Introduction
Cholangiocarcinoma is defined as an epithelial tumor
with features of cholangiocyte differentiation. According to the
anatomic location, it is categorized as either intrahepatic or
extrahepatic cholangiocarcinoma. Although cholangiocarcinoma
accounts for 3% of all gastrointestinal tumors, it is the most
common biliary malignancy and the second most common hepatic
malignancy after hepatocellular carcinoma (1). The overall survival rate of
cholangiocarcinoma of less than 5% at 5 years has not significantly
changed over the past 3 decades (2). Surgical treatment is the preferred
option for all subtypes. Yet, surgical intervention is limited due
to the lack of effective markers for early diagnosis, thus only a
small number of patients benefit from surgery. Furthermore, a high
postoperative recurrence rate and low sensitivity to
chemotherapeutics are critical factors which contribute to the poor
prognosis of cholangiocarcinoma (3,4).
Several risk factors including primary sclerosing cholangitis,
liver fluke infestation and hepatolithiasis have been described.
However, little is known concerning the mechanisms of
carcinogenesis. Therefore, there is a dire need for improving our
understanding of the biology of cholangiocarcinoma to develop
effective early diagnostic and therapeutic options.
Apoptosis-related protein-1 (Apr-1), also known as
melanoma-associated antigen (MAGE)-H1 and restin, was first cloned
by our group from the apoptotic tumor cell line HL-60 when induced
by all-trans retinoic acid (ATRA) (5). It belongs to the MAGE gene superfamily
based on the bioinformatic analysis of its gene structure compared
with other homologues in the GenBank (6). According to their expression patterns,
MAGE genes can be divided into two groups. Type I MAGE genes are
expressed in tumors of various histological origins, but are
completely silent in normal tissues, with the exception of male
germ cells and placenta; thus, the corresponding proteins represent
attractive targets for cancer immunotherapy (6). On the contrary, type II MAGE genes are
ubiquitously expressed in somatic cells, both in tumors and normal
adult tissues, which suggests that they may play an important role
in biological processes (7).
Although the activation and expression of MAGEs were reported in
various human cancers including cholangiocarcinoma, the
physiological function of MAGEs remains largely unknown (8,9).
However, the involvement of MAGEs in the regulation of cell cycle
progression (10) and apoptosis
(11,12) has been suggested. Thus, the
identification of the mechanisms responsible for the biological
functions of MAGEs may shed new light on the understanding of the
cause and development of cholangiocarcinoma.
According to our previous study, the expression
pattern of Apr-1 indicates it is a type II MAGE gene and is
involved in cancer cell proliferation and survival (13,14).
In the present study, we first examined the expression pattern of
Apr-1 in human cholangiocarcinoma tissues, and then investigated
the effects of induced overexpression of Apr-1 on cell growth and
cell cycle regulation in human cholangiocarcinoma cell line QBC939.
Furthermore, we analyzed the cell cycle-specific gene expression
profile in QBC939 cells upon Apr-1 expression, as well as the
expression of several key cell cycle regulatory proteins in human
cholangiocarcinoma tissues. These studies facilitate a better
understanding of the fundamental aspects of Apr-1, as a type II
MAGE gene, in the tumorigenesis and tumor development of
cholangiocarcinoma.
Materials and methods
Tissues
Four fresh-frozen cholangiocarcinoma samples and
matched tumor-adjacent tissues obtained from Xijing Hospital
(Fourth Military Medical University, Xi'an, Shaanxi Province,
China) were collected and stored at −70°C. Formalin-fixed
paraffin-embedded (FFPE) human cholangiocarcinoma tissue and
tumor-adjacent tissue samples were collected from the Department of
Pathology, Xijing Hospital. Ethical approval was obtained from the
Xijing Hospital Ethics Committee.
Reagents
Anti-Apr-1 (anti-MAGE-H1) polyclonal antibody
(HPA011324) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Anti-Cdk2 (ab6538), anti-Cks1/2 (ab54643) and anti-β-actin (ab8227)
rabbit polyclonal antibodies were purchased from Abcam (Shanghai,
China). Restriction enzymes BamHI, XbaI, SalI,
ScaI, ExTaq polymerase, DNA marker DL2000,
λDNA/EcoRI+HindIII marker were purchased from Takara
(Dalian, China). Liposome™ reagent, Dulbecco's modified Eagle's
medium (DMEM) and fetal bovine serum (FBS) were obtained from
Gibco-BRL (Gaithersburg, MD, USA). RNasin was purchased from
Promega (Madison, WI, USA). G418 and TRIzol were from Invitrogen
(Carlsbad, CA, USA). High-capacity cDNA reverse transcription kit
was from Applied Biosystems (Carlsbad, CA, USA).
Plasmid
A BamHI site was introduced to the 5′-end and
a SalI site was introduced to the 3′-end of the primers for
cloning the open reading frame of Apr-1. The sequences were as
follows: sense BamHI, 5′-TAGGATCCggagacatgcctcggg-3′;
antisense SalI, 5′-ACGCGTCGACgatctacttaaggggc-3′. The
purified PCR products and pcDNA3.0 vector were cut by
BamHI/SalI and BamHI/XhoII,
respectively. The digested fragments were harvested and cloned into
pcDNA3.0 between the same sites of SalI/XhoI to yield
pcDNA3.0-Apr-1. The recombinant plasmid was confirmed by digestion
of SalI/XhoI and sequencing.
Cell culture
Human cholangiocarcinoma QBC939 cells were stocked
in our laboratory. Cells were cultured in DMEM containing 10% FBS,
50 IU/ml penicillin and 50 µg/ml gentamycin at 37°C under an
atmosphere of 5% CO2. For gene transfection, QBC939
cells in optimal growth conditions were divided into 3 groups: the
control (QBC939 cells), blank vector group (pcDNA3.0-transfected
QBC939 cells) and experimental group (pcDNA3.0-Apr-1-transfected
QBC939 cells). Transfection procedures were performed according to
the manufacturer's instructions. The resistant cells were screened
by G418. G418-resistant clones were obtained after a 2-week
selection.
Quantitative RT-PCR
Total RNA from the frozen tissues or QBC939 cells
was extracted using TRIzol following the manufacturer's
instructions. Reverse transcription reactions were conducted using
a high-capacity cDNA reverse transcription kit. All primers were
optimized for amplification under reaction conditions as follows:
95°C for 10 min, followed by 30 cycles of 95°C for 15 sec and 60°C
for 1 min. Melt curve analysis was performed for all samples after
completion of the amplification protocol. β-actin was used as the
housekeeping gene expression control. Listed below are the primer
sequences used for quantitative PCR: Cdk2 forward,
5′-TCTGCCATTCTCATCGGGTC-3′ and Cdk2 reverse,
5′-ATTTGCAGCCCAGGAGGATTT-3′; Cks1 forward,
5′-AGCAAACCGAGCGATCATGT-3′ and Cks1 reverse,
5′-TGCTGAACGCCAAGATTCCT-3′; Cks2 forward, 5′-TCTTCGCGCTCTCGTTT
CAT-3′ and Cks2 reverse, 5′-TGGACACCAAGTCTCCT CCA-3′.
