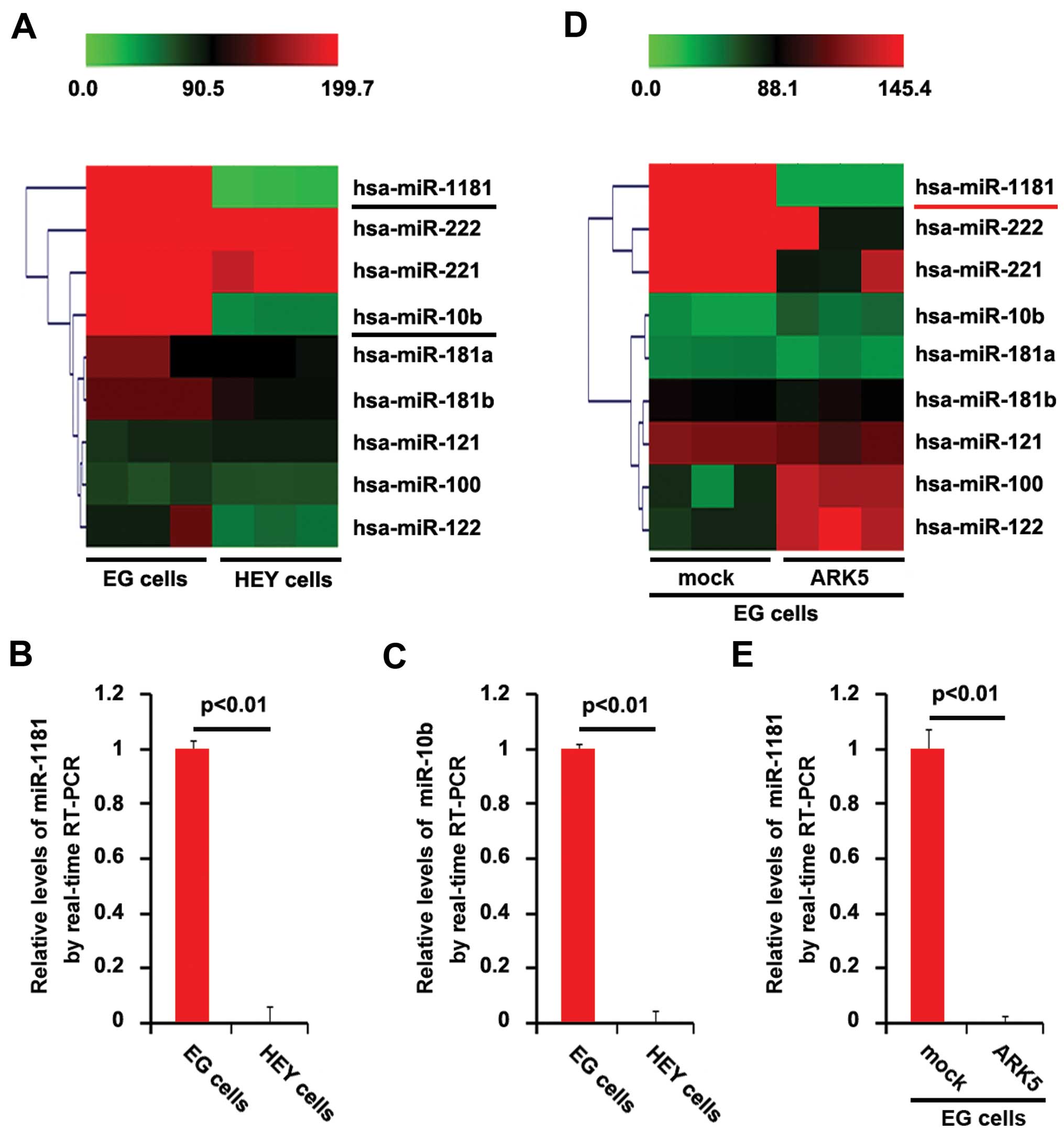
miRNA microarrays were performed. RNAs isolated from
EG and HEY cells were hybridized to a custom miRNA microarray
platform. After hybridization, quantification and normalization
were carried out three times, a number of miRNAs, particularly
miR-1181 and miR-10b, were downregulated >150-fold in HEY cells
(ARK5-positive), compared with EG cells (ARK5-negative) (Fig. 3A). We also performed RT-PCR to
confirm the results of miRNA microarray. Consistent with miRNA
microarray, the results of RT-PCR showed that miR-1181 and miR-10b
were significantly downregulated in HEY cells, compared with EG
cells (Fig. 3B and C). The results
suggested that miR-1181 and miR-10b inhibition may be associated
with the overexpression of ARK5 in ovarian cancer cells. To
identify the association between ARK5 and the two miRNAs, we
transfected EG cells (ARK5-negative) with ARK5-expressing plasmids
or empty vectors and then miRNA microarrays were performed. We
found that ARK5 downregulated miR-1181 expression >100-fold,
although it did not affect miR-10b expression in EG cells (Fig. 3D). Moreover, the results of RT-PCR
confirmed that ARK5 significantly inhibited miR-1181 expression in
EG cells (Fig. 3E).
