Introduction
Osteosarcoma is a primary mesenchymal tumor
characterized histologically by malignant tumor cells that directly
produce osteoid or immature bone. It is the most commonly diagnosed
primary malignancy of the bone, particularly among children and
adolescents (1). As the disease
results in active bone growth and repair (2), it is highly destructive and
metastatic. In the last several decades, therapies utilizing
surgery and chemical agents have improved the 5-year survival rate
of these patients to ~80% (3).
However, the cure rate of patients bearing osteosarcoma is still
very poor, and the majority eventually die of metastases (4). Thus, an understanding of the genes and
molecules involved in osteosarcoma is particularly critical for
developing new therapies and improving patient outcomes.
miR-603 has been reported as a tumor suppressor in
thyroid cell tumors (5), laryngeal
squamous cell carcinoma (6) and GH
adenoma (7). miR-603 has also been
previously reported to be overexpressed in pancreatic (8), ovarian (9) and colorectal cancer (10). However, its function in osteosarcoma
is not yet known.
Breast cancer cell 2 (BRCC2) is unique among the
proto-oncogenes, being localized to mitochondria and extending cell
survival by blocking programmed cell death (11). BRCC2 expression is reported to
prevent apoptosis in a wide variety of cell types, including
T-lymphocytes, prostate cancer cells and embryonic sensory neurons
(12). Previous studies have
reported that expression of BRCC2 suppresses cellular proliferation
and is associated with less aggressive biological behavior and
better prognosis (13,14).
In the present study, we found that miR-603 is
overexpressed in osteosarcoma, as compared to normal non-cancer
tissue and a normal human osteoblastic cell line. miR-603
significantly promoted osteosarcoma cell growth in vitro and
in vivo. We further demonstrated that BRCC2 is a direct
target of miR-603. miR-603 appears to be a potential therapeutic
target for use in osteosarcoma treatment.
Materials and methods
Ethical approval
All procedures performed in the studies involving
human participants were in accordance with the ethical standards of
the Institutional and/or National Research Committee and with the
1964 Helsinki Declaration and its later amendments or comparable
ethical standards. All applicable international, national and/or
institutional guidelines for the care and use of animals were
followed.
Tissue samples
From January 2010 to March 2014, a total of 50
primary osteosarcoma and corresponding non-cancerous bone tissue
samples were collected from the Fourth Affiliated Hospital of the
China Medical University. All specimens were obtained after
procuring written informed consent according to a protocol approved
by the Institutional Review Board of the Fourth Affiliated Hospital
of the China Medical University. Specimens were snap-frozen in
liquid nitrogen and stored at −80°C for quantitative real-time
reverse-transcriptase polymerase chain reaction (qRT-PCR) analysis.
All of the 50 patients received follow-up. The median follow-up was
60 months (range 4–62 months).
Cell lines and maintenance
Human osteosarcoma cell lines (MG-63, Saos-2, U2OS
and 143B) and human normal osteoblastic cell line hFOB1.19 were
obtained from the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). The cell lines mentioned above were
cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen,
USA) supplemented with 10% (w/v) fetal bovine serum (FBS) and
incubated at 37°C with 5% (v/v) CO2.
RNA extraction and quantitative real-time
PCR
Total RNAs were extracted from the cultured human
tissue specimens and cells using TRIzol reagent (Invitrogen)
according to the manufacturer's instructions. For the detection of
mature miR-603 and relative mRNA, RNA was reverse transcribed using
a TaqMan reverse transcription kit (Applied Biosystems, Foster
City, CA, USA). The reaction was incubated at 94°C for 4 min
followed by 35 cycles of 20 sec at 94°C, 30 sec at 60°C and 30 sec
at 72°C. All PCRs were carried out in triplicate on an ABI 7500
Real-Time PCR system (Applied Biosystems), and miRNA and mRNA
expression was normalized to U6 snRNA and GAPDH, respectively,
using the 2−ΔΔCt method, and at least 3 independent
experiments were performed to generate each data set.
Tumor formation in nude mice
Nude mice (4-weeks old, 18.0–22.0 g in weight) were
subcutaneously injected with 2×106 cells transfected
with either miR-603 mimics or its inhibitor. Every 2 days
post-inoculation, the length and width of the implanted tumors were
measured with a Vernier caliper. After 3 weeks, the mice were
sacrificed to measure tumor weights. The mice were manipulated and
housed according to protocols approved by the Local Medical
Experimental Animal Care Commission.
Transfection
miRNA-603 mimics and its inhibitor were purchased
from GenePharma Co. Ltd. (Shanghai, China). For transfection, MG-63
and U2OS cells were grown to 90% confluency, and transfected with
miR-603 mimics or its inhibitor using Lipofectamine 2000
(Invitrogen) by incubation in Opti-Mem I media for 4 h according to
the manufacturer's protocols. Cells were harvested 48 or 72 h
post-transfection. The transfection efficiency was confirmed by
quantitative real-time PCR analysis. For detecting BRCC2, the cells
were transfected again with BRCC2 or its silence gene.
Luciferase reporter assays
Cells cultured into 6-well plates were transfected
with 0.05 µg of the pRL-TK vector (Promega, Madison, WI,
USA) containing Renilla luciferase along with 30 nM miR-603
mimics or inhibitor. Cells cultured for 24 h were then transfected
with BRCC2-wild-type (WT) or BRCC2-mutant reporter plasmid
containing firefly luciferase using Lipofectamine 2000. Luciferase
activity was measured using a dual luciferase assay system
(Promega) 48 h post-transfection. Renilla activity was
normalized to firefly activity to control for transfection
efficiency.
MTT assay
The transfected cells were seeded into 96-well
plates at a density of 3×103 cells/well. MTT solution
(20 µl of 5 mg/ml MTT) was added to each well (for a total
volume of 250 µl), and the plates were incubated for 4 h at
37°C. Following the removal of the culture medium, the remaining
crystals were dissolved in dimethylsulfoxide (DMSO), and the
absorbance was measured at 570 nm using a microplate reader. Cell
proliferation was assessed daily for 4 consecutive days.
Colony formation assay
Different cells were seeded in triplicate at 500
cells/6-cm dishes in complete medium. The cells were then cultured
in DMEM containing 10% FBS and DMEM was changed every 2 days. After
incubation for 15 days, the cells were fixed with methanol and
stained with 0.1% crystal violet. Visible colonies were manually
counted. Triplicate wells were measured for each group.
Immunohistochemistry
All steps were carried out at room temperature as
previously described (15). Cells
transfected with miR-603 mimics or its inhibitor were plated on
poly-D-lysine-coated glass coverslips 72 h post-transfection and
were fixed in 4% (w/v) paraformaldehyde (Electron Microscopy
Sciences) for 1 h, washed in phosphate-buffered saline (PBS)
(Invitrogen) (6 times, 5 min each), incubated for 1 h in blocking
buffer (PBS, 0.8% Triton X-100, 10% normal goat serum), and then
incubated overnight in blocking buffer with anti-BRCC2 (1:100)
antibodies. The cells were washed in PBS (6 times, 5 min each)
before the addition of goat anti-mouse secondary antibodies
(Invitrogen, Molecular Probes) at 1:250 for 60 min followed by
washing in PBS (6 times, 5 min each). Finally, the slides were
incubated with 3,3′-diaminobenzidine (BioGenex Laboratories, San
Ramon, CA, USA) and color development was closely monitored under a
microscope. The slides were counterstained with hematoxylin.
Western blotting
Western blotting was used to detect the expression
of BRCC2 at the protein level as previously described (15). Anti-β-actin (Abcam) was used as the
protein-loading control. The protein complex was detected with
enhanced chemiluminescence reagents. Digital images were visualized
using the electrochemiluminescence detection system
(Invitrogen).
