Introduction
HSPC238, also known as RNF181 (ring finger
protein 181), is a tumor suppressor gene located at chromosome
2p11.2 (1). It encodes a 153 amino
acid protein that contains a RING finger domain in its first 40 to
60 amino acid residues. It has been reported that the HSPC238
protein has ubiquitin ligase E3 enzyme activity (2). In a previous study, our group found
that HSPC238 was significantly downregulated in HCC when compared
with adjacent tissues. Further studies showed that HSPC238 could
significantly inhibit the proliferation and invasion of hepatoma
cells in vivo and in vitro, and that it played a
tumor suppressor role in the development of HCC (3). Previous studies also showed that
HSPC238 inhibited the growth of HepG2 and SMMC7721 cells through
the ERK/MAPK pathway, but the underlying molecular mechanism is
unclear.
The zinc finger proteins are a class of important
transcription factors that are deregulated in many human diseases,
including cancer (4). Moreover, the
zinc finger domain may be involved in many biological processes via
binding of Zn2+ and interactions with other proteins.
However, it is unclear with which proteins HSPC238 interacts. In
the present study, to provide a foundation for further subsequent
study of HSPC238's tumor suppressor function, we performed a yeast
two-hybrid assay to identify the proteins that interact with the
HSPC238, followed by validation with confocal microscopy,
co-immunoprecipitation and pull-down assays.
Materials and methods
Reagents and cells
The yeast two-hybrid system and the human fetal
liver cDNA library were purchased from Clontech Laboratories, Inc.,
(Mountain View, CA, USA). 293T, HepG2 and SMMC7721 cell lines were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Cells were grown in RPMI-1640 medium (Gibco,
Life Technologies, Grand Island, NY, USA) containing 10% fetal
bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml
streptomycin and were incubated at 37°C in a 5% CO2
atmosphere. Lipofectimine 2000 transfection reagents, and the
plasmids pGBKT7, pGADT7 and pCDNA3.1 (+) were purchased from Life
Technologies. DNA gel purification kits and a small plasmid
extraction kits were purchased from Axygen (Tewksbury, MA, USA).
Anti-HSPC238 rabbit antibody was obtained from Proteintech Group
Inc. (Chicago, IL, USA); anti-HMOX1, anti-MT2A, anti-RPS27A and
anti-ubiquitin B (UBB) antibodies were purchased from Abclonal
(Cambridge, MA, USA); and anti-HIS mouse antibody and anti-Flag
mouse antibody were purchased from Zoonbio Tech Co., Ltd. (Nanjing,
China). The secondary CY3-labeled goat anti-rabbit and FITC-labeled
goat anti-mouse antibodies were purchased from Rockland
Immunochemicals Inc. (Gilbertsville, PA, USA).
Yeast two-hybrid assay
The ORF of HSPC238 gene was amplified by PCR and
subcloned into the yeast two-hybrid bait vector pGBKT7 to get
vector pGBKT7-HSPC238. The vectors pGBKT7-HSPC238 and pGADT7 were
co-transformed into the yeast strain AH109 in SD-Trp-Leu medium.
Three reporter genes (LacZ, HIS and ADE2) were
used to detect self-activation of yeast transformants
(pGBKT7-HSPC238 + pGADT7). Transformants with no self-activation of
the three reporter genes were used for subsequent library
screening. Matchmaker Human Fetal Liver Library plasmids (cat. no.
638805; Clontech) were transformed into yeast AH109 that had been
co-transformed with pGBKT7-HSPC238 and pGADT7. The transformants
were plated on SD-3 + 3AT agar medium and incubated for one to two
weeks. One hundred and twenty-four transformants were further
tested with the three reporter genes (LacZ, HIS and
ADE2). Positive clone strains were inoculated in liquid
SD-Leu medium. Yeast plasmids were extracted and further
transformed into E. coli Top10 fresh competent cells for
amplification. Following extraction from E. coli, the
plasmids were subjected to DNA sequencing and BLAST alignment
analysis.
Confocal laser location detection
HSPC238 and target genes (HMOX1,
RPS27A, UBB and MT2A) were cloned into
pcDNA3.1(+) to get the recombinant plasmids HSPC238-pcDNA3.1(+),
HMOX1-pcDNA3.1(+), UBB-pcDNA3.1(+), RPS27A-pcDNA3.1(+) and
MT2A-pcDNA3.1(+). 293T or SMMC7721 cells were transformed with the
HSPC238-pcDNA3.1(+) plasmid together with pcDNA3.1(+)-target
plasmids. Colocalization of HSPC238 with the four proteins in 293T
and SMMC7721 cells was observed by confocal microscopy using
antibodies against the recombinant target protein and HSPC238.
Co-immunoprecipitation
Cells were washed with pre-chilled PBS twice,
re-suspended in cold RIPA lysis buffer (20 mmol/l Tris-HCl, pH 7.5;
150 mmol/l NaCl; 1 mmol/l Na2EDTA; 1 mmol/l EGTA; 1%
Triton; 2.5 mmol/l sodium pyrophosphate; 1 mmol/l
β-glycerophosphate; 1 mmol/l Na3VO4; 1 mg/ml
leupeptin; and 1 mmol/l phenylmethyl-sulfonylfluoride) and lysed on
ice for 15 min at 4°C. Protein A/G-agarose beads were washed twice
with PBS and diluted to 50% protein A/G agarose in PBS. After
centrifuging at 14,000 × g and 4°C for 15 min, the cell lysate
supernatant was immediately transferred to new tubes and added to
the 50% protein A/G agarose at a ratio of 100 µl/1 ml sample
solution. After shaking on a horizontal shaker at 4°C for 10 min,
the mixture was centrifuged at 14,000 × g for 15 min at 4°C. The
supernatant was transferred to new tubes and the concentration of
the total protein was determined with a BCA assay. Total protein
(500 µg) was mixed with 10 µg primary antibody in 500
µl of solution and shaken on a rotating shaker at 4°C
overnight. The next day, the mixture was centrifuged at 14,000 × g
for 5 sec. The pellet was washed with pre-chilled PBS three times,
re-suspended in 15 µl 2X SDS sample buffer and boiled at
100°C for 5 min. The sample was then subjected to western blot
analysis.