miR-1181 promotes MET in ovarian cancer
cells
Given that ARK5 promoted EMT in ovarian cancer cells
and inhibited miR-1181 expression in the cells, we examined whether
miR-1181 inhibited EMT and promoted MET, while ARK5 promoted EMT by
inhibiting miR-1181 expression in ovarian cancer cells. To identify
the role of miR-1181 in regulating EMT in ovarian cancer cells, HEY
cells were transfected with pre-miR-1181 and control miR. Following
transfection, miR-1181 expression was detected by RT-PCR and the
results showed that miR-1181 was increased by pre-miR-1181 in HEY
cells (Fig. 4A). Additionally, its
overexpression caused significant changes in HEY cell morphology
(MET) (Fig. 4B). To identify
whether miR-1181 promoted MET, we performed western blotting to
detect mesenchymal and epithelial markers. The results demonstrated
that the expression of vimentin and α-SMA were significantly
suppressed in HEY cells transfected with pre-miR-1181, compared
with the cells transfected with control miR (Fig. 4C). We also identified that
E-cadherin was upregulated by pre-miR-1181 in HEY cells (Fig. 4C). Wound-healing, migration and
invasion assays were performed to detect the migration and invasion
of HEY cells transfected with pre-miR-1181 or control miR. Ectopic
miR-1181 significantly inhibited motility (Fig. 4D and E) and invasion (Fig. 4E).
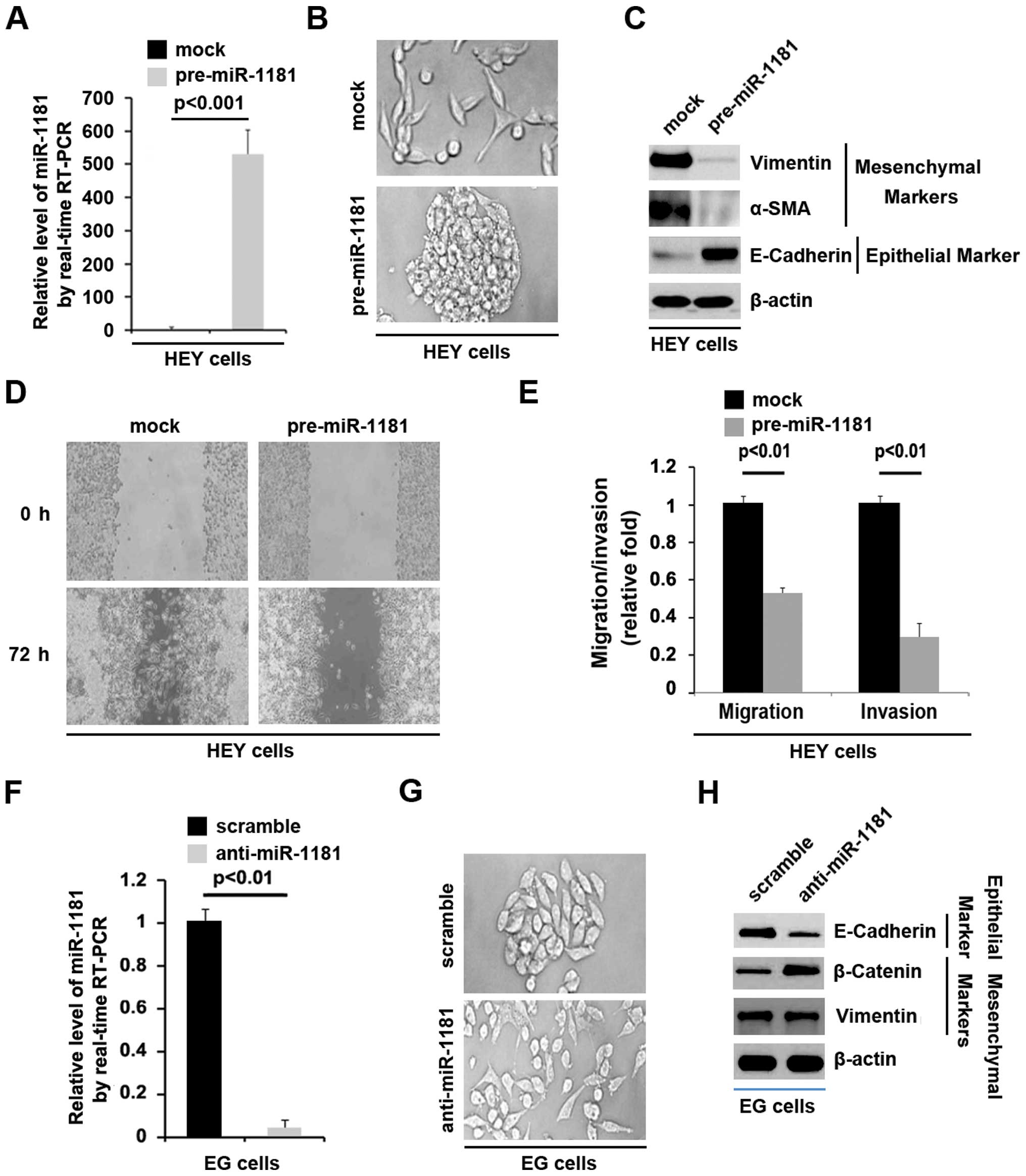 | Figure 4miR-1181 promotes MET in ovarian
cancer. (A) RT-PCR for miR-1181 in HEY cells transfected as
indicated. Mock groups were transfected with control miR. U6 served
as a loading control, n=3. (B) HEY cells were transfected as
indicated. Mock groups were transfected with control miR. Cells
were then photographed, n=3. (C) Western blotting for vimentin,
