Introduction
Epstein-Barr virus (EBV) is a human lymphotrophic
double-stranded γ-DNA herpes virus. EBV infection is common within
the global population, with ~90% of adults testing positive for
serum viral capsid antigen (VCA)-IgG antibody. Latent EBV usually
persists after infection, and may lead to malignant transformation
of host cells. EBV has been found to be involved in the occurrence
and progression of nasopharyngeal carcinoma, lymphoma, gastric
cancer, and other malignancies, and is considered as a human tumor
virus (1). EBV infection of resting
B lymphocytes in vitro may induce cell activation and
proliferation, potentially resulting in the establishment of
immortalized lymphoblastoid cell lines (LCLs). These LCLs express
virally-encoded proteins, including 6 EBV nuclear proteins (EBNAs),
EBNA-1, -2, -3A, -3B, -3C and -LP, and 3 latent membrane proteins
(LMPs), LMP-1, -2A and -2B (2,3),
associated with cell proliferation and tumorigenesis.
We previously confirmed that transplantation of
peripheral blood lymphocytes (PBLs) from healthy human subjects
with latent EBV infection into severe combined immunodeficiency
(SCID) mice induced EBV-associated human-origin B-cell lymphoma
(4,5). However, there has been no
comprehensive and systematic evaluation of viral LMP and EBNA gene
products in EBV-associated lymphomas. In the present study, we
evaluated EBV gene and protein expression levels in EBV-induced
lymphoma cells in SCID mice using quantitative real-time polymerase
chain reaction (qRT-PCR) and western blotting, and compared them
with matched lymphocytes from healthy human blood donors with
latent EBV infection to reveal the changes in EBV gene expression
profiles associated with EBV-induced lymphoma.
Materials and methods
Materials
Peripheral venous blood (300–400 ml) from nine
healthy blood donors was provided by Hengyang Blood Center.
SCID-Beige mice were purchased from Beijing Weitong Lihua
Experimental Animal Technology Co., Ltd. (Beijing, China). The
present study protocol was approved by the Medical Ethics Committee
of the University of South China.
Methods
Detection of the EBV infection status
of blood donors
Plasma EBV VCA-IgG was detected using an EBV-VCA-IgG
ELISA kit (ADL Embedded Solutions Inc., San Diego, CA, USA). The
presence of EBV DNA in the donor blood cells was detected by
extraction of DNA from whole blood, followed by PCR amplification
of the 82-bp LMP-1 gene sequence (GI: 896226, forward,
5′-CTGCTCATCGCTCTCTGGAA-3′ and reverse,
5′-AGACAAGTAAGCACCCGAAGATG-3′) (2,3). The
PCR reaction conditions were as follows: a 4-min pre-denaturation
at 94°C, a 30-sec denaturation at 94°C, a 30-sec annealing at 52°C,
a 30-sec extension at 72°C, for 30 cycles, followed by a 5-min
extension at 72°C. The size of the amplified product was 82 bp. The
PCR products were identified using 2% agarose gel electrophoresis,
with LMP-1 amplification results observed under UV using a
gel-imaging and analysis system.
Construction of Hu-PBL/SCID chimeric
mice
Hu-PBL/SCID chimeric mice were constructed by
inoculation of SCID mice with lymphocytes isolated from the
peripheral blood of human donors with latent EBV infection. PBLs
were isolated using lymphocyte separation medium (Tianjin Hao Yang
Biological Manufacture Co., Ltd., Tianjin, China) from peripheral
venous blood of healthy adults with latent EBV infection, diluted
to 8–10×107/ml using RPMI-1640 culture medium without
fetal bovine serum. Each SCID mouse was inoculated
intraperitone-ally with 1 ml PBL suspension under aseptic
conditions. PBLs from each blood donor were used to inoculate 3 or
4 SCID mice. After inoculation, each SCID mouse was administered
cyclosporin A (Sandoz Co., Novartis, Switzerland) via
intra-peritoneal injection at 10 mg/kg/day for 2 consecutive days.
The dose was adjusted to 15 mg/kg every other day from the third
day, for a total of 11 doses.
Development and pathological
examination of EBV-induced tumors in SCID mice
Hu-PBL/SCID chimeric mice were maintained in a
laminar-air-flow rack under specific pathogen-free conditions. The
mice were euthanized to reduce suffering in the event of death or
sickness, and surviving mice were sacrificed 4 months after PBL
inoculation. All mice were subjected to detailed autopsy. The
abdominal and mediastinal cavities and vital organs were examined,
and tumor shape, size, color, texture, and invasion of adjacent
organs were examined. Each induced tumor was divided into two
parts: one part was frozen in liquid nitrogen for RNA, DNA, or
protein extraction; the other part was fixed in 10% neutral
formalin, embedded in paraffin, and sliced into 4-µm serial
sections for routine hematoxylin and eosin or immunohistochemical
staining.
Immunohistochemical staining was performed using
antibodies against human leukocyte common antigen CD45 (LCA),
B-cell markers (CD20 and CD79a), and T-cell markers (CD45RO and
CD3) (Maixin Biotech. Co., Ltd., Fuzhou, China). The SP
immunohistochemistry and DAB kits were purchased from Fuzhou Maixin
Biotechnology Co., Ltd. (China). Staining was carried out according
to the manufacturer's instructions. Phosphate-buffered saline
replaced the primary antibody as a negative control.
PCR detection of human-specific Alu
sequence in induced tumors in SCID mice
DNA was extracted from tumors induced in SCID mice
and the 221-bp human-specific Alu sequence was amplified using PCR
(5). The sequence of the human Alu
primer was as follows: forward, 5′-CACCTGTAATCCCAGCAGTTT-3′ and
reverse, 5′-CGCGATCTCGGCTCACTGCA-3′. The PCR product was identified
by 1.5% agarose gel electrophoresis and observed under UV light
using a gel-imaging analysis system.
