Introduction
Breast cancer is the most common malignancy in women
around the world, and its incidence and mortality rates are
increasing (1,2). While surgery, radiotherapy,
chemotherapy, hormone and/or biological methods are available
clinical therapies for breast cancer, their therapeutic efficacy is
still limited. Furthermore, the detailed pathogenesis of breast
cancer is still unclear. Therefore, it is of great importance to
gain a better understanding of the molecular mechanisms underlying
breast cancer pathogenesis, which will help to identify an
effective and specific molecular target for the development of
novel and promising therapeutic strategies for breast cancer.
The SOX genes, located on the sex-determining region
of the Y chromosome, are a highly conserved family of genes
encoding various transcription factors. All family members share a
high-mobility group (HMG-box) DNA-binding domain (3,4).
According to their HMG-box homology, the SOX family is divided into
ten groups (A–J) (4–7). SOX18, a member of group F (SOX F),
affects vascularization, neolymphangiogenesis, and tumor growth.
SOX18 mutations are characterized by defective blood and lymphatic
vessel formation, resulting in
hypotrichosis-lymphodema-teleangiectasia syndrome (8–10). In
addition, loss of function of SOX18 results in vascular and coat
anomalies in ragged (Ra) mutant mice (11). Furthermore, SOX18 overexpression has
recently been detected in several cancer-derived cell lines,
including those from melanoma, hepatic carcinoma, and breast,
gastric, lung, and pancreatic cancers (12–15).
However, mechanistic insight into how SOX18 exerts its function in
human breast cancer cells is still very limited.
Considering the important role of SOX18 in
tumorigenesis and tumor progression, we investigated the functional
role of SOX18 in breast cancer and explored the potential
underlying mechanism of SOX18 in regulating breast cancer cell
proliferation and invasion. We found that the mRNA and protein
levels of SOX18 in human breast cancer cells were significantly
increased, as compared with normal human breast epithelial cells.
Then, by siRNA-mediated silencing of SOX18 in two breast cancer
cell lines, BT474 and MCF-7, we demonstrated that SOX18 was
involved in the regulation of cell proliferation, apoptosis, and
invasion. Moreover, we found that SOX18 was associated with the
regulation of several tumorigenic genes. Taken together, the
results of our study provide evidence that SOX18 is a novel and
promising molecular target for human breast cancer therapy.
Materials and methods
Cell culture
Human breast cancer cell lines, including BT-474,
MCF-7, Hs578T, T-47D, and SK-BR-3, and the normal human breast
epithelial cell line, MCF-10 were from the Cell Bank of the
Shanghai Institutes for Biological Sciences, Chinese Academy of
Sciences (Shanghai, China). MCF-10A cells were maintained in
Dulbecco's modified Eagle's medium: F12 (DMEM: F12) medium
supplemented with 5% horse serum (Hyclone, Logan, UT, USA), 10
μg/ml insulin (Sigma-Aldrich, St. Louis, MO, USA), 100 ng/ml
cholera toxin (Sigma-Aldrich), 20 ng/ml epidermal growth factor
(EGF; Peprotech, Rocky Hill, NJ, USA), 0.5 μg/ml
hydrocortisone (Sigma-Aldrich), 100 U/ml penicillin, and 100 mg/ml
streptomycin. All other cell lines (human breast cancer cell lines)
were grown in RPMI-1640 medium containing 10% fetal bovine serum
(FBS; Gibco, Rockville, MD, USA), 100 U/ml penicillin, and 100
mg/ml streptomycin. These cell lines were maintained in a
humidified incubator with 5% CO2 at 37°C.
Silencing of SOX18 by small interfering
RNA (siRNA)
siRNAs directed against either human SOX18 or
negative control (NC) were purchased from Ambion (Austin, TX, USA).
BT-474 and MCF-7 cells were grown in 6-well plates and transfected
with SOX18 siRNA or NC (at a final concentration, 125 nM) using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to
the manufacturer's instructions. At 48 h after transfection, cells
were harvested for RT-PCR or western blot analyses.
CCK-8 assay
As described in detail previously (16), cell proliferation was assessed using
the Cell Counting Kit-8 (CCK-8) (Beyotime, Nantong, China) method.
Briefly, cells were seeded onto 96-well culture plates, and then
transfected with SOX18 siRNA, or negative control (NC) using
Lipofectamine 2000 according to manufacturer's instructions. At 48
h after siRNA transfection, CCK-8 solution was added to each well
and incubated for 2 h, and the absorbance was read at 450 nm. The
assay was performed in triplicate and repeated three times.
Protein extraction and western
blotting
Total protein from treated and untreated BT-474 and
MCF-7 cells was extracted using RIPA lysis buffer containing
phenylmethylsulfonyl fluoride (PMSF). Protein concentrations were
determined using the bicinchoninic acid (BCA) protein assay kit
(Boster Biology Co., Wuhan, China). Equal amounts of protein were
separated on SDS-polyacrylamide gels and electrotransferred to a
PVDF (polyvinylidene fluoride) membrane. Membranes were blocked
with 5% skim milk for 1 h at room temperature, and probed at 4°C
overnight with primary antibodies. The appropriate secondary
antibody was applied at room temperature for 1 h. An ECL detection
method (Pierce Biotechnology, Rockford, IL, USA) was subsequently
used to visualize the protein band of interest. Densitometric
analysis was performed using ImageJ software. Primary antibodies
were purchased from the following companies: i) SOX18, RhoA, PDGFB,
IGF1R and MMP7 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); ii)
Bcl-2, Bax and β-actin (ProteinTech, Chicago, IL, USA).
Reverse transcription and real-time PCR
(RT-PCR)
Total RNA was extracted from treated and untreated
cells using TRIzol reagent (Invitrogen) according to the
manufacturer's protocol. cDNA was then synthesized using the
PrimeScript RT reagent kit (Takara, Dalian, China) according to the
manufacturer's instructions. Next, real-time PCR was performed
using the LightCycler 480 and SYBR green master mix (Roche
Diagnostics Ltd., Lewes, UK). β-actin served as an internal
control. Results were normalized to the endogenous control
(β-actin). All RT-PCR amplifications were performed in triplicate.