MTT cell proliferation assay
Each group of QBC939 cells was seeded at
1×104 cells/well into a 96-well plate. For cell growth
analysis, 20 µl freshly made MTT (5 mg/ml) was added into
each well and incubated for 4 h at 37°C. Then, cell culture media
were removed and replaced with 150 µl of dimethyl-sulfoxide
(DMSO) for a further 10-min incubation with gentle shaking until
the crystals were dissolved. The optical density (OD) value of each
well was measured using a microculture plate reader (Coulter
American) with a test wavelength of 490 nm. Three duplicate wells
for each group were measured per day.
Cell cycle analysis
The floating and adherent cells were harvested and
washed twice with phosphate-buffered saline (PBS), then
re-suspended in 300 µl PBS and fixed by adding 700 µl
cold ethanol in 70% ethanol at 4°C overnight. After washing twice
with PBS, 100 µl of fixed cells (~1×106 cells)
were stained with 300 µl propidium iodide (PI) for 15 min.
Cell cycle analysis by flow cytometry was performed. The
percentages of cells at the G1, G2 and S phases were measured.
Cell cycle gene expression array
analysis
To determine the expression of cell cycle-specific
genes, a GEArray Q series human cell cycle gene array kit
(SuperArray Biosciences, Frederick, MD, USA) was used. Total RNA
was isolated from QBC939 cells transfected for 48 h in 100-mm
dishes with 10.0 µg of either empty vector pcDNA3.0 or
pcDNA3.0-Apr-1. Both RNA samples were reverse-transcribed to
produce 32P-labeled probes following the manufacturer's
protocol. Cell cycle-specific genes in the nylon membranes were
hybridized to heat denatured radiolabeled probes at 55°C for 16 h
in a hybridization buffer provided by the manufacturer. After
washing twice in 2X SSC/1% SDS, followed by two additional washes
in 0.1X SSC/1% SDS at 55°C for 15 min each, the membranes were
exposed to X-ray film. The spots were detected by autoradiography,
and the intensities of the corresponding spots in the two membranes
were compared for two RNA populations used to generate the
probes.
Western blotting
Immunoblot analysis was performed according to
standard procedures using the following antibodies and dilutions:
anti-Apr-1 1:500, anti-Cdk2 1:1,000, anti-Cks1/2 1:1,000 and
anti-β-actin 1:1,000. Equal amounts of protein from the cells and
tissues were separated by SDS-PAGE and transferred to
polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Shanghai,
China). The membranes were blocked with milk, and primary antibody
incubations were performed at room temperature for 2 h. Secondary
antibody HRP-conjugated anti-rabbit (1:5,000) was used and signals
were detected with SuperSignal West Pico Substrate (Thermo
Scientific, Shanghai, China). The visualization of bands was
performed by exposure to high-performance autoradiography film.
Immunohistochemistry
Paraffin-embedded tissue sections on microscopic
slides were processed through a graded series of alcohols and
rehydrated in distilled water. Heat-induced antigen retrieval was
performed by citrate buffer (10 mmol/l concentration, pH 6.0), and
standard indirect biotin-avidin immunohistochemical analysis was