Pull-down assay
The purified 6xHis-tagged HSPC238 protein (6–10 mg)
in 100 µl buffer B [50 mM sodium phosphate buffer, pH 7.5,
100 mM NaCl, 5 mM MgCl2, 2 mM β-mercaptoethanol and 100
µg bovine serum albumin (BSA)/ml] was incubated with 1 ml of
a 50% slurry of Ni-NTA-agarose resin (Qiagen Inc., Valencia, CA,
USA) that was pre-equilibrated in buffer B. After 90-min incubation
at room temperature, the beads were washed three times with 400
µl buffer B and twice with 400 µl buffer B with 50 mM
imidazole. After an additional wash with 100 µl buffer B
with 50 mM imidazole, proteins bound to the beads were released by
adding 100 µl buffer B with 350 mM imidazole. The presence
of HSPC238 in different fractions was analyzed by western
blotting.
Western blotting
Equal amounts of protein from each sample were
denatured in SDS sample buffer and separated on a 12% SDS
polyacrylamide gel. Proteins were transferred to polyvinylidene
difluoride (PVDF) membranes in buffer containing 25 mM Tris, 192 mM
glycine and 20% methanol. Membranes were blocked with Tris-buffered
saline (TBS; 137 mM NaCl, 20 mM Tris-HCl, pH 7.5) containing 0.05%
Tween-20 and 5% dried milk powder, incubated first with primary
antibodies and then with horseradish peroxidase-conjugated
secondary antibodies. Signals were developed with the enhanced
chemiluminescence detection system (Pierce, Thermo Fischer
Scientific, Bonn, Germany).
Results
Yeast two-hybrid screening results
To identify proteins that potentially physically
interact with HSPC238, we performed a yeast two-hybrid screen with
HSPC238 as bait against Matchmaker Human Fetal Liver Library. One
hundred and twenty-four positive clones were picked from the first
screen and tested with LacZ, HIS and ADE2
selective media. Twelve genes were found that potentially
interacted with HSPC238 (Table I)
after sequencing plasmid DNA from clones that passed through
successive screenings. After reviewing the literature, we finally
selected HMOX1, MT2A, RPS27A and UBB
