Introduction
Autophagy is a highly conserved metabolic pathway
that is required for intracellular degradation of long-lived
proteins or damaged organelles. Initially, the autophagic process
includes the formation of double-membraned autophagosomes, and
these subsequently fuse with lysosomes to degrade the contents
inside (1,2). In cancer cells, autophagy clears
damaged organelles and unfolded proteins, and also provides
cellular energy to enhance cell survival; while chronic or
excessive autophagy can contribute to cell death (3).
Currently, breast cancer is the most prevalent
cancer diagnosed in women, with an estimated 1.8 million cases
reported worldwide in 2013 (4).
Radiation is commonly adopted as an adjuvant therapy for the
management of breast cancer (5).
However, there is growing evidence that autophagy is induced by
ionizing radiation, and this induction plays a crucial role in
radiosensitivity (6,7). Furthermore, the regulatory effect of
autophagy in radiation-induced cell death remains controversial,
and the underlying molecular mechanisms remain to be fully
characterized.
Several autophagy-related genes (Atgs) have been
identified in relation to the autophagy machinery. For example,
Beclin 1 (the mammalian orthologue of yeast Atg6) and
Atg5 are required for the biogenesis of autophagosomes
(8). The autophagy pathway is also
associated with multiple intracellular signaling pathways. In
particular, damage-regulated autophagy modulator (DRAM), a p53
signaling effector, is a lysosomal protein that contributes to
p53-regulated autophagy induction (9). The findings of the present study
suggest that autophagy is induced by DRAM which leads to increased
expression of beclin 1 and production of p53 (10). In addition, p53 has been shown to
have a critical role in inducing DRAM-mediated autophagy in normal
hepatocytes (7702) and hepatocellular carcinoma HepG2 cells in
response to starvation, while p53 overexpression induces
DRAM-mediated autophagy (11).
Recent evidence also indicates that the p53/DRAM-autophagy axis
contributes to anticancer reagents that induce cytotoxicity in
breast cancer cells (12,13). However, the potential involvement of
the p53/DRAM signaling pathway in radiation-induced autophagic cell
death remains unknown.
In this study, activation of autophagy in MCF-7
breast cancer cells was investigated following ionizing radiation
treatment. Various assays were performed to characterize the
radiation-induced autophagy that was achieved. In particular, the
gene, DRAM, was identified in a gene microarray analysis,
thereby recognizing a potential role for this gene in
radiation-induced autophagic cell death.
Materials and methods
Reagents
The following reagents were purchased as indicated:
Dulbecco's modified Eagle's medium (DMEM) culture medium and fetal
bovine serum (FBS) (Life Technologies, Grand Island, NY, USA);
trypan blue solution (Sigma-Aldrich, St. Louis, MO, USA); protease
inhibitor cocktail (Roche Diagnostics, Basel, Switzerland); siRNAs
targeting Atg5 (sc-41445) and Beclin 1 (sc-29797) and
control siRNA-A (sc-37007) (Santa Cruz Biotechnology, Santa Cruz,
CA, USA); anti-Atg5 (#2630) and anti-Beclin 1 (#3738) primary
antibodies (Cell Signaling Technology, Beverly, MA, USA); mouse
monoclonal anti-p53 antibody (sc-126; Santa Cruz Biotechnology);
rabbit monoclonal anti-TIGAR (ab37910) and anti-DRAM (ab68987)
antibodies (Abcam, Cambridge, UK); rabbit anti-LC3 (no. 12741P)
antibody (Cell Signaling Technology); mouse anti-GAPDH (no. A3853)
and anti-actin (no. A5441) primary antibodies (Sigma-Aldrich);
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L,
no. 170-6515) and goat anti-mouse IgG (H+L, no. 170-6516)
antibodies (Bio-Rad Laboratories, Hercules, CA, USA); and primers
used for PCR were synthesized by Takara (Dalian, China).
Cell culture and radiation
MCF-7 breast cancer cells were maintained in DMEM
supplemented with 5% FBS, 100 U/ml of penicillin, and 100
µg/ml of streptomycin in a humidified 5% CO2
incubator at 37°C. The cells were exposed to 2, 4, 6, or 8 Gy
radiation at a rate of 0.40 Gy/min using an X-ray generator (X-RAD
320ix; Precision X-Ray Inc., North Branford, CT, USA). Untreated
cells were used as a control.
Monodansylcadaverine (MDC) staining
assay
MDC staining was used to determine the presence of
autophagic vacuoles. Briefly, MCF-7 cells were pre-seeded onto
glass cover slips and 24 h later the cells were irradiated. After
an additional 24 h, the irradiated cells were washed twice with
cold phosphate-buffered saline (PBS) and then were subsequently
incubated with 0.05 mM MDC solution in DMEM for 1 h at 37°C. After
three washes with PBS, the cells were fixed in 4% paraformaldehyde
for 15 min and then were examined with a confocal scanning
microscope (OLYMPUS-FA500; Olympus, Japan). Fluorescence intensity
was quantified by using a FACSCanto flow cytometer (BD Biosciences,
Franklin Lakes, NJ, USA).
CCK-8 assay for cell viability
Cells were seeded in 96-well plates
(5×103 cells/well) and were maintained in complete
culture medium for 24 h. Forty-eight hours after treatment, CCK-8
(Dojindo) solution was added to each well (10 µl). After 2
h, absorbance values for each plate were measured at 450 nm using a
microplate reader (Synergy HT; Bio-Tek, Winooski, VT, USA).
Plasmid transfection
A pcDNA3.1-RFP-LC3 (or RFP-LC3) plasmid expressing
the autophagy-related gene, lc3, was constructed by
inserting the lc3 cassette into the EcoRI and
BamHI sites of the pcDNA3.1-RFP vector (VPY0003; Changsha
YRBio, Hunan, China). Following sequencing confirmation of the
resulting ligated vector, MCF-7 cells were transfected with RFP-LC3
using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) as
described previously (14,15). For each transfection, 30 fields were
randomly selected and the number of cells expressing RFP-labeled
LC3 puncta were calculated in each field. Three independent
experiments were conducted for each group.