performed as previously described (15,16).
Statistical analysis
The data are presented as mean ± SEM. Statistical
analysis was performed using SPSS 21.0. Statistical evaluation of
the data was performed by two-way analysis of variance (ANOVA) or
unpaired t-test (two groups). A value of p<0.05 was considered
to indicate a statistically significant result.
Results
Apr-1 expression is reduced in human
cholangiocarcinoma tissues
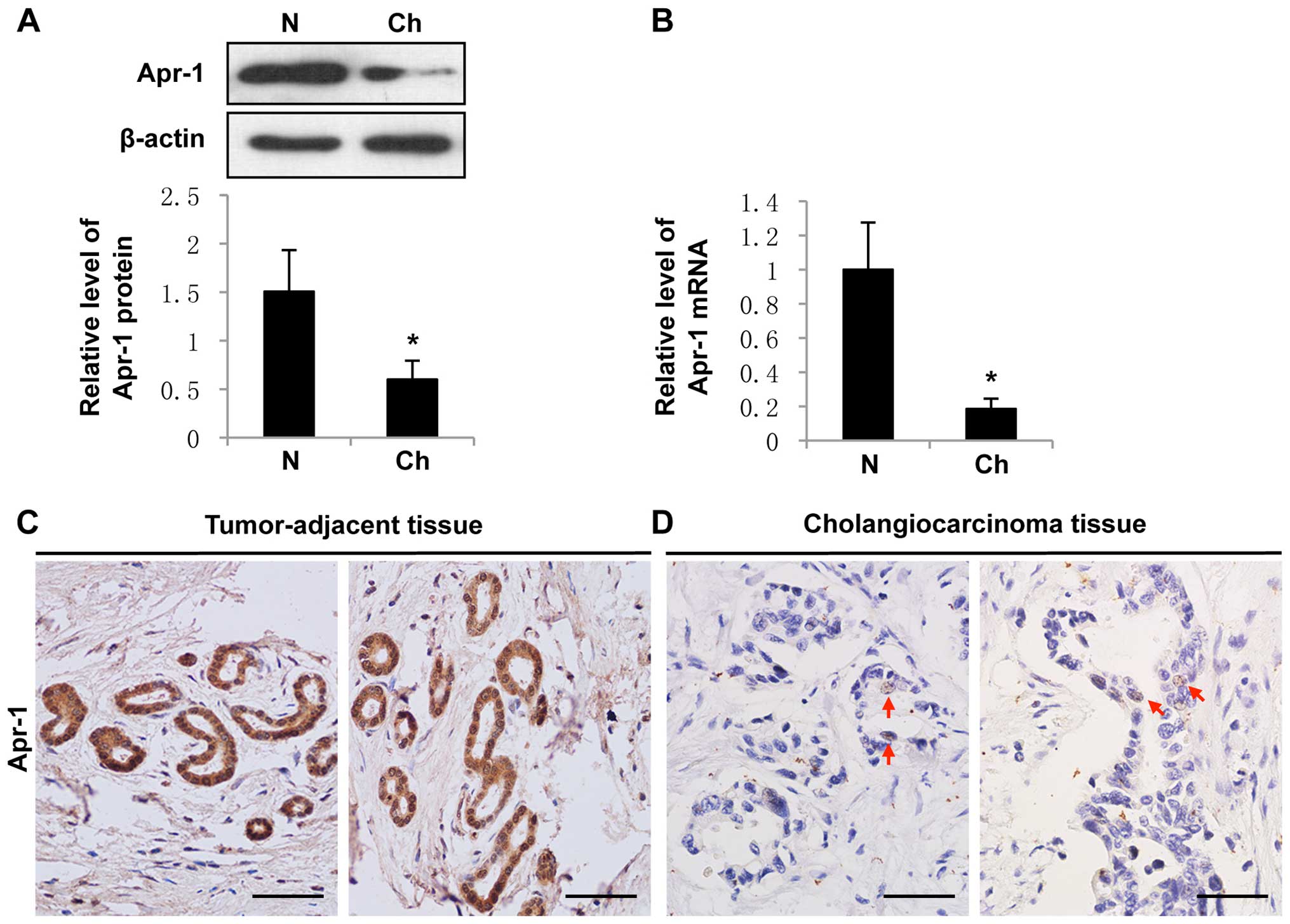
We performed western blotting and qRT-PCR assay to
evaluate the Apr-1 expression in pooled samples of 4 human
intrahepatic cholangiocarcinoma and paired tumor-adjacent hepatic
tissues. The results showed that Apr-1 expression was significant
lower in the cholangiocarcinoma tissues than that in the
tumor-adjacent hepatic tissues (Fig. 1A
and B). We next examined Apr-1 expression in 6 pathologically
graded (moderate to well differentiated) human cholangiocarcinoma
FFPE samples by immunohistochemistry (IHC). All normal or
tumor-adjacent bile ducts exhibited medium to strong positive
staining of Apr-1 in both the cytoplasm and nuclei (Fig. 1C). In contrast, Apr-1 protein was
undetectable in most of the cholangiocarcinoma tissues. Only rare
carcinoma cells showed very weak trace nuclear expression of Apr-1
(Fig. 1D). We did not find any
Apr-1 expression in the samples derived from human
cholangiocarcinoma cell line QBC939 (data not shown). Therefore, it
appears that Apr-1 expression is essential for normal cell function
and is downregulated during cholangiocarcinoma development.
Apr-1 inhibits cell proliferation of
QBC939 cells by inducing G2/M phase arrest
Given the fact that QBC939 cells do not express
Apr-1, we sought to investigate the effects of induced
overexpression of Apr-1 on cholangiocarcinoma cell proliferation
and survival. QBC939 cells were transfected with pcDNA3.0-Apr-1,
and stable cells were screened by G418. MTT assay was carried out
to analyze the viability of the QBC939 cells with Apr-1 expression.
The results showed that the growth of the QBC939-pcDNA3.0-Apr-1
cells was significantly inhibited (p<0.001, by two-way ANOVA)
(Fig. 2A). Subsequently, we
analyzed changes in the cell cycle distribution in the QBC939,
QBC939-pcDNA3.0 and QBC939-pcDNA3.0-Apr-1 cells. The results
demonstrated that QBC939-pcDNA3.0-Apr-1 cells were promoted to
enter into the following phase from the G1 phase, which resulted in
4.2% more cells arrested in the G2/M phase, coinciding with 2.5%
more cells accumulated in the S phase when compared with the
QBC939-pcDNA3.0 group (Fig. 2B).
Meanwhile, cell death was not observed in these three cell lines.
These data indicate that Apr-1 may play a role in
cholangiocarcinoma cell proliferation by negatively regulating the
cell cycle at the G2/M point.
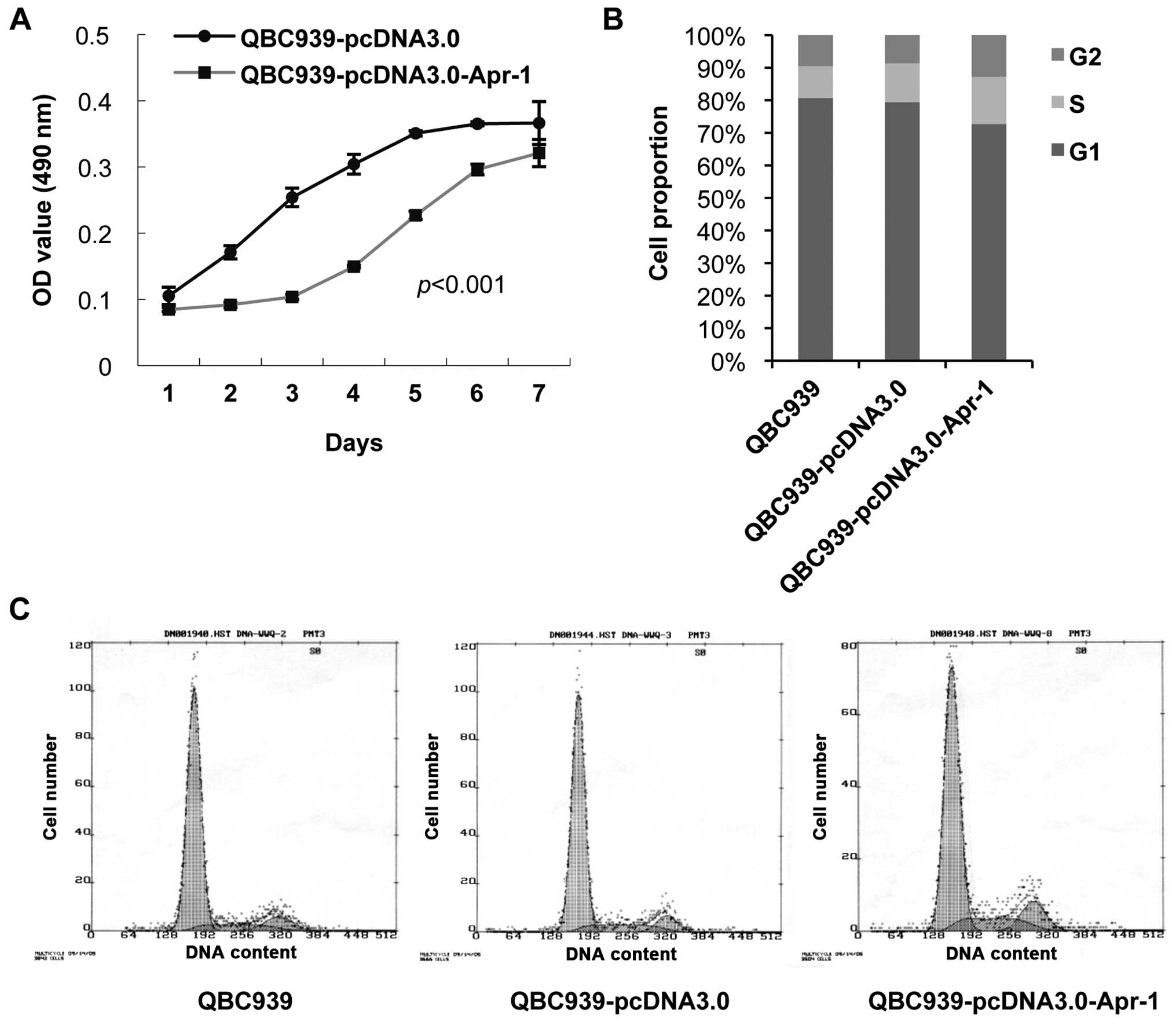 | Figure 2Effects of Apr-1 expression on the
cell growth and cell cycle progression of QBC939 cells. (A) Growth
curve of QBC939-pcDNA3.0 (transfected with pcDNA3.0) and
QBC939-pcDNA3.0-Apr-1 (transfected with pcDNA3.0-Apr-1) cells by
MTT cell proliferation assay. Data represent the mean ± SEM.