RNA interference for BRCC2
One siRNA lentivirus against BRCC2 and a
non-targeting siRNA (both from Sigma-Aldrich) were transfected into
U2OS-anti-miR-603 cells in 48-well plates according to the
manufacturer's instructions. The multiplicity of infection (MOI;
number of transducing lentiviral particles/cell) was 5. Puromycin
selection was performed at a concentration of 0.5 µg/ml for
10 days.
Statistical analysis
All data are presented as the mean ± standard error
of the mean (SEM). Statistical significance was determined using
t-test or analysis of variance (ANOVA) using the SPSS 18.0 program.
p<0.05 was considered to indicate a statistically significant
difference.
Results
miR-603 is upregulated in human
osteosarcoma carcinoma tissues and cell lines
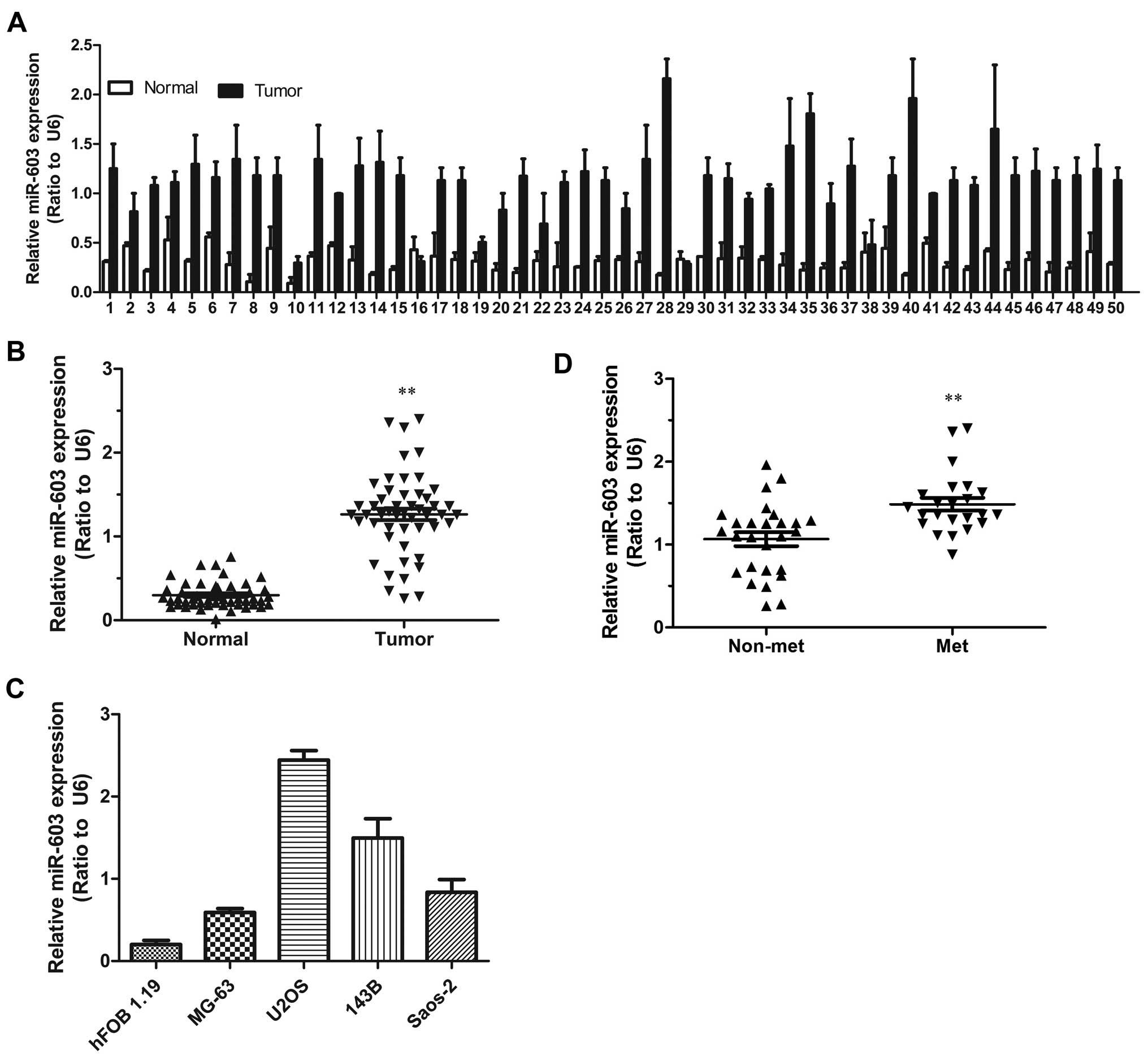
To explore the expression level of miR-603 in human
osteosarcoma carcinoma development, miR-603 expression was assessed
in 50 paired osteosarcoma carcinoma and adjacent non-tumor tissues.
According to the qRT-PCR analysis, miR-603 was strongly upregulated
in the 50 tumor tissues by 1.5- to 6-fold when compared with that
in the matched non-tumor tissues (Fig.
1A and B). We next examined miR-6–3 expression levels in a
panel of 4 widely used human osteosarcoma cell lines in comparison
to levels in the non-malignant cell line hFOB 1.19.
Correspondingly, miR-603 expression levels were consistently
increased in the osteosarcoma cell lines (Fig. 1C). To investigate the clinical
impact of elevated miR-603 expression in osteosarcoma, we assessed
the association between miR-603 expression levels and clinical
outcome. miR-603 expression levels were statistically increased in
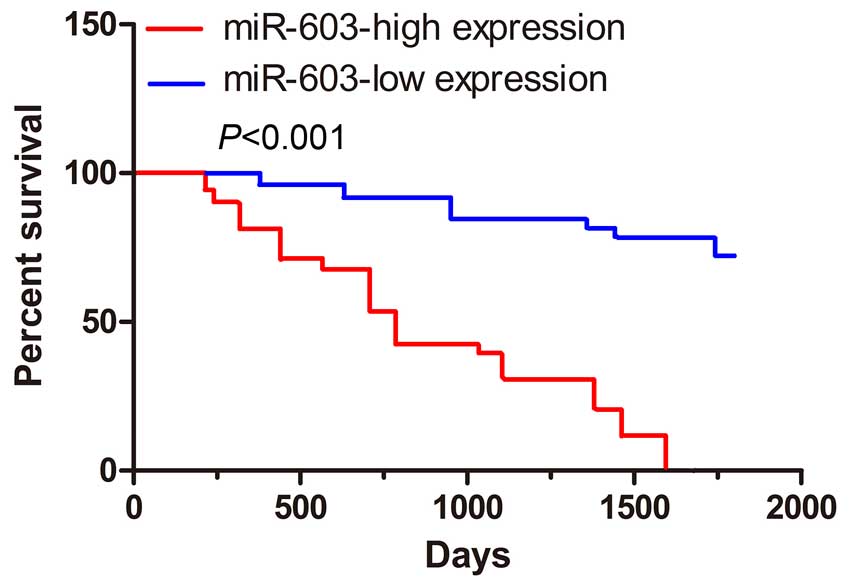
patients of stage III-IV in comparison to the stage I–II (Fig. 1D). Furthermore, in order to
determine the prognostic impact of miR-603 expression in
osteosarcoma, we categorized osteosarcoma patients into two groups
based on miR-603 expression levels. Patients with tumors displaying
high miR-603 expression levels had a significantly reduced rate of
survival compared to those with low miR-603 (Fig. 2).
