Introduction
Ovarian carcinoma is a leading cause of gynecologic
malignancies (1). It is estimated
that 5-year survival rates could be more than 90% in ovarian
cancers experiencing early detection of the malignancy;
nevertheless, due to the anatomical location the primary ovarian
cancer is insidious, fewer than 20% of ovarian cancers could be
detected at their early stages, this is also the main reason for
the high lethality of this malignancy (2,3). On
the other hand, ovarian carcinoma is comprised of histologically
diverse subtypes but still commonly treated as a single disorder
with limited stratification based on histological characteristics
or molecular genetic alterations, the 5-year survival rates of this
malignancy remains under 50% (4,5). This
prompted us to seek better understanding of the molecular genetic
alterations contributing to the initiation and progression of
ovarian cancer.
RAS/RAF/MEK/ERK cascade is a key signaling pathway
regulating diverse biological processes such as cell proliferation,
survival and programmed cell death (6,7).
Multiple lines of evidence have suggested that this signaling
pathway is frequently deregulated in human cancers as a result of
either genetic aberrations of their components or over-activation
of upstream cell-surface receptors (7,8).
Amongst these reported genetic alterations, mutations in the BRAF
and three RAS members (KRAS, NRAS and HRAS) were prevalent in human
cancers, while mutations in other components such as MAPK1 (ERK2)
were relatively infrequently detected (9-12). Of
note, a large-scale sequencing study have identified a high
frequency of MAPK1 mutations (6/79, 7.6%) in primary cervical
squamous cell carcinomas, among the 6 patients with MAPK1
mutations, 4 samples harbored MAPK1 p.E322K mutation and the
remaining 2 samples harbored either MAPK1 p.E81K or p.E220K
mutation; thus MAPK1 p.E322 mutation was considered as the
potential mutational hot spot (4/6, 66.7%) in cervical cancer
(13). Till now, the mutational
statues of MAPK1 mutations in other cancer types, such as ovarian
cancer, remain largely unexplored.
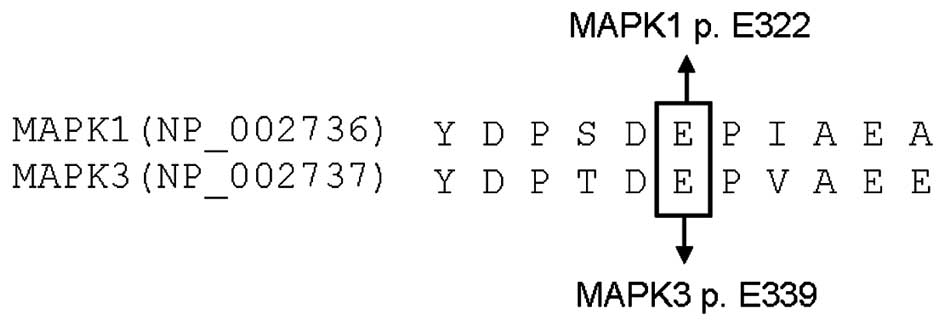
Considering the fact that there existed some
overlaps of molecular genetic aberrations between ovarian and
cervical carcinomas, such as prevalent PIK3CA and TP53 mutations
(13,14), it raises the possibility that
ovarian carcinoma may also harbor MAPK1 mutations. On the other
hand, homologous residues of paralogous genes are frequently
mutated in certain cancer types, thus we wanted to explore the
possibility that MAPK3 p.E339 residue, the homologue residue of
MAPK1 p.E322, would be also mutated in ovarian carcinoma.
In the present study, we analyzed a cohort of 263
Chinese ovarian cancer samples with distinct subtypes for the
presence of MAPK1 and MAPK3 mutations. Furthermore, the potential
hotspot mutations in several newly-identified cancer-related genes
were also analyzed in our samples previously and here, including
PPP2R1A, RNF43, POLE1, DICER1, CTCF, RPL22, DNMT3A, TRRAP, IDH1 and
IDH2 (15-18), with the aim of exploring the
possibility that these mutations would play synergistic role with
MAPK1 mutation in the development of this malignancy.