Determination of EBV gene expression
in induced tumors by qRT-PCR
Total RNA was extracted and purified and used to
synthesize cDNA. An RNA extraction kit (Omega Bio-Tek Inc.,
Doraville, GA, USA) was used to extract total RNA from induced
tumor tissues and matched lymphocytes from 'normal' donors, with
B95–8 cells as a positive control (B95–8
cells were derived from marmoset leukocytes transformed by EBV).
The quality and quantity of the RNA samples were determined using a
UV spectrophotometer (Perkin-Elmer, Fremont, CA, USA), followed by
storage at −80°C. In accordance with the instructions of the
reverse transcription kit (Promega Corporation, Madison, WI, USA),
2 µg RNA from each sample was used for cDNA synthesis. mRNA
expression was detected by qRT-PCR using SYBR Premix Ex Taq reagent
(Takara, Dalian, China). qRT-PCR was carried out in a 96-well plate
using an ABI 7700 Real-Time PCR system (Applied Biosystems, Foster
City, CA, USA). The primers used are listed in Table I.
 | Table IPrimer sequences of qRT-PCR
(5′-3′). |
Table I
Primer sequences of qRT-PCR
(5′-3′).
| Gene name | Forward primer | Reverse primer | Size (bp) |
|---|
| LMP-1 |
CTGCTCATCGCTCTCTGGAA |
AGACAAGTAAGCACCCGAAGATG | 82 |
| LMP-2A |
CGTCACTCGGACTATCAACCAC |
CTTCCTCTGCCCGCTTCTT | 149 |
| LMP-2B |
CGCCGTTTGACTGTTTGTG |
AGCAGCAGCGTCATGGAA | 125 |
| EBNA-1 |
GTTCCTCGCCTTAGGTTGTA |
AGCTCTCCTGGCTAGGAGTC | 124 |
| EBNA-2 |
GTCTGGCACATGCAAGACA |
TCTGCCACCTGCAACACTAA | 154 |
| EBNA-3A |
CTAATGGCCTGTCGAATGG |
TTTCAGCGCATCGACACA | 103 |
| EBNA-3B |
GGATCGTCACCACCATTGT |
GGTGGGATCTGAGCCTATTT | 159 |
| EBNA-3C |
GGCACATTGTCTTCCGTGTC |
TACAGACTACCGGCGAGCAT | 220 |
| EBNA-LP |
TCCCCTCGGACAGCTCCTA |
CCACTTACCACCTCCCCTTCT | 117 |
| GAPDH |
GCACCGTCAAGGCTGAGAAC |
TGGTGAAGACGCCAGTGGA | 138 |
RNA from B95–8 cells was used as a
template to obtain cDNA for qRT-PCR. A standard curve was
established based on the initial cDNA duplication number and CT
value for real-time fluorescent quantitative detection of
B95–8 cells. The template content in the sample was
determined by relative quantitation using the ΔΔCT method,
standardized by the duplication number of the housekeeping gene
glyceraldehyde 3-phosphate dehydrogenase. Each sample was tested in
triplicate.
Determination of EBV protein
expression in the induced tumors by western blotting
Total protein was extracted from sodium dodecyl
sulfate (SDS) lysates (SDS lysis buffer; Beyotime, China) of
induced tumor tissues and matched 'normal' lymphocytes. Proteins
were quantified using a BCA kit (Enhanced BCA protein assay kit;
Beyotime). Protein samples (50 µg) were separated using
8–12% SDS-polyacrylamide gel electrophoresis and electrotransferred
to polyvinylidene difluoride membranes (Millipore Corporation,
Boston, MA, USA). Membranes were blocked in Tris-buffered saline
containing 5% skimmed milk (TBST; 25 mm Tris-HCl, 150 mM NaCl, pH
7.5 and 0.05% Tween-20) for 4 h, followed by the addition of the
following diluted primary antibodies containing 5% skimmed milk:
EBNA-1 (mouse monoclonal anti-EBNA-1; Santa Cruz Biotechnology,
Santa Cruz, CA, USA), EBNA-2 (rat monoclonal anti-EBNA-2; Millipore
Corporation) and LMP-1 (mouse monoclonal anti-EBV LMP-1; Dako,
Glostrup, Denmark), with rat anti-human β-actin (anti-β-actin mouse
monoclonal antibody; Beijing ComWin Biotech, China) as an internal
control, at 4°C overnight. The TBST membrane was rinsed four times
for 10 min each. The corresponding secondary antibodies were then
added (Beijing ComWin Biotech), followed by incubation for 2 h at
room temperature. The membrane was then rinsed another four times
for 10 min each. ECL Plus luminescence reagent (Pierce ECL Western
Blotting Substrate; Pierce, Rockford, IL, USA) was then dripped
onto the membrane, followed by pressing, image developing, and
fixing in an X-ray machine, and image collection using a
fluorescence image-analysis system. Each sample was tested in
triplicate.
Statistical analysis
All experimental data are presented as mean ±
standard deviation. Induced lymphoma cells and matched 'normal'
lymphocytes were compared by t-tests, using SPSS13.0 software. A
value of P<0.05 was considered to indicate statistical
significance.