Transfection of small interfering RNA
(siRNA)
To knockdown expression levels of Atg5 and
Beclin 1, MCF-7 cells were transfected with siRNAs targeting
Atg5 and Beclin 1, as described in our previous study
(16). In addition, a scrambled
siRNA was used as a control. Briefly, one day prior to
transfection, MCF-7 cells were seeded into 100-mm tissue culture
plates. Forty-eight hours later when the cells reached 30–50%
confluency, the cells were transfected with each of the three types
of siRNAs (siAtg5, siBeclin1 and sicon, respectively) diluted 1:5
with Oligofectamine reagent (Invitrogen) and then diluted with 40
µl of serum-free DMEM. After 5–10 min at room temperature
(RT), another tube containing 10 µl of 20 µM siRNA
added into 440 µl of serum-free DMEM was added to each
diluted Oligofectamine mixture. After 15–20 min at RT, the
siRNA-Oligofectamine-reagent complex solution was added to 2.5 ml
of serum-free DMEM and this mixture was added to each dish of cells
that had been washed once with serum-free DMEM. The final
concentration of each siRNA in medium was 40 nM. As a control, 2.5
ml serum-free DMEM with only the sicon siRNA was added onto the
cells. After the transfected cells were incubated for 4 h at 37°C
in a 5% CO2 incubator, 2.5 ml serum-free DMEM and 400
µl serum (FBS) were added to each plate. On the fourth day
after transfection, 2×106 cells from each transfection
were divided equally into the wells of a 6-well plate. On the sixth
or seventh day following transfection, each set of cells was
harvested for analysis.
Western blot analysis
Total protein extracts were collected from MCF-7
cells in RIPA lysis buffer [HEPES (50 mM), NaCl (150 mM), EDTA (1
mM), EGTA (2.5 mM), NaF (10 mM), DTT (1 mM), sodium orthovanadate
(1 mM), PMSF (1 mM), NP-40 (1%), and SDS (0.1%)]. Proteins were
heated to 95°C for 5 min and 40 µg of the extracted proteins
was prepared in 2-ml aliquots and were mixed with 20 µl of
protease inhibitor cocktail. After a 5-min incubation on ice, the
samples were sonicated and centrifuged at 12,000 rpm for 10 min.
The resulting supernatants were transferred to new tubes, and each
were mixed with 5X SDS before being loaded onto a 10% SDS-PAGE gel.
Following transfer of the gels to nitrocellulose membranes, the
membranes were blocked with 5% non-fat dried milk in Tris-buffered
saline (TBS) containing 10 mM Tris (pH 7.5), 100 mM NaCl, and 0.1%
Tween-20 at RT. After 1.5 h, the membranes were incubated with the
appropriate primary antibodies overnight at 4°C. The dilutions used
for the primary antibodies were: anti-Beclin 1 (1:1,000), anti-LC3
(1:1,000), anti-GAPDH (1:1,000), anti-Atg5 (1:1,000), anti-actin
(1:1,000), anti-p53 (1:500), anti-TIGAR (1:1,000) and anti-DRAM
(1:300). After the membranes were washed, the membranes were
incubated with the appropriate HRP-conjugated secondary antibodies
at RT. After 1 h, bound antibodies were visualized with a
chemiluminescence detection system according to the manufacturer's
instructions (Pierce, Burlingame, CA, USA). Detection of GAPDH and
actin were used as loading controls.
Determination of cell death and
apoptosis
MCF-7 cells were plated onto 6-well plates and were
pre-incubated with or without the autophagy inhibitor,
3-methyladenine (3-MA), for 1 h followed by exposure to 4-Gy
radiation. At the indicated time-points after irradiation, the
cells were collected and washed with PBS three times.
To evaluate cell death, cells were stained with
trypan blue dye which is excluded from live cells yet penetrates
into dead cells and produces a red fluorescent signal that can be
quantified by flow cytometry (FACSCanto; BD Biosciences).
To detect cell apoptosis, collected cells were
stained with an Annexin V-FITC Apoptosis Detection kit I according
to the manufacturer's recommendation (BD Biosciences). After the
cells were counted by flow cytometry (FACSCanto; BD Biosciences),
they were analyzed with FCS Express v2.0 software (De Novo
Software, Thornhill, ON, Canada).
Short hairpin RNA (shRNA) constructs and
transfection
shRNAs were designed according to 'www.idtdna.com' and were synthesized, denatured,
annealed, and ligated to the pSUPER vector within the BglII
and HindIII sites. The shRNA sequences for targeting DRAM
were: sense,
5′-AGCTTGCCACATACGGATGGTCATTTCAAGAGAATGACCATCCGTATGTGGCTTTTTA-3′
and antisense,
5′-GATCTAAAAAGCCACATACGGATGGTCATTCTCTTGAAATGACCATCCGTATGTGGCA-3′.
The shRNA sequences for targeting p53 were: sense,
5′-GATCCCCGGAGGTTGTGAGGCACTGCTTCAAGAGAGCAGTGCCTCACAACCTCCTTTTTA-3′
and antisense,
5′-AGCTTAAAAAGGAGGTTGTGAGGCACTGCTCTCTTGAAGCAGTGCCTCACAACCTCCGGG-3′.
A pSUPER construct expressing a scrambled sequence with no
significant homology to any known mammalian mRNA was used as a
control. All of the plasmids were constructed in our laboratory.
The plasmids were transfected into 293T packaging cells by calcium
phosphate co-precipitation (Ampho Pack plasmid 10 µg,
pSUPER-shRNA plasmid 10 µg, 2 M CaCl2 31
µl, ddH2O to 250 µl, and 250 µl 2X
HEPES buffer salt solution) and supernatants containing pseudovirus
particles were collected after 72 h and applied to MCF-7 cells in
the presence of Polybrene (8 µg/ml). Stable cell clones were
selected in the presence of puromycin (0.8 µg/ml) for 7
days.
Gene microarray analysis
MCF-7 cells were exposed to 0, 4, and 8 Gy of
radiation for 4 h and then total RNA samples were collected using
an mRNA isolation kit (Ambion, Austin, TX, USA). All of the RNA
samples were labeled with Cy5/Cy3 and then were hybridized to a
Human Whole Genome OneArray (Phalanx Biotech Group, Taiwan). The
hybridized chips were scanned with an Axon 4000 scanner (Molecular
Devices, Sunnyvale, CA, USA) and spot quantification was performed
by using GenePix 4.1 software (Molecular Devices). Hierarchical
clustering was performed by Cluster 3.0 software (Molecular
Devices) for the expression profiles obtained. Differentially
expressed genes that exhibited greater than a 2-fold change were
further analyzed by Pathway-Express software (Onto-Tools, Wayne
State University, Detroit, MI, USA).