Fold changes were calculated in relation to reference control cDNA
as previously described (17). The
sequences of PCR primers for SOX18, RhoA, PDGFB, IGF1R, MMP7,
Bcl-2, Bax, and β-actin are listed in Table I.
 | Table IThe primer sequences of PCR. |
Table I
The primer sequences of PCR.
| Primer | Sequence |
|---|
| β-actin | F:
5′-GCGCGGCTACAGCTTCA-3′ |
| R:
5′-TCTCCTTAATGTCACGCACGAT-3′ |
| SOX18 | F:
5′-CGCGTGTATGTTTGGTTC-3′ |
| R:
5′-ATGTAACCCTGGCAACTC-3′ |
| Bcl-2 | F: 5′-
ATGTGTGTGGAGAGCGTCAACC-3′ |
| R: 5′-
GCATCCCAGCCTCCGTTATC-3′ |
| Bax | F:
5′-CCTTTTCTACTTTGCCAGCAAAC-3′ |
| R: 5′-
GAGGCCGTCCCAACCAC-3′ |
| RhoA | F:
5′-GAGTGTTCAGCAAAGACCAAAG-3′ |
| R:
5′-TTGCAGCAAGGTTTCACAAG-3′ |
| PDGFB | F:
5′-CTCGATCCGCTCCTTTGATG-3′ |
| R:
5′-AGGAAGTTGGCGTTGGTG-3′ |
| IGF1R | F:
5′-GAGCCTCCTGTGAAAGTG-3′ |
| R:
5′-GCATCCTGCCCATCATAC-3′ |
| MMP-7 | F:
5′-GAGTGCCAGATGTTGCAGAA-3′ |
| R: 5
AAATGCAGGGGGATCTCTTT-3′ |
Cell invasion assay
As described in detail previously (18), transwell insert assays were used to
assess BT-474 and MCF-7 cell invasion ability. Briefly, for the
invasion assay, siRNA-transfected cells in serum-free RPMI-1640
were seeded in the upper chambers of Matrigel-coated Transwell
plates. To the lower chamber was added RPMI-1640 containing 10% FBS
(600 μl) as a chemotactic factor. All of the Transwell
chambers were then incubated at 37°C for 48 h. Each subclone was
seeded in triplicate. The migrated cells were observed under a
Leica inverted microscope (Deerfield, IL, USA) and counted.
Caspase-3 activity assay
The caspase-3 colorimetric assay is based on the
hydrolysis of the peptide substrate
acetyl-Asp-Glu-Val-Asp-p-nitroanilide (Ac-DEVD-pNA) by caspase-3,
resulting in the release of the p-nitroaniline (pNA) moiety. pNA
has a high absorbance at 405 nm. Cell lysates were prepared using
the Caspase-3 activity kit (EMD Millipore Corp., Billerica, MA,
USA). The concentration of pNA released from the substrate is
calculated from the absorbance values at 405 nm. The rest of the
detailed procedure was performed according to the manufacturer's
instructions. This assay was performed in triplicate and repeated
three times.
Statistical analysis
Data are summarized as mean ± SD from at least three
independent experiments. Statistical analysis was carried out using
SPSS 17.0 software. Statistical differences were processed using
one-way analysis of variance (ANOVA). A P-value of <0.05 was
considered to be statistically significant.
Results
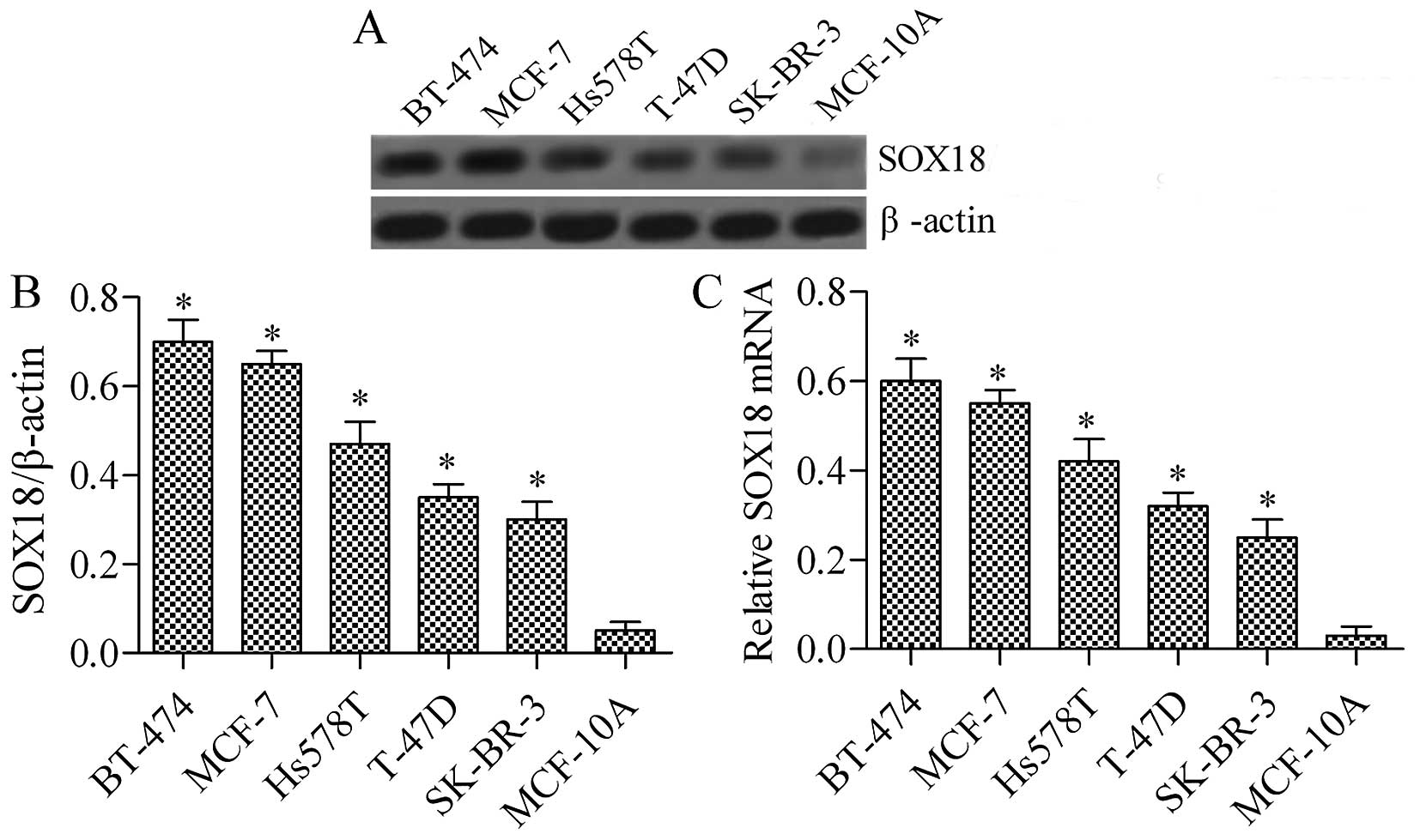
SOX18 is highly expressed in various
human breast cancer cell lines
To investigate the function of SOX18 in human breast
cancer cell lines, we evaluated its expression in four breast
cancer cell lines (BT-474, MCF-7, Hs578T, T-47D, and SK-BR-3) and a
normal human breast epithelial cell line (MCF-10A) using RT-PCR and
western blotting. As shown in Fig.