Results
EBV-infection status of the blood
donors
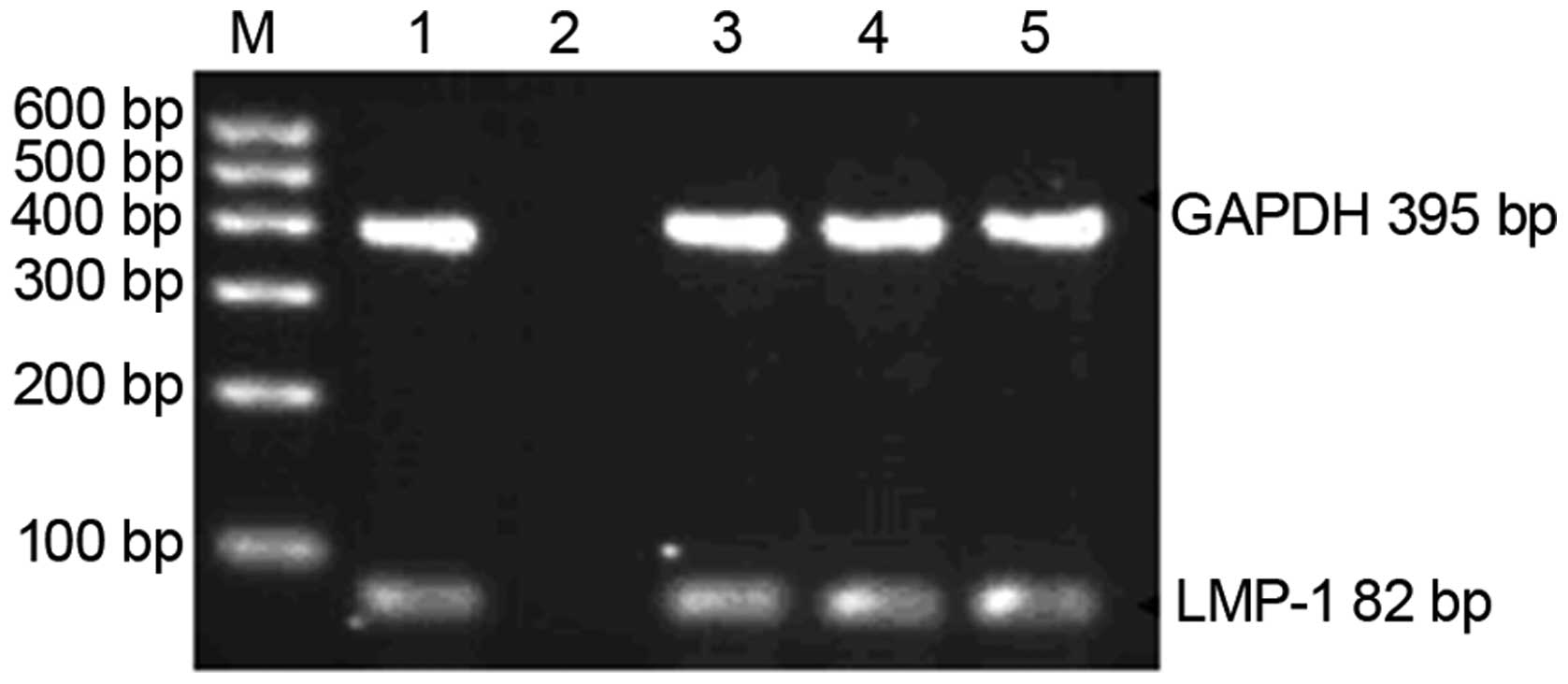
All 9 blood donors tested positive for latent EBV
infection according to both EBV-VCA-IgG detection and PCR
amplification of the LMP-1 gene of EB virus (Fig. 1).
Tumorigenesis in the SCID mice
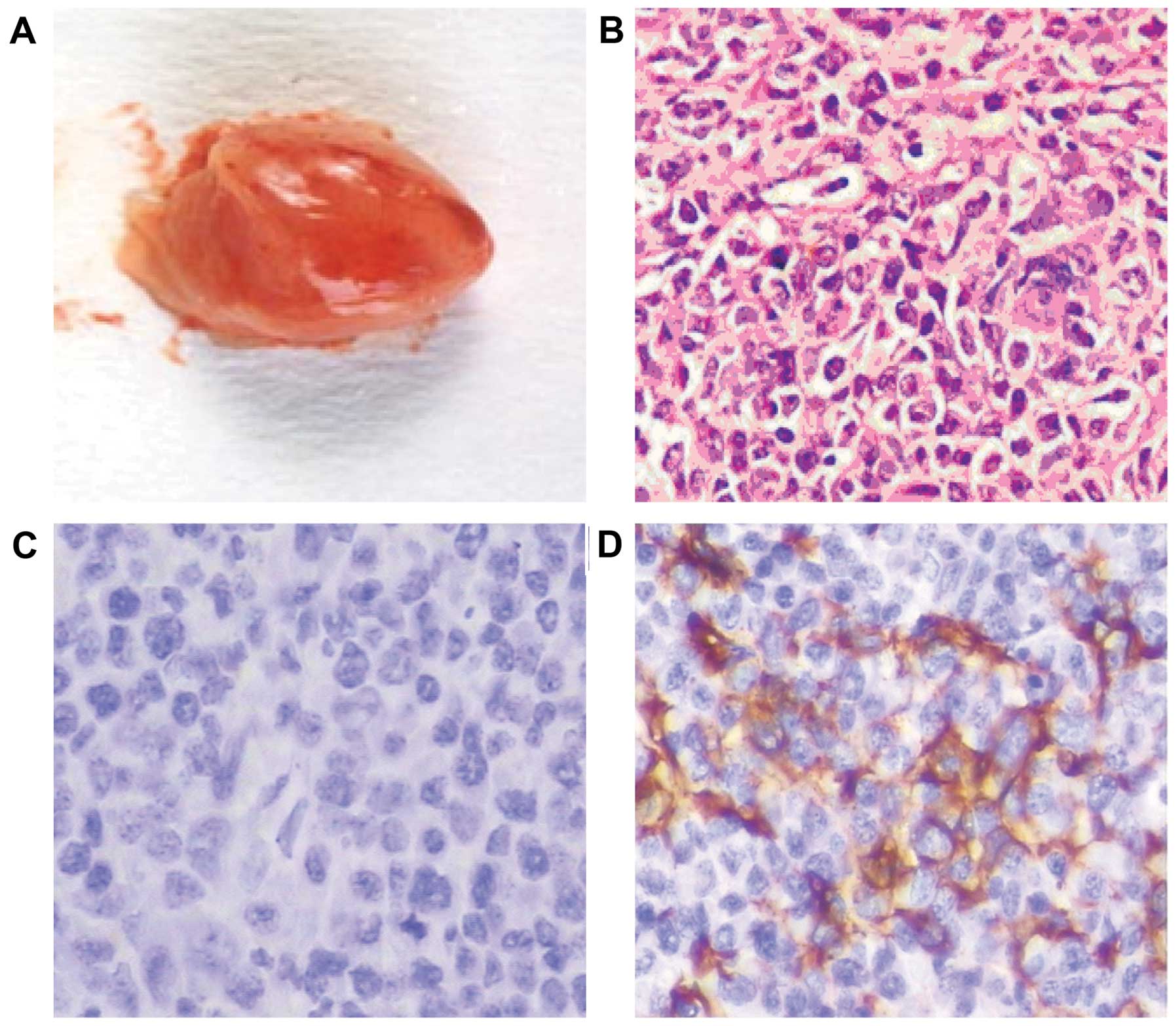
Tumors were formed in the mediastinum and
intraperitoneal cavity of the SCID mice. The induced tumors
appeared nodular, gray, or gray-red to the naked eye (Fig. 2A), with a fish-like texture in
cross-section. Under light microscopy, the tumor cells demonstrated
mixed morphology resembling plasmacytoid lymphocytes, centroblastic
cells and immunoblasts (Fig. 2B).