p<0.001, as determined by two-way ANOVA. (B and C) Cell cycle
analysis of QBC939 (G1, 80.7%; G2, 9.5%; S, 9.8%), QBC939-pcDNA3.0
(G1, 79.4%; G2, 8.6%; S, 12.0%) and QBC939-pcDNA3.0-Apr-1 cells
(G1, 72.7%; G2, 12.8%; S, 14.5%) by flow cytometry. |
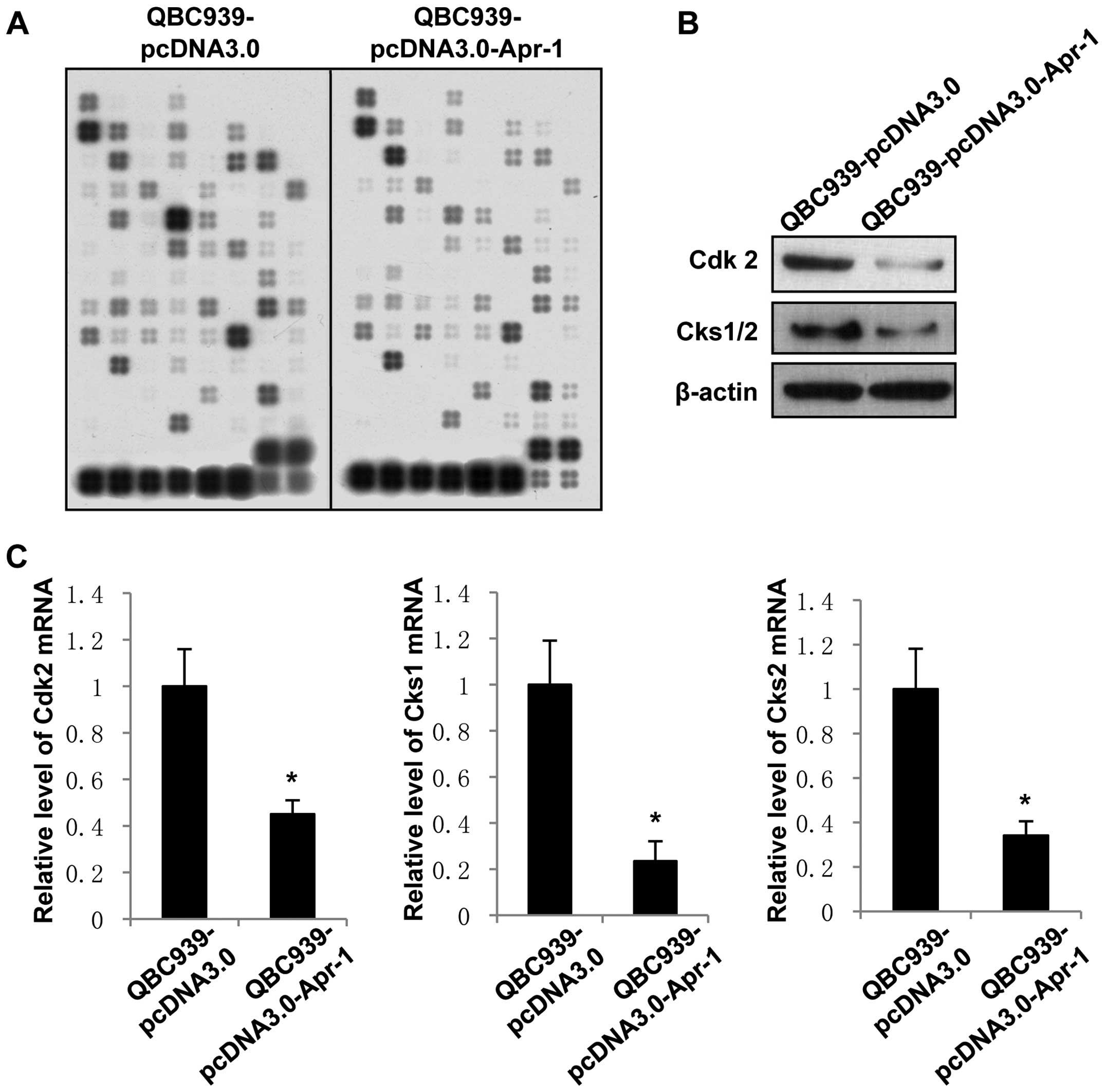
Cdk2, Cks1 and Cks2 are involved in
Apr-1-induced cell cycle arrest
Based on the above data, we further investigated
whether the transcription levels of cell cycle-regulatory genes are
affected by expression of Apr-1. Cell cycle-specific gene
expression array was used to determine the differences in gene
expression between the QBC939-pcDNA3.0-Apr-1 and QBC939-pcDNA3.0
cells. The results showed that 23 cell cycle-specific genes
including several cyclins and cyclin-dependent kinases
(Cdks) were downregulated >2-fold in the
QBC939-pcDNA3.0-Apr-1 cells compared with the levels in the
QBC939-pcDNA3.0 group (Fig. 3A and
Table I). There were also two
genes, S phase kinase-associated protein 2 (Skp2) and
ubiquitin-activating enzyme (UBE1) which are ubiquitin
signaling pathway factors associated with the cell cycle, that were
found to be upregulated upon Apr-1 expression (Table I). Notably, Cdk1 and
Cdk2, cyclin-dependent kinase catalytic subunits that are
important regulators of cell cycle transition between different
cell cycle stages, were downregulated >4-fold by Apr-1.