Materials and methods
Patients and ethics statement
A collection of 263 formalin-fixed,
paraffin-embedded (FFPE) samples with distinct subtypes of ovarian
carcinomas were recruited from the archives of Department of
Pathology, Jiangxi Provincial Maternal and Child Health Hospital
from March 2007 to May 2015. Each sample was reviewed by two
experienced pathologists, all of the recruited samples contained
>70% cancerous cells and did not undergo chemotherapy and/or
radiotherapy. Among these patients, 251 patients were described
previously (15–18) and the additional 12 patients
diagnosed with ovarian mixed germ cell tumor were newly recruited
(Table I). In addition, among these
cases, 238 paired adjacent non-cancerous samples were also taken
from archival blocks of these oophorectomy samples where no
cancerous cells were identified by hematoxylin-eosin staining. The
Institutional Review Board of Jiangxi Provincial Maternal and Child
Health Hospital approved this study and an informed consent was
obtained from each patient prior to this study. The study was
conducted according to the Declaration of Helsinki.
 | Table IThe mutations of the MAPK1 and MAPK3
genes in 263 samples with distinct subtypes of ovarian tumors. |
Table I
The mutations of the MAPK1 and MAPK3
genes in 263 samples with distinct subtypes of ovarian tumors.
| Subtype | No. | MAPK1 p.D321N | MAPK3 p.E339 |
|---|
| Epithelial | | | |
| Serous | 76 | 0/76 | 0/76 |
| Clear cell | 43 | 0/43 | 0/43 |
| Endometrioid | 37 | 0/37 | 0/37 |
| Mucinous | 15 | 0/15 | 0/15 |
|
Undifferentiated | 3 | 0/3 | 0/3 |
| Unclassified | 4 | 0/4 | 0/4 |
| Transitional
cell | 3 | 0/3 | 0/3 |
| Mixed | 2 | 0/2 | 0/2 |
| Non-epithelial germ
cell tumor | | | |
| Yolk sac | 11 | 0/11 | 0/11 |
| Dysgerminoma | 7 | 0/7 | 0/7 |
| Teratoma | 9 | 0/9 | 0/9 |
| Mixed | 18 | 2/18 | 0/18 |
| Gender
cord-stromal | | | |
| Granulosa
cell | 16 | 0/16 | 0/16 |
|
Sertoli-Leydig | 2 | 0/2 | 0/2 |
| Krukenberg
tumor | 17 | 0/17 | 0/17 |
MAPK1 and MAPK3 hotspot mutation
analyses
The genomic DNA of each sample was isolated by
commercially available kits (OMEGA Bio-Tek Inc., Doraville, GA,
USA) and the quantity and quality of DNA was determined
spectrophotometrically. For sequence analysis of the potential
MAPK1 and MAPK3 mutations, a 246- and a 231-bp PCR fragment
covering MAPK1 p.E322 and the paralogous MAPK3 p.E339 residue was
amplified by PCR, respectively, with the following primer pairs:
forward, 5′-CTGCTCTCACTACTGCAAAACC-3′ and reverse,
5′-TGGCAGCAGGTATATCTCAGG-3′ for MAPK1; forward,
5′-CTGACTCCTGCCCTTCCATA-3′ and reverse, 5′-GGGTGGTAGAGACAGCAAGG-3′
for MAPK3. A total of 200 ng of genomic DNA was used for each
amplification reaction in a total volume of 30 µl, after an
initial denaturation step at 94°C for 3 min, 35 cycles were run
with the following conditions: denaturation at 94°C for 30 sec,
annealing at 55 or 60°C for 30 sec, extension at 72°C for 30 sec;
finally followed by a final extension at 72°C for 10 min. All PCR
reactions were performed in a Thermal Cycler 2720 (Applied
Biosystems, Foster City, CA, USA). After purification, the PCR
products were subjected to DNA sequencing on an ABI Prism 3730 DNA
sequencer (Applied Biosystems). An independent PCR and
bidirectional sequencing was used to verify the identified
mutations. The somatic status of these identified MAPK1 mutations
were determined by sequencing the MAPK1 gene in correspondingly
paired adjacent normal tissues.
PPP2R1A, RNF43, POLE1, DICER1, CTCF,
RPL22, DNMT3A, TRRAP, IDH1 and IDH2 hotspot mutation analyses
The mutational status of several potential ovarian
cancer-related genes were analyzed in our samples previously
(15–18) and here (ovarian mixed germ cell
tumor, n=12), including PPP2R1A, RNF43, POLE1, DICER1, CTCF, RPL22,
DNMT3A, TRRAP, IDH1 and IDH2, the PCR and DNA sequencing reactions
were performed as previously described (15–18).