1, the expression of SOX18 was significantly upregulated in
breast cancer cell lines when compared with that of matched normal
human breast epithelial cells, both at the protein (Fig. 1A) and mRNA expression levels
(Fig. 1B). The data revealed that
SOX18 is overexpressed in breast cancer cell lines, indicating that
increased SOX18 protein expression is clearly involved in human
breast cancer development.
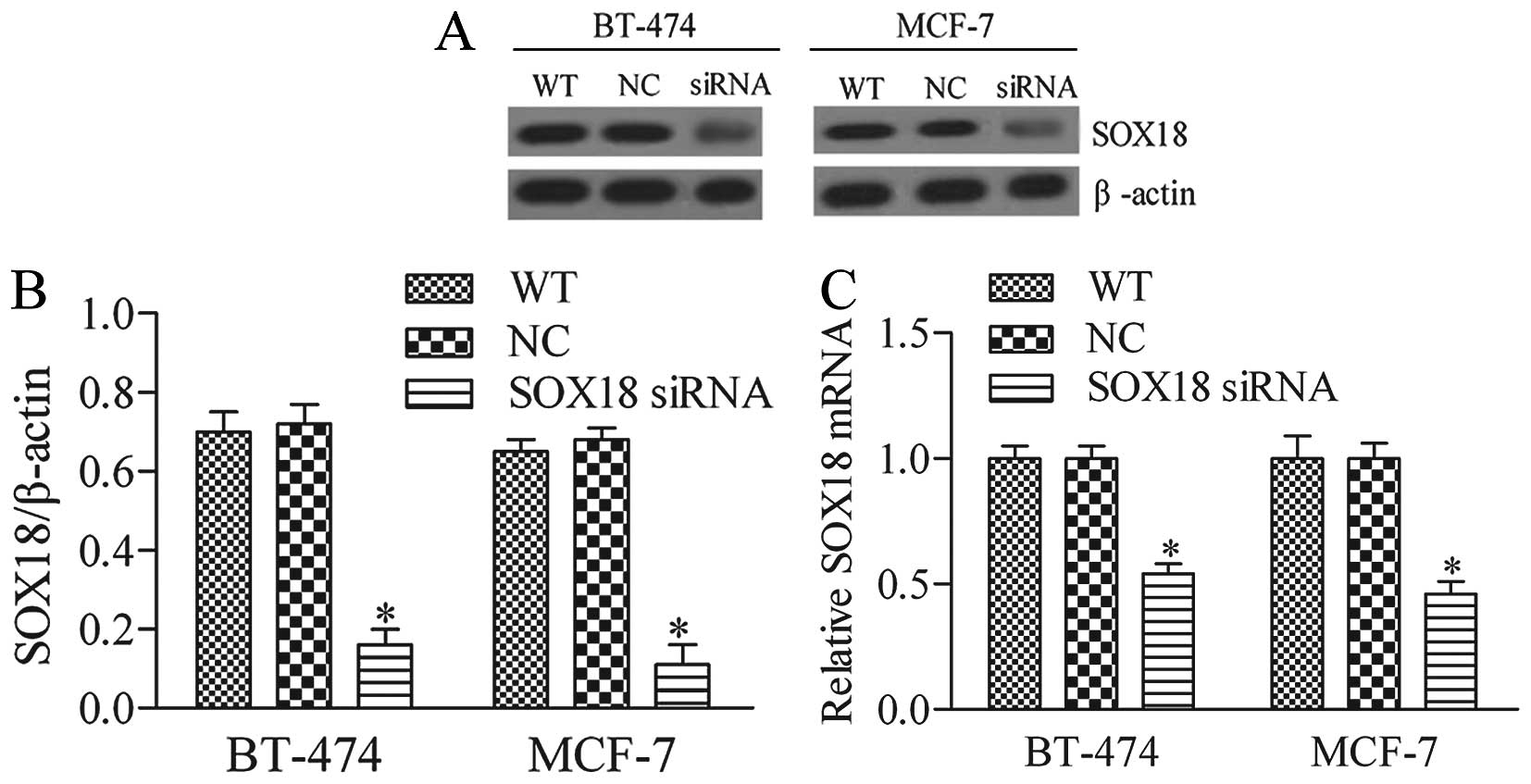
Silencing of SOX18 by RNAi
To investigate the potential function of SOX18, the
breast cancer cell lines, BT-474 and MCF-7 were selected for the
RNAi experiment and transfected with one siRNA targeting human
SOX18 (SOX18 siRNA) and a negative control (NC) siRNA. The
silencing efficiency of the siRNA on SOX18 expression was then
detected by western blot and RT-PCR analysis. Our results showed
that SOX18 siRNA was able to efficiently and significantly suppress
endogenous SOX18 expression in BT-474 and MCF-7 cells (Fig. 2A and B).