Moreover, the expression levels of Cdk-interacting protein
Cks1 and cyclin-dependent kinase subunit (Cks2),
which are tightly associated with Cdks and mitotic entry, were
decreased ~3-fold upon Apr-1 expression in the QBC939 cells.
 | Table IApr-1 regulates transcription of cell
cycle-related genes in QBC939 cells (downregulated or upregulated
>2-fold). |
Table I
Apr-1 regulates transcription of cell
cycle-related genes in QBC939 cells (downregulated or upregulated
>2-fold).
| Gene name | Description | Fold-change | GenBank |
|---|
| Cks1 | Cyclin-dependent
kinase subunit 1 | −2.934 | NM_001826 |
| Cks2 | Cyclin-dependent
kinase subunit 2 | −3.126 | NM_001827 |
| Cdk2 | Cyclin-dependent
kinase 2 | −4.477 | NM_001798 |
| Cdk1 | Cell division cycle
2, G1 to S and G2 to M | −4.648 | NM_001786 |
| Cyclin H | Cyclin H | −2.139 | NM_001239 |
| MRE11A | Homolog A
(Cerevisiae) | −3.049 | NM_005590 |
| CDK8 | Cyclin-dependent
kinase 8 | −3.258 | NM_001260 |
| CDC45 | CDC45 cell division
cycle 45-like | −3.298 | NM_003504 |
| P21/Waf1 | Cyclin-dependent
kinase inhibitor 1A | −3.632 | NM_000389 |
| p16/INK4 | Cyclin-dependent
kinase inhibitor 2A | −3.782 | NM_000077 |
| CHK2/RAD53 | CHK2 checkpoint
homolog | −3.872 | NM_007194 |
| Chk1 | CHK1 checkpoint
homolog | −3.936 | NM_001274 |
| PRC1 | Protein regulator
of cytokinesis 1 | −4.022 | NM_003981 |
| E2F | E2F transcription
factor 1 | −4.061 | NM_005225 |
| P55CDC | CDC20 cell division
cycle 20 homolog | −4.224 | NM_001255 |
| Cyclin F | Cyclin F | −4.353 | NM_001761 |
| Cdk4 | Cyclin-dependent
kinase 4 | −4. 441 | NM_000075 |
| Cdc16 | CDC16 cell division
cycle 16 | −4.445 | NM_003903 |
| GADD45 | Cell division cycle
2, G1 to S | −4.612 | NM_001924 |
| SUMO-1 | Ubiquitin-like 1
(sentrin) | −4.848 | NM_003352 |
| Apaf-1 | Apoptotic protease
activating factor | −4.925 | NM_001160 |
| Cyclin D3 | Cyclin D3 | −4.942 | NM_001760 |
| p18/INK4C | p18, inhibits
CDK4 | −4.989 | NM_078626 |
| Skp2 | S phase
kinase-associated protein 2 | +2.427 | NM_005983 |
| UBE1 |
Ubiquitin-activating enzyme E1 | +2.012 | NM_003334 |
To confirm the negative-regulatory effects of Apr-1
on Cdk2, Cks1 and Cks2 expression, we used western blotting and
qRT-PCR assay to identify the changes in expression of these genes
in the pcDNA3.0-Apr-1-transfected QBC939 cells. As shown in
Fig. 3B and C, pcDNA3.0-Apr-1
transfection inhibited the expression of Cdk2 and Cks1/2 in the
QBC939 cells at both the protein and mRNA levels. The results
indicated that expression of Apr-1 induces G2/M phase arrest of
cholangiocarcinoma cells potentially by inhibiting the
transcription of Cdk2, Cks1 and Cks2.
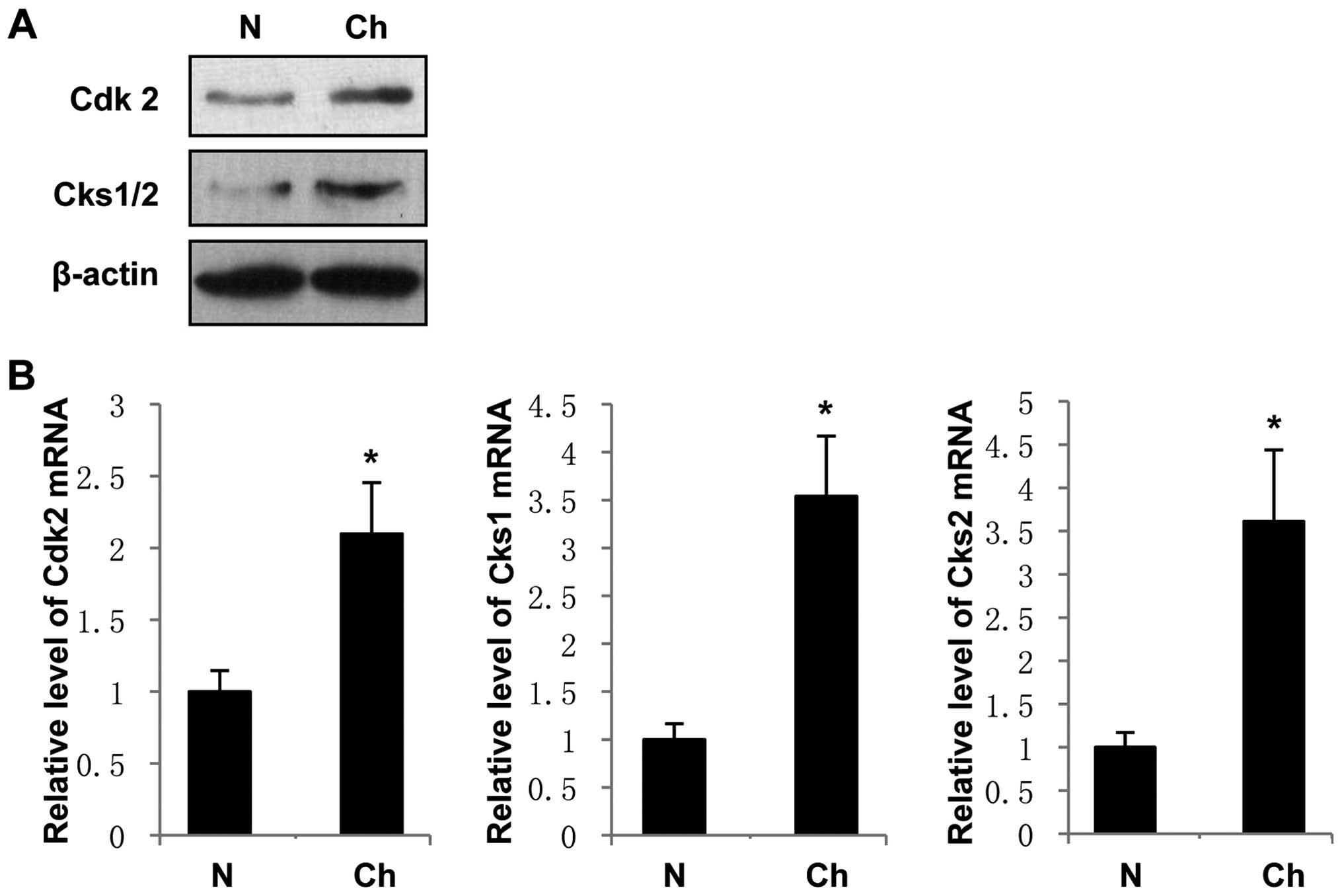
Cdk2, Cks1 and Cks2 expression levels are
elevated in human cholangiocarcinoma tissues
To further characterize the correlation between
Apr-1 and Cdk2, Cks1 and Cks2 expression in cholangiocarcinoma, we
detected Cdk2, Cks1 and Cks2 protein and mRNA levels in tumor and
tumor-adjacent hepatic tissues. Western blotting showed that Cdk2
and Cks1/2 were accumulated in the samples of cholangiocarcinoma
tissues compared with matched tumor-adjacent hepatic tissues
(Fig. 4A), which was consistent
with the results of qRT-PCR that Cdk2, Cks1 and Cks2 levels were
significantly higher in cholangiocarcinoma tissues than that in
tumor-adjacent hepatic tissues (Fig.