α-SMA and E-cadherin in HEY cells transfected with pre-miR-1181.
Mock groups were transfected with control miR. β-actin served as a
loading control, n=3. (D) Wound-healing assays for HEY cells
transfected as indicated. Mock groups were transfected with control
miR. The cell layer was photographed, n=3. (E) Invasion and
migration assays for HEY cells transfected as indicated. Mock
groups were transfected with control miR, n=3. (F) RT-PCR for
miR-1181 in EG cells transfected as indicated. U6 served as a
loading control, n=3. (G) EG cells were transfected as indicated.
Cells were then photographed, n=3. (H) Western blotting for
E-cadherin, β-catenin and vimentin in EG cells transfected as
indicated. β-actin served as a loading control, n=3. MET,
mesenchymal-epithelial transition. |
As shown above miR-1181 promoted MET in ovarian
cancer cells. Thus, to provide further evidence that the roles of
miR-1181 were involved in MET of ovarian cancer cells, we studied
the effects of an inhibitor of miR-1181 and anti-miR-1181. After
stable transfection, miR-1181 expression was detected by RT-PCR in
EG cells. The results showed that anti-miR-1181 significantly
downregulated miR-1181 expression in EG cells (Fig. 4F). We also observed the morphology
of EG cells transfected with anti-miR-1181, compared with the cells
transfected with scramble. The results showed that contrary to
miR-1181, silencing miR-1181 significantly changed EMT (Fig. 4G). Western blotting was performed to
detect whether epithelial and mesenchymal markers were affected by
anti-miR-1181 in EG ovarian cancer cells. The results showed that
E-cadherin was significantly inhibited in the cells transfected
with anti-miR-1181, whereas β-catenin was promoted by the
transfection (Fig. 4H).