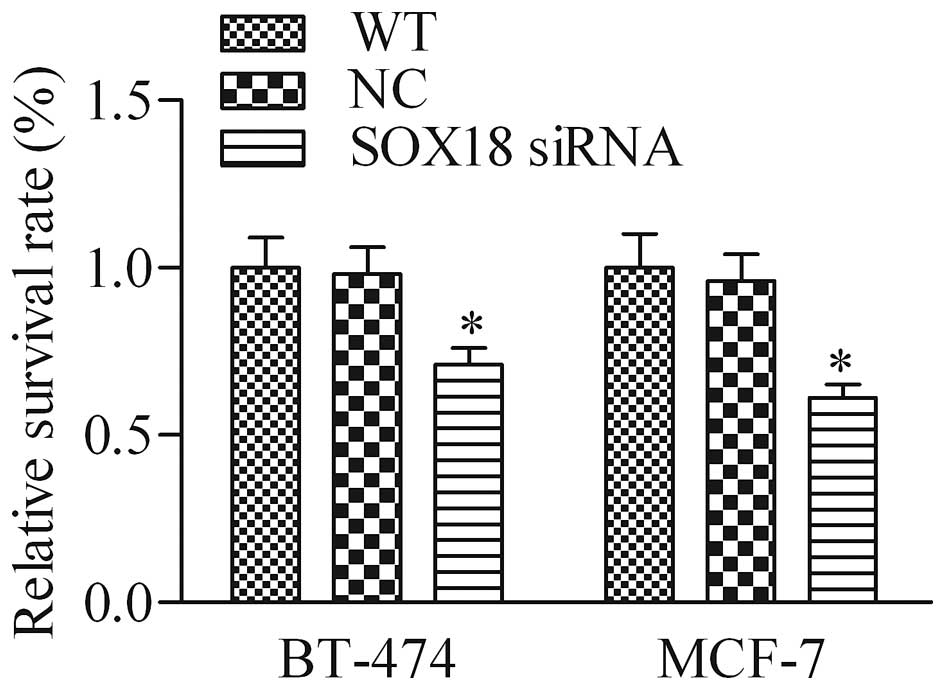
SOX18 knockdown inhibits human breast
cancer cell proliferation
To test whether the silencing effect of SOX18
suppresses the proliferation of human breast cancer cells, BT-474
and MCF-7, the CCK-8 assay was performed. As shown in Fig. 3, cell proliferation of BT-474 and
MCF-7 transfected with SOX18 siRNA was notably impaired when
compared to the corresponding WT and NC cells. These results
suggest that knockdown of SOX18 represses breast cancer cell
proliferation.
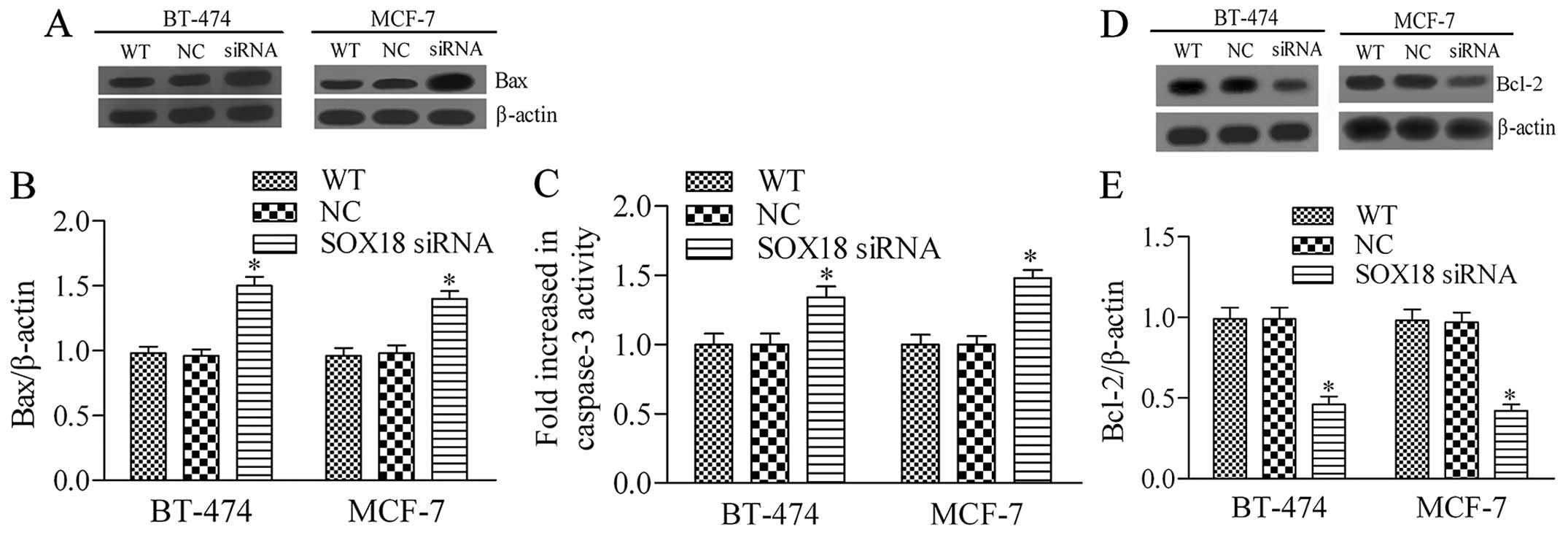
Silencing of SOX18 in human breast cancer
induces cell apoptosis
To further investigate the role of SXO18 in breast
cancer cells, we next detected the effect of SOX18 silencing on
breast cancer cell apoptosis. The results showed that knockdown of
SOX18 significantly increased the expression of Bax and the
activity of caspase-3 (Fig. 4A–C)
in both BT-474 and MCF-7 cells. Furthermore, the protein expression
level of Bcl-2, an anti-apoptotic protein, was markedly decreased
by SOX18 siRNA transfection. These results imply that knockdown of
SOX18 promotes breast cancer cell apoptosis.