4B). These data suggest that Apr-1 expression, which is reduced
in cholangiocarcinoma, is negatively correlated with Cdk2, Cks1 and
Cks2 expression.
Discussion
Cholangiocarcinoma is a highly malignant cancer with
a poor prognosis. Abnormalities in various signaling cascades,
molecules and genetic mutations are involved in the pathogenesis of
cholangiocarcinoma. Recurrent mutations including KRAS,
BRAF, TP53, Smad and p16 (INK4a)
are characteristic of cholangiocarcinoma. KRAS and
BRAF mutant cholangiocarcinomas have been associated with a
worse long-term survival. Meanwhile, disruption of the RAF/MEK/MAPK
pathway by RAS or BRAF mutation has been found in >60% of
cholangiocarcinomas, indicating that these pathways are important
in cholangiocarcinoma carcinogenesis (17). Wnt signaling also plays an important
role in cholangiocarcinoma carcinogenesis through activation of
downstream target genes such as cyclin D1 and c-myc (18,19).
Although extensive efforts have been made to explore the
transformation mechanism of cholangiocarcinoma in the past decades,
detailed molecular and cellular mechanisms remain unclear. More
comprehensive understanding of the mechanisms involved in the
pathogenesis of cholangiocarcinoma is critical for the development
of effective therapies.
Apr-1 belongs to the MAGE superfamily for which over
30 members have been identified. Although the first MAGE family
gene was discovered in 1991, MAGEs have been well studied for
>20 years in melanomas (20),
yet their functions still remain unclear.
Based on our previous study, Apr-1 protein was found
to be localized in the nucleus and is believed to be an
apoptosis-related gene since its transcripts were upregulated
during all-trans-retinoic acid-induced apoptosis in human
promyelocytic leukaemia cells (5,14).
In situ hybridization assay on tissue microarrays showed
that Apr-1 is expressed in esophageal carcinoma, normal hepatic
tissue and hepatic tissue adjacent to hepatocellular carcinoma, but
is absent in normal esophageal mucosa and hepatocellular carcinoma
(data not shown). The expression pattern suggests that Apr-1 is a
type II MAGE gene. Recently, type II MAGE proteins are under
increasing attention due to their roles in the regulation of cell
cycle progression and apoptosis.
In the present study, we found that Apr-1 triggered
cell cycle arrest in QBC939 cells. QBC939-pcDNA3.0-Apr-1 cells
showed a decrease in the G1 phase and were dominantly arrested in
the G2/M phase after Apr-1 overexpression. Using gene expression
array, we identified that Apr-1 induced G2/M phase arrest of QBC939
cells via a mechanism mediated by downregulation of the cell cycle
checkpoint-related genes, in which Cdk2, Cks1 and Cks2 expression
levels played critical roles during this course.
Cdks are a family of protein kinases that drive the
cell cycle progression through their periodic activation.
Cyclin-Cdk complexes phosphorylate specific substrates in a
particular cell cycle phase (21).
The cyclin B-Cdk1 complex is vital for entering mitosis (M phase),
while Cdk2 is involved with cyclin A and E and activated from late
G1 until the onset of mitosis (22). Moreover, deregulation of Cdk2 can
result in DNA damage accumulation and loss of DNA damage checkpoint
control (23–25). In the present study, we found that
Cdk1 and Cdk2 were inhibited by Apr-1 expression. Thus suggests
that Apr-1 induces cholangiocarcinoma cell cycle arrest in the G2
phase by impairing the expression of Cdk1 and Cdk2.
Given their essential role in cell cycle
progression, Cdk1 and Cdk2 are highly regulated by, among others,
cyclin dependent-kinase subunit Cks1 and Cks2. The human cyclin
kinase subunit family consists of two well-conserved members, Cks1
and Cks2, both of which were identified based on the sequence
homology to yeast suc1 and Cks1 (also named Cdc28 kinase subunit 1)
that are essential for cell cycle control (26). Accumulative evidence shows that the
two Cks members have distinct regulatory functions in mammalian
cells although 81% of protein products are identical and share one
or more redundant functions in both humans and mice (27). Cks1 can specifically regulate cell
cycle G1/S transition by promoting the process of
SCFSkp2-mediated ubiquitination and p27kip1
(a Cdk2 inhibitor) degradation (28,29).
Therefore, cells lacking Cks1 have elevated p27 and reduced Cdk2
activity (29,30). Cks2 has been shown to be essential
for the first metaphase/anaphase transition in mammalian meiosis;
however, its function is not clearly identified in the cell cycle
(23,24,31,32).
Recent study has shown that overexpression of Cks1 or Cks2 can
maintain Cdk2 in an active state (33). Therefore, in the present study,
decreased Cdk2 level upon Apr-1 expression could directly result
from Apr-1 regulation and/or be mediated by Cks1 and Cks2
inhibition.
Numerous studies report abnormal Cks1/2 expression
in various malignant tumors including hepatocellular carcinoma,
bladder, gastric and breast cancer, and meningioma (34–38).