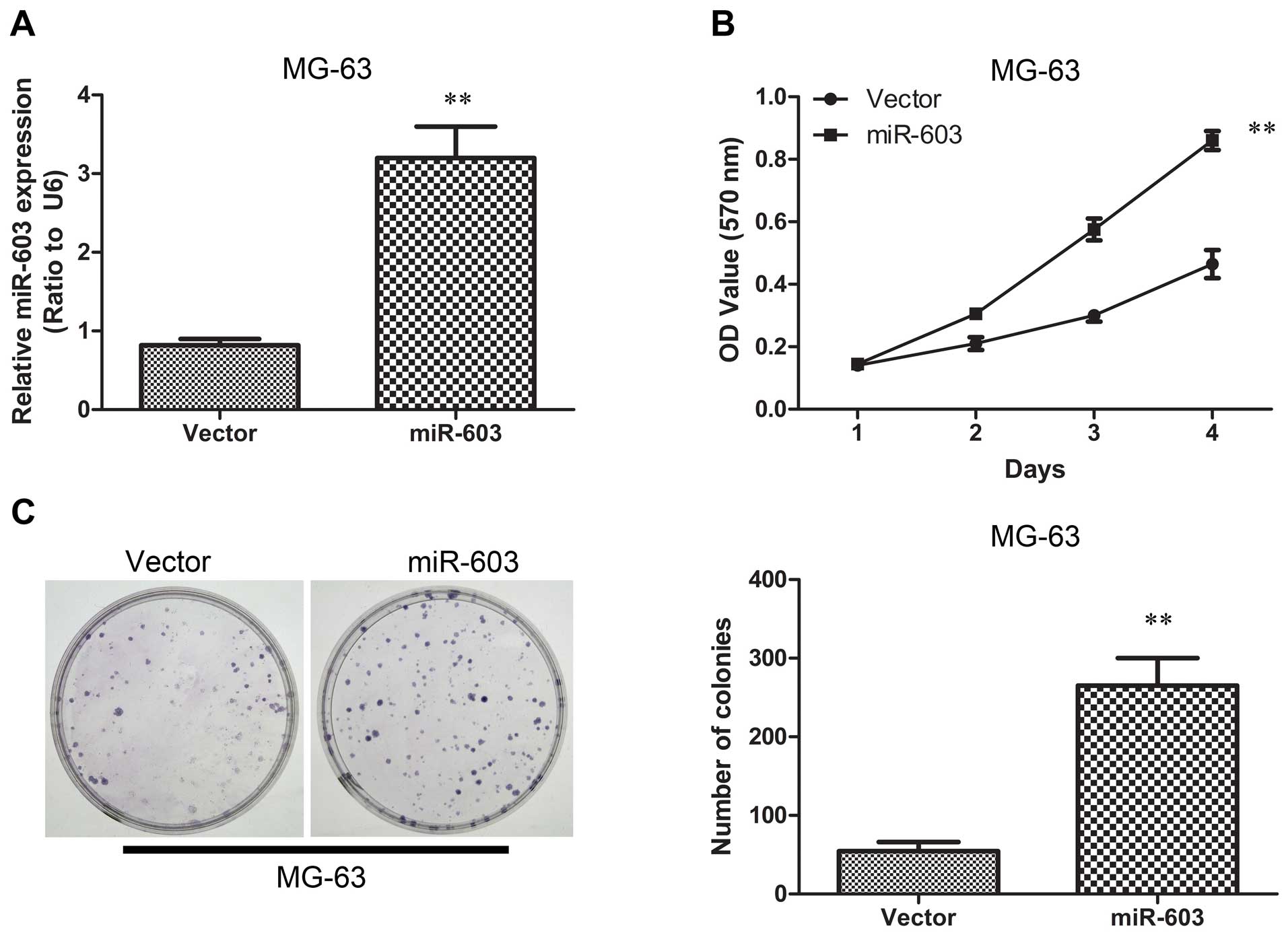
Overexpression of miR-603 in MG-63 cells
enhances tumorigenesis in vitro and in vivo
To evaluate a possible involvement of miR-603 in
osteosarcoma tumorigenesis, we examined its expression level in
human osteosarcoma. We established MG-63 cells overexpressing
miR-603 by introducing miR-603 mimics using lentiviral vectors. The
miR-603 expression levels were tested using qRT-PCR, and the
results showed that the miR-603 group expressed higher miR-603
levels as compared with the vector group (Fig. 3A). We next tested the hypothesis
that overexpression of miR-603 in MG-63 cells promotes cell growth
and colony formation ability. We performed MTT and colony formation
assays. The MTT assay showed that high expression of miR-603
resulted in more rapid proliferation compared with the vector group
(Fig. 3B). Furthermore,
transfection with miR-603 mimics resulted in significantly promoted
colony formation under serum starvation (Fig. 3C). The two results indicated that
miR-603 is potentially oncogenic. The miR-603-mediated tumorigenic
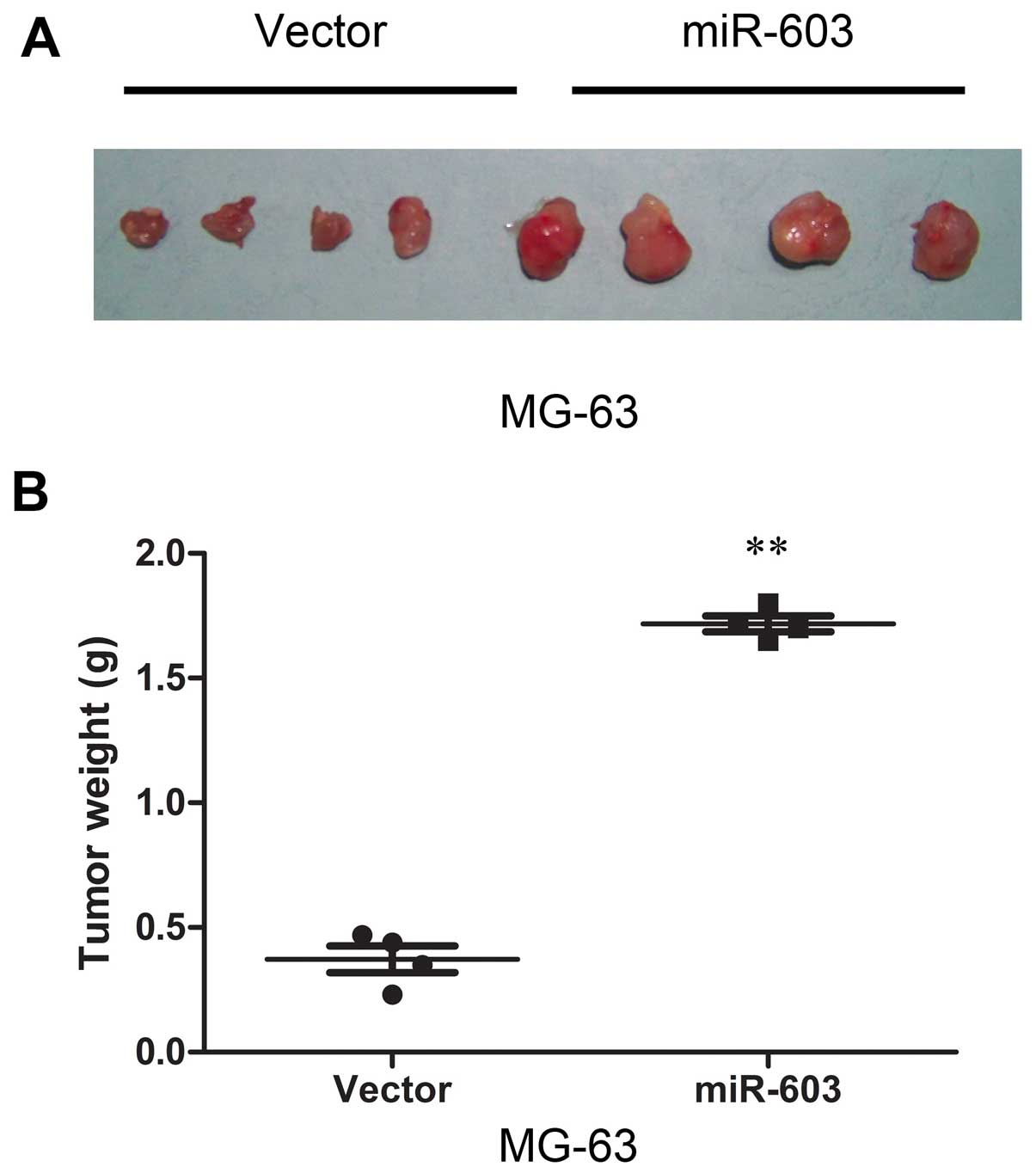
effects were confirmed in a nude mouse subcutaneous tumor model.