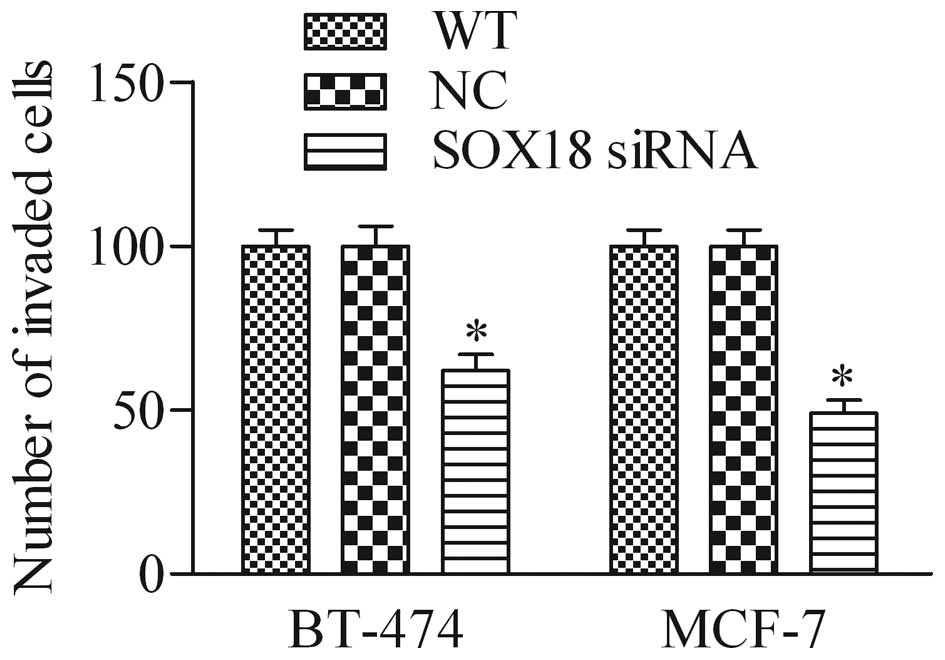
Silencing of SOX18 affects cell invasion
in human breast cancer cells
To further investigate the biological role of SOX18
in breast cancer cells, we detected the effect of SOX18 siRNA on
breast cancer cell invasion using the Transwell invasion assay. As
shown in Fig. 5, the number of
invaded cells in SOX18 siRNA-knockdown cells was significantly
decreased in BT-474 and MCF-7 cells when compared with that of the
WT and NC cells, suggesting that depletion of SOX18 markedly
decreased the cell invasive ability of breast cancer cells.
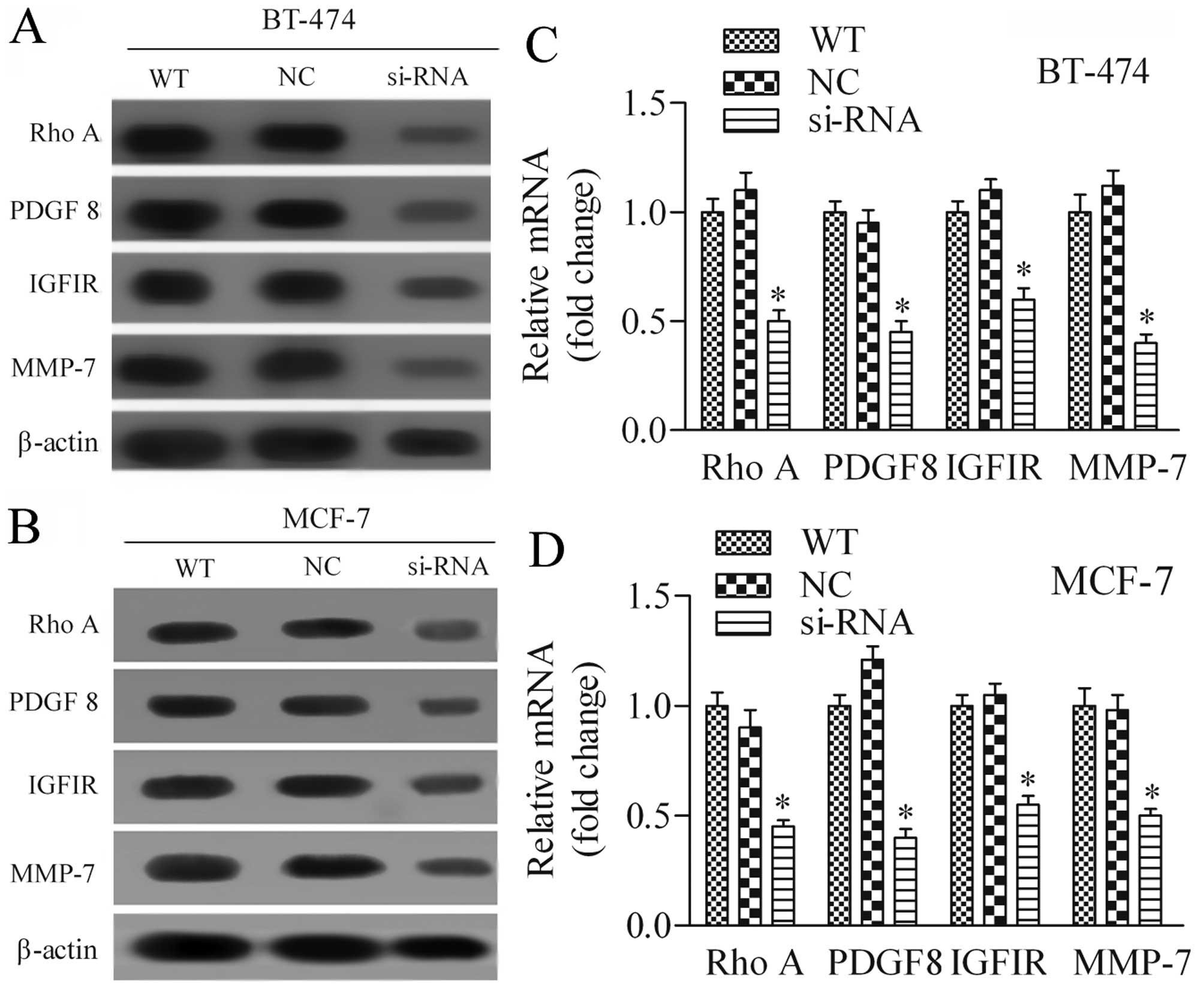
Silencing of SOX18 in human breast cancer
cells modulates the mRNA and protein expression of RhoA, PDGFB,
IGF1R, and MMP-7
To explore the potential underlying molecular basis
of SOX18 in regulating breast cancer cell proliferation and
invasion, we examined the expression of several oncogenic proteins,
including RhoA, PDGFB, IGF1R and MMP-7 by western blot analysis and
RT-PCR. Results showed that both protein (Fig. 6A) and mRNA (Fig. 6B) expression levels of these
detected genes were markedly decreased after inhibition of SOX18
expression in BT-474 cells in comparison with control cells.