Immunohistochemical staining of the induced tumors was negative for
T-cell markers (CD3 and CD45RO) (Fig.
2C) and positive for LCA B-cell markers (CD20 and CD79a)
(Fig. 2D), and consistent with the
pathological diagnosis of diffuse large B-cell lymphoma.
Determination of the induced tumor origin
using Alu PCR
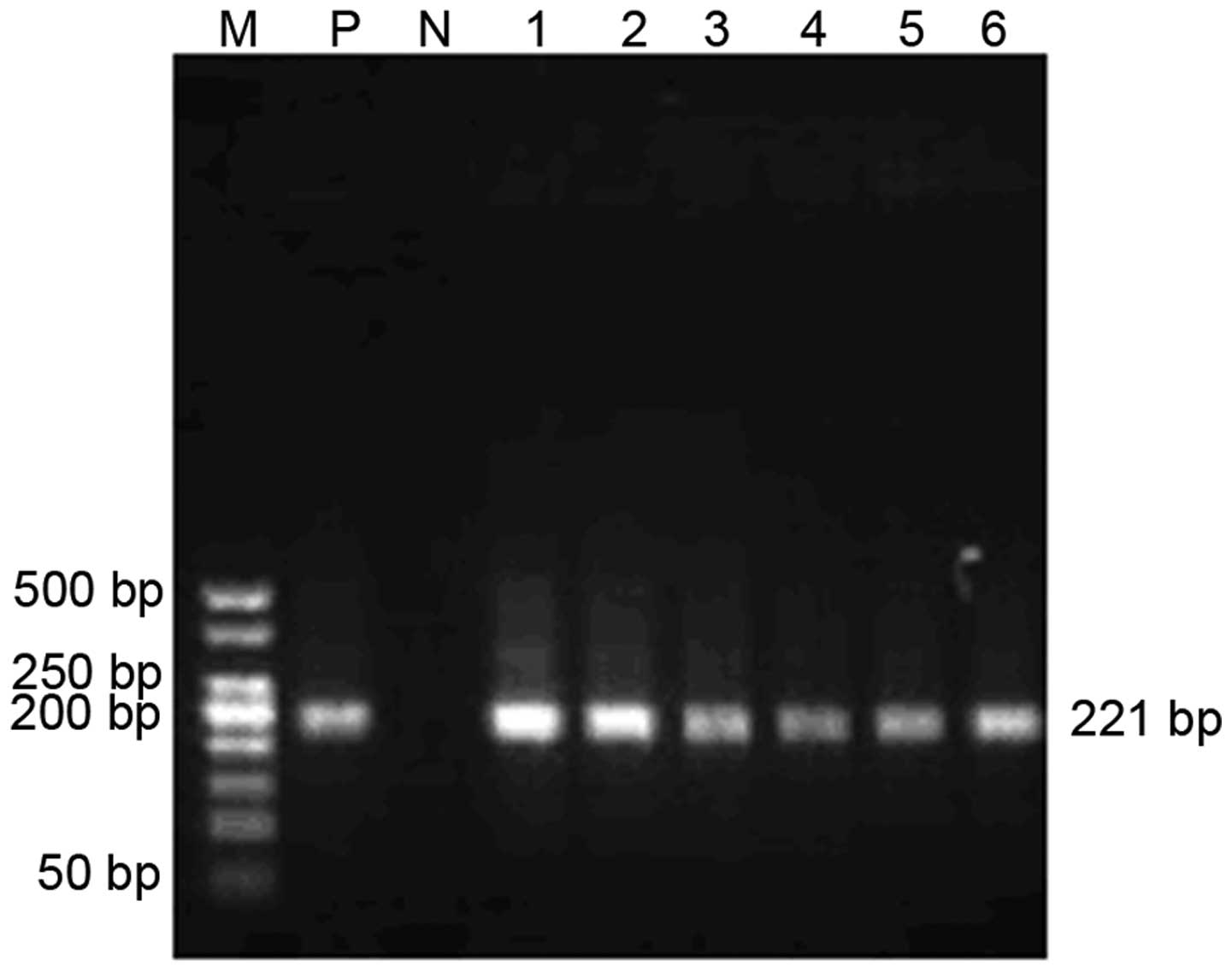
DNA was extracted from the tumor tissues, followed
by PCR amplification of the 221-bp human-specific Alu sequence,
which confirmed that the induced lymphomas were human in origin
(Fig. 3).