for further validation.
 | Table IProteins that potentially interact
with HSPC238. |
Table I
Proteins that potentially interact
with HSPC238.
| Name | Accession
number | Description |
|---|
| UBB | NM_018955.2 | Homo sapiens
ubiquitin B (UBB), mRNA |
| MT2A | NM_005953.3 | Homo sapiens
metallothionein 2A (MT2A), mRNA |
| HBA1 | NM_000558.3 | Homo sapiens
hemoglobin, α 1 (HBA1), mRNA |
| RPS27A | NM_001177413.1 | Homo sapiens
ribosomal protein S27a (RPS27A), transcript variant 3, mRNA |
| MT1E | NM_175617.3 | Homo sapiens
metallothionein 1E (MT1E), mRNA |
| NDUFB10 | NM_004548.2 | Homo sapiens
NADH dehydrogenase (ubiquinone) 1 β subcomplex, 10, 22 kDa
(NDUFB10), nuclear gene encoding mitochondrial protein, mRNA |
| MCM7 | NM_182776.1 | Homo sapiens
minichromosome maintenance complex component 7 (MCM7), transcript
variant 2, mRNA |
| ALB | NM_000477.5 | Homo sapiens
albumin (ALB), mRNA |
| ELL2 | NM_012081.5 | Homo sapiens
elongation factor, RNA polymerase II, 2 (ELL2), mRNA |
| MT1H | NM_005951.2 | Homo sapiens
metallothionein 1H (MT1H), mRNA |
| RPL5 | NM_000969.3 | Homo sapiens
ribosomal protein L5 (RPL5), mRNA |
| FGB | NM_001184741.1 | Homo sapiens
fibrinogen β chain (FGB), transcript variant 2, mRNA |
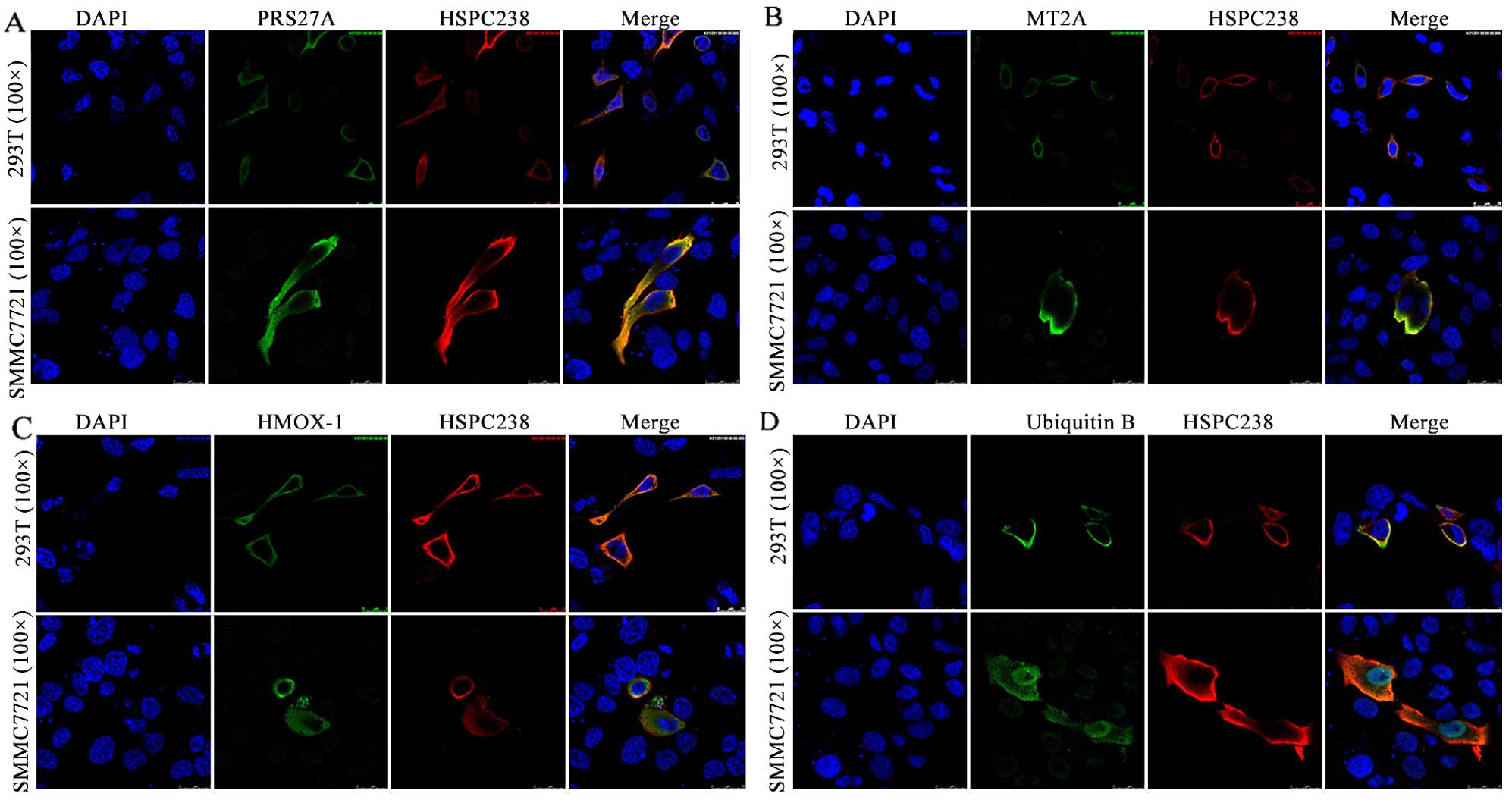
HMOX1, MT2A, RPS27A and UBB localize in
the cytoplasm and co-localize with HSPC238
We transfected 293T or SMMC7721 cells with
pcDNA3.1-HSPC238-Flag together with either pcDNA3.1-HMOX1–6xHis,
pcDNA3.1s-RPS27A-6xHis, pcDNA3.1-UBB-6xHis or pcDNA3.1-MT2A-6xHis,
and determined the co-localization of HSPC238 with the
corresponding candidate protein by confocal microscopy. In both
293T and SMMC7721 cells, RPS27A was found to co-localize with
HSPC238 (Fig. 1A). Similar
co-localization of HSPC238 with MT2A (Fig. 1B), HMOX1 (Fig. 1C) and UBB (Fig. 1D) were observed in both 293T and
SMMC7721 cells. Moreover, the results showed that HSPC238, HMOX1,
MT2A, RPS27A and UBB all localized in the cytoplasm of 293T and
SMMC7721 cells (Fig. 1).