Furthermore, similar data were observed using MCF-7 cells (Fig. 6C and D). These data suggest that
SOX18 is an important transcription factor in breast cancer that
modulates the expression of several oncogenic proteins.
Discussion
It is now clear that SOX genes encoding
transcription factors are involved in embryonic development and
oncogenesis (19). While the role
and functions of SOX18 in various cancers are proundly reseached,
the specific molecular functions of SOX18 in human breast cancer
remain obscure. Here, we evaluated SOX18 expression in human breast
cancer cell lines (BT-474, MCF-7, Hs578T, T-47D, and SK-BR-3) using
RT-PCR and western blot analysis, and we found that SOX18 was
highly expressed in breast cancer cell lines. Furthermore,
knockdown of SOX18 by SOX18 siRNA suppressed cell proliferation and
invasion of breast cancer cells, but promoted breast cancer cell
apoptosis. Most importantly, several oncogenic proteins, including
RhoA, PDGFB, IGF1R, and MMP-7, were found to be regulated by SOX18.
These results suggested that SOX18 was a critical regulator
involved in human breast cancer.
Previous studies showed that SOX18 mRNA was
expressed in pancreatic and breast cancer cells (13); in non-small cell lung cancer, SOX18
expression in mRNA and protein was significantly lower than in
non-malignant lung tissue (12);
Also, in ovarian cancer, SOX18 expression was detected in the cell
nuclei as well as the cytoplasm using immunohistochemical methods
(20). In the current study, the
expression level of SOX18 mRNA and protein was significantly
upregulated in human breast cancer cell lines when compared with a
normal human breast epithelial cell line (Fig. 1A), which is consistent with the
results obtained by immunohistochemistry showing that SOX18 was
rarely expressed in non-malignant breast duct cells and frequently
in invasive ductal breast carcinoma (21). Taken together, these findings
suggest that SOX18 may serve as a novel oncogene and a potential
therapeutic molecular target.
Previous studies have suggested the promoting effect
of SOX18 on cell proliferation of vascular smooth muscle cells
(22) and MCF-7 breast cancer cells
(15). In line with these findings,
knockdown of SOX18 in human breast cancer cells significantly
impaired cell proliferation (Fig.
3). Moreover, we assessed the apoptotic function of SOX18 in
SOX18 siRNA-treated breast cancer cells (Fig. 4A). Our data showed that the
silencing of SOX18 significantly downregulated the expression of
Bcl-2 and increased the activity of caspase-3 and expression of Bax
in BT-474 and MCF-7 cells (Fig. 4),
which might have contributed to the inhibition of proliferation in
SOX18-knockdown cells.
In addition, the expression of SOX18 is mainly
involved in tumor metastatic processes in lung and gastric cancer,
where it plays an important role in regulating vascular formation
and degradation of extracellular matrix tumor metastasis (23,24).
Consistent with these studies, we found that SOX18 siRNA treatment
significantly decreased the invasive capabilities of breast cancer
cells (Fig. 5), suggesting that
tumor metastasis can be inhibited by SOX18 silencing in human
breast cancer cells.
To elucidate the underlying mechanisms involved in
cell proliferation, apoptosis and invasion induced by SOX18
silencing, we investigated the expression of several downstream
signaling molecules of SOX18, including RhoA, PDGFB, IGF1R, and
MMP-7, in SOX18 siRNA-knockdown breast cancer cells. RhoA, a small
GTP-binding protein, has been reported to be associated with the
occurrence, invasion, and metastasis of various tumors (25–27).
PDGFB, a platelet-derived growth factor, is a member of the PDGF
family (PDGFA, B, C, and D) that is involved in multiple
tumor-associated processes, including tumor growth and metastasis
(autocrine or paracrine) (28),
tumor angio genesis (29), and
tumor fibroblasts (30). IGF1R is
reported to be overexpressed in some types of human cancer,
including lung, breast, pancreatic, prostate and glioma cancers
(31–36). Moreover, IGF1R has been confirmed to
play critical roles in multiple biological processes of tumor
diseases, including malignant transformation, proliferation,
anti-apoptosis, vascularization, and invasion (37). MMP-7 can cleave plasminogen and
collagen XVIII to generate angiostatin and a 28 kDa
endostatin-spanning fragment, respectively (38). Previous studies have also suggested
that SOX18 RNAi significantly downregulated the expression of
detected genes in SOX18 siRNA-knockdown hepatocellular carcinoma
cells (14) and in human
endothelial cells (39). Our
findings further proved that the expression of detected genes is
significantly downregulated in SOX18 siRNA-treated breast cancer
cells (Fig. 6), indicating that
tumor progression can be inhibited by SOX18 silencing.