miR-1181 degrades HOXA10 by targeting its
3′UTR in ovarian cancer
miRNAs are a new class of small (~22 nucleotide)
non-coding RNAs that negatively regulate protein-coding gene
expression by targeting mRNA degradation or translation inhibition
(19–21). Thus, a search for downstream targets
of miR-1181 in silico was conducted. The commonly used
prediction algorithm-miRanda (http://www.microrna.org/microrna/home.do) was utilized
to analyze the predicted target genes of miR-1181. A number of
target genes was identified, including HOXA10, which was
overexpressed and promoted migration, and invasion in human ovarian
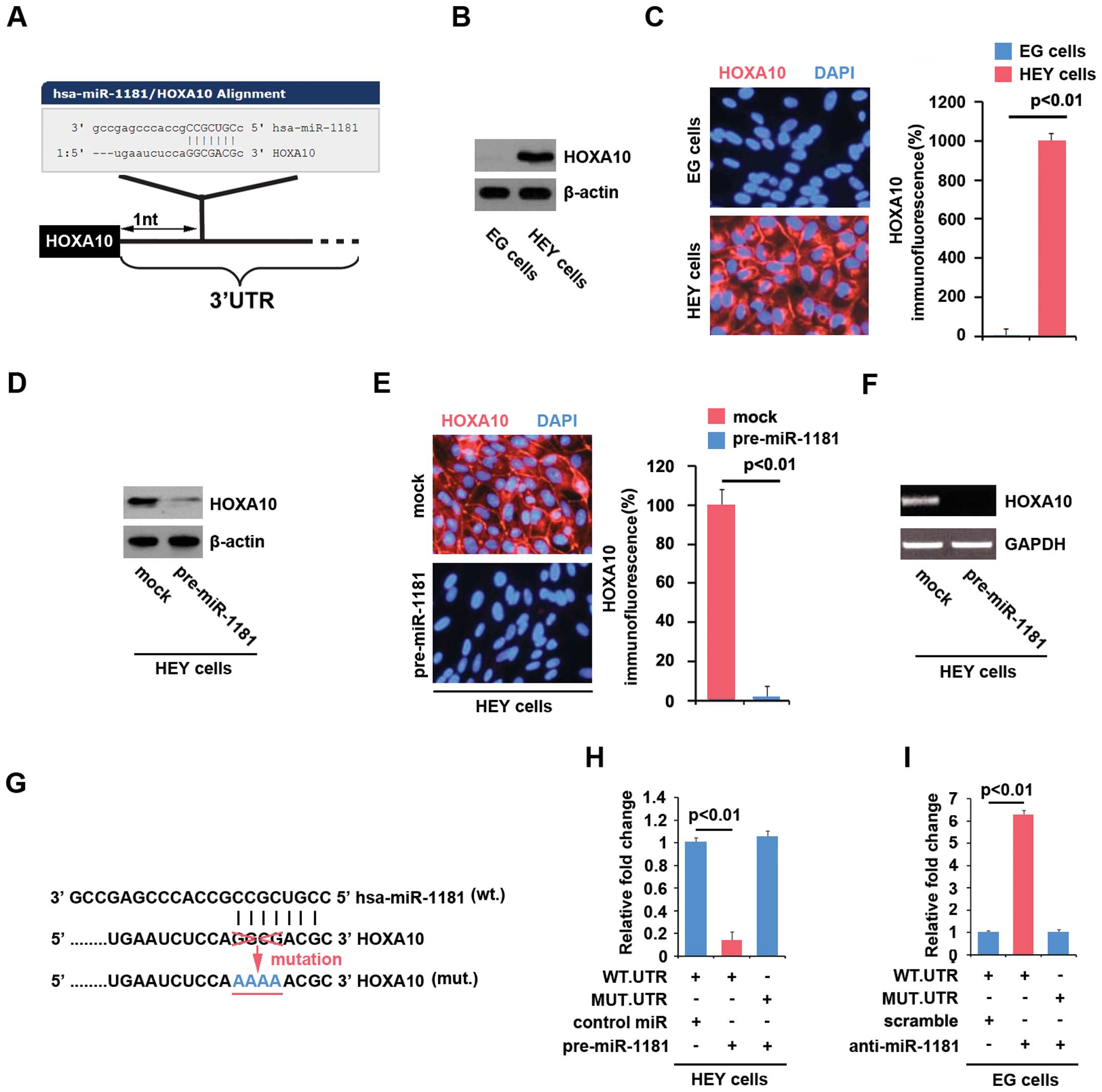
cancer and correlated with poor survival (36). A schematic of predicted
miR-1181-binding sites in the 3′UTR of HOXA10 mRNA is shown in
Fig. 5A. miR-1181 was found to
promote MET in ovarian cancer cells by targeting HOXA10 in ovarian
cancer.
Given that miR-1181 expression was significantly
upregulated in EG cells compared with HEY cells, we performed
western blotting to detect HOXA10 protein in the two cell lines.
The results showed that HOXA10 was detected in HEY cells, but not
in EG cells (Fig. 5B). We also
performed immunofluorescence analyses in EG and HEY cells. The
results showed that HOXA10 protein was evidently suppressed in the
EG cells (Fig. 5C). The results
suggested that HOXA10 was negatively associated with miR-1181
expression.
To confirm this finding, we performed western
blotting in HEY cells transfected with pre-miR-1181 or control miR.
The results showed that HOXA10 protein was evidently suppressed in
the cells transfected with pre-miR-1181 (Fig. 5D). We also performed
immunofluorescence analyses in the cells. Consistent with the
results of western blotting, immunofluorescence analyses
demonstrated that HOXA10 was reduced in HEY cells transfected with
pre-miR-1181, compared with the control miR-transfected groups
(Fig. 5E). We then performed RT-PCR
to detect HOXA10 mRNA expression in HEY cells transfected with
pre-miR-1181 or control miR. The results showed that HOXA10 mRNA
(Fig. 5F) was significantly
suppressed in the cells transfected with pre-miR-1181. To
demonstrate the direct regulation of HOXA10 by miR-1181 through its
3′UTR, we constructed luciferase reporters with the targeting
sequences of wild-type (HOXA10-WT-luc) and mutated HOXA10 3′UTRs
(HOXA10-MUT-luc) (Fig. 5G). Both
the wild-type and mutant reporters were introduced into HEY cells.