The results demonstrated that cells in the miR-603 group formed
progressively growing solid tumors. By contrast, cells in the
vector group produced much smaller tumors (Fig. 4A). Finally, as shown in Fig. 4A and B, overexpression of miR-603
promoted tumor growth in vivo. All of these findings above
demonstrated that miR-603 induces a more aggressive phenotype of
osteosarcoma carcinoma.
Inhibition of miR-603 in U2OS cells
reduces tumorigenesis in vitro and in vivo
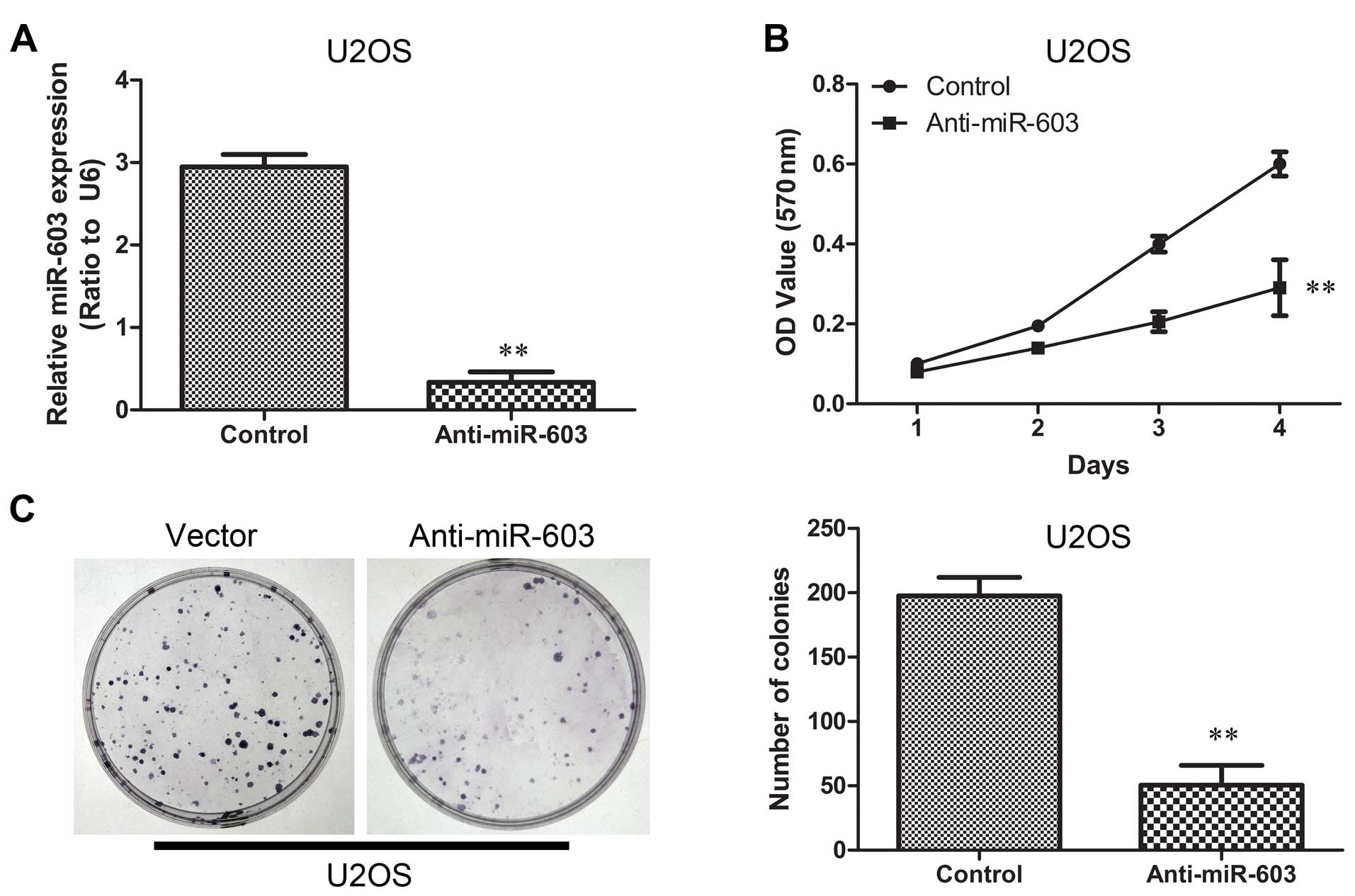
We suppressed miR-603 expression in U2OS cells by
introducing anti-miR-603 mimics using lentiviral vectors. The
suppression of miR-603 expression was confirmed using qRT-PCR
(Fig. 5A). To corroborate this
hypothesis, we investigated whether downregulation of miR-603 in
U2OS cells reduces cell growth and colony formation ability. MTT
and colony formation assays were also carried out. The
proliferation curve showed that cells in the anti-miR-603 group
exhibited significant decline in proliferation when compared to
cells in the vector group (Fig.