LMP and EBNA mRNA expression in induced
lymphomas
mRNA expression levels of the three LMPs were
detected in induced-lymphoma cells from nine Hu-PBL/SCID chimeric
mice and matched PBLs from donors by fluorescence quantitative PCR
(Table II). LMP-1 gene expression
in the induced-lymphoma cells was increased 256-fold compared with
the expression level in the 'normal' lymphocytes (P<0.05),
LMP-2A was upregulated 38-fold (P<0.05), and LMP-2B was
upregulated 331-fold (P<0.05).
 | Table IIExpression of LMPs in the EBV-induced
lymphoma cells and normal human lymphocytes (relative mRNA). |
Table II
Expression of LMPs in the EBV-induced
lymphoma cells and normal human lymphocytes (relative mRNA).
| Normal lymphocytes
(mean ± SD)×10−4 | EBV-induced
lymphoma (mean ± SD)×10−4 | P-value |
|---|
| LMP-1 | 2.414±1.080 |
617.402±122.101 | <0.05 |
| LMP-2A | 2.975±1.640 | 112.101±8.382 | <0.05 |
| LMP-2B | 0.640±0.304 | 211.603±48.420 | <0.05 |
mRNA expression levels of the six EBNA genes were
also detected by fluorescence quantitative PCR in induced-lymphoma
cells and matched PBLs (Table
III). EBNA-1 expression in the induced-lymphoma cells was
upregulated 1,157-fold compared with the expression level in the
'normal' lymphocytes (P<0.05), and EBNA-3A was upregulated
1,154-fold (P<0.05). EBNA-2, EBNA-3B, EBNA-3C and EBNA-LP were
not expressed in the 'normal' lymphocytes, but were significantly
expressed in the induced-lymphoma cells.
 | Table IIIExpression of EBNAs in the
EBV-induced lymphoma cells and normal human lymphocytes (relative
mRNA). |
Table III
Expression of EBNAs in the
EBV-induced lymphoma cells and normal human lymphocytes (relative
mRNA).
| Normal lymphocytes
(mean ± SD)×10−4 | EBV-induced
lymphoma (mean ± SD)×10−4 | P-value |
|---|
| EBNA-1 | 0.318±0.034 | 368.041±33.502 | <0.05 |
| EBNA-2 | 0.000±0.011 | 56.680±19.867 | <0.05 |
| EBNA-3A | 0.324±0.046 |
374.033±116.041 | <0.05 |
| EBNA-3B | 0.000±0.012 |
248.107±193.501 | <0.05 |
| EBNA-3C | 0.000±0.007 |
339.413±195.020 | <0.05 |
| EBNA-LP | 0.000±0.003 | 322.105±84.801 | <0.05 |
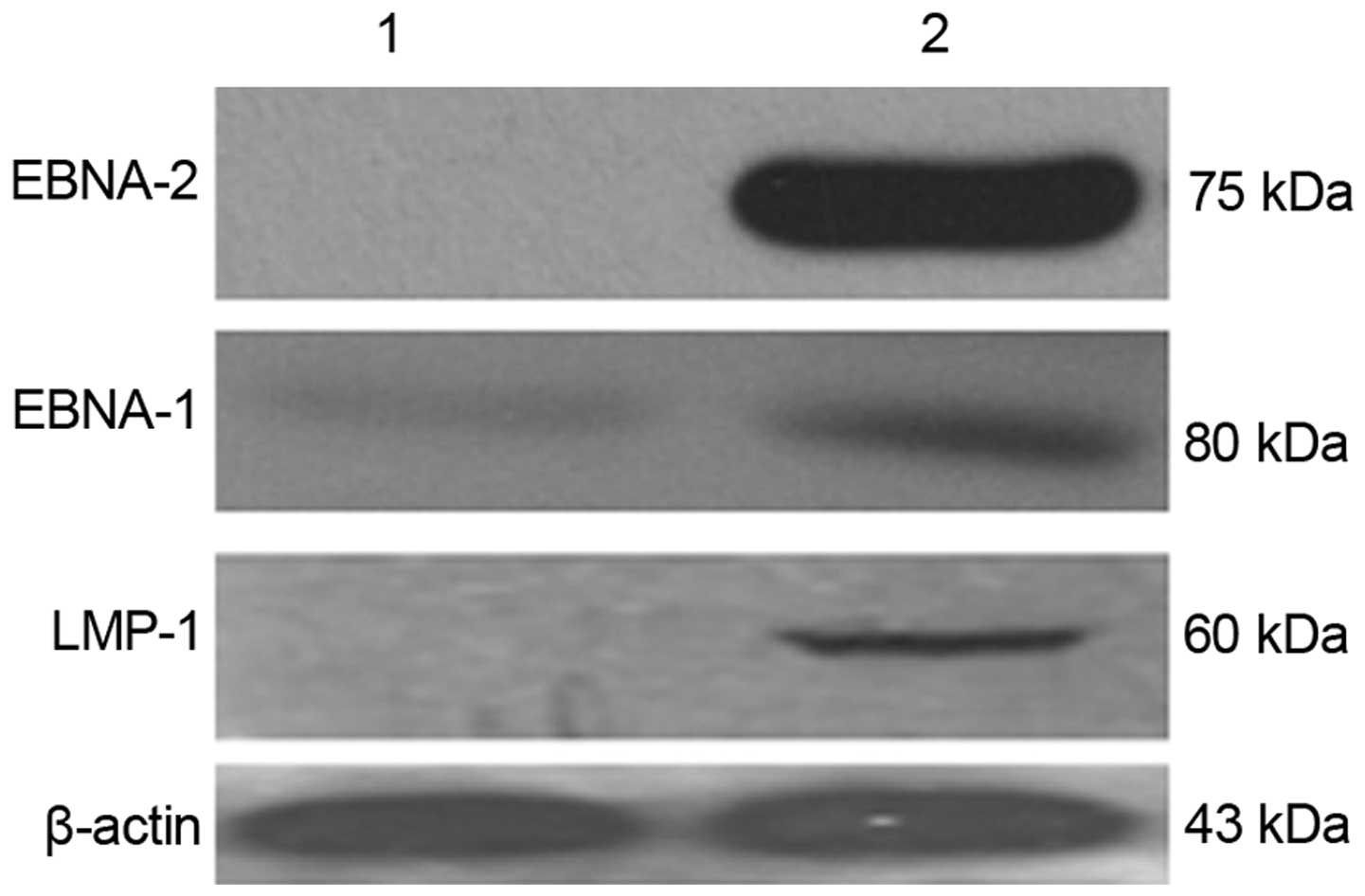
Detection of LMP-1, EBNA-1 and EBNA-2
protein expression by western blotting
LMP-1, EBNA-1 and EBNA-2 protein expression levels
in the EBV-induced lymphoma cells and matched 'normal' lymphocytes
were detected by western blotting. LMP-1, EBNA-1 and EBNA-2 protein
levels were significantly upregulated in the EBV-induced lymphomas
(Fig. 4), consistent with the mRNA
expression trends demonstrated by fluorescence quantitative PCR.