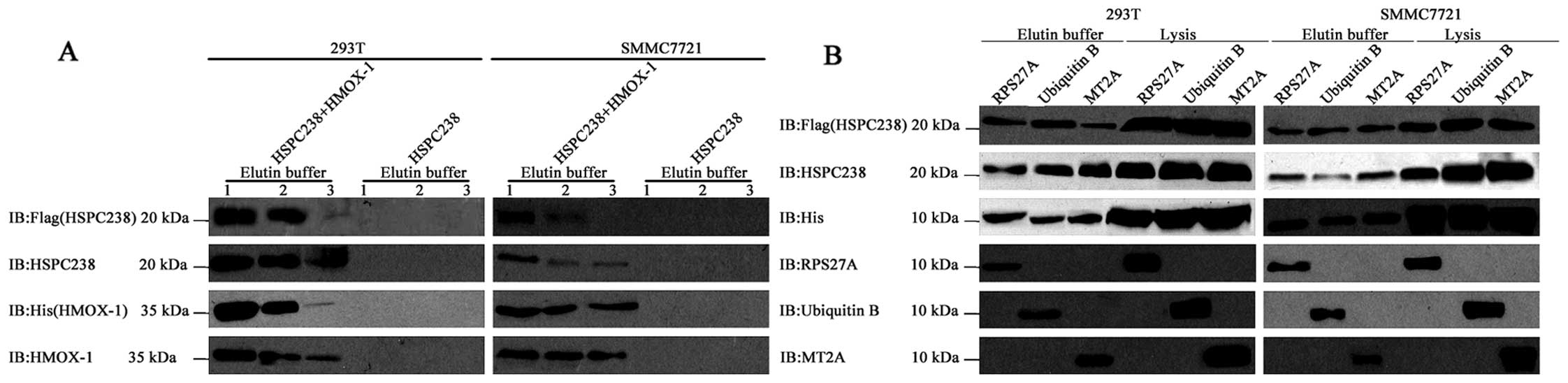
HMOX1, MT2A, RPS27A and UBB can be
co-immunoprecipitated with HSPC238
We performed co-immunoprecipitation assays to
confirm the physical interaction of HSPC238 with HMOX1, MT2A,
RPS27A and UBB. No endogenous Flag or His was observed in either
293T or SMMC7721 cells, while transfection of pcDNA3.1-HSPC238-Flag
(Fig. 2A, left panel) and
pcDNA3.1-HMOX1-His (Fig. 2A, right
panel) vectors resulted in high levels of Flag and His signals,
respectively. Similarly, strong His signals were detected in both
cell lines when they were transfected with pcDNA3.1-RPS27A-His,
pcDNA3.1-UBB-His or pcDNA3.1-MT2A-His (Fig. 2B). In the HSPC238 single
transfection, no specific HMOX1 band was detected with either
anti-His or anti-HMOX1 antibody in the IP precipitate or the cell
lysates (Fig. 2C). In contrast,
following co-transfection of pcDNA3.1-HSPC238-Flag and
pcDNA3.1-HMOX1-His, HMOX1 was detected with both anti-His and
anti-HMOX1 antibody in both the IP precipitate and the cell lysates
(Fig. 2C). Similarly, RPS27A, UBB
and MT2A were detected by either anti-His or anti-RPS27A, UBB or
MT2A antibody, respectively, in both the IP precipitate and the
cell lysates (Fig. 2D). These
results indicated that HMOX-1, RPS27A, UBB and MT2A could be
co-immunoprecipitated with HSPC238.
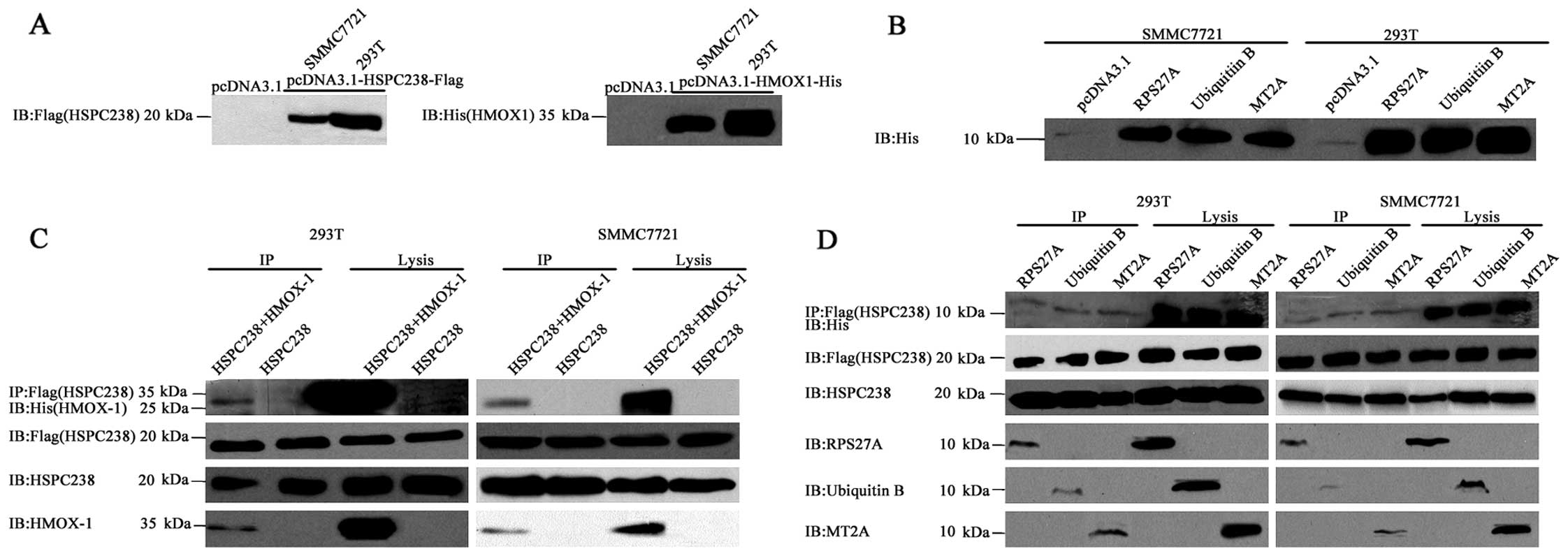 | Figure 2HMOX1, MT2A, RPS27A and UBB
co-immunoprecipitate with HSPC238. (A) 293T or SMMC7721 cells were
transfected with pcDNA3.1, pcDNA3.1-HSPC238-Flag or
pcDNA3.1-HMOX1-6xHis. Total proteins were extracted for
immunoblotting of His and Flag tag. (B) 293T or SMMC7721 cells were
transfected with pcDNA3.1, pcDNA3.1-RPS27A-6xHis,
pcDNA3.1-UBB-6xHis or pcDNA3.1-MT2A-6xHis. Total proteins were
extracted for immunoblotting of the His tag. (C) 293T or SMMC7721
cells were co-transfected with pcDNA3.1-HSPC238-Flag, or
pcDNA3.1-HSPC238-Flag together with pcDNA3.1-HMOX1-6xHis
pcDNA3.1-RPS27A-6xHis, pcDNA3.1-UBB-6xHis or pcDNA3.1-MT2A-6xHis.