Quantitative real-time PCR (qPCR)
Total RNA was isolated using RNAiso Plus reagent
according to the manufacturer's instructions (Takara). The quality
and quantity of the RNA samples collected were analyzed by
measuring the A260/A280 ratio for each with an ultraviolet
spectrophotometer (Beckman Coulter, Miami, FL, USA). Each RNA
sample (2 µg) was subjected to reverse transcription using a
PrimeScript RT reagent kit (Takara). All of the primers were
designed with Primer5.0 (Premier, Canada) and their specificity was
verified by Blast NCBI. The forward and reverse primer sequences
for the target genes included: 5′-CAAGTGTGGGCTGCTGAGGA-3′ and
5′-AGCCTGGGTACAGGTTGTTGATG-3′ for DRAM;
5′-CAGTGATCTCATGAGGACAAAGCA-3′ and 5′-CCATGGCCCTCAGCTCACTTA-3 for
TIGER; and 5′-GCACCGTCAAGGCTGAGAAC-3′ and 5′-TGGTGAAGACGCCAGTGGA-3′
for GAPDH, respectively in each case. To perform qPCR (Stratagene
MX3000P; Agilent Technologies, Santa Clara, CA, USA), the SYBR
Premix Ex TaqII reagent (Takara) was used to amplify the resulting
cDNAs. The conditions for the qPCR cycles included: an activation
step at 95°C for 10 sec, 40 cycles of denaturation at 95°C for 20
sec, and an annealing step at 60°C for 20 sec. The formula,
2−ΔΔCT, where ΔCT is the value from the threshold cycle
(CT) of the treated sample subtracted from the CT value of the
untreated or zero time-point control sample was used to calculate
mRNA levels (17). The relative
amount of mRNA was normalized to the levels of GAPDH
mRNA.
Statistical analysis
All of the experiments described were performed in
triplicate. For the analysis of cell death, each experiment was
performed with 3–6 replicates. Data are presented as the mean ±
standard deviation (SD). Student's t-test was performed for
statistical analyses and a p-value <0.05 was considered
statistically significant.
Results
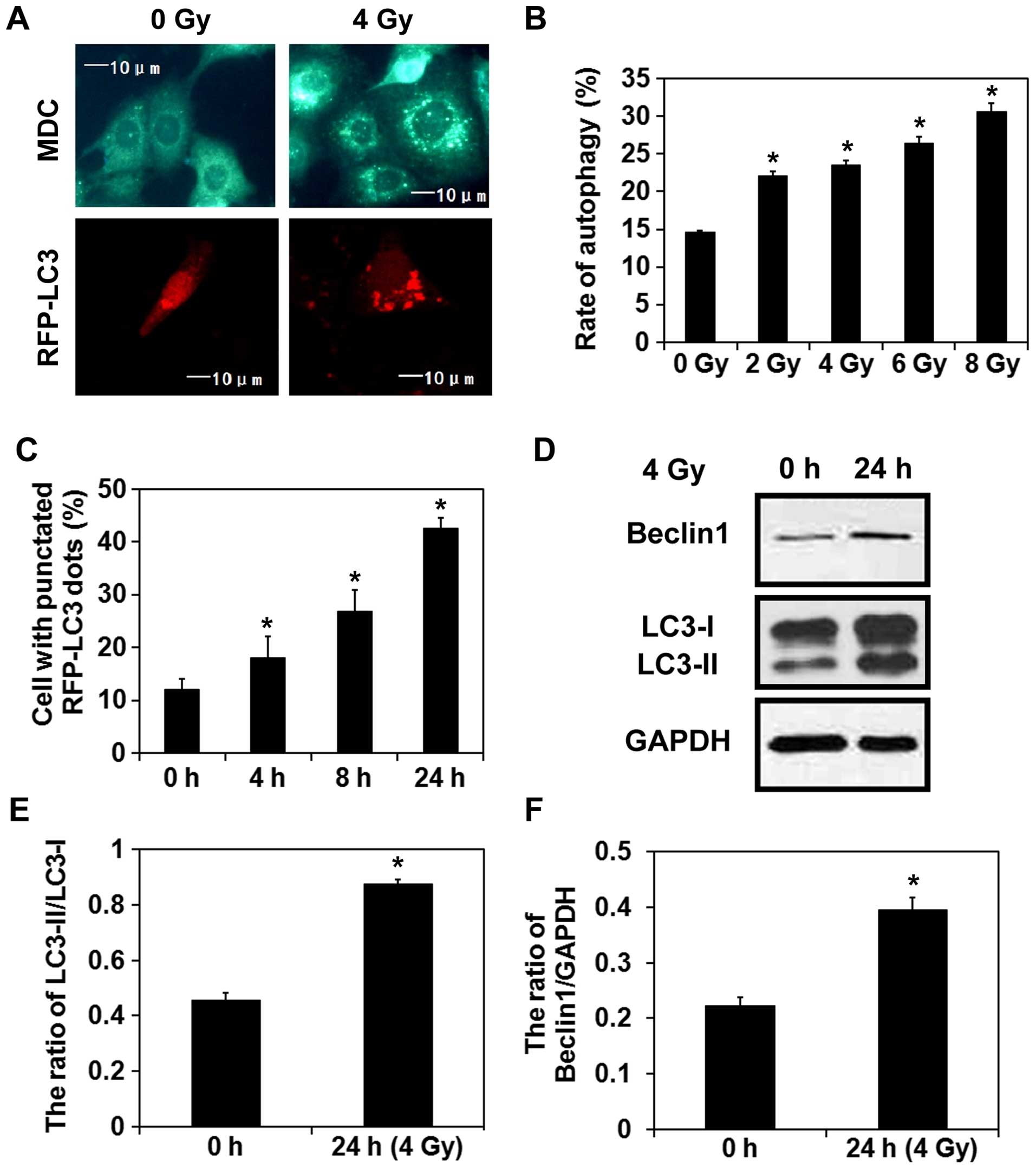
Radiation induces autophagy in breast
cancer cells
MDC is a lysosomotropic compound that is commonly
used for the detection of autophagic vacuoles in cells (18). Thus, staining with MDC was initially
performed to investigate the induction of autophagy in MCF-7 breast
cancer cells exposed to 4 Gy of radiation. Twenty-four hours later,
a higher number of MDC-labeled vacuoles was observed in the
irradiated cells than that detected in the control cells (Fig. 1A). Moreover, a dose-dependent
increase in the number of positively-stained cells was observed
within 24 h after irradiation. RFP-LC3 is a fluorescent protein
which labels both autophagosomes and autolysosomes (19), and a higher number of LC3-labeled
puncta were also observed in the irradiated cells compared with the
control cells (Fig. 1A). In
addition, the percentage of cells expressing LC3-positive puncta
gradually increased with time after irradiation (Fig. 1C). In accordance with these results,
western blot analysis demonstrated that expression of the
autophagy-related proteins, Beclin 1 and LC3-II, were upregulated
24 h after the exposure of the MCF-7 cells to 4-Gy radiation
(Fig. 1D–F). Taken together, these
results suggest that radiation induces autophagy in MCF-7
cells.