The EBNA-2 protein level was also increased in the EBV-induced
lymphoma cells compared with that in the matched 'normal'
lymphocytes before transplantation.
Discussion
EBV is an important human tumor virus, the oncogenic
effect of which is mainly realized through viral gene transcription
and the effects of the encoded proteins on the biological behavior
of the infected cells. It is therefore necessary to understand the
process of EBV genome transcription and expression during
tumorigenesis, as well as its functional activity in its host cells
(6–8).
LMP-1 has been considered to be the most important
oncogenic EBV gene, able to mediate cell proliferation and inhibit
apoptosis. Using qRT-PCR and western blotting, we showed that LMP-1
expression was upregulated in the EBV-induced lymphoma cells
compared with the expression level in the original 'normal'
lymphocytes with latent EBV infection, confirming and supporting
the role of LMP-1 in EBV-associated lymphomas. Zhang et al
constructed transgenic mice expressing LMP-1 and confirmed its
important role in B-cell proliferation and transformation (9). Increased expression of LMP-1 protein
promoted cell proliferation in NK/T cell lymphoma (10). Previous studies also showed that
LMP-1 activated β-catenin through the phosphatidylinositol
3-kinase/Akt signaling pathway, associated with the proliferation
of EBV-infected B cells (11).
LMP-1 could regulate DAPK1 expression and activate NF-κB signaling
in LCLs (12). Transcription
products of LMP-2A could be sustainably detected in EBV-associated
malignant tumors, suggesting that LMP-2A may play an important role
in persistent in vivo viral infection and EBV-associated
diseases (13). LMP-2A gene
expression was upregulated 38-fold in EBV-induced lymphoma cells
compared with 'normal' lymphocytes, according to qRT-PCR
(P<0.05), while LMP-2B was upregulated 331-fold (P<0.05).
These results suggest that LMP-2A and LMP-2B are also involved in
the occurrence and development of EBV-induced lymphomas. Previous
studies reported that LMP-2A could promote the malignant
transformation of cells and the survival and activation of B cells
by increasing the expression of genes associated with cell cycle
induction and apoptosis inhibition, together with LMP-1 (14,15).
Through regulating tumor necrosis factor receptor-associated factor
2 expression, LMP-2A regulates NF-κB signaling pathway activation
mediated by LMP-1 (16,17), thus helping lymphoma cells to escape
apoptosis. LMP-2B regulates the function of LMP-2A (13), preventing the potential lysis of
EBV, while high levels of LMP-2B expression also accelerate the
transition from latent to proliferative EBV infection.
EBNA-1 appears to be expressed in all EBV-associated
tumors, with significant implications for the stability of the EBV
genome in the host cells (18),
which is in turn crucial for the maintenance, replication, and
transcription of the EBV episome (19). EBNA-1, LMP-1 and LMP-2A may
participate in the progression of a variety of human malignant
tumors induced by EBV, and expression of these genes in the
infected cells thus indicates a potential risk of cancer (20). Our qRT-PCR and western blotting
results showed that EBNA-1 was upregulated in EBV-induced lymphoma
cells compared with the level in the 'normal' lymphocytes. In
addition to helping to maintain the viral episome, EBNA-1 can also
control viral replication and gene expression. Acting via a hidden
cis-acting mechanism, EBNA1 can help virus-carrying host
cells to escape the host immune system (21).
EBNA-2 is one of first viral gene products to be
expressed in host cells after EBV infection and plays a key role in
the immortalization of infected B cells (22). EBNA-2 is directly responsible for
initiating the transcription of EBV-related proteins during the
type III incubation period, resulting in excessive growth of LCLs
(23). After binding to
CBF1/RBP-JK, EBNA-2 promotes the expression of EBNA genes and LMP-1
protein, thus promoting cell growth and proliferation (24). EBNA-2 can also activate the signal
transduction pathway mediated by JAK-STAT, leading to continuous
proliferation of lymphocytes (25).
The present study demonstrated high levels of EBNA-2 expression in
induced-lymphoma cells, suggesting that it may play an important
role in the pathogenesis of EBV-associated lymphomas.