Total proteins were extracted for co-immunoprecipitation (IP) with
anti-Flag antibodies, followed by immunoblotting (IB) with anti-His
tag, anti-HSPC238, anti-HMOX-1, anti-RPS27A, anti-UBB and anti-MT2A
antibodies. (D) 293T or SMMC7721 cells were co-transfected with
pcDNA3.1-HSPC238-Flag or pcDNA3.1-HSPC238-Flag together with
pcDNA3.1-HMOX1-6xHis. Total proteins were extracted for
co-immunoprecipitation (IP) with anti-Flag antibodies, followed by
immunoblotting (IB) with anti-His tag, HSPC238, HMOX-1, RPS27A, UBB
and MT2A antibodies. |
HSPC238 can be pulled down by HMOX-1,
RPS27A, UBB or MT2A
We performed protein pull-down assays to examine the
interaction of HSPC238 with HMOX1, MT2A, RPS27A and UBB. In the
HSPC238 single transfection, no specific HSPC238 bands were
detected with either anti-Flag or anti-HSPC238 antibody in the
eluents (Fig. 3A). In the
co-transfection of pcDNA3.1-HSPC238-Flag and pcDNA3.1-HMOX1-His,
HMOX1 was detected in the eluent by both anti-His and anti-HMOX1
antibody and HSPC238 was detected by both anti-Flag and
anti-HSPC238 antibody. In the co-transfection of
pcDNA3.1-HSPC238-Flag with all three of the other plasmids, RPS27A,
UBB and MT2A were detected with anti-His as well as anti-RPS27A,
UBB or MT2A antibody, respectively, in both the eluents and the
cell lysates. In this co-transfection, HSPC238 was detected by both
anti-Flag and anti-HSPC238 antibody in eluents and cell lysates
(Fig. 3B). These results indicate
that either HMOX-1, RPS27A, UBB or MT2A can pull down HSPC238.
Discussion
In the present study, we successfully screened 12
proteins that interact with HSPC238 using a yeast two-hybrid assay.
Among these proteins, HMOX1, RPS27A, UBB and MT2A were further
validated by co-localization, co-immunoprecipitation and pull down
assays. Our results provide new insights concerning the molecular
functions of HSPC238.
HSPC238 is a tumor suppressor gene that is
known to have a special RING finger domain (1). The RING finger domain, which is a
novel zinc finger protein domain defined by Lovering in 1994, may
be involved in many biological processes via binding of
Zn2+ and interaction with other proteins. The zinc
finger proteins are a class of important transcription factors that
are deregulated in many human diseases, including cancers (4). Brophy et al (2) confirmed that HSPC238 has ubiquitin
ligase E3 enzyme activity. Because the ubiquitin ligase E3 enzymes
are the key enzymes in the ubiquitin-proteasome pathway, HSPC238
may play a tumor suppressor role by promoting the degradation of
tumor-promoting proteins or by interfering with the degradation of
tumor suppressors.
Previous studies (2–4) showed
that HSPC238 inhibited the growth of HepG2 and SMMC7721 cells
through the ERK/MAPK pathway, but the underlying molecular
mechanism remains unclear. In the present study, we demonstrate the
physical interaction of HMOX-1, RPS27A, UBB and MT2A with HSPC238
with yeast two-hybrid, confocal laser, co-immunoprecipitation and
pull-down techniques. The physical interactions we report here
between these proteins may lead to the discovery of novel
mechanisms through which HSPC238 regulates the cellular
processes.
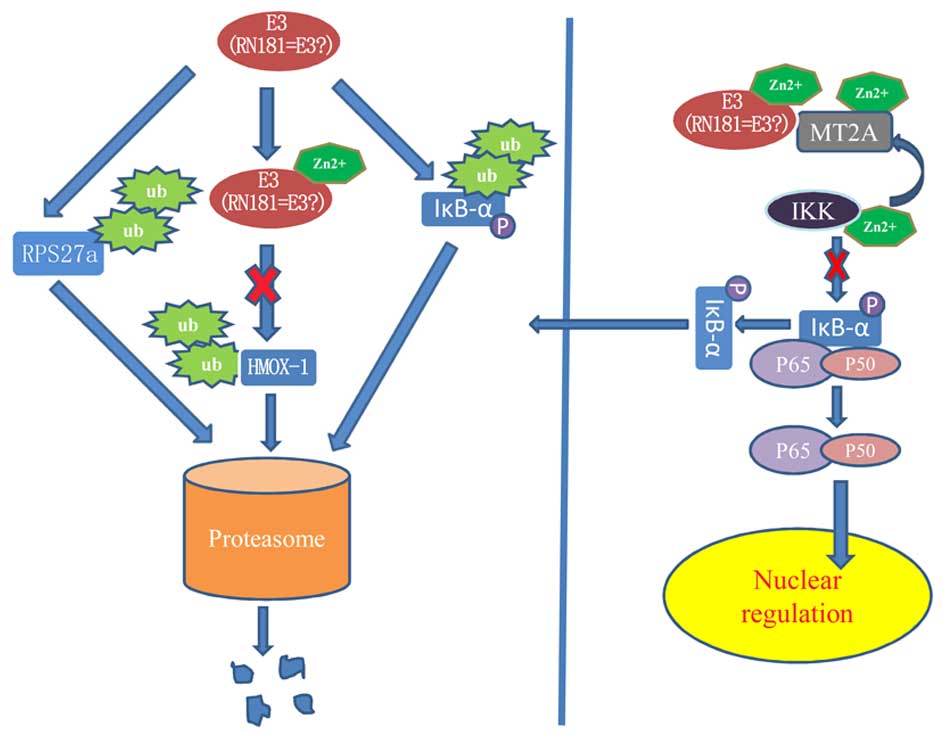
HSPC238 with RPS27a
RPS27a is a ribosomal protein that plays an
important role in protein synthesis. Accumulating evidence
indicates that ribosomal proteins have other functions, such as the
regulation of DNA replication and repair, mRNA transcription and
processing, cell apoptosis, malignant transformation of normal
cells and developmental regulation. All of these functions have
significance for tumorigenesis and anticancer drug resistance.