The luciferase reporter assay showed that the luciferase activities
of HOXA10-WT-luc plasmids were significantly suppressed in the
cells transfected with pre-miR-1181, suggesting that miR-1181
targeted 3′UTR of HOXA10 mRNA (Fig.
5H). To determine whether miR-1181 targeted 3′UTR of HOXA10 at
the predicted sites, we mutated four bases in the predicted sites
(Fig. 5G). Mutant reporters were
subsequently introduced into HEY cells as expected. The luciferase
activities of HOXA10-MUT-luc were not suppressed by miR-1181 in HEY
cells (Fig. 5H).
Given that miR-1181 overexpression inhibited
HOXA10-WT-luc plasmids at the predicted sites, we investigated
whether silencing miR-1181 affected activity of the HOXA10-WT-luc
plasmids. Thus, the luciferase reporter assay was performed again
and the results showed that contrary to pre-miR-1181, anti-miR-1181
significantly promoted the luciferase activity of HOXA10-WT-luc in
EG cells (Fig. 5I). Moreover,
mutant reporters were introduced into EG cells, although the
luciferase activities of HOXA10-MUT-luc were not affected by
anti-miR-1181 in EG cells (Fig.
5I). The results suggested that miR-1181 degraded HOXA10 by
targeting the specific sites predicted in silico in ovarian
cancer cells.
HOXA10 promotes EMT in ovarian cancer
cells
As shown above, miR-1181 degrades HOXA10 by
targeting its 3′UTR in ovarian cancer cells. Thus, we examined the
roles of HOXA10 in EMT of ovarian cancer.
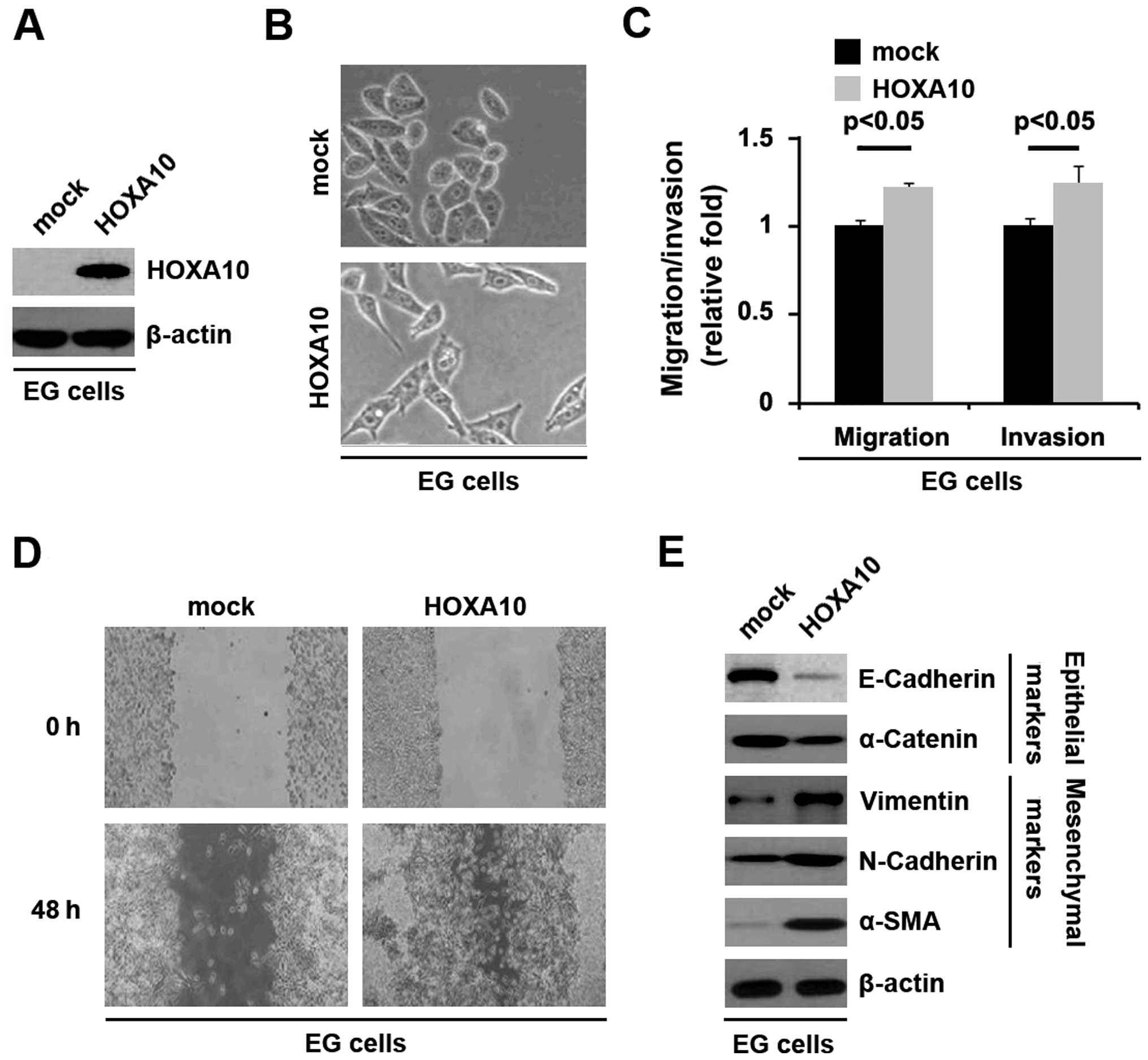
To identify the role of HOXA10 in regulating EMT in
ovarian cancer, the cells were transfected with HOXA10-expressing
plasmids. After stable transfection, HOXA10 protein expression was
detected by western blotting and the results showed that HOXA10
protein was increased by HOXA10-expressing plasmids in the cells
(Fig. 6A). HOXA10 overexpression
caused significant changes in EMT (Fig.
6B). In addition, to identify whether HOXA10 affected invasion
and migration, we performed invasion, migration, and wound-healing
assays. HOXA10 resulted in increased cell invasion (Fig. 6C) and migration (Fig. 6C and D) in EG cells.
To verify that the changes in cell morphology and
characteristics of invasion and migration were caused by EMT, the
expression levels of epithelial and mesenchymal markers were
compared in EG cells transfected with HOXA10-expressing plasmids or
empty vectors. The results revealed that the epithelial markers