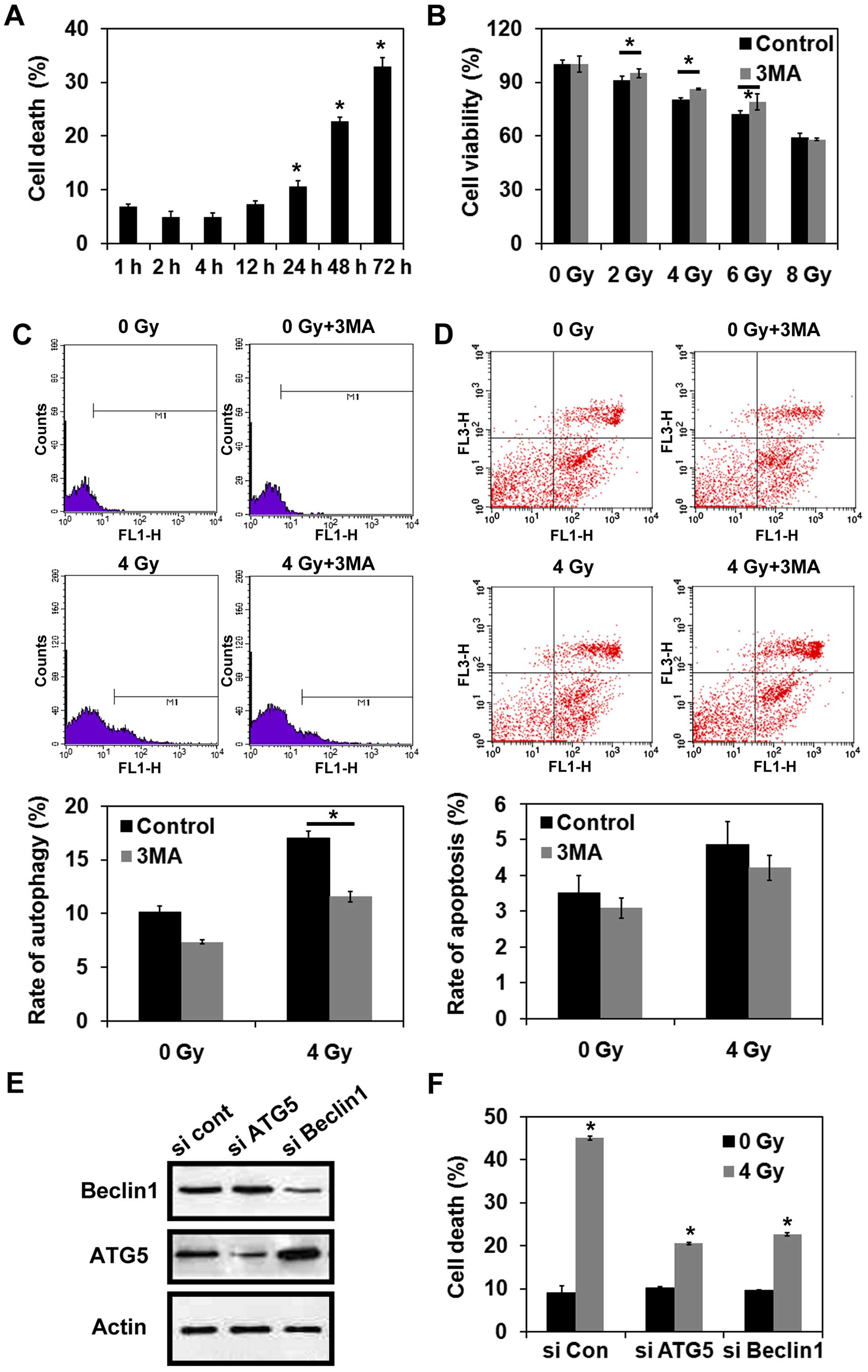
Radiation induces autophagic cell death
in MCF-7 cells
Trypan blue dye exclusion assays were performed to
detect cell death following the exposure of MCF-7 cells to 4-Gy
radiation. As shown in Fig. 2A, the
percentage of cells undergoing cell death increased in a
time-dependent manner following irradiation. Next, the effect of
inhibiting autophagy prior to radiation on cell viability was
examined. For this purpose, MCF-7 cells were treated with or
without 3-MA, and cell viability was examined in CCK-8 assays
following the exposure of MCF-7 cells to various doses of
radiation. As shown in Fig. 2B,
treatment with 3-MA reversed the reduction in cell viability that
was observed for the cells that were exposed to 2, 4, or 6 Gy
radiation alone (P<0.05). These results suggest that autophagy
may be the predominant pathway that mediates cell death following
irradiation. However, inhibition of autophagy did not prevent cell
death following the exposure of MCF-7 cells to 8-Gy radiation
(P>0.05) (Fig. 2B). MDC staining
further indicated that treatment with 3-MA significantly reduced
the induction of autophagy in MCF-7 cells exposed to 4-Gy radiation
(P<0.05) (Fig. 2C). However,
there was no significant difference in the levels of apoptosis
detected for the cells that were irradiated with or without 3-MA
pretreatment (P>0.05) (Fig. 2D).
When the autophagy-related genes, Atg5 and Beclin 1,
were each knocked down in MCF-7 cells, both sets of cells were
protected against cell death following exposure to 4-Gy radiation
(Fig. 2E and F). In combination,
these data indicate that radiation can lead to autophagic cell
death in MCF-7 cells.