Wong et al (5) reported that the RPS27a gene is
overexpressed in human colon cancer cells. Ganger et al
(6) showed that RPS27a was
expressed at low levels in chronic hepatitis cells, but was highly
expressed in cirrhotic and hepatocellular carcinoma cells.
Similarly, Fatima et al (7)
found that RPS27a may be an important factor during the processes
of HB virus-mediated liver cancer. These facts suggest that RPS27a
might play a key role in the tumorigenesis of liver cancer. Our
previous studies (3) have shown
that HSPC238 had ubiquitin ligase E3 activity and significantly
inhibited the growth of hepatoma cells in vivo and in
vitro. The physical interaction of HSPC238 with RPS27a suggests
that HSPC238 may function as a tumor suppressor by promoting the
degradation of RPS27a through the ubiquitin-proteasome pathway
(Fig. 4).
HSPC238 with HMOX-1
HMOX-1 is an isoform of heme oxygenase, a key heme
degradation enzyme that decomposes hemoglobin into carbon monoxide,
ferrous ion and biliverdin (8).
HMOX-1 plays an important role in the self-protection of cells;
however, HMOX-1 also plays key roles during tumorigenesis through
its anti-oxidative and anti-apoptotic functions. In a chemical
carcinogenesis liver cancer experiment, Cabalero et al
(9) found that decreased expression
of HMOX-1 is associated with malignant tumor progression. However,
HMOX-1 expression in the Kuffer cells of macrophages and tumor
nodules in the periphery of necrotic tissue is increased,
indicating that HMOX-1 may have a protective role in chronic liver
injury and suggesting that HMOX-1 is a potential agent for the
treatment of primary liver cancer. Zou et al (10) reported that HMOX-1 inhibited the
migration and growth of hepatocellular carcinoma cells by
inhibiting the expression of IL-6 in vitro and in
vivo. Carbon monoxide generation by heme oxygenase also
inhibits the migration of cancer cells by inhibiting the activity
of IL-6 and p38MAPK. These studies indicate that HMOX-1 might
prevent the tumorigenesis of liver cancer via a mechanism similar
to heme oxygenase. Lin et al (11) reported that HMOX-1 degradation was
mediated by a Zn2+ dependent RING-E3 enzyme.
Zn2+ chelating agents significantly inhibited the
ubiquitination of HMOX-1, whereas ubiquitination was significantly
enhanced by ZnCl2. Because HSPC238 contains a
Zn2+ binding zinc finger structure, the interaction of
HSPC238 with HMOX-1 suggest that HSPC238 may inhibit tumorigenesis
by preventing the ubiquitination and degradation of HMOX-1
(Fig. 4).
HSPC238 with MT2A
Of all the MT-II protein subtypes, MT-2A is the only
one with a known biological effect (12). Not only does MT-2A have
anti-radiation, heavy metal detoxification, free radical
scavenging, tissue damage repair, trace element balance and
anti-aging effects, but it is also associated with cell
proliferation, apoptosis, tumor development and drug resistance.
MT2A is overexpressed in breast, lung, bladder and ovarian cancer,
but downregulated in liver and gastric cancer. It was found that
hepatitis B virus infection was associated with downregulation of
MT2A expression in hepatoma cells (13). Mao et al (14) reported that exogenous MT1M inhibited
the growth of hepatoma cells in vitro and in vivo by
reducing TNF-α-induced activation of NF-κB. Pan et al
(15) showed that MT2A inhibited
the activation of the NF-κB pathway by upregulating IκB, thereby
inhibiting the growth of gastric cancer cells. Similarly, restoring
the expression of MT2A in vitro and in vivo could
inhibit the growth of gastric cancer cells. Habel et al
(16) found that overexpression of
MT2A reduced the intracellular activity of zinc ions and the growth
kinetics of cells, thereby inhibiting the growth of osteosarcoma
cells. In addition, Zn2+ was found to enhance the
phosphorylation of IκB, which in turn promoted the activity of
NF-κB (17). Various signals, such
as virus infections, lead to the phosphorylation and degradation of
IκB via the ubiquitin-proteasome pathway. These facts suggest that
in the NF-κB pathway, MT2A inhibits the activation of IKK via
chelating zinc ions, thereby inhibiting the phosphorylation,
ubiquitination and degradation of IκB and, therefore, preventing
the activation of NF-κB. HSPC238 can interact with Zn2+,
and it may inhibit the activation of NF-κB by a process similar to
that described above. Our present study shows that HSPC238
interacts with MT2A (Fig. 4). We
speculate that a HSPC238 and MT2A complex may exert a synergistic
antitumor effect.