Interestingly, SOX18 expression also plays a crucial
role in vascularization and lymphangiogenesis. Previous studies
showed that vascular cell growth was inhibited by antisense SOX18
in endothelial cells and vascular smooth muscle cells (22); the expression of dominant-negative
SOX18 in human umbilical vein endothelial cells impaired capillary
tube formation (15). Additionally,
knockdown of SOX18 selectively impaired lymphatic sprouting,
resulted in defective lymphatic thoracic duct formation, and caused
lymphedema (40). SOX18 expression
is not responsible for the maintenance of lymphatic identity in the
normal organism, whereas under pathological conditions, such as
tumor growth, loss of SOX18 function impairs tumor-induced
angiogenesis and lymphangiogenesis and decreases cancer cell
metastasis (41). However,
knowledge of whether knockdown of SOX18 expression decreases cancer
metastasis through inhibition of angiogenesis and lymphangiogenesis
in breast cancer would be required for further research.
In conclusion, this study showed for the first time
that SOX18 silencing by siRNA inhibited proliferation and invasion
and promoted apoptosis in human breast cancer cell lines in
vitro. Furthermore, our findings showed that knockdown of SOX18
significantly downregulated the expression of associated
tumorigenic genes (RhoA, PDGFB, IGF1R, and MMP-7) in SOX18
siRNA-knockdown breast cancer cells. Our results suggest that SOX18
is a promising therapeutic molecular target for the treatment of
breast cancer.
References
|
1
|
Anaya-Ruiz M and Perez-Santos M:
Innovation status of gene therapy for breast cancer. Asian Pac J
Cancer Prev. 16:4133–4136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang GQ, He C, Tao L and Liu F: Role of
DJ-1 siRNA in reverse sensitivity of breast cancer cells to
chemotherapy and its possible mechanism. Int J Clin Exp Pathol.
8:6944–6951. 2015.PubMed/NCBI
|
|
3
|
Harley VR, Lovell-Badge R and Goodfellow
PN: Definition of a consensus DNA binding site for SRY. Nucleic
Acids Res. 22:1500–1501. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wegner M: From head to toes: The multiple
facets of Sox proteins. Nucleic Acids Res. 27:1409–1420. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dunn TL, Mynett-Johnson L, Wright EM,
Hosking BM, Koopman PA and Muscat GE: Sequence and expression of
Sox-18 encoding a new HMG-box transcription factor. Gene.
161:223–225. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanai Y, Kanai-Azuma M, Noce T, Saido TC,
Shiroishi T, Hayashi Y and Yazaki K: Identification of two Sox17
messenger RNA isoforms, with and without the high mobility group
box region, and their differential expression in mouse
spermatogenesis. J Cell Biol. 133:667–681. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taniguchi K, Hiraoka Y, Ogawa M, Sakai Y,
Kido S and Aiso S: Isolation and characterization of a mouse
SRY-related cDNA, mSox7. Biochim Biophys Acta. 1445:225–231. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Downes M, François M, Ferguson C, Parton
RG and Koopman P: Vascular defects in a mouse model of
hypotrichosis-lymphedematelangiectasia syndrome indicate a role for
SOX18 in blood vessel maturation. Hum Mol Genet. 18:2839–2850.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
François M, Caprini A, Hosking B, Orsenigo
F, Wilhelm D, Browne C, Paavonen K, Karnezis T, Shayan R, Downes M,
et al: Sox18 induces development of the lymphatic vasculature in
mice. Nature. 456:643–647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irrthum A, Devriendt K, Chitayat D,
Matthijs G, Glade C, Steijlen PM, Fryns JP, Van Steensel MA and
Vikkula M: Mutations in the transcription factor gene SOX18
underlie recessive and dominant forms of
hypotrichosis-lymphedematelangiectasia. Am J Hum Genet.
72:1470–1478. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pennisi D, Gardner J, Chambers D, Hosking
B, Peters J, Muscat G, Abbott C and Koopman P: Mutations in Sox18
underlie cardiovascular and hair follicle defects in ragged mice.
Nat Genet. 24:434–437. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jethon A, Pula B, Olbromski M, Werynska B,
Muszczynska-Bernhard B, Witkiewicz W, Dziegiel P and
Podhorska-Okolow M: Prognostic significance of SOX18 expression in
non-small cell lung cancer. Int J Oncol. 46:123–132. 2015.
|
|
13
|
Saitoh T and Katoh M: Expression of human
SOX18 in normal tissues and tumors. Int J Mol Med. 10:339–344.
2002.PubMed/NCBI
|
|
14
|
Wang G, Wei Z, Jia H, Zhao W, Yang G and
Zhao H: Knockdown of SOX18 inhibits the proliferation, migration
and invasion of hepatocellular carcinoma cells. Oncol Rep.
34:1121–1128. 2015.PubMed/NCBI
|
|
15
|
Young N, Hahn CN, Poh A, Dong C, Wilhelm
D, Olsson J, Muscat GE, Parsons P, Gamble JR and Koopman P: Effect
of disrupted SOX18 transcription factor function on tumor growth,
vascularization, and endothelial development. J Natl Cancer Inst.
98:1060–1067. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di J, Huang H, Qu D, Tang J, Cao W, Lu Z,
Cheng Q, Yang J, Bai J, Zhang Y, et al: Rap2B promotes
proliferation, migration, and invasion of human breast cancer
through calcium-related ERK1/2 signaling pathway. Sci Rep.
5:123632015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
McGarry T, Veale DJ, Gao W, Orr C, Fearon
U and Connolly M: Toll-like receptor 2 (TLR2) induces migration and
invasive mechanisms in rheumatoid arthritis. Arthritis Res Ther.
17:1532015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castillo SD and Sanchez-Cespedes M: The
SOX family of genes in cancer development: Biological relevance and
opportunities for therapy. Expert Opin Ther Targets. 16:903–919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pula B, Kobierzycki C, Solinski D,
Olbromski M, Nowak-Markwitz E, Spaczynski M, Kedzia W, Zabel M and
Dziegiel P: SOX18 expression predicts response to platinum-based
chemotherapy in ovarian cancer. Anticancer Res. 34:4029–4037.