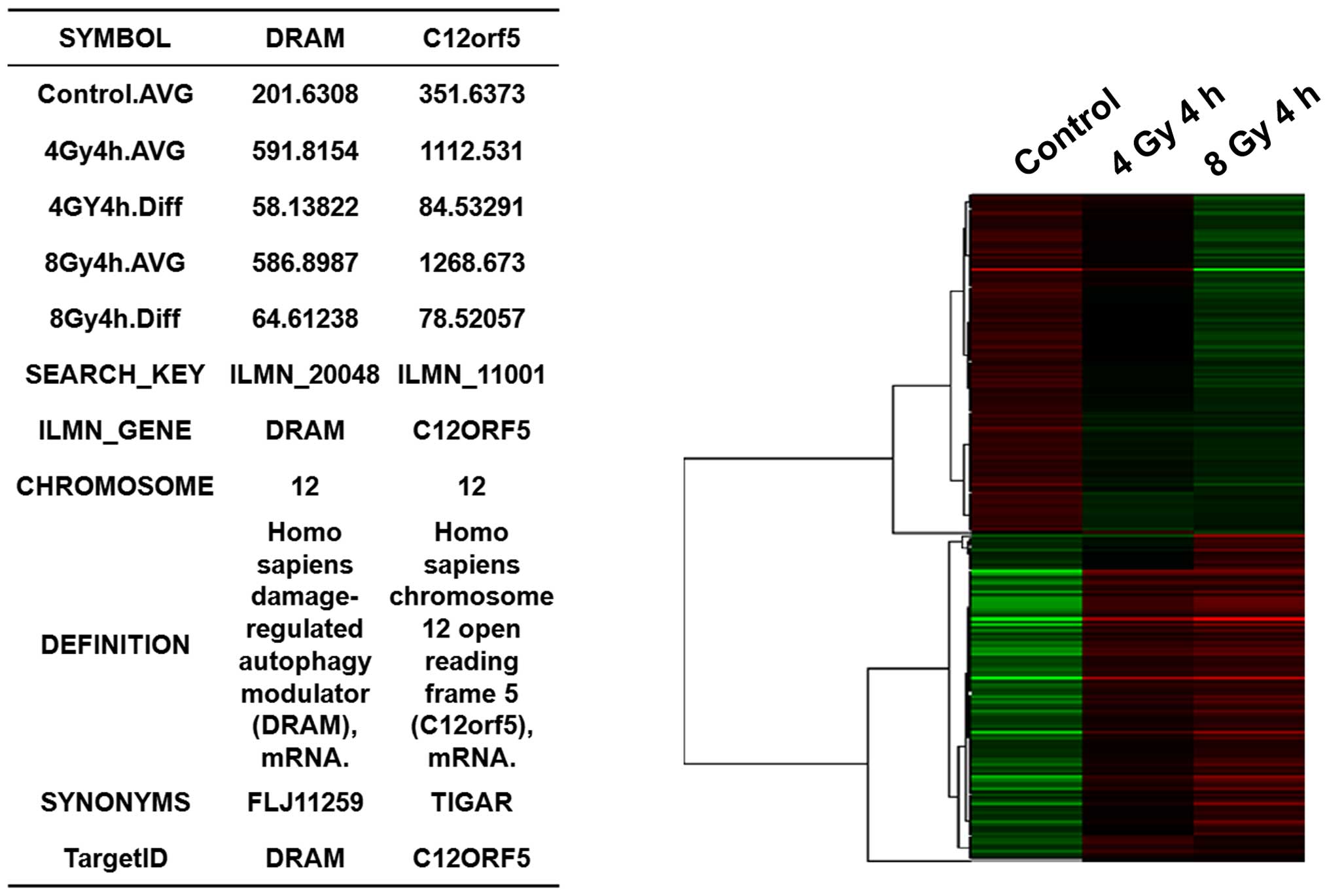
Gene microarray analysis of irradiated
MCF-7 cells
To identify regulatory genes that participate in
radiation-induced autophagic cell death, MCF-7 cells were exposed
to 0, 4, or 8 Gy radiation and then were subjected to a gene
microarray analysis 4 h later. Compared with the non-irradiated
control cells, there were 160 differentially expressed genes that
were detected after the exposure of the cells to 4 Gy radiation,
including 93 genes that were upregulated (indicated in red in the
row of 4 Gy 4 h) and 67 genes that were downregulated (indicated in
green in the row of 4 Gy 4 h) (Fig.
3). Overall, these differentially expressed genes were
associated with 10 signaling pathways. For the MCF-7 cells that
were exposed to 8 Gy of radiation, a total of 236 differentially
expressed genes were detected [including 121 upregulated genes
(indicated in red in the row of 8 Gy 4 h) and 115 downregulated
genes (indicated in green in the row of 8 Gy 4 h)], and these were
associated with 9 signaling pathways (Fig. 3). Furthermore, significant
alterations in the expression of the autophagy-related genes,
DRAM and TIGAR (also known as C12orf5), were detected
following the exposure of the cells to 8-Gy radiation. Therefore,
it appears that DRAM and TIGAR contribute to
radiation-induced autophagic cell death in MCF-7 cells.
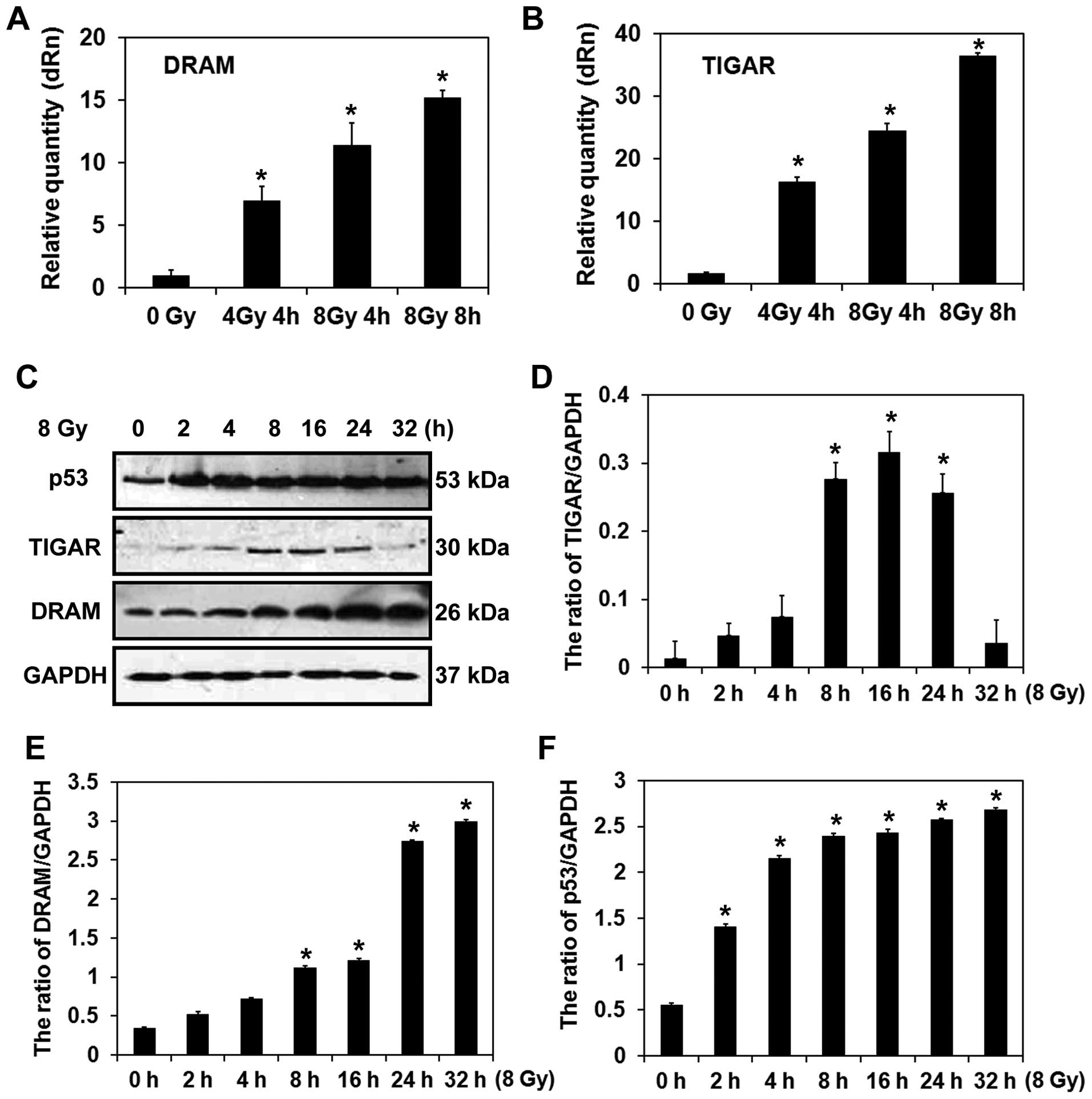
Characterization of DRAM and TIGAR in