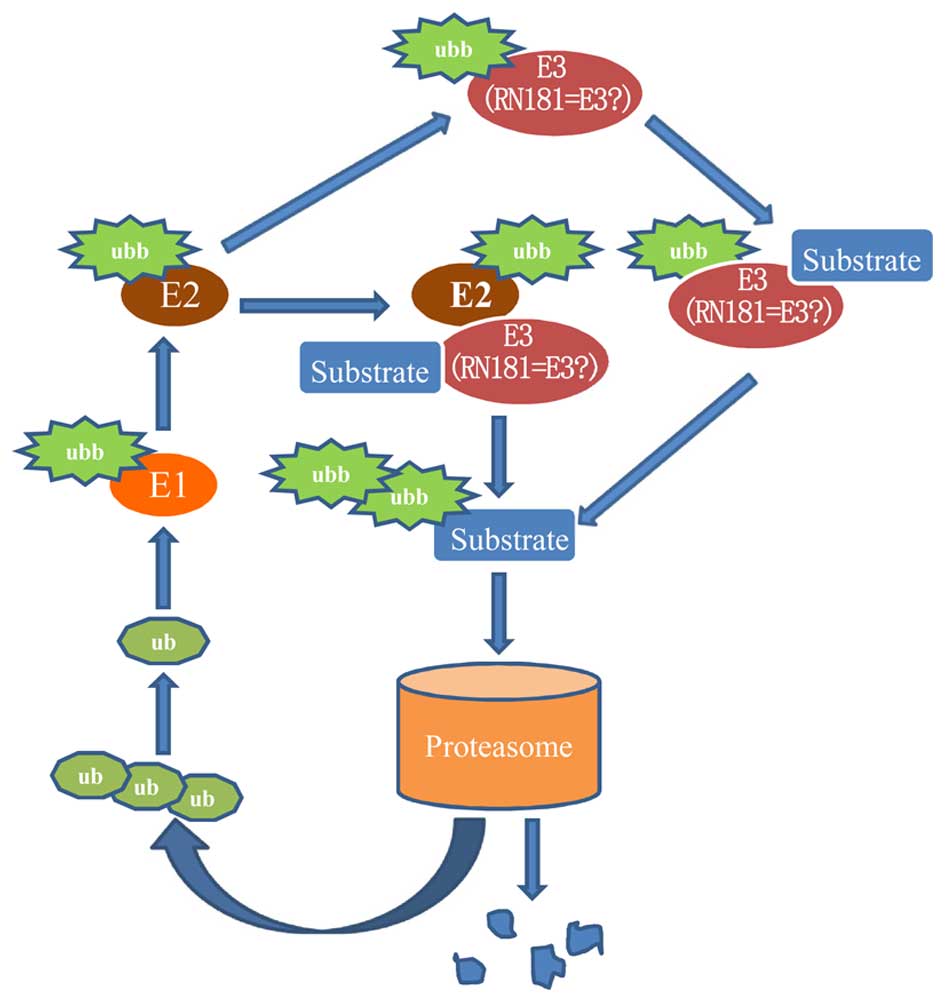
HSPC238 with UBB
Ubiquitin B is a ubiquitin protein, but its function
is largely unknown. Tian et al (18) reported that UBB might promote the
apoptosis of cervical cancer cells. Our observation that UBB
interacts with HSPC238 implies that UBB may be involved in the
multi-step processes of tumorigenesis through the
ubiquitinproteasome pathway (Fig.
5).
In summary, the four proteins screened in the
present study have both direct and indirect relationships with the
ubiquitin-proteasome pathway. The dysregulation of the
ubiquitinproteasome pathway is closely associated with the
development of a variety of diseases, including diseases of the
cardiovascular system (19–21), lymphoma (22), kidney disease (23,24),
nervous system disorders (25,26),
gastrointestinal malignancies (27–29)
and has an especially strong association with the development of
many cancers. HSPC238 has a RING-finger domain and E3 enzyme
activity. Since E3 enzyme activity is a key component of the
ubiquitin-proteasome pathway, we speculate that HSPC238 may
interact with HO-1, MT2A, RPS27a and UBB in the
ubiquitin-proteasome pathway. Thus, HSPC238 may suppress
tumorigenesis by: i) indirectly promoting the ubiquitination of the
cancer-promoting protein RPS27a; ii) inhibiting the NF-κB pathway
through the binding of Zn2+ and the formation of a
complex with MT2A; iii) preventing the ubiquitination of tumor
suppressor protein HO-1 by binding Zn2+; and iv)
combining and regulating the function of the small molecule
ubiquitin UBB.
Acknowledgments
This research was supported by the Natural Science
Foundation of China (no. 81101534); the Natural Science Foundation
of Guangdong Province (no. S2012010010824); and the Medical
Research Foundation of Guangdong Province (no. A2013875).
References
|
1
|
Joazeiro CA, Wing SS, Huang H, Leverson
JD, Hunter T and Liu YC: The tyrosine kinase negative regulator
c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase.
Science. 286:309–312. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brophy TM, Raab M, Daxecker H, Culligan
KG, Lehmann I, Chubb AJ, Treumann A and Moran N: RN181, a novel
ubiquitin E3 ligase that interacts with the KVGFFKR motif of
platelet integrin alpha(IIb)beta3. Biochem Biophys Res Commun.
369:1088–1093. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang S, Huang X, Li Y, Lao H, Zhang Y,
Dong H, Xu W, Li JL and Li M: RN181 suppresses hepatocellular
carcinoma growth by inhibition of the ERK/MAPK pathway. Hepatology.