However, the mechanistic link between Cks protein deregulation and
oncogenesis remains to be elucidated. Liberal et al found
that human mammary epithelial cells with Cks1 or Cks2
overexpression became resistant to DNA damage response mediated by
cell cycle checkpoint that was triggered by oncoproteins, thus,
allowing cells to continue proliferating even under replicative
stress (33). More recently, Cks2
was found to be significantly elevated in cholangiocarcinoma
tissues and its downregulation inhibited cholangiocarcinoma cell
proliferation in vitro and in vivo; particularly,
Cks2 knockdown induced cholangiocarcinoma cell cycle arrest in the
G2/M phase by facilitating cell apoptosis, which suggests that Cks2
may serve as an independent prognostic factor in patients with
cholangiocarcinoma (39).
Consistently, we found that Cks1 and Cks2 were accumulated in human
cholangiocarcinoma samples. Most importantly, we demonstrated that
Apr-1 expression was downregulated in cholangiocarcinoma and
induced overexpression of Apr-1 in cholangiocarcinoma cells
inhibited cell proliferation, mechanistically mediated by Cks1 and
Cks2 deficiency, indicating that Apr-1 can regulate the expression
of Cks1 and Cks2 in cholangiocarcinoma. Two cell cycle-related
genes, Skp2 and UBE1, showed >2-fold increase at
the transcription level after overexpression of Apr-1. Skp2 is a
member of the F-box family of substrate-recognition subunits of SCF
ubiquitin-protein ligase complexes that has been implicated in
ubiquitin-mediated degradation (40); UBE1, also known as an E1 enzyme,
catalyzes the first step in the ubiquitination reaction (41). Both Skp2 and UBE1 function to
regulate other cell cycle molecules by ubiquitination. It has been
recognized that Skp2 can interact with Cks1 to degrade
p27Kip1 protein, which serves as a negative cell cycle
regulator exemplifying a class of tumor suppressors that controls
cell cycle progression (42).
Recent findings indicate that high expression of Skp2 is associated
with aggressiveness and a poor prognosis in a large variety of
cancers, including cholangiocarcinoma (43). The present study did not address
whether there is a feedback loop leading to upregulation of
Skp2 upon Apr-1-induced cell cycle arrest. Additional
studies are required to clarify the regulatory mechanisms involved
in elevated Skp2 and UBE1 transcription induced by
Apr-1 overexpression.
In conclusion, our data strongly suggest that Apr-1
has a tumor-suppressor function in cholangiocarcinoma cells,
mechanistically by inducing cell cycle arrest through regulating a
series of cell cycle regulators. Thus, our study lays a foundation
for further investigation of the underlying mechanisms of Apr-1
downregulation during the development and progression of
cholangiocarcinoma in order to explore potential therapeutic
targets for cholangiocarcinoma treatment.
Acknowledgments
The present study was supported by the National
Nature Science Foundation of China (no. 81472299).
References
|
1
|
Olnes MJ and Erlich R: A review and update
on cholangiocarcinoma. Oncology. 66:167–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sandhu DS, Shire AM and Roberts LR:
Epigenetic DNA hypermethylation in cholangiocarcinoma: Potential
roles in pathogenesis, diagnosis and identification of treatment
targets. Liver Int. 28:12–27. 2008. View Article : Google Scholar
|
|
3
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rizvi S and Gores GJ: Pathogenesis,
diagnosis, and management of cholangiocarcinoma. Gastroenterology.
145:1215–1229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu F, Yan W, Zhao ZL, Chai YB, Lu F, Wang
Q, Peng WD, Yang AG and Wang CJ: Improved PCR-based subtractive
hybridization strategy for cloning differentially expressed genes.
Biotechniques. 29:310–313. 2000.PubMed/NCBI
|
|
6
|
Chomez P, De Backer O, Bertrand M, De
Plaen E, Boon T and Lucas S: An overview of the MAGE gene family
with the identification of all human members of the family. Cancer
Res. 61:5544–5551. 2001.PubMed/NCBI
|
|
7
|
Barker PA and Salehi A: The MAGE proteins:
Emerging roles in cell cycle progression, apoptosis, and
neurogenetic disease. J Neurosci Res. 67:705–712. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bertrand M, Huijbers I, Chomez P and De
Backer O: Comparative expression analysis of the MAGED genes during
embryogenesis and brain development. Dev Dyn. 230:325–334. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sang M, Wang L, Ding C, Zhou X, Wang B,
Wang L, Lian Y and Shan B: Melanoma-associated antigen genes - an
update. Cancer Lett. 302:85–90. 2011. View Article : Google Scholar
|
|
10
|
Saburi S, Nadano D, Akama TO, Hirama K,
Yamanouchi K, Naito K, Tojo H, Tachi C and Fukuda MN: The trophinin
gene encodes a novel group of MAGE proteins, magphinins, and
regulates cell proliferation during gametogenesis in the mouse. J
Biol Chem. 276:49378–49389. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salehi AH, Roux PP, Kubu CJ, Zeindler C,
Bhakar A, Tannis LL, Verdi JM and Barker PA: NRAGE, a novel MAGE
protein, interacts with the p75 neurotrophin receptor and
facilitates nerve growth factor-dependent apoptosis. Neuron.
27:279–288. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Selimovic D, Sprenger A, Hannig M, Haïkel
Y and Hassan M: Apoptosis related protein-1 triggers melanoma cell
death via interaction with the juxtamembrane region of p75
neurotrophin receptor. J Cell Mol Med. 16:349–361. 2012. View Article : Google Scholar
|
|
13
|
Yan W, Li Q and Zhu F: Apoptosis-related
genes cloned by improved subtractive hybridization. Zhonghua Zhong
Liu Za Zhi. 23:193–195. 2001.In Chinese.
|
|
14
|
Yan W, Wang W-L, Zhu F, Cheng SQ, Li QL,
Wang L and Wang CJ: Cloning and subcellular localization of apr-1 -
a new gene of tumor specific antigen family. Ai Zheng. 24:129–134.
2005.In Chinese. PubMed/NCBI
|
|
15
|
Zhang Y, Li Q, Zhu F, Cui J, Li K, Li Q,
Wang R, Wang W, Wang W and Yan W: Subcellular localization of
APMCF1 and its biological significance of expression pattern in
normal and malignant human tissues. J Exp Clin Cancer Res.
28:1112009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Li Q, Huang W, Zhang J, Han Z,
Wei H, Cui J, Wang Y and Yan W: Increased expression of
apoptosis-related protein 3 is highly associated with tumorigenesis
and progression of cervical squamous cell carcinoma. Hum Pathol.