5B). Furthermore, transfection with anti-miR-603 mimics
obviously restrained colony formation under serum starvation
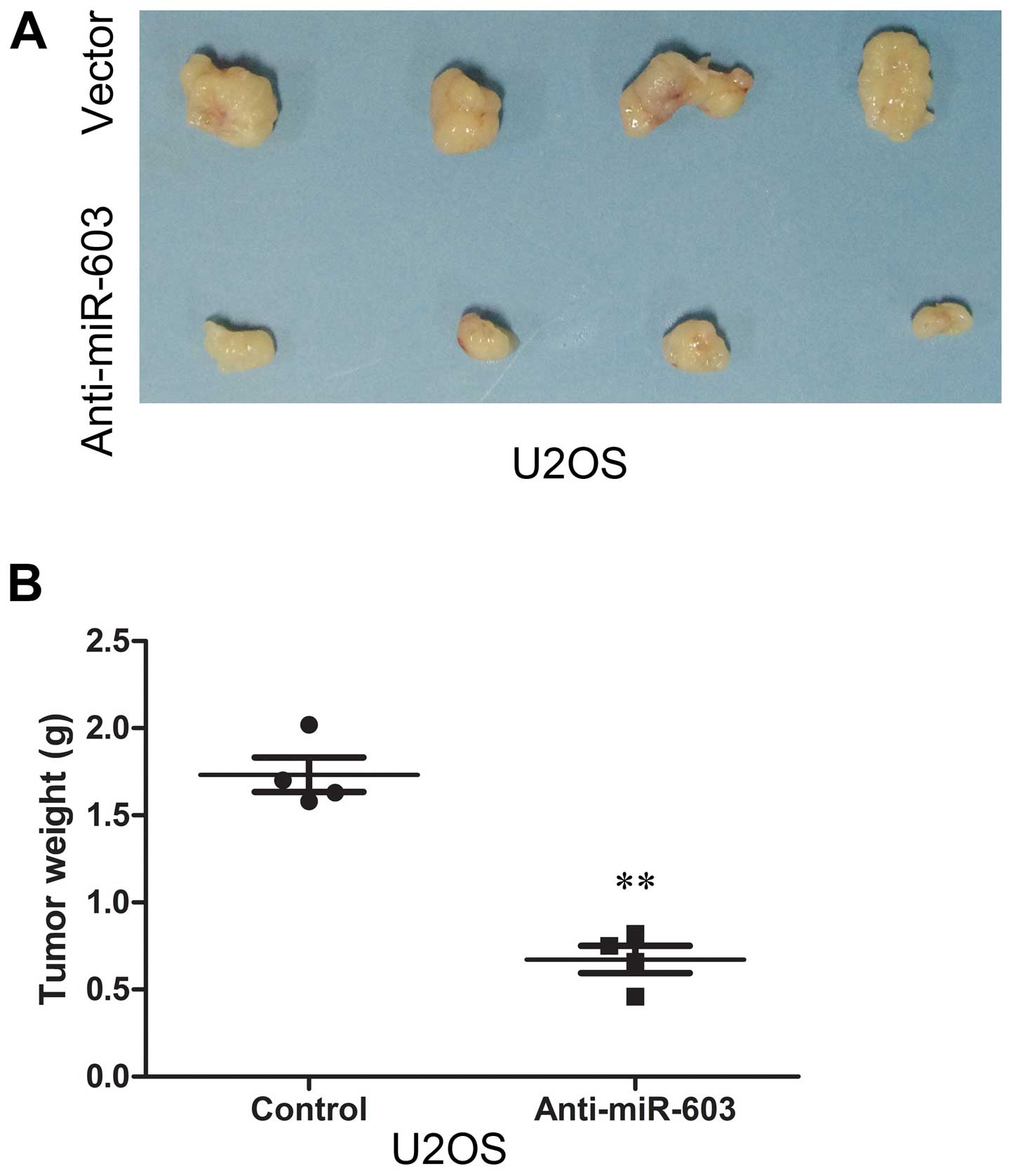
(Fig. 5C). Animal studies were
conducted to further evaluate the effect of anti-miR-603 on
osteosarcoma tumor growth in a nude mouse subcutaneous tumor model.
The results demonstrated that cells in the vector group formed
progressively growing solid tumors. By contrast, in the
anti-miR-603 group much smaller tumors were formed (Fig. 6A). Finally, the mean weight of
tumors in the control group was significantly higher than that in
the anti-miR-603 group (Fig.
6B).
miR-603 reduces the protein levels of
BRCC2 by inhibiting translation
In order to explore the mechanisms by which miR-603
promotes osteosarcoma tumorigenesis, in silico prediction
models were employed to search for genes regulated by miR-603. We
chose to investigate signal transducer and activator of BRCC2 as a
possible target of miR-603 since it is a known tumor-suppressor
gene that has been previously reported to be a direct target of
miR-603. To confirm that BRCC2 is a target of miR-603, the RNA
levels of BRCC2 were assessed in MG-63 cells overexpressing miR-603
(Fig. 7C) and U2OS cells with
silenced miR-603 (Fig. 7D).
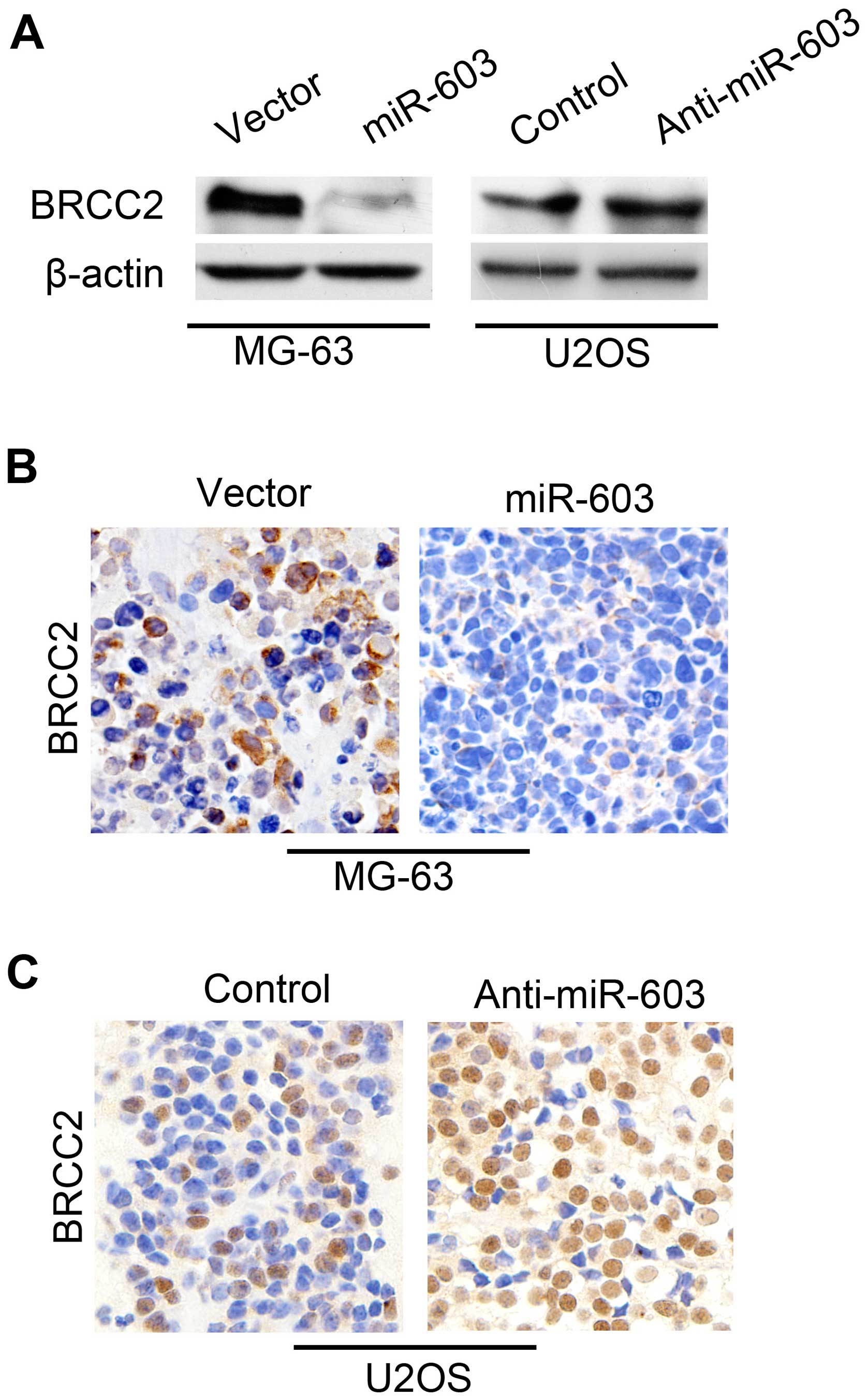
Correlation between miR-603 and BRCC2 expression was assessed by
western blot analysis (Fig. 8A) and
the immunohistochemical analysis in vivo (Fig. 8B and C). We found that BRCC2 was
downregulated in the miR-603-overexpressing cells, whereas BRCC2
was increased by the miR-603-specific inhibitor compared with that
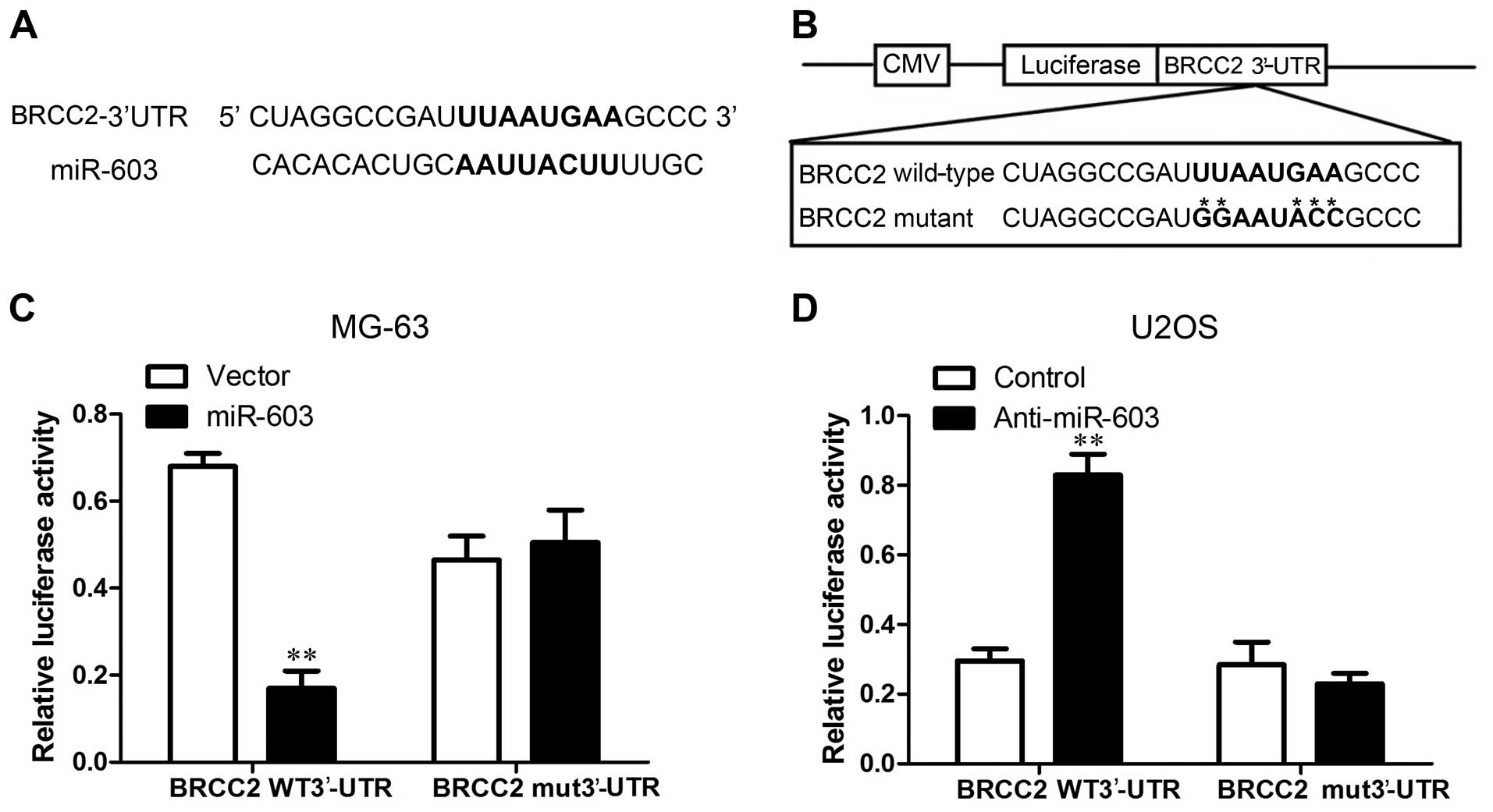
observed in the vector cells. We also repeated the luciferase assay
to confirm BRCC2 as a target of miR-603 in osteosarcoma cells, and
assessed whether miR-603 inhibits the translation of BRCC2. One
potential binding site for miR-603 was located on the 3′-UTR of
BRCC2 mRNA (Fig. 7A and B). To
further validate whether a miR-603 binding site in the BRCC2 3′-UTR
mediated this suppression, we inserted the BRCC2 3′-UTR transcript