radiation-induced autophagic cell death in MCF-7 cells
In the qPCR analysis, mRNA levels of DRAM and TIGAR
were found to be significantly elevated following the exposure of
MCF-7 cells to radiation (Fig. 4A and
B). As shown by western blot results, the levels of p53
gradually increased, as did the levels of DRAM, with time following
radiation (Fig. 4C and E). However,
while the expression of TIGAR gradually increased and peaked 16 h
after the irradiation treatment, the levels decreased thereafter
(Fig. 4C and D). Previously, DRAM
was identified as a crucial effector of p53-activated autophagy.
However, more recently, the lysosomal protein, DRAM has also been
found to be required for p53-induced autophagy and cell death
(20). Consistent with the DRAM
data, expression levels of p53 were higher after the MCF-7 cells
were exposed to radiation compared with the control cells (Fig. 4C and F). These data suggest that
regulation of DRAM by p53 may mediate autophagy in MCF-7 cells
following exposure to radiation.
p53/DRAM mediates radiation-induced
autophagic cell death
To confirm the potential regulatory role of the
p53/DRAM signaling pathway in radiation-induced autophagic cell
death, a stable p53-deficient MCF-7 cell line and
DRAM-deficient MCF-7 cell line were established following
the infection of MCF-7 cells with a pSUPER-DRAM shRNA and a
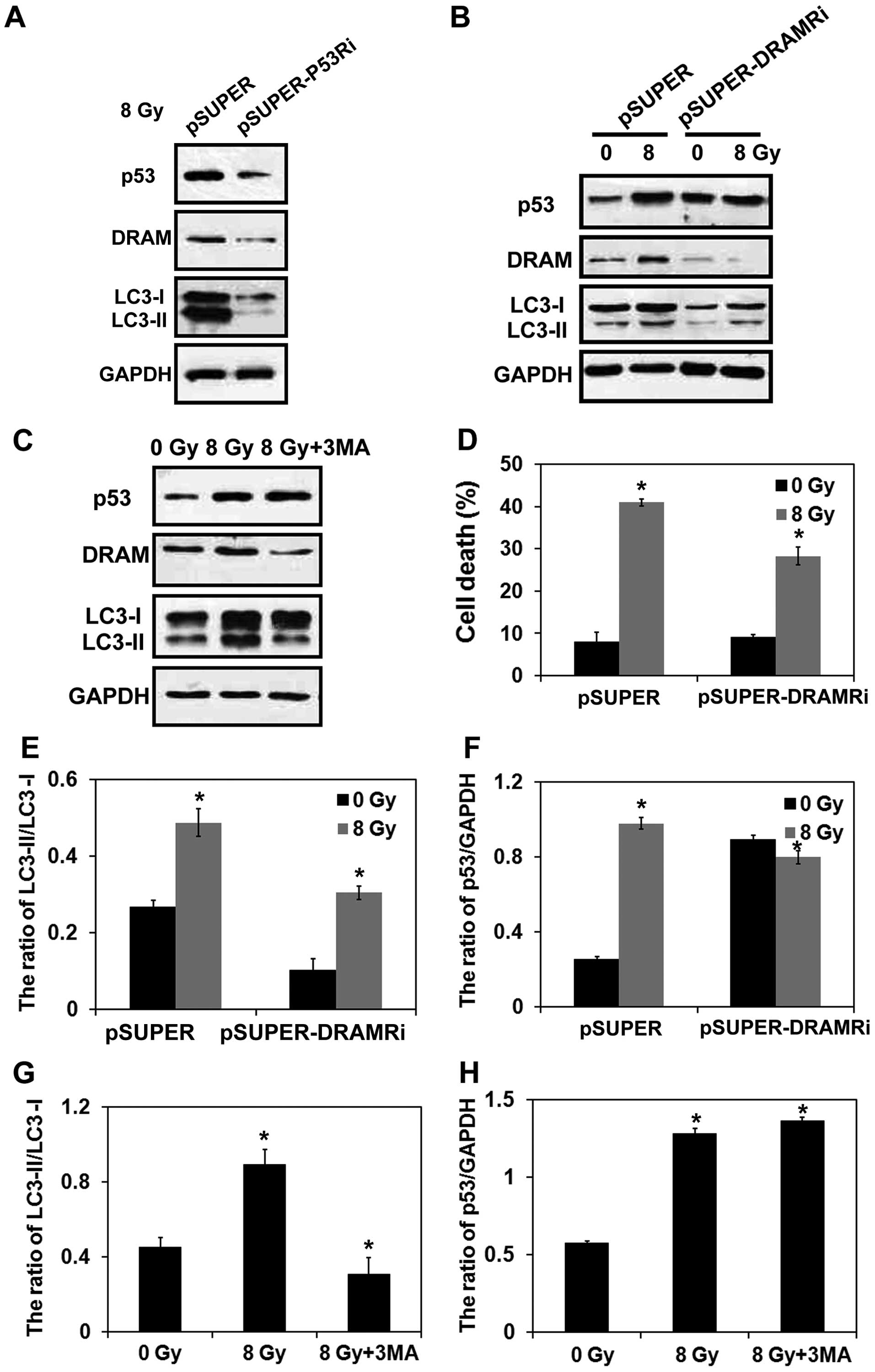
pSUPER-p53 shRNA. Western blot analysis confirmed that silencing of
p53 and DRAM were achieved (Fig.