(E-cadherin and α-catenin) were consistently repressed, whereas
mesenchymal markers (vimentin, N-cadherin and α-SMA) were induced
by HOXA10 overexpression in EG cells (Fig. 6E).
Discussion
EMT is a crucial developmental program in which
immotile epithelial cells acquire mesenchymal features. Activation
of EMT triggers tumor cell invasion and dissemination and is thus
considered as the initiating step of cancer metastasis (37,38).
Epithelial cells are characterized by several features including,
cohesive interactions among cells, facilitating the formation of
continuous cell layers; existence of three membrane domains:
apical, lateral and basal; presence of tight junctions between
apical and lateral domains; apicobasal polarized distribution of
the various organelles and cytoskeleton components; expression of
epithelial markers such as E-cadherin and α-catenin; lack of
mobility of individual epithelial cells with respect to their local
environment (39–41). Mesenchymal architectures are
different from epithelial ones including features such as loose or
no interactions among cells; no clear apical and lateral membranes;
no apico-basal polarized distribution of organelles and
cytoskeleton components; and expression of mesenchymal markers,
such as N-cadherin, vimentin, β-catenin, α-SMA, SNAI1, TWIST,
TGFB1, ZEB1 and TGFB2; motile cells having invasive properties
(39,42–44).
During EMT, cell-cell junctions are altered, cells lose epithelial
polarity, express the mesenchymal markers and the resulting
reorganization of the actin cytoskeleton supports cell migration.
We found that ARK5 protein expression was positively associated
with mesenchymal markers in ovarian cancer cell lines. Moreover,
its overexpression caused significant changes in EG cell morphology
(EMT) and unregulated mesenchymal markers and downregulated
epithelial markers. Contrary to its overexpression, silencing ARK5
caused significant changes in HEY cell morphology (MET) and
suppressed expression of mesenchymal markers, such as vimentin,
SNAI1, TWIST, TGFB1, ZEB1 and vimentin as well as upregulated
epithelial marker E-cadherin.
miRNAs, the small non-coding RNA molecules that
suppress gene expression by interacting with the 3′ untranslated
regions (3′UTRs) of target mRNAs, have also been associated with
EMT and cancer (4,32–34).
Epithelial-mesenchymal transition (EMT) has recently been
associated with stem-cell phenotype (27,45).