Evolutionary conservation analysis
Twenty-six vertebrate species were selected from
GenBank to analyze the evolutionary conservation status of MAPK1
mutation, including Homo sapiens (NP_002736), Pan
troglodytes (XP_003317171), Mus musculus (NP_036079),
Rattus norvegicus (NP_446294), Cricetulus griseus
(XP_007641645), Odobenus rosmarus divergens (XP_004400572),
Oryctolagus cuniculus (XP_008270402), Tupaia
chinensis (XP_006140283), Ovis aries (XP_0040177265),
Canis lupus familiaris (NP_001104270), Sus scrofa
(NP_001185851), Bos taurus (NP_786987), Vicugna pacos
(XP_006213368), Equus caballus (XP_005612442), Alligator
mississippiensis (XP_006269381), Chelonia mydas
(XP_007054992), Lipotes vexillifer (XP_007451077),
Xenopus laevis (NP_001083548), Python bivittatus
(XP_007422500), Columba livia (XP_005515231), Zonotrichia
albicollis (XP_005488954), Eptesicus fuscus
(XP_0081408655), Gallus gallus (NP_989481), Astyanax
mexicanus (XP_007229493), Poecilia reticulata
(XP_008417574) and Danio rerio (NP_878308).
Protein structural modeling
The protein structural modeling was performed by
DeepView Swiss-PdbViewer 4.0 software (19). Three PDB structures of human MAPK1
were available (3sa0.1.A, 1wzy.1.A and 4qte.1.A) in the ExPASy
database (http://www.expasy.org). Based on these
structures, by displaying 'show backbone oxygen', 'show dots
surface' and 'sender in solid 3D', wild-type MAPK1 were built
firstly and the p.D321N mutant MAPK1 was subsequently modeled by
changing aspartic acid 321 to asparagine. Additionally, p.D321V
mutant MAPK1 was also modeled according to the same procedure.
Results
MAPK1 and MAPK3 mutations in ovarian
carcinoma
The clinical information of the sample cohort has
been described previously (15–17). A
total of 263 ovarian carcinomas were screened for the potential
mutations in the MAPK1 p.E322 and paralogous MAPK3 p.E339 residues
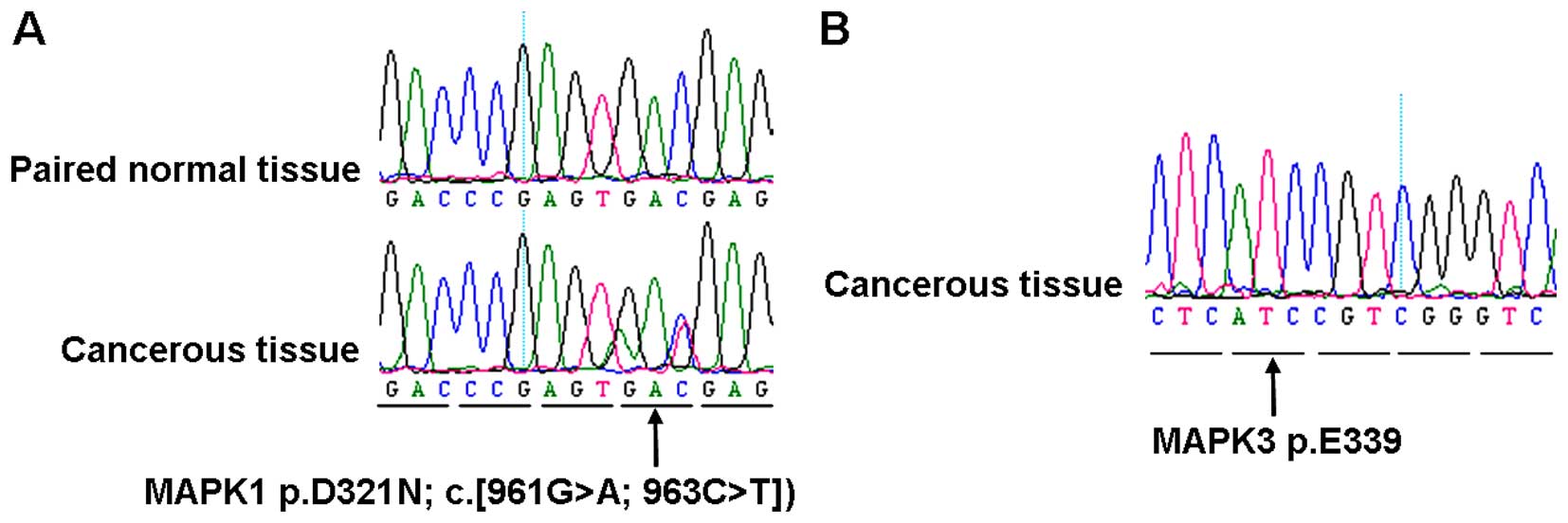
(Table I and Fig. 1). Although MAPK1 p.E322 mutations
were not detected in these samples, intriguingly, a novel,
previously unreported mutation in the 321st residue [p.D321N, c.