2014.PubMed/NCBI
|
|
21
|
Pula B, Olbromski M, Wojnar A,
Gomulkiewicz A, Witkiewicz W, Ugorski M, Dziegiel P and
Podhorska-Okolow M: Impact of SOX18 expression in cancer cells and
vessels on the outcome of invasive ductal breast carcinoma. Cell
Oncol (Dordr). 36:469–483. 2013. View Article : Google Scholar
|
|
22
|
García-Ramírez M, Martínez-González J,
Juan-Babot JO, Rodríguez C and Badimon L: Transcription factor
SOX18 is expressed in human coronary atherosclerotic lesions and
regulates DNA synthesis and vascular cell growth. Arterioscler
Thromb Vasc Biol. 25:2398–2403. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Azhikina T, Kozlova A, Skvortsov T and
Sverdlov E: Heterogeneity and degree of TIMP4, GATA4, SOX18, and
EGFL7 gene promoter methylation in non-small cell lung cancer and
surrounding tissues. Cancer Genet. 204:492–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma LJ, Wang J, Deng L and Yu S: Expression
of SOX18, VEGF-C and VEGFR-3 and their clinical significance in
gastric carcinoma. J Clin Exp Pathol. 29:1310–1316. 2013.
|
|
25
|
Huang KH, Lan YT, Chen MH, Chao Y, Lo SS,
Li AF, Wu CW, Chiou SH, Yang MH, Shyr YM, et al: The Correlation
Between RhoA Expression and Clinicopathological Characteristics in
Gastric Cancer Patients After Curative Surgery. World J Surg.
39:2289–2299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li XR, Ji F, Ouyang J, Wu W, Qian LY and
Yang KY: Overexpression of RhoA is associated with poor prognosis
in hepatocellular carcinoma. Eur J Surg Oncol. 32:1130–1134. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang M, Wang XJ and Liu BR: Effect of
shRNA targeted against RhoA on proliferation and migration of human
colonic cancer cells. Int J Clin Exp Pathol. 8:7040–7044.
2015.PubMed/NCBI
|
|
28
|
Kuzmanov A, Hopfer U, Marti P,
Meyer-Schaller N, Yilmaz M and Christofori G: LIM-homeobox gene 2
promotes tumor growth and metastasis by inducing autocrine and
paracrine PDGF-B signaling. Mol Oncol. 8:401–416. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kryza T, Achard C, Parent C, Marchand-Adam
S, Guillon-Munos A, Iochmann S, Korkmaz B, Respaud R, Courty Y and
Heuzé-Vourc'h N: Angiogenesis stimulated by human
kallikrein-related peptidase 12 acting via a platelet-derived
growth factor B-dependent paracrine pathway. FASEB J. 28:740–751.
2014. View Article : Google Scholar
|
|
30
|
Pietras K, Sjöblom T, Rubin K, Heldin CH
and Ostman A: PDGF receptors as cancer drug targets. Cancer Cell.
3:439–443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deng WY, Li N, Wan XB, Luo SX and Zhang
YW: Phosphorylated insulin-like growth factor-1 receptor expression
predicts poor prognosis of Chinese patients with gastric cancer.
Med Oncol. 31:1412014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Farabaugh SM, Boone DN and Lee AV: Role of
IGF1R in breast cancer subtypes, stemness, and lineage
differentiation. Front Endocrinol (Lausanne). 6:592015.
|
|
33
|
Furukawa J, Wraight CJ, Freier SM, Peralta
E, Atley LM, Monia BP, Gleave ME and Cox ME: Antisense
oligonucleotide targeting of insulin-like growth factor-1 receptor
(IGF-1R) in prostate cancer. Prostate. 70:206–218. 2010.
|
|
34
|
Hirano H, Lopes MB, Laws ER Jr, Asakura T,
Goto M, Carpenter JE, Karns LR and VandenBerg SR: Insulin-like
growth factor-1 content and pattern of expression correlates with
histopathologic grade in diffusely infiltrating astrocytomas. Neuro
Oncol. 1:109–119. 1999.
|
|
35
|
Ning XH, Wang YZ, Bai CM and Li J:
Clinical significance of insulin-like growth factor-1 receptor in
platinum-based chemotherapy for non-small cell lung cancer.
Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 32:366–370. 2010.In Chinese.
PubMed/NCBI
|
|
36
|
Zhao S, Qiu Z, He J, Li L and Li W:
Insulin-like growth factor receptor 1 (IGF1R) expression and
survival in non-small cell lung cancer patients: A meta-analysis.
Int J Clin Exp Pathol. 7:6694–6704. 2014.PubMed/NCBI
|
|
37
|
Pollak M: The insulin and insulin-like
growth factor receptor family in neoplasia: An update. Nat Rev
Cancer. 12:159–169. 2012.PubMed/NCBI
|
|
38
|
Patterson BC and Sang QA:
Angiostatin-converting enzyme activities of human matrilysin
(MMP-7) and gelatinase B/type IV collagenase (MMP-9). J Biol Chem.
272:28823–28825. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hoeth M, Niederleithner H, Hofer-Warbinek
R, Bilban M, Mayer H, Resch U, Lemberger C, Wagner O, Hofer E,
Petzelbauer P, et al: The transcription factor SOX18 regulates the
expression of matrix metalloproteinase 7 and guidance molecules in
human endothelial cells. PLoS One. 7:e309822012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cermenati S, Moleri S, Neyt C, Bresciani
E, Carra S, Grassini DR, Omini A, Goi M, Cotelli F, François M, et
al: Sox18 genetically interacts with VegfC to regulate
lymphangiogenesis in zebrafish. Arterioscler Thromb Vasc Biol.
33:1238–1247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Duong T, Proulx ST, Luciani P, Leroux JC,
Detmar M, Koopman P and Francois M: Genetic ablation of SOX18
function suppresses tumor lymphangiogenesis and metastasis of
melanoma in mice. Cancer Res. 72:3105–3114. 2012. View Article : Google Scholar : PubMed/NCBI
|