It has been reported that miR-1181 can inhibit stem cell-like
phenotypes and suppresses SOX2 and STAT3 in human pancreatic cancer
(26). Consistent with previous
studies, we confirmed that miR-1181 promoted MET in ovarian cancer
cells and its expression was significantly downregulated by ARK5.
However, whether ARK5 and miR-1181 are linked to the stem-cell
phenotype of ovarian cancer remains to be determined.
An increased expression of HOXA10 was detected in
almost all ovarian carcinomas (46). In the present study, we confirmed
that HOXA10 promoted EMT in ovarian cancer. Elucidating the
mechanism that miR-1181 promotes MET in ovarian cancer by
downregulating HOXA10 may be useful to understand the mechanism of
EMT. Thus, targeting ARK5 and restoration of miR-1181 may be a
promising therapeutic approach to suppress HOXA10-mediated EMT.
However, the role of the ARK5/miR-1181/HOXA10 axis remains to be
confirmed in vivo.
Abbreviations:
|
EMT
|
epithelial-mesenchymal transition
|
|
MET
|
mesenchymal-epithelial transition
|
References
|
1
|
Nieto MA: The ins and outs of the
epithelial to mesenchymal transition in health and disease. Annu
Rev Cell Dev Biol. 27:347–376. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Savagner P, Yamada KM and Thiery JP: The
zinc-finger protein slug causes desmosome dissociation, an initial
and necessary step for growth factor-induced epithelial-mesenchymal
transition. J Cell Biol. 137:1403–1419. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang CJ, Chao CH, Xia W, Yang JY, Xiong
Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al: p53 regulates
epithelialmesenchymal transition and stem cell properties through
modulating miRNAs. Nat Cell Biol. 13:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gravdal K, Halvorsen OJ, Haukaas SA and
Akslen LA: A switch from E-cadherin to N-cadherin expression
indicates epithelial to mesenchymal transition and is of strong and
independent importance for the progress of prostate cancer. Clin
Cancer Res. 13:7003–7011. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hader C, Marlier A and Cantley L:
Mesenchymal-epithelial transition in epithelial response to injury:
The role of Foxc2. Oncogene. 29:1031–1040. 2010. View Article : Google Scholar :
|
|
7
|
Suzuki A, Kusakai G, Kishimoto A, Lu J,
Ogura T, Lavin MF and Esumi H: Identification of a novel protein
kinase mediating Akt survival signaling to the ATM protein. J Biol
Chem. 278:48–53. 2003. View Article : Google Scholar
|
|
8
|
Suzuki A, Kusakai G, Kishimoto A, Lu J,
Ogura T and Esumi H: ARK5 suppresses the cell death induced by
nutrient starvation and death receptors via inhibition of caspase 8
activation, but not by chemotherapeutic agents or UV irradiation.
Oncogene. 22:6177–6182. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki A, Kusakai G, Kishimoto A, Shimojo
Y, Miyamoto S, Ogura T, Ochiai A and Esumi H: Regulation of
caspase-6 and FLIP by the AMPK family member ARK5. Oncogene.
23:7067–7075. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kusakai G, Suzuki A, Ogura T, Kaminishi M
and Esumi H: Strong association of ARK5 with tumor invasion and
metastasis. J Exp Clin Cancer Res. 23:263–268. 2004.PubMed/NCBI
|
|
11
|
Kusakai G, Suzuki A, Ogura T, Miyamoto S,
Ochiai A, Kaminishi M and Esumi H: ARK5 expression in colorectal
cancer and its implications for tumor progression. Am J Pathol.
164:987–995. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki A, Lu J, Kusakai G, Kishimoto A,
Ogura T and Esumi H: ARK5 is a tumor invasion-associated factor
downstream of Akt signaling. Mol Cell Biol. 24:3526–3535. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen P, Li K, Liang Y, Li L and Zhu X:
High NUAK1 expression correlates with poor prognosis and involved
in NSCLC cells migration and invasion. Exp Lung Res. 39:9–17. 2013.
View Article : Google Scholar
|
|
14
|
Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y,
Li H, Wang L, Wang X and Zhao C: MiR-204 inhibits human NSCLC
metastasis through suppression of NUAK1. Br J Cancer.
111:2316–2327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bell RE, Khaled M, Netanely D, Schubert S,
Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R,
et al: Transcription factor/microRNA axis blocks melanoma invasion
program by miR-211 targeting NUAK1. J Invest Dermatol. 134:441–451.
2014. View Article : Google Scholar
|
|
16
|
Lu S, Niu N, Guo H, Tang J, Guo W, Liu Z,
Shi L, Sun T, Zhou F, Li H, et al: ARK5 promotes glioma cell
invasion, and its elevated expression is correlated with poor
clinical outcome. Eur J Cancer. 49:752–763. 2013. View Article : Google Scholar
|
|
17
|
Chang XZ, Yu J, Liu HY, Dong RH and Cao
XC: ARK5 is associated with the invasive and metastatic potential
of human breast cancer cells. J Cancer Res Clin Oncol. 138:247–254.