EBNA-3 consists of three subtypes EBNA-3A, EBNA-3B
and EBNA-3C associated with gene regulation. Young et al
(26) found that EBNA-3A induced
the re-distribution of heat shock protein 70 in immortalized
EBV-induced B lymphocytes in vitro, promoting the expression
of chaperone and co-chaperone proteins in the host cells. Although
EBNA-3B is not essential for B-cell immortalization, its expression
plays an important role in the proliferation and initiation of
EBV-infected B lymphocytes (27).
No previous studies have reported on the expression of EBNA-3 and
EBNA-LP genes in tumor cells, and their roles and functions remain
unknown. However, the present study found that EBNA-3A gene
expression was upregulated in EBV-induced tumor cells compared with
this level in the 'normal' lymphocytes. EBNA-3B, EBNA-3C and
EBNA-LP genes were not expressed in 'normal' cells, but were
positively expressed in EBV-induced lymphoma cells. Further studies
are needed to clarify the roles of EBNA-3 and EBNA-LP. Tursiella
et al (28) inhibited
EBNA-3A expression by transfection of EBNA-3A-specific shRNA and
observed that the cell cycle in Wp-R BL Sal cells and LCLs stopped
at G0/G1, with growth inhibition and
increased apoptosis, together with upregulation of
p21WAF1/CIP1 and Bim expression. Wp-R BL Sal cells may
suppress the expression of Bim through EBNA-3A, inhibiting
Bim-mediated apoptosis and thus affecting the oncogenic potential.
EBNA-3C is one of the essential antigens required for in
vitro primary B-cell transformation. EBNA-3C acts as a
transcriptional co-regulator by interacting with various cellular
and viral factors (29). EBNA-LP
and EBNA-2 were found to be co-expressed in EBV-infected B
lymphocytes and to play a critical role in the survival and growth
of lymphoblastoids (30). EBNA-LP
is also a co-activator of EBNA-2, which can enhance B-cell
transformation mediated by EBNA-2 (31). Furthermore, EBNA-LP and EBNA-2 can
induce the activation of cellular and viral genes, including LMP-1
and cyclin D2 (32). The above
results suggest that EBNA-3 and EBNA-LP may be important oncogenic
factors involved in the malignant transformation of lymphocytes by
EBV.
The results of the present study showed that LMP and
EBNA gene expression levels were upregulated in an EBV-associated
lymphoma model, suggesting that their products may play important
roles in the malignant transformation of lymphocytes. Further
studies are needed to explore the mechanisms responsible for the
actions of these EBV genes.
Acknowledgments
This research was supported by the National Natural
Science Foundation of China (grant nos. 81272182 and 81372134), and
the Construct Program of the Key Discipline in Hunan Province
(2011–76), and Hunan Province Cooperative Innovation Center for
Molecular Target New Drug Study (2014–405).
References
|
1
|
Lan K, Verma SC, Murakami M, Bajaj B and
Robertson ES: Epstein-Barr virus (EBV): Infection, propagation,
quantitation, and storage. Curr Protoc Microbiol Chapter 14. Unit
14E.2. 2007. View Article : Google Scholar
|
|
2
|
Rasul AE, Nagy N, Sohlberg E, Ádori M,
Claesson HE, Klein G and Klein E: Simultaneous detection of the two
main proliferation driving EBV encoded proteins, EBNA-2 and LMP-1
in single B cells. J Immunol Methods. 385:60–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tang Y, Luo C, Cheng A, Lu S, Xu J, Fu T
and Gan R: Expression of latent membrane proteins in Epstein Barr
virus-transformed lymphocytes in vitro. Mol Med Rep. 10:1117–1121.
2014.PubMed/NCBI
|
|
4
|
Gan R, Yin Z, Liu T, Wang L, Tang Y and
Song Y: Cyclosporine A effectively inhibits graft-versus-host
disease during development of Epstein-Barr virus-infected human B
cell lymphoma in SCID mouse. Cancer Sci. 94:796–801. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gan R, Xie X, He J, Liu X, Hong L, Tang Y,
Liu F and Xie H: Gene analysis of Epstein-Barr virus-associated
lymphomas in Hu-Pbl/SCID chimeras. Tumori. 96:465–472.
2010.PubMed/NCBI
|
|
6
|
Klein E, Kis LL and Klein G: Epstein-Barr
virus infection in humans: From harmless to life endangering
virus-lymphocyte interactions. Oncogene. 26:1297–1305. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Canaan A, Haviv I, Urban AE, Schulz VP,
Hartman S, Zhang Z, Palejev D, Deisseroth AB, Lacy J, Snyder M, et
al: EBNA1 regulates cellular gene expression by binding cellular
promoters. Proc Natl Acad Sci USA. 106:22421–22426. 2009.
View Article : Google Scholar
|
|
8
|
Klein G, Klein E and Kashuba E:
Interaction of Epstein-Barr virus (EBV) with human B-lymphocytes.
Biochem Biophys Res Commun. 396:67–73. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang B, Kracker S, Yasuda T, Casola S,
Vanneman M, Hömig-Hölzel C, Wang Z, Derudder E, Li S, Chakraborty
T, et al: Immune surveillance and therapy of lymphomas driven by
Epstein-Barr virus protein LMP1 in a mouse model. Cell.
148:739–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ramakrishnan R, Donahue H, Garcia D, Tan
J, Shimizu N, Rice AP and Ling PD: Epstein-Barr virus BART9 miRNA
modulates LMP1 levels and affects growth rate of nasal NK T cell
lymphomas. PLoS One. 6:e272712011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomita M, Dewan MZ, Yamamoto N, Kikuchi A
and Mori N: Epstein-Barr virus-encoded latent membrane protein 1
activates beta-catenin signaling in B lymphocytes. Cancer Sci.