(961G>A; 963 C>T)] adjacent to MAPK1 p.E322, was detected in
2 out of 18 (11.1%) samples with ovarian mixed germ cell tumor, and
the somatic status of these mutation was confirmed by sequencing
the correspondingly adjacent normal tissues (Table I and Fig. 2A). In addition, no mutations were
detected in the remaining samples (Table I). The age of the patients with
mutations (OCC-44 and OCC-115) was 28 and 30 years, and affected
the right and bilateral ovaries, respectively; while the 16 ovarian
mixed germ cell tumors with wild-type MAPK1 affected 8 left, 3
bilateral and 5 right ovaries, respectively (17). Of note, OCC-115, the sample with
bilateral ovaries affected, whose bilateral cancerous ovaries were
obtained and MAPK1 p.D321N mutation was detected only in the right
ovary while absent in the left cancerous ovary. Moreover, no MAPK3
mutation was detected in our samples (Table I and Fig. 2B), including the MAPK3 p.D338 and
p.E339 residues, which were the corresponding paralogous residues
of MAPK1 p.D321 and p.E322, respectively.
Association of MAPK1 mutation with other
genetic alterations in ovarian mixed germ cell tumor
We screened our samples for the presence of PPP2R1A,
RNF43, POLE1, DICER1, CTCF, RPL22, DNMT3A, TRRAP, IDH1 and IDH2
mutations in our prior (15-18)
and the present study. Nevertheless, no mutation was detected in
these genes.
Evolutionary conservation analysis and
protein structural modeling
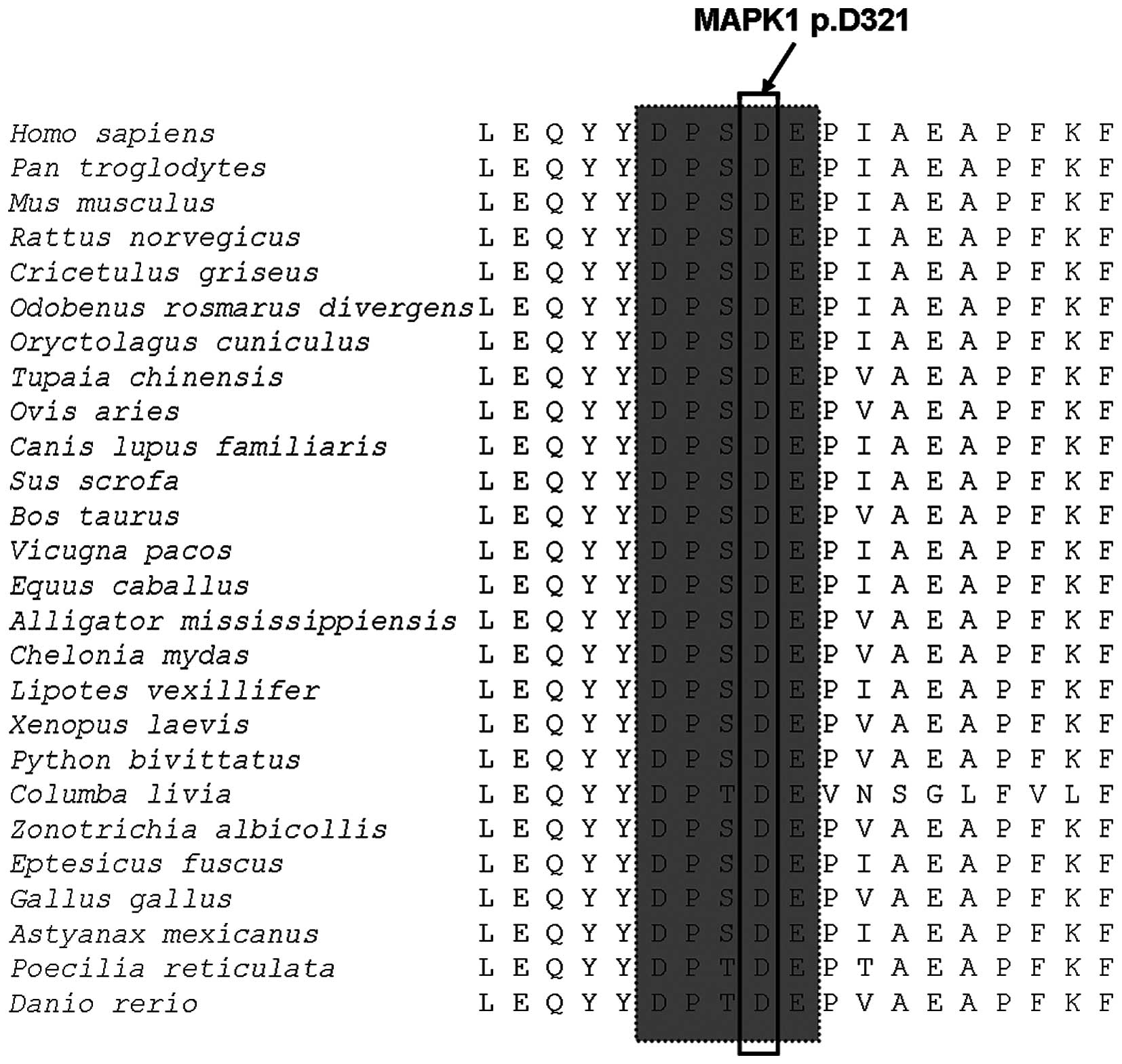
MAPK1 p.D321 residue is located in the cytoplasmic
retention motif (http://www.uniprot.org/) and the result of
evolutionary conservation analysis suggested that this residue was
highly conserved in vertebrate from Homo sapiens to Danio
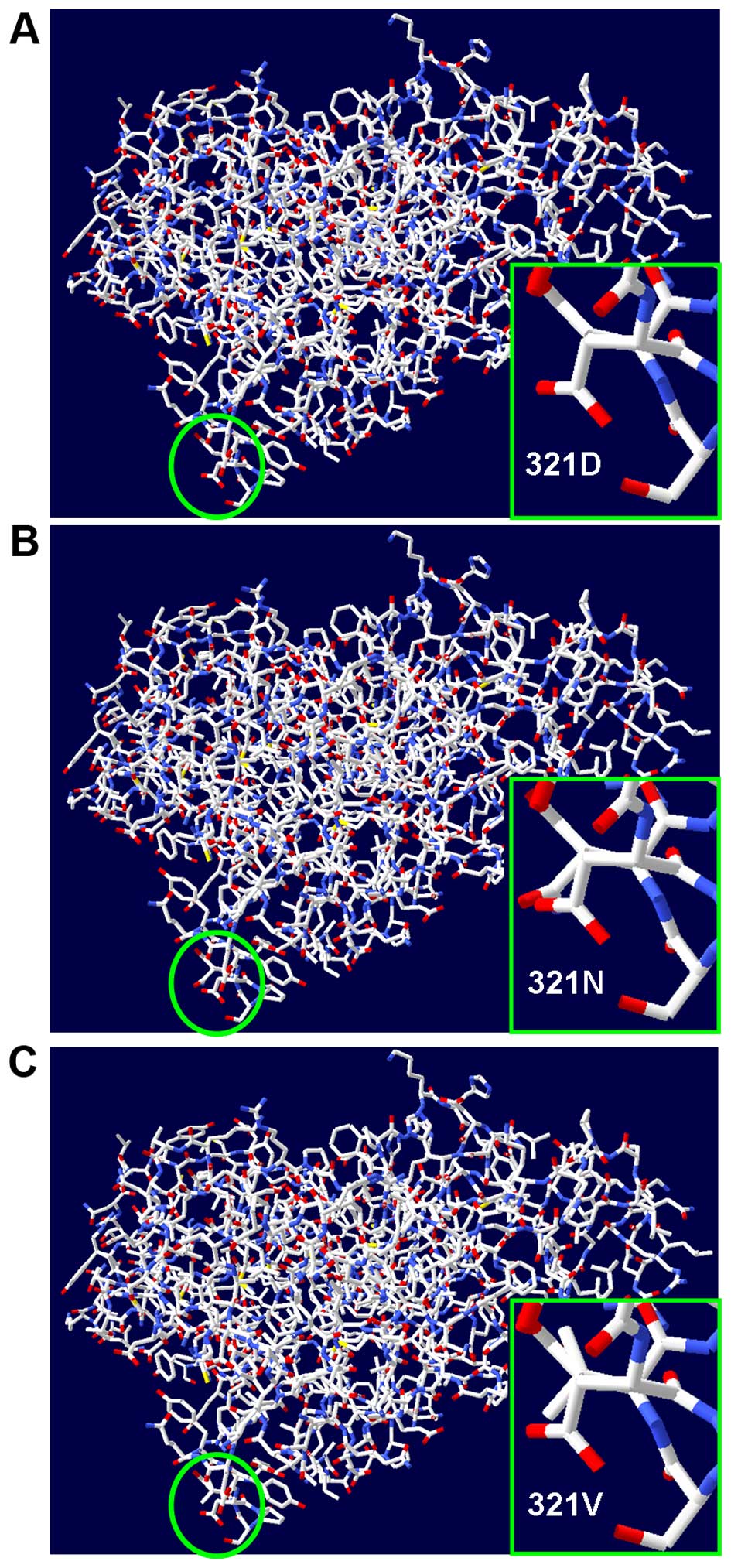
rerio (Fig. 3). Protein
structural modeling results suggested that the MAPK1 p.D321N mutant
and another p.D321 mutant (p.D321V) which was identified in
malignant melanomas (20),
exhibited structural changes in all of the 3 PDB versions of MAPK1
protein. Herein, one of the PDB structures (3sa0.1.A) used in the
present study is displayed (Fig.