2012. View Article : Google Scholar
|
|
18
|
Cui J, Yu Y, Lu GF, Liu C, Liu X, Xu YX
and Zheng PY: Overexpression of ARK5 is associated with poor
prognosis in hepatocellular carcinoma. Tumour Biol. 34:1913–1918.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang J, Li Z, Yu C, Chen M, Tian S and
Sun C: MiR-1181 inhibits stem cell-like phenotypes and suppresses
SOX2 and STAT3 in human pancreatic cancer. Cancer Lett.
356:962–970. 2015. View Article : Google Scholar
|
|
27
|
Polyak K and Weinberg RA: Transitions
between epithelial and mesenchymal states: Acquisition of malignant
and stem cell traits. Nat Rev Cancer. 9:265–273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar
|
|
29
|
Zuo JH, Zhu W, Li MY, Li XH, Yi H, Zeng
GQ, Wan XX, He QY, Li JH, Qu JQ, et al: Activation of EGFR promotes
squamous carcinoma SCC10A cell migration and invasion via inducing
EMT-like phenotype change and MMP-9-mediated degradation of
E-cadherin. J Cell Biochem. 112:2508–2517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jung H, Lee KP, Park SJ, Park JH, Jang YS,
Choi SY, Jung JG, Jo K, Park DY, Yoon JH, et al: TMPRSS4 promotes
invasion, migration and metastasis of human tumor cells by
facilitating an epithelial-mesenchymal transition. Oncogene.
27:2635–2647. 2008. View Article : Google Scholar
|
|
31
|
Christiansen JJ and Rajasekaran AK:
Reassessing epithelial to mesenchymal transition as a prerequisite
for carcinoma invasion and metastasis. Cancer Res. 66:8319–8326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bracken CP, Gregory PA, Kolesnikoff N,
Bert AG, Wang J, Shannon MF and Goodall GJ: A double-negative
feedback loop between ZEB1-SIP1 and the microRNA-200 family
regulates epithelial-mesenchymal transition. Cancer Res.
68:7846–7854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gregory PA, Bracken CP, Bert AG and
Goodall GJ: MicroRNAs as regulators of epithelial-mesenchymal
transition. Cell Cycle. 7:3112–3118. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma L, Young J, Prabhala H, Pan E, Mestdagh
P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S,
et al: miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin
and cancer metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
36
|
Li B, Jin H, Yu Y, Gu C, Zhou X, Zhao N
and Feng Y: HOXA10 is overexpressed in human ovarian clear cell
adenocarcinoma and correlates with poor survival. Int J Gynecol
Cancer. 19:1347–1352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsuji T, Ibaragi S and Hu GF:
Epithelial-mesenchymal transition and cell cooperativity in
metastasis. Cancer Res. 69:7135–7139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gavert N and Ben-Ze'ev A:
Epithelial-mesenchymal transition and the invasive potential of
tumors. Trends Mol Med. 14:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vasioukhin V, Bauer C, Degenstein L, Wise
B and Fuchs E: Hyperproliferation and defects in epithelial
polarity upon conditional ablation of alpha-catenin in skin. Cell.
104:605–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schlegelmilch K, Mohseni M, Kirak O,
Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J,
Brummelkamp TR, et al: Yap1 acts downstream of α-catenin to control
epidermal proliferation. Cell. 144:782–795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Masszi A, Di Ciano C, Sirokmány G, Arthur
WT, Rotstein OD, Wang J, McCulloch CA, Rosivall L, Mucsi I and
Kapus A: Central role for Rho in TGF-beta1-induced alpha-smooth
muscle actin expression during epithelial-mesenchymal transition.
Am J Physiol Renal Physiol. 284:F911–F924. 2003. View Article : Google Scholar
|
|
43
|
Cano A, Pérez-Moreno MA, Rodrigo I,
Locascio A, Blanco MJ, del Barrio MG, Portillo F and Nieto MA: The
transcription factor snail controls epithelial-mesenchymal
transitions by repressing E-cadherin expression. Nat Cell Biol.
2:76–83. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng W, Jiang Y, Liu C, Shen O, Tang W
and Wang X: Identification of aberrant promoter hypomethylation of
HOXA10 in ovarian cancer. J Cancer Res Clin Oncol. 136:1221–1227.
2010. View Article : Google Scholar : PubMed/NCBI
|