53:1932–1942. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lange R, Christoph A, Thiesen HJ, Vopper
G, Johnson KR, Lemaire L, Plomann M, Cremer H, Barthels D and
Heinlein UA: Developmentally regulated mouse gene NK10 encodes a
zinc finger repressor protein with differential DNA-binding
domains. DNA Cell Biol. 14:971–981. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wong JM, Mafune K, Yow H, Rivers EN,
Ravikumar TS, Steele GD Jr and Chen LB: Ubiquitin-ribosomal protein
S27a gene overexpressed in human colorectal carcinoma is an early
growth response gene. Cancer Res. 53:1916–1920. 1993.PubMed/NCBI
|
|
6
|
Ganger DR, Hamilton PD, Klos DJ, Jakate S,
McChesney L and Fernandez-Pol JA: Differential expression of
metallopanstimulin/S27 ribosomal protein in hepatic regeneration
and neoplasia. Cancer Detect Prev. 25:231–236. 2001.PubMed/NCBI
|
|
7
|
Fatima G, Mathan G and Kumar V: The HBx
protein of hepatitis B virus regulates the expression,
intracellular distribution and functions of ribosomal protein S27a.
J Gen Virol. 93:706–715. 2012. View Article : Google Scholar
|
|
8
|
Wu ML, Layne MD and Yet SF: Heme
oxygenase-1 in environmental toxin-induced lung disease. Toxicol
Mech Methods. 22:323–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caballero F, Meiss R, Gimenez A, Batlle A
and Vazquez E: Immunohistochemical analysis of heme oxygenase-1 in
preneoplastic and neoplastic lesions during chemical
hepatocarcinogenesis. Int J Exp Pathol. 85:213–222. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zou C, Zhang H, Li Q, Xiao H, Yu L, Ke S,
Zhou L, Liu W, Wang W, Huang H, et al: Heme oxygenase-1: a
molecular brake on hepatocellular carcinoma cell migration.
Carcinogenesis. 32:1840–1848. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin PH, Chiang MT and Chau LY:
Ubiquitin-proteasome system mediates heme oxygenase-1 degradation
through endoplasmic reticulum-associated degradation pathway.
Biochim Biophys Acta. 1783:1826–1834. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coyle P, Philcox JC, Carey LC and Rofe AM:
Metallothionein: the multipurpose protein. Cell Mol Life Sci.
59:627–647. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tao X, Zheng JM, Xu AM, Chen XF and Zhang
SH: Downregulated expression of metallothionein and its
clinicopathological significance in hepatocellular carcinoma.
Hepatol Res. 37:820–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mao J, Yu H, Wang C, Sun L, Jiang W, Zhang
P, Xiao Q, Han D, Saiyin H, Zhu J, et al: Metallothionein MT1M is a
tumor suppressor of human hepatocellular carcinomas.
Carcinogenesis. 33:2568–2577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan Y, Huang J, Xing R, Yin X, Cui J, Li
W, Yu J and Lu Y: Metallothionein 2A inhibits NF-kappaB pathway
activation and predicts clinical outcome segregated with TNM stage
in gastric cancer patients following radical resection. J Transl
Med. 11:1732013. View Article : Google Scholar
|
|
16
|
Habel N, Hamidouche Z, Girault I,
Patiño-García A, Lecanda F, Marie PJ and Fromigué O: Zinc
chelation: a metallothionein 2A's mechanism of action involved in
osteosarcoma cell death and chemotherapy resistance. Cell Death
Dis. 4:e8742013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bao B, Prasad AS, Beck FW and Sarkar FH:
Zinc upregulates NF-kappaB activation via phosphorylation of
IkappaB in HUT-78 (Th0) cells. FEBS Lett. 581:4507–4511. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian Y, Ding W, Wang Y, Ji T, Sun S, Mo Q,
Chen P, Fang Y, Liu J, Wang B, et al: Ubiquitin B in cervical
cancer: critical for the maintenance of cancer stem-like cell
characters. PLoS One. 8:e844572013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cacciapuoti F: Role of
ubiquitin-proteasome system (UPS) in left ventricular hypertrophy
(LVH). Am J Cardiovasc Dis. 4:1–5. 2014.PubMed/NCBI
|
|
20
|
Powell SR, Herrmann J, Lerman A, Patterson
C and Wang X: The ubiquitin-proteasome system and cardiovascular
disease. Prog Mol Biol Transl Sci. 109:295–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schlossarek S and Carrier L: The
ubiquitin-proteasome system in cardiomyopathies. Curr Opin Cardiol.
26:190–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suh KS, Tanaka T, Sarojini S, Nightingale
G, Gharbaran R, Pecora A and Goy A: The role of the ubiquitin
proteasome system in lymphoma. Crit Rev Oncol Hematol. 87:306–322.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fukasawa H: The role of the
ubiquitin-proteasome system in kidney diseases. Clin Exp Nephrol.
16:507–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thomas SS and Mitch WE: Mechanisms
stimulating muscle wasting in chronic kidney disease: the roles of
the ubiquitin-proteasome system and myostatin. Clin Exp Nephrol.
17:174–182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rogers N, Paine S, Bedford L and Layfield
R: Review: the ubiquitin-proteasome system: contributions to cell
death or survival in neurodegeneration. Neuropathol Appl Neurobiol.
36:113–124. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying Z, Wang H and Wang G: The ubiquitin
proteasome system as a potential target for the treatment of
neurodegenerative diseases. Curr Pharm Des. 19:3305–3314. 2013.
View Article : Google Scholar
|
|
27
|
Dawson SP: Hepatocellular carcinoma and
the ubiquitin-proteasome system. Biochim Biophys Acta.
1782:775–784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grande E, Earl J, Fuentes R and Carrato A:
New targeted approaches against the ubiquitin-proteasome system in
gastrointestinal malignancies. Expert Rev Anticancer Ther.
12:457–467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Voutsadakis IA: The ubiquitin-proteasome
system in colorectal cancer. Biochim Biophys Acta. 1782:800–808.
2008. View Article : Google Scholar : PubMed/NCBI
|