or a mutated version into the luciferase system. The detection of
luciferase activity showed that the activity of luciferase combined
with wild-type BRCC2 3′-UTR was decreased in the MG-63-miR-603
cells and increased in the U2OS-anti-miR-603 cells, whereas the
luciferase activity was not altered by the vector when BRCC2 3′-UTR
possessing a mutation in the putative miR-603 binding site. As no
significant differences in the BRCC2 mRNA levels were observed
between the MG-63 vector as well as U2OS controls, miR-603 did not
appear to degrade BRCC2 mRNA (Fig. 7C
and D). These results together suggest that miR-603
downregulates BRCC2 expression in osteosarcoma via translational
inhibition.
BRCC2 is responsible for the
colony-forming and proliferative ability of MG-63 and U2OS
cells
In order to investigate the correlation between
BRCC2 expression and colony-forming and proliferativen ability of
the osteosarcoma cells, we constructed MG-63-miR-603 cells
transfected with BRCC2 and U2OS-anti-miR-603 cells transfected with
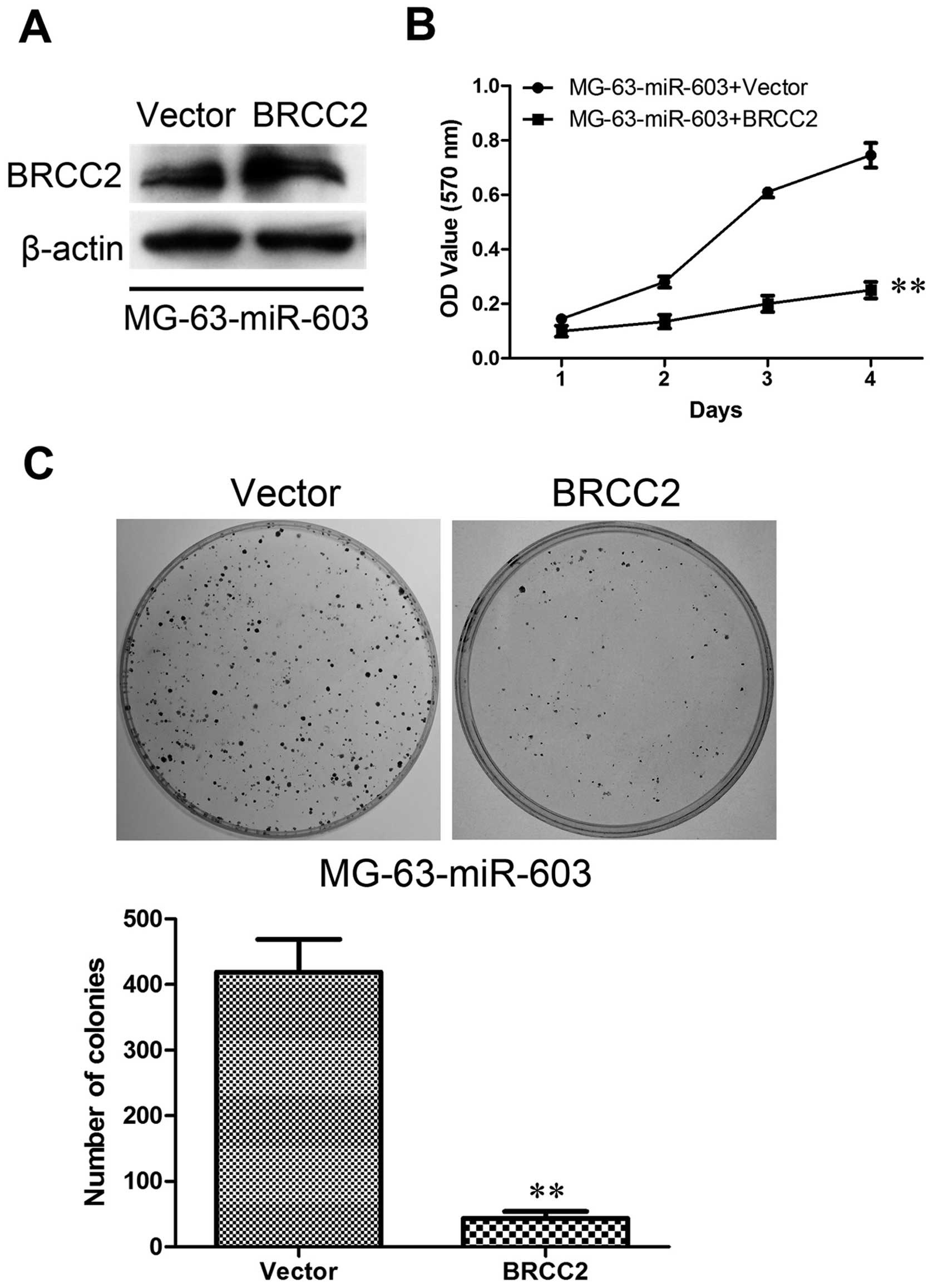
specific siRNAs against BRCC2. The expression of BRCC2 at the
protein level was analyzed by western blotting. The results
confirmed that BRCC2 expression was significantly upregulated in
the MG-63-miR-603 cells (Fig. 9A)
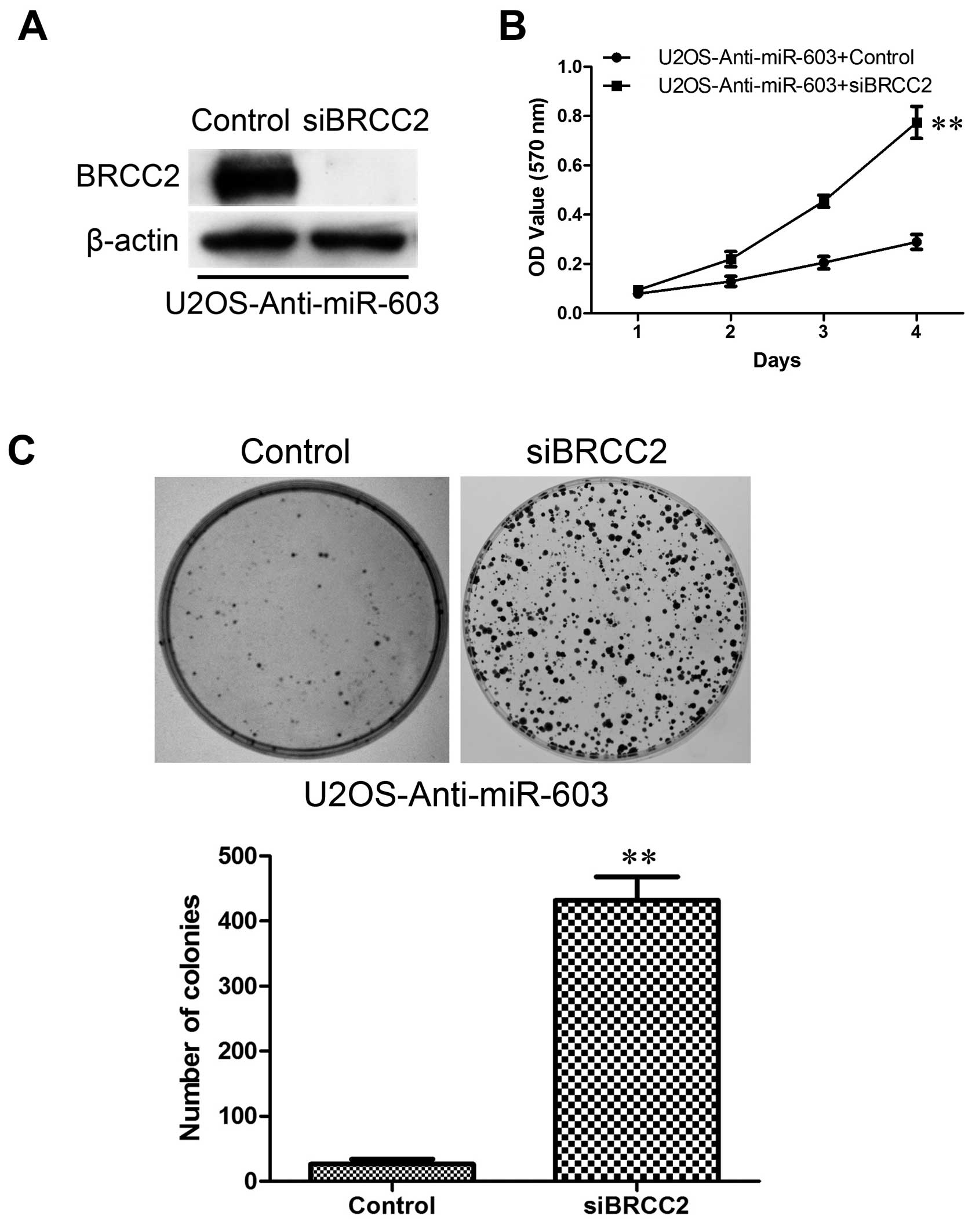
and silenced in U2OS-anti-miR-603 cells (Fig. 10A). Then, the treated cells were
evaluated for tumorigenesis using MTT and colony formation assays.
As expected, overexpression of BRCC2 strongly suppressed cell
proliferation compared to the vector cells (Fig. 9B) and clearly decreased colony
formation ability (Fig. 9C).
Inhibition of BRCC2 expression strongly promoted cell proliferation
compared to the control cells (Fig.
10B) and increased colony formation (Fig. 10C). Collectively, these results
indicate that reduced BRCC2 expression induced by miR-603 is
responsible for colony-forming and proliferation ability of the
MG-63 and U2OS cells.
Discussion
miRNAs are a type of small-molecule non-coding RNAs,