4).
Discussion
Previous studies have suggested that MAPK1 mutations
were either rare or absent in human cancers (21-23)
(http://www.sanger.ac.uk/cosmic).
However, a recent integrated genomic characterization study
identified a high frequency of MAPK1 mutations in primary cervical
squamous cell carcinomas (13).
Ovarian mixed germ cell tumor is an ovarian germ
cell tumor containing two or more types of germ cell components
with extremely low incidence worldwide (24). Up to date, the detailed molecular
aberrations underlying ovarian mixed germ cell tumor remains
largely unknown. In the present study, a novel MAPK1 p.D321N
somatic mutation was detected in 2 out of 18 ovarian mixed germ
cell tumors but not in other subtypes of ovarian carcinoma,
implying this mutation may play an active role specifically in the
pathogenesis of ovarian mixed germ cell tumor. Intriguingly,
OCC-115, the MAPK1-mutated sample with both ovaries affected,
harbored MAPK1 mutation in the right ovary only but not in the
contralateral cancerous ovary. The discrepant mutational status of
MAPK1 between the paired cancerous ovaries in the same patient
indicated that the genetic alterations underlying ovarian mixed
germ cell tumor may be more complicated than we thought, even in
samples with similar genetic backgrounds and tumor
microenvironments. To our knowledge, this is the first report
revealing a novel MAPK1 mutation in ovarian mixed germ cell tumor,
and that this mutation may be actively involved in the
tumorigenesis process and may be a potential molecular therapy
target for this disorder.
Multiple genetic alterations are necessary for the
development of human cancers, it thus would be crucial for the
diagnosis and therapy of cancer patients to understand their
underlying combined events of genetic alterations (13,14,22).
Nevertheless, we failed to detect any mutations in the 10 novel
cancer-related genes in our ovarian mixed germ cell tumors
previously (15-18) and here, including PPP2R1A, RNF43,
POLE1, DICER1, CTCF, RPL22, DNMT3A, TRRAP, IDH1 and IDH2. There
results implicated that these genetic alterations may not play
synergistic roles with MAPK1 p.D321N mutation in the development of
the ovarian mixed germ cell tumor. In addition, no MAPK3 mutation
was detected in our sample cohort, indicating that MAPK3 mutations
may be not actively involved in the pathogenesis of ovarian
carcinoma.
In contrast to the mixed subtype, MAPK1 mutations
were not detected in other subtypes of germ cell tumors, including
patients with yolk sac (n=11), dysgerminoma (n=7) and teratoma
(n=9) subtypes. The inconsistent mutational status of MAPK1 in
various subtypes of germ cell tumor further indicated that germ
cell tumor of ovary was heterogeneous and the potential molecular
genetic alterations underlying these tumor subtypes may be quite
complex (25).
In addition, MAPK1 mutations were not found in the
76 ovarian serous or 15 mucinous carcinomas. This was consistent
with several previous observations based on whole-exome or genome
sequencing projects, where MAPK1 mutations were absent in 32 small
cell (26), 15 mucinous (27), 2 serous borderline (28) and 997 serous subtypes of the ovary
(14) (http://www.sanger.ac.uk/cosmic). Together, these
combined data show that MAPK1 mutations may be rare in ovarian
serous or mucinous carcinomas. Similarly, MAPK1 mutations were not
detected in other primary and secondary ovarian carcinomas in our
samples, implying that MAPK1 mutations may not be involved in
tumorigenesis of these tumor subtypes.