5A and B). This in vitro model was then exposed to 8-Gy
radiation. We found that radiation induced downregulation of DRAM
and LC3-II in the p53-deficient MCF-7 cells (Fig. 5A). This in vitro model was
then exposed to 8-Gy radiation, and radiation-induced upregulation
of p53, DRAM, and LC3-II were reversed compared with wild-type
MCF-7 cells that were exposed to the same radiation in a
DRAM-deficient MCF-7 cell line (Fig. 5B). These results are in agreement
with the results of the statistical analysis in the graph shown in
Fig. 5E and F). Previously, it was
reported that siRNA-mediated knockdown of p53 was able to
completely block DRAM expression and significantly reduce the level
of autophagy (11). However, in the
present study, silencing of DRAM only partially blocked autophagy,
which might be due to the upregulated p53 level after irradiation.
It is also possible as p53 may induce autophagy by mediating other
autophagy related genes. As shown in Fig. 5C, inhibition of autophagy by 3-MA
further reduced the radiation-induced upregulation of LC3-II as
well as DRAM, while no significant difference was detected in the
expression of p53 in the wild-type MCF-7 cells that were irradiated
(Fig. 5G and H). In trypan blue
exclusion assays, DRAM-deficient MCF-7 cells also exhibited a
marked reduction in cell death rate after irradiation (Fig. 5D). Thus, the present findings
suggest that the p53/DRAM signaling pathway mediates
radiation-induced autophagic cell death in MCF-7 cells.
Discussion
Ionizing radiation is an efficient adjuvant therapy
for the management of breast cancer after surgery as it reduces
local recurrence and has been shown to prolong the long-term
survival of patients (21).
However, breast cancer cells can develop radioresistance, and the
ability to promote radiosensitivity could potentially improve the
therapeutic outcome in these cases (22). In recent studies, autophagy has been
shown to play a key role in the radiosensitivity and
radioresistance of various cancer therapies (6). However, the precise role of autophagy
in radiation-induced cytotoxicity of breast cancer cells has not
been fully characterized. In the present study, radiation triggered
autophagic cell death in MCF-7 breast cancer cells, and we provide
evidence to indicate this process is modulated by the p53/DRAM
signaling pathway.
Increased autophagic activity in irradiated MCF-7
cells was demonstrated with the presence of enhanced MDC staining,
an accumulation of LC3-positive puncta, and upregulated expression
of autophagy-related genes. Moreover, these observations were
accompanied by enhanced cytotoxicity, suggesting the involvement of
autophagy in radiation-induced cell injury. Similarly, enhanced
autophagy has been observed in other studies of breast cancer cells
that were subjected to ionizing radiation (14,23).
However, it should be noted that autophagy can act as a
double-edged sword in modulating radiation-triggered cancer cell
death (24). For example, Han et
al reported that blockage of autophagy overcame the
radioresistance of breast cancer cells (25), and Bristol et al reported
that an inhibition of autophagic activity in MCF-7 cells aggravated
cytotoxicity and a cytoprotective role was indicated for autophagy
induced by radiation (26). Thus,
it appears that autophagy may favor cancer cell survival and
radioadaption by contributing to the maintenance of intracellular
homeostasis in cells (27).
However, radiation has also been shown to induce autophagic cell
death in MCF-7 cells (28). In
accordance with this finding, the results of the present study
demonstrate that autophagy can act as a pro-death mechanism.
Specifically, it was demonstrated in the present study that
inhibition of the autophagic pathway by pharmacological
interference or with the knockdown of autophagy-related genes could
prevent cell death-induced radiation. Therefore, it is hypothesized
that augmentation of autophagy may sensitize breast cancer cells to
radiotoxicity.
In the gene microarray analysis that was performed,
marked changes in expression of the autophagy-related genes,
DRAM and TIGAR, were observed. DRAM plays a crucial
role in mediating a cell's response to DNA damage (9), and is also a downstream target of p53.
Furthermore, activation of the p53/DRAM signaling pathway has been
found to mediate the induction of autophagy in breast cancer cells
by anticancer reagents such as doxorubicin (29) and acetazolamide (13). Consistent with these findings,
overexpression of DRAM and p53 were detected
following the irradiation of MCF-7 breast cancer cells in the
present study. Conversely, silencing of p53 decreased
DRAM and LC3-II. Silencing of DRAM reversed
radiation-induced of the autophagy-related gene, LC3-II and
increased the expression levels of p53. The latter observation may
involve a compensatory mechanism by which radiation-induced
upregulation of p53 is reversed, and thus, a positive feedback loop
may exist between p53 and DRAM. It has been reported that
DRAM-induced autophagy was also accompanied by an increased number
of autophagic vacuoles and upregulated expression of LC3 (10). Knockdown of DRAM attenuates
the ability of wild-type p53 to induce autophagy (30). Furthermore, knockdown of DRAM
in MDA-MB-231 cells, which represent a p53-mutated breast cancer
cell line, did not induce DRAM-1 expression or autophagy (29). It was also observed in the present
study that DRAM silencing partially prevented radiation-induced
cell death in MCF-7 cells, and this result indicates a potential
role for the p53/DRAM-autophagy axis in radiation-induced
cytotoxicity of breast cancer cells.
In conclusion, the results of the present study
demonstrate that ionizing radiation induces autophagic cell death
of MCF-7 cells via the p53/DRAM signaling pathway, and they also
identify potential targets for improving radiosensitivity in the
treatment of breast cancer.
References
|
1
|
Galluzzi L, Vicencio JM, Kepp O, Tasdemir
E, Maiuri MC and Kroemer G: To die or not to die: That is the
autophagic question. Curr Mol Med. 8:78–91. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xie Z and Klionsky DJ: Autophagosome
formation: Core machinery and adaptations. Nat Cell Biol.