which play an important role in the regulation of
post-transcriptional gene expression levels. The functions of
miRNAs vary in various clinical diseases and may regulate all
aspects of bioactivities, including differentiation and
development, metabolism, proliferation, apoptotic cell death, viral
infection and tumorigenesis (16).
Abnormal cellular development, as occurs in cancer, has also been
associated with miRNAs (17). The
miRNAs with increased expression levels in tumors may function as
oncogenes by negatively regulating tumor-suppressor genes. In
contrast, the miRNAs downregulated in cancer may function as tumor
suppressors and inhibit cancer development by downregulating
responding oncogenes (18). To
summarize, miRNAs participate in all stages of tumor
development.
Osteosarcoma is one of the most common tumors that
threats human life. The incidence of osteosarcoma accounts for 20%
of all primary malignant bone tumors, behind multiple myeloma.
Typical osteosarcoma is a highly malignant intra-medullary tumor,
accounting for 85% of all bone sarcoma (19). Study of the relationships between
miRNAs and osteosarcoma may provide a new direction for the
diagnosis and treatment of osteosarcoma. In previous studies,
miR-143, miR-145, miR-195, miR-133a, miR-218 and miR-34a were
reported to inhibit osteosarcoma cell proliferation and to suppress
tumorigenicity (20–25); miR-221, miR-214, miR-128, miR-181,
miR-33a and miR-27a were reported to induce osteosarcoma cell
survival and promote cell proliferation (26–31).
It appears that miR-603 functions differentially in
various types of tumors. Our results found that it acts as an
oncogene by suppressing BRCC2. BRCC2 expression is reported to
prevent apoptosis in a wide variety of cell types and as a target
of miRNAs. Suppression or induction of BRCC2 affects tumorigenesis.
miR-195, miR-24–2 and miR-365–2 were reported as negative
regulators of BRCC2 through direct binding to their respective
binding sites in the 3′-UTR of the human BRCC2 gene (32). While, miR-15 and miR-16 induce
apoptosis by targeting BRCC2 (33).