Functionally, it is not yet clear whether MAPK1
p.D321N mutation would play substantial role in the pathogenesis of
ovarian mixed germ cell tumor. Prior large-scale sequencing efforts
have detected MAPK1 p.D321N (c.961G>A) mutation in 1 out of 35
(2.9%) patients with head and neck squamous cell carcinoma
(29) (http://www.sanger.ac.uk/cosmic), and MAPK1 p.D321V
(c.962A>T) mutation in 1 out of 147 malignant melanomas
(20). In addition, the results of
protein sequence homology and evolutionary conservation analyses
indicated that MAPK1 p.D321 residue located in the cytoplasmic
retention motif (http://www.uniprot.org/) and was highly conserved in
26 vertebrate species from Homo sapiens to Danio
rerio. On the other hand, the results of protein structure
modeling indicated that the mutations in the MAPK1 p.D321 residue
(p.D321N and p.D321V) would lead to protein structure changes and
may affect the activity of MAPK1. Altogether, we speculated that
MAPK1 p.D321N mutation may promote the development of ovarian mixed
germ cell tumor, via change of MAPK1 activity or subcellular
localization. However, further functional assays would be necessary
to confirm these speculations.
A main limitation of the present study was that we
have screened only a short DNA fragment spanning the potential
mutational hotspot of MAPK1 and MAPK3, it could be more informative
to detect the entire coding region of the MAPK1 and MAPK3 genes.
However, this is limited mainly by our available DNA materials
isolated from FFPE tissues and we thus failed to test this aspect.
Furthermore, we have analyzed the mutational status of MAPK1 in a
total of 18 ovarian mixed germ cell tumors, further analysis using
larger sample sizes will be helpful for validating the accurate
frequency of MAPK1 p.D321N mutation in this specific subtype of
ovarian carcinoma.
In conclusion, our study reveals a novel mutation
associated with ovarian mixed germ cell tumor but not other
subtypes, thus, this mutation may play active role in the
progression of ovarian mixed germ cell tumor and may be a promising
therapeutic target for this disorder. Moreover, absence of MAPK3,
PPP2R1A, RNF43, POLE1, DICER1, CTCF, RPL22, DNMT3A, TRRAP, IDH1 and
IDH2 mutations in MAPK1-mutated ovarian mixed germ cell tumors
indicated that these mutations may not cooperate with MAPK1
mutation in the development of this disorder.
Acknowledgments
We thank all participants involved in the present
study. The present study was supported by grants from the National
Natural Science Foundation of China (no. 81260384), the Natural
Science Foundation of Jiangxi (no. 20142BAB215003) and Basic
Equipment Grant for Research Institute of Jiangxi (no.
20151BBA13051).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Munkarah A, Chatterjee M and Tainsky MA:
Update on ovarian cancer screening. Curr Opin Obstet Gynecol.
19:22–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tinelli A, Vergara D, Martignago R, Leo G,
Pisanò M and Malvasi A: An outlook on ovarian cancer and borderline
ovarian tumors: Focus on genomic and proteomic findings. Curr
Genomics. 10:240–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lowe KA, Chia VM, Taylor A, O'Malley C,
Kelsh M, Mohamed M, Mowat FS and Goff B: An international
assessment of ovarian cancer incidence and mortality. Gynecol
Oncol. 130:107–114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davis A, Tinker AV and Friedlander M:
'Platinum resistant' ovarian cancer: What is it, who to treat and
how to measure benefit? Gynecol Oncol. 133:624–631. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Luca A, Maiello MR, D'Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Targets. 16(Suppl 2):
S17–S27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Steelman LS, Franklin RA, Abrams SL,
Chappell W, Kempf CR, Bäsecke J, Stivala F, Donia M, Fagone P,
Nicoletti F, et al: Roles of the Ras/Raf/MEK/ERK pathway in
leukemia therapy. Leukemia. 25:1080–1094. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M,
Tafuri A, et al: Roles of the Raf/MEK/ERK pathway in cell growth,
malignant transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar
|
|
9
|
Holderfield M, Deuker MM, McCormick F and
McMahon M: Targeting RAF kinases for cancer therapy: BRAF-mutated
melanoma and beyond. Nat Rev Cancer. 14:455–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Kaiser CE, Frett B and Li HY:
Targeting mutant KRAS for anticancer therapeutics: A review of
novel small molecule modulators. J Med Chem. 56:5219–5230. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tsai JH, Huang WC, Jhuang JY, Jeng YM,
Cheng ML, Chiu HY, Kuo KT and Liau JY: Frequent activating HRAS
mutations in trichilemmoma. Br J Dermatol. 171:1073–1077. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trietsch MD, Spaans VM, ter Haar NT, Osse
EM, Peters AA, Gaarenstroom KN and Fleuren GJ: CDKN2A(p16) and HRAS
are frequently mutated in vulvar squamous cell carcinoma. Gynecol
Oncol. 135:149–155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bell DBA, Birrer M, Chien J, Cramer D, Dao
F, Dhir R, DiSaia P, Gabra H, Glenn P, Godwin A, et al Cancer
Genome Atlas Research Network: Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar
|
|
15
|
Wang F, Zou Y, Liu FY, Yu XH, Huang H,
Zhang N, Qi YY, Liu RF, Liu XY, Chen J, et al: Infrequent mutations
of the PPP2R1A and PPP2R1B genes in patients with ovarian cancer.