9:1102–1109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dalby KN, Tekedereli I, Lopez-Berestein G
and Ozpolat B: Targeting the prodeath and prosurvival functions of
autophagy as novel therapeutic strategies in cancer. Autophagy.
6:322–329. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fitzmaurice C, Dicker D, Pain A, Hamavid
H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R,
Wolfe C, et al Global Burden of Disease Cancer Collaboration: The
Global Burden of Cancer 2013. JAMA Oncol. 1:505–527. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsoutsou PG, Koukourakis MI, Azria D and
Belkacémi Y: Optimal timing for adjuvant radiation therapy in
breast cancer: A comprehensive review and perspectives. Crit Rev
Oncol Hematol. 71:102–116. 2009. View Article : Google Scholar
|
|
6
|
Yang Y, Yang Y, Yang X, Zhu H, Guo Q, Chen
X, Zhang H, Cheng H and Sun X: Autophagy and its function in
radiosensitivity. Tumour Biol. 36:4079–4087. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gewirtz DA: The four faces of autophagy:
Implications for cancer therapy. Cancer Res. 74:647–651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Juenemann K and Reits EA: Alternative
macroautophagic pathways. Int J Cell Biol. 2012:1897942012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crighton D, Wilkinson S, O'Prey J, Syed N,
Smith P, Harrison PR, Gasco M, Garrone O, Crook T and Ryan KM:
DRAM, a p53-induced modulator of autophagy, is critical for
apoptosis. Cell. 126:121–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu BS, Zhao K, Jia X, Wu YY and Xing CG:
Effects of damage-regulated autophagy regulator gene on the SGC7901
human gastric cancer cell line. Oncol Lett. 8:657–662.
2014.PubMed/NCBI
|
|
11
|
Liu K, Shi Y, Guo XH, Ouyang YB, Wang SS,
Liu DJ, Wang AN, Li N and Chen DX: Phosphorylated AKT inhibits the
apoptosis induced by DRAM-mediated mitophagy in hepatocellular
carcinoma by preventing the translocation of DRAM to mitochondria.
Cell Death Dis. 5:e10782014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim TH and Kim HS, Kang YJ, Yoon S, Lee J,
Choi WS, Jung JH and Kim HS: Psammaplin A induces Sirtuin
1-dependent autophagic cell death in doxorubicin-resistant
MCF-7/adr human breast cancer cells and xenografts. Biochim Biophys
Acta. 1850:401–410. 2015. View Article : Google Scholar
|
|
13
|
Mohammadpour R, Safarian S, Ejeian F,
Sheikholya-Lavasani Z, Abdolmohammadi MH and Sheinabi N:
Acetazolamide triggers death inducing autophagy in T-47D breast
cancer cells. Cell Biol Int. 38:228–238. 2014. View Article : Google Scholar
|
|
14
|
Yi H, Liang B, Jia J, Liang N, Xu H, Ju G,
Ma S and Liu X: Differential roles of miR-199a-5p in
radiation-induced autophagy in breast cancer cells. FEBS Lett.
587:436–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang N, Jia L, Liu Y, Liang B, Kong D,
Yan M, Ma S and Liu X: ATM pathway is essential for ionizing
radiation-induced autophagy. Cell Signal. 25:2530–2539. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang C, Tang X, Guo X, Niikura Y, Kitagawa
K, Cui K, Wong ST, Fu L and Xu B: Aurora-B mediated ATM serine 1403
phosphorylation is required for mitotic ATM activation and the
spindle checkpoint. Mol Cell. 44:597–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Vázquez CL and Colombo MI: Assays to
assess autophagy induction and fusion of autophagic vacuoles with a
degradative compartment, using monodansylcadaverine (MDC) and
DQ-BSA. Methods Enzymol. 452:85–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kimura S, Noda T and Yoshimori T:
Dissection of the autophagosome maturation process by a novel
reporter protein, tandem fluorescent-tagged LC3. Autophagy.
3:452–460. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crighton D, Wilkinson S and Ryan KM: DRAM
links autophagy to p53 and programmed cell death. Autophagy.
3:72–74. 2007. View Article : Google Scholar
|
|
21
|
Lee LJ and Harris JR: Innovations in
radiation therapy (RT) for breast cancer. Breast. 18(Suppl 3):
S103–S111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kaidar-Person O, Lai C, Kuten A and
Belkacemi Y; AROME: 'The Infinite Maze' of breast cancer, signaling
pathways and radioresistance. Breast. 22:411–418. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chaachouay H, Ohneseit P, Toulany M,
Kehlbach R, Multhoff G and Rodemann HP: Autophagy contributes to
resistance of tumor cells to ionizing radiation. Radiother Oncol.
99:287–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hönscheid P, Datta K and Muders MH:
Autophagy: Detection, regulation and its role in cancer and therapy
response. Int J Radiat Biol. 90:628–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han MW, Lee JC, Choi JY, Kim GC, Chang HW,
Nam HY, Kim SW and Kim SY: Autophagy inhibition can overcome
radio-resistance in breast cancer cells through suppression of TAK1
activation. Anticancer Res. 34:1449–1455. 2014.PubMed/NCBI
|
|
26
|
Bristol ML, Di X, Beckman MJ, Wilson EN,
Henderson SC, Maiti A, Fan Z and Gewirtz DA: Dual functions of
autophagy in the response of breast tumor cells to radiation:
Cytoprotective autophagy with radiation alone and cytotoxic
autophagy in radio-sensitization by vitamin D 3. Autophagy.
8:739–753. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szumiel I: Radiation hormesis: Autophagy
and other cellular mechanisms. Int J Radiat Biol. 88:619–628. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhong R, Xu H, Chen G, Zhao G, Gao Y, Liu
X, Ma S and Dong L: The role of hypoxia-inducible factor-1α in
radiation-induced autophagic cell death in breast cancer cells.
Tumour Biol. 36:7077–7083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gomes LR, Vessoni AT and Menck CF:
Three-dimensional microenvironment confers enhanced sensitivity to
doxorubicin by reducing p53-dependent induction of autophagy.
Oncogene. 34:5329–5340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takahashi M, Kakudo Y, Takahashi S,
Sakamoto Y, Kato S and Ishioka C: Overexpression of DRAM enhances
p53-dependent apoptosis. Cancer Med. 2:1–10. 2013. View Article : Google Scholar : PubMed/NCBI
|