The definition of osteosarcoma is deceptively
straightforward. It is a malignant tumor of connective tissue
origin within which tumor cells produce bone or osteoid (34). Our results revealed that miR-603
enhanced tumor growth by downregulation of BRCC2 expression via
translational inhibition in osteosarcoma cell lines and clinical
samples. Increased miR-603 expression was associated with
aggressive clinicopathological features and poor prognosis. The
suppression of miR-603 exhibited an antitumor effect both in
vitro and in vivo. Our findings demonstrate that miR-603
may be a potential novel target for the gene therapy of
osteosarcoma.
In summary, the present study showed that miR-603 is
significantly upregulated in osteosarcoma, meanwhile it is related
with poor patient overall survival. This demonstrates that miR-603
has powerful oncogenic, proliferative and invasive regulatory
effects that are mediated by BRCC2. The study indicates that
miR-603 acts as an oncogene and is a promising therapeutic target
in osteosarcoma.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geller DS and Gorlick R: Osteosarcoma: A
review of diagnosis, management, and treatment strategies. Clin Adv
Hematol Oncol. 8:705–718. 2010.
|
|
3
|
Zhang Y, Zhang L, Zhang G, Li S, Duan J,
Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al: Osteosarcoma
metastasis: Prospective role of ezrin. Tumour Biol. 35:5055–5059.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Farrill JS and Gordon N: Autophagy in
osteosarcoma. Adv Exp Med Biol. 804:147–160. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mussnich P, D'Angelo D, Leone V, Croce CM
and Fusco A: The High Mobility Group A proteins contribute to
thyroid cell transformation by regulating miR-603 and miR-10b
expression. Mol Oncol. 7:531–542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ayaz L, Görür A, Yaroğlu HY, Ozcan C and
Tamer L: Differential expression of microRNAs in plasma of patients
with laryngeal squamous cell carcinoma: Potential early-detection
markers for laryngeal squamous cell carcinoma. J Cancer Res Clin
Oncol. 139:1499–1506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
D'Angelo D, Palmieri D, Mussnich P, Roche
M, Wierinckx A, Raverot G, Fedele M, Croce CM, Trouillas J and
Fusco A: Altered microRNA expression profile in human pituitary GH
adenomas: Down-regulation of miRNA targeting HMGA1, HMGA2, and
E2F1. J Clin Endocrinol Metab. 97:E1128–E1138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu J, Li A, Hong SM, Hruban RH and Goggins
M: MicroRNA alterations of pancreatic intraepithelial neoplasias.
Clin Cancer Res. 18:981–992. 2012. View Article : Google Scholar :
|
|
9
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang FJ, Ding Y, Mao YY, Jing FY, Zhang
ZY, Jiang LF, Guo JF, Sun XJ, Jin MJ and Chen K: Associations
between hsa-miR-603 polymorphism, lifestyle-related factors and
colorectal cancer risk. Cancer Biomark. 14:225–231. 2014.PubMed/NCBI
|
|
11
|
Zutter M, Hockenbery D, Silverman GA and
Korsmeyer SJ: Immunolocalization of the Bcl-2 protein within
hematopoietic neoplasms. Blood. 78:1062–1068. 1991.PubMed/NCBI
|
|
12
|
Pietenpol JA, Papadopoulos N, Markowitz S,
Willson JK, Kinzler KW and Vogelstein B: Paradoxical inhibition of
solid tumor cell growth by bcl2. Cancer Res. 54:3714–3717.
1994.PubMed/NCBI
|
|
13
|
EI-Emshaty HM, Saad EA, Toson EA, Abdel
Malak CA and Gadelhak NA: Apoptosis and cell proliferation:
Correlation with BCL-2 and P53 oncoprotein expression in human
hepatocellular carcinoma. Hepatogastroenterology. 61:1393–1401.
2014.PubMed/NCBI
|
|
14
|
Garewal J, Garewal R and Sircar K:
Expression of Bcl-2 and MIB-1 markers in oral squamous cell
carcinoma (OSCC) - A comparative study. J Clin Diagn Res.
8:QC01–QC04. 2014.PubMed/NCBI
|
|
15
|
Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang
P, He X, Wang Q, Huang Y, Jen KY, Labarge MA, et al: CUL4A induces
epithelial-mesenchymal transition and promotes cancer metastasis by
regulating ZEB1 expression. Cancer Res. 74:520–531. 2014.
View Article : Google Scholar :
|
|
16
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar
|
|
17
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
19
|
Unni KK: Chordoma. Dahlin's Bone Tumors:
General Aspects and Data on 11,087 cases. 5th edition.
Lippincott-Raven; New York, NY: pp. 291–305. 1996
|
|
20
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fan L, Wu Q, Xing X, Wei Y and Shao Z:
MicroRNA-145 targets vascular endothelial growth factor and
inhibits invasion and metastasis of osteosarcoma cells. Acta
Biochim Biophys Sin. 44:407–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao JH, Zhou RP, Peng AF, Liu ZL, Huang
SH, Long XH and Shu Y: microRNA-195 suppresses osteosarcoma cell
invasion and migration in vitro by targeting FASN. Oncol Lett.
4:1125–1129. 2012.PubMed/NCBI
|
|
23
|
Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y,
Wang Z, Wang Z, Cheng P, Tong D, et al: MicroRNA-133a,
downregulated in osteosarcoma, suppresses proliferation and
promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 56:220–226.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin J, Cai L, Liu ZM and Zhou XS:
miRNA-218 inhibits osteosar-coma cell migration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar
|
|
25
|
Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q
and Ma B: MicroRNA-34a inhibits the proliferation and metastasis of
osteosarcoma cells both in vitro and in vivo. PLoS One.
7:e337782012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Z and Cai H, Lin L, Tang M and Cai H:
Upregulated expression of microRNA-214 is linked to tumor
progression and adverse prognosis in pediatric osteosarcoma.
Pediatr Blood Cancer. 61:206–210. 2014. View Article : Google Scholar
|
|
28
|
Shen L, Chen XD and Zhang YH: MicroRNA-128
promotes proliferation in osteosarcoma cells by downregulating
PTEN. Tumour Biol. 35:2069–2074. 2014. View Article : Google Scholar
|
|
29
|
Jianwei Z, Fan L, Xiancheng L, Enzhong B,
Shuai L and Can L: MicroRNA 181a improves proliferation and
invasion, suppresses apoptosis of osteosarcoma cell. Tumour Biol.
34:3331–3337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Y, Huang Z, Wu S, Zang X, Liu M and
Shi J: miR-33a is up-regulated in chemoresistant osteosarcoma and
promotes osteosarcoma cell resistance to cisplatin by
down-regulating TWIST. J Exp Clin Cancer Res. 33:122014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pan W, Wang H, Jianwei R and Ye Z:
MicroRNA-27a promotes proliferation, migration and invasion by
targeting MAP2K4 in human osteosarcoma cells. Cell Physiol Biochem.
33:402–412. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh R and Saini N: Downregulation of
BCL2 by miRNAs augments drug-induced apoptosis - a combined
computational and experimental approach. J Cell Sci. 125:1568–1578.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, et al: miR-15 and miR-16 induce apoptosis by
targeting BCL2. Proc Natl Acad Sci USA. 102:13944–13949. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Klein MJ and Siegal GP: Osteosarcoma:
Anatomic and histologic variants. Am J Clin Pathol. 125:555–581.
2006. View Article : Google Scholar : PubMed/NCBI
|