Mol Med Rep. 7:1826–1830. 2013.PubMed/NCBI
|
|
16
|
Zou Y, Wang F, Liu FY, Huang MZ, Li W,
Yuan XQ, Huang OP and He M: RNF43 mutations are recurrent in
Chinese patients with mucinous ovarian carcinoma but absent in
other subtypes of ovarian cancer. Gene. 531:112–116. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zou Y, Liu FY, Liu H, Wang F, Li W, Huang
MZ, Huang Y, Yuan XQ, Xu XY, Huang OP, et al: Frequent POLE1
p.S297F mutation in Chinese patients with ovarian endometrioid
carcinoma. Mutat Res. 761:49–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou Y, Huang MZ, Liu FY, Yang BC, Wang LQ,
Wang F, Yu XH, Wan L, Wan XD, Xu XY, et al: Absence of DICER1,
CTCF, RPL22, DNMT3A, TRRAP, IDH1 and IDH2 hotspot mutations in
patients with various subtypes of ovarian carcinomas. Biomed Rep.
3:33–37. 2015.
|
|
19
|
Guex N and Peitsch MC: SWISS-MODEL and the
Swiss- PdbViewer: An environment for comparative protein modeling.
Electrophoresis. 18:2714–2723. 1997. View Article : Google Scholar
|
|
20
|
Krauthammer M, Kong Y, Ha BH, Evans P,
Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et
al: Exome sequencing identifies recurrent somatic RAC1 mutations in
melanoma. Nat Genet. 44:1006–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Muzny DMBM, Chang K, Dinh HH, Drummond JA,
Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Reid JG, et al
Cancer Genome Atlas Network: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar
|
|
22
|
Biankin AV, Waddell N, Kassahn KS, Gingras
MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J,
et al Australian Pancreatic Cancer Genome Initiative: Pancreatic
cancer genomes reveal aberrations in axon guidance pathway genes.
Nature. 491:399–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landau DA, Carter SL, Stojanov P, McKenna
A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A,
Wang L, et al: Evolution and impact of subclonal mutations in
chronic lymphocytic leukemia. Cell. 152:714–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goyal LD, Kaur S and Kawatra K: Malignant
mixed germ cell tumour of ovary - an unusual combination and review
of literature. J Ovarian Res. 7(91)2014. View Article : Google Scholar
|
|
25
|
Heravi-Moussavi A, Anglesio MS, Cheng SW,
Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE,
et al: Recurrent somatic DICER1 mutations in nonepithelial ovarian
cancers. N Engl J Med. 366:234–242. 2012. View Article : Google Scholar
|
|
26
|
Witkowski L, Carrot-Zhang J, Albrecht S,
Fahiminiya S, Hamel N, Tomiak E, Grynspan D, Saloustros E, Nadaf J,
Rivera B, et al: Germline and somatic SMARCA4 mutations
characterize small cell carcinoma of the ovary, hypercalcemic type.
Nat Genet. 46:438–443. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ryland GL, Hunter SM, Doyle MA, Rowley SM,
Christie M, Allan PE, Bowtell DD, Gorringe KL and Campbell IG;
Australian Ovarian Cancer Study Group: RNF43 is a tumour suppressor
gene mutated in mucinous tumours of the ovary. J Pathol.
229:469–476. 2013. View Article : Google Scholar
|
|
28
|
Boyd J, Luo B, Peri S, Wirchansky B,
Hughes L, Forsythe C and Wu H: Whole exome sequence analysis of
serous borderline tumors of the ovary. Gynecol Oncol. 130:560–564.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pickering CR, Zhang J, Yoo SY, Bengtsson
L, Moorthy S, Neskey DM, Zhao M, Ortega Alves MV, Chang K, Drummond
J, et al: Integrative genomic characterization of oral squamous
cell carcinoma identifies frequent somatic drivers. Cancer Discov.
3:770–781. 2013. View Article : Google Scholar : PubMed/NCBI
|