44:388–393. 2013. View Article : Google Scholar
|
|
17
|
Robertson S, Hyder O, Dodson R, Nayar SK,
Poling J, Beierl K, Eshleman JR, Lin MT, Pawlik TM and Anders RA:
The frequency of KRAS and BRAF mutations in intrahepatic
cholangiocarcinomas and their correlation with clinical outcome.
Hum Pathol. 44:2768–2773. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yothaisong S, Dokduang H, Techasen A,
Namwat N, Yongvanit P, Bhudhisawasdi V, Puapairoj A, Riggins GJ and
Loilome W: Increased activation of PI3K/AKT signaling pathway is
associated with cholangiocarcinoma metastasis and PI3K/mTOR
inhibition presents a possible therapeutic strategy. Tumour Biol.
34:3637–3648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maemura K, Natsugoe S and Takao S:
Molecular mechanism of cholangiocarcinoma carcinogenesis. J
Hepatobiliary Pancreat Sci. 21:754–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Bruggen P, Traversari C, Chomez P,
Lurquin C, De Plaen E, Van den Eynde B, Knuth A and Boon T: A gene
encoding an antigen recognized by cytolytic T lymphocytes on a
human melanoma. Science. 254:1643–1647. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morgan DO: Cyclin-dependent kinases:
Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol.
13:261–291. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pagano M, Pepperkok R, Lukas J, Baldin V,
Ansorge W, Bartek J and Draetta G: Regulation of the cell cycle by
the cdk2 protein kinase in cultured human fibroblasts. J Cell Biol.
121:101–111. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Merrick KA, Larochelle S, Zhang C, Allen
JJ, Shokat KM and Fisher RP: Distinct activation pathways confer
cyclin-binding specificity on Cdk1 and Cdk2 in human cells. Mol
Cell. 32:662–672. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tane S and Chibazakura T: Cyclin A
overexpression induces chromosomal double-strand breaks in
mammalian cells. Cell Cycle. 8:3900–3903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wheeler LW, Lents NH and Baldassare JJ:
Cyclin A-CDK activity during G1 phase impairs MCM chromatin loading
and inhibits DNA synthesis in mammalian cells. Cell Cycle.
7:2179–2188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hayles J, Beach D, Durkacz B and Nurse P:
The fission yeast cell cycle control gene cdc2: Isolation of a
sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet.
202:291–293. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Richardson HE, Stueland CS, Thomas J,
Russell P and Reed SI: Human cDNAs encoding homologs of the small
p34Cdc28/Cdc2-associated protein of Saccharomyces
cerevisiae and Schizosaccharomyces pombe. Genes Dev. 4:1332–1344.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ganoth D, Bornstein G, Ko TK, Larsen B,
Tyers M, Pagano M and Hershko A: The cell-cycle regulatory protein
Cks1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nat
Cell Biol. 3:321–324. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spruck C, Strohmaier H, Watson M, Smith
AP, Ryan A, Krek TW and Reed SI: A CDK-independent function of
mammalian Cks1: Targeting of SCFSkp2 to the CDK
inhibitor p27Kip1. Mol Cell. 7:639–650. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Frontini M, Kukalev A, Leo E, Ng YM,
Cervantes M, Cheng CW, Holic R, Dormann D, Tse E, Pommier Y, et al:
The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2
activity in maintaining replicative fidelity and neurodevelopment.
Dev Cell. 23:356–370. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Spruck CH, de Miguel MP, Smith APL, Ryan
A, Stein P, Schultz RM, Lincoln AJ, Donovan PJ and Reed SI:
Requirement of Cks2 for the first metaphase/anaphase transition of
mammalian meiosis. Science. 300:647–650. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rother K, Dengl M, Lorenz J, Tschöp K,
Kirschner R, Mössner J and Engeland K: Gene expression of
cyclin-dependent kinase subunit Cks2 is repressed by the tumor
suppressor p53 but not by the related proteins p63 or p73. FEBS
Lett. 581:1166–1172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liberal V, Martinsson-Ahlzén HS, Liberal
J, Spruck CH, Widschwendter M, McGowan CH and Reed SI:
Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression
overrides the DNA damage response barrier triggered by activated
oncoproteins. Proc Natl Acad Sci USA. 109:2754–2759. 2012.
View Article : Google Scholar :
|
|
34
|
Shen DY, Fang ZX, You P, Liu PG, Wang F,
Huang CL, Yao XB, Chen ZX and Zhang ZY: Clinical significance and
expression of cyclin kinase subunits 1 and 2 in hepatocellular
carcinoma. Liver Int. 30:119–125. 2010. View Article : Google Scholar
|
|
35
|
Chen R, Feng C and Xu Y: Cyclin-dependent
kinase-associated protein Cks2 is associated with bladder cancer
progression. J Int Med Res. 39:533–540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang MA, Kim J-T, Kim JH, Kim SY, Kim YH,
Yeom YI, Lee Y and Lee HG: Upregulation of the cycline kinase
subunit CKS2 increases cell proliferation rate in gastric cancer. J
Cancer Res Clin Oncol. 135:761–769. 2009. View Article : Google Scholar
|
|
37
|
Tanaka F, Matsuzaki S, Mimori K, Kita Y,
Inoue H and Mori M: Clinicopathological and biological significance
of CDC28 protein kinase regulatory subunit 2 overexpression in
human gastric cancer. Int J Oncol. 39:361–372. 2011.PubMed/NCBI
|
|
38
|
Menghi F, Orzan FN, Eoli M, Farinotti M,
Maderna E, Pisati F, Bianchessi D, Valletta L, Lodrini S, Galli G,
et al: DNA microarray analysis identifies CKS2 and LEPR as
potential markers of meningioma recurrence. Oncologist.
16:1440–1450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen DY, Zhan YH, Wang QM, Rui G and Zhang
ZM: Oncogenic potential of cyclin kinase subunit-2 in
cholangiocarcinoma. Liver Int. 33:137–148. 2013. View Article : Google Scholar
|
|
40
|
Cardozo T and Pagano M: The SCF ubiquitin
ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol.
5:739–751. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Weissman AM: Themes and variations on
ubiquitylation. Nat Rev Mol Cell Biol. 2:169–178. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Frescas D and Pagano M: Deregulated
proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the
scales of cancer. Nat Rev Cancer. 8:438–449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hashimoto N, Yachida S, Okano K,
Wakabayashi H, Imaida K, Kurokohchi K, Masaki T, Kinoshita H,
Tominaga M, Ajiki T, et al: Immunohistochemically detected
expression of p27Kip1 and Skp2 predicts survival in
patients with intrahepatic cholangiocarcinomas. Ann Surg Oncol.
16:395–403. 2009. View Article : Google Scholar
|