100:807–812. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee CW, Leu SJ, Tzeng RY, Wang SF, Tsai
SC, Sun KH, Chen RH and Huang JC: Latent membrane protein 1 of
Epstein-Barr virus regulates death-associated protein kinase 1 in
lymphoblastoid cell line. Virology. 413:19–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rechsteiner MP, Berger C, Zauner L,
Sigrist JA, Weber M, Longnecker R, Bernasconi M and Nadal D: Latent
membrane protein 2B regulates susceptibility to induction of lytic
Epstein-Barr virus infection. J Virol. 82:1739–1747. 2008.
View Article : Google Scholar :
|
|
14
|
Bultema R, Longnecker R and
Swanson-Mungerson M: Epstein-Barr virus LMP2A accelerates
MYC-induced lymphomagenesis. Oncogene. 28:1471–1476. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wasil LR, Tomaszewski MJ, Hoji A and Rowe
DT: The effect of Epstein-Barr virus latent membrane protein 2
expression on the kinetics of early B cell infection. PLoS One.
8:e540102013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guasparri I, Bubman D and Cesarman E: EBV
LMP2A affects LMP1-mediated NF-kappaB signaling and survival of
lymphoma cells by regulating TRAF2 expression. Blood.
111:3813–3820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vrazo AC, Chauchard M, Raab-Traub N and
Longnecker R: Epstein-Barr virus LMP2A reduces hyperactivation
induced by LMP1 to restore normal B cell phenotype in transgenic
mice. PLoS Pathog. 8:e10026622012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Do NV, Ingemar E, Phi PT, Jenny A, Chinh
TT, Zeng Y and Hu L: A major EBNA1 variant from Asian EBV isolates
shows enhanced transcriptional activity compared to prototype
B95.8. Virus Res. 132:15–24. 2008. View Article : Google Scholar
|
|
19
|
Chen YL, Liu CD, Cheng CP, Zhao B, Hsu HJ,
Shen CL, Chiu SJ, Kieff E and Peng CW: Nucleolin is important for
Epstein-Barr virus nuclear antigen 1-mediated episome binding,
maintenance, and transcription. Proc Natl Acad Sci USA.
111:243–248. 2014. View Article : Google Scholar
|
|
20
|
Liu X, Tang J, Wang M, Ma Q and Wang Y:
Visual detection and evaluation of latent and lytic gene expression
during Epstein-Barr virus infection using one-step reverse
transcription loop-mediated isothermal amplification. Int J Mol
Sci. 14:23922–23940. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Daskalogianni C, Pyndiah S, Apcher S,
Mazars A, Manoury B, Ammari N, Nylander K, Voisset C, Blondel M and
Fåhraeus R: Epstein-Barr virus-encoded EBNA1 and ZEBRA: Targets for
therapeutic strategies against EBV-carrying cancers. J Pathol.
235:334–341. 2015. View Article : Google Scholar
|
|
22
|
Pagès F, Galon J, Karaschuk G, Dudziak D,
Camus M, Lazar V, Camilleri-Broët S, Lagorce-Pagès C, Lebel-Binay
S, Laux G, et al: Epstein-Barr virus nuclear antigen 2 induces
interleukin-18 receptor expression in B cells. Blood.
105:1632–1639. 2005. View Article : Google Scholar
|
|
23
|
Rowe M, Raithatha S and Shannon-Lowe C:
Counteracting effects of cellular Notch and Epstein-Barr virus
EBNA2: Implications for stromal effects on virus-host interactions.
J Virol. 88:12065–12076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yue W, Gershburg E and Pagano JS:
Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase
suppresses trans-activation of the LMP1 promoter. J Virol.
79:5880–5885. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Konforte D, Simard N and Paige CJ:
Interleukin-21 regulates expression of key Epstein-Barr virus
oncoproteins, EBNA2 and LMP1, in infected human B cells. Virology.
374:100–113. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Young P, Anderton E, Paschos K, White R
and Allday MJ: Epstein-Barr virus nuclear antigen (EBNA) 3A induces
the expression of and interacts with a subset of chaperones and
co-chaperones. J Gen Virol. 89:866–877. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen A, Zhao B, Kieff E, Aster JC and Wang
F: EBNA-3B- and EBNA-3C-regulated cellular genes in Epstein-Barr
virus-immortalized lymphoblastoid cell lines. J Virol.
80:10139–10150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tursiella ML, Bowman ER, Wanzeck KC, Throm
RE, Liao J, Zhu J and Sample CE: Epstein-Barr virus nuclear antigen
3A promotes cellular proliferation by repression of the
cyclin-dependent kinase inhibitor p21WAF1/CIP1. PLoS Pathog.
10:e10044152014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saha A and Robertson ES: Impact of EBV
essential nuclear protein EBNA-3C on B-cell proliferation and
apoptosis. Future Microbiol. 8:323–352. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Portal D, Zhou H, Zhao B, Kharchenko PV,
Lowry E, Wong L, Quackenbush J, Holloway D, Jiang S, Lu Y, et al:
Epstein-Barr virus nuclear antigen leader protein localizes to
promoters and enhancers with cell transcription factors and EBNA2.
Proc Natl Acad Sci USA. 110:18537–18542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Portal D, Zhao B, Calderwood MA,
Sommermann T, Johannsen E and Kieff E: EBV nuclear antigen EBNALP
dismisses transcription repressors NCoR and RBPJ from enhancers and
EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci
USA. 108:7808–7813. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yokoyama A, Tanaka M, Matsuda G, Kato K,
Kanamori M, Kawasaki H, Hirano H, Kitabayashi I, Ohki M, Hirai K,
et al: Identification of major phosphorylation sites of
Epstein-Barr virus nuclear antigen leader protein (EBNA-LP):
Ability of EBNA-LP to induce latent membrane protein 1
cooperatively with EBNA-2 is regulated by phosphorylation. J Virol.
75:5119–5128. 2001. View Article : Google Scholar : PubMed/NCBI
|