Introduction
Breast cancer (BC) is the most commonly diagnosed
female cancer (1). BC ranks first
in Saudi females as well, accounting for 26% of all newly diagnosed
female cancers as reported by the National Cancer Registry, Saudi
Arabia (2). In the US and Western
Europe, the median age at presentation is ~63 years in comparison
to 48 years in Saudi Arabia (2). BC
is clinically a heterogeneous disease depending on genetic
variation between tumor and healthy tissue (3). Studies characterizing transcriptomics
and molecular genetics of cancers including BC have been conducted
on European or US patients. However, variability in the molecular
signature of cancers from different ethnic groups has been
reported, and clear differences in the patterns of p53 mutations in
BC have been found among different ethnic groups (4,5).
In the recent decade, a high throughput
transcriptomic genome-wide approach has had a major impact on BC
research (6). Studies have been
conducted to classify clinically distinct subclasses of tumors and
treatment prediction (6–8). To date, gene expression profiling
studies have been reported mainly from the Caucasian population,
while less data are available for non-Caucasian and Arabian
populations (9,10). Saudi society is different from
Western countries and common BC risk factors such as nulliparity,
low parity, first time pregnancy at late age, and no history of
breast feeding, are usually not applicable in the present study,
yet, the high incidence of BC is alarming among Saudi women
(11,12).
LEP, formerly depicted as a circulating
hormone, is predominantly produced by adipocytes along with other
adipocytokines such as adiponectin, tumor necrosis factor-α
(TNF-α), vascular endothelial growth factor (VEGF)
and interleukin-6 (IL-6). LEP activates the leptin receptor
in different tissues and basically acts as a mitogen, survival
factor and regulator of metabolism, feeding behavior and energy
homeostasis. Research has demonstrated that the function of LEP
goes far beyond metabolism and it has been postulated to be a key
mediator in obesity-associated cancers such as that of breast,
colorectal and prostrate cancer (13). Researchers have demonstrated the
overexpression of LEP and its receptors in BC using
immunohistochemistry or RT-PCR, and LEP has been proposed as a
prospective molecular target for cancer prediction, prevention and
therapeutics (14,15). In the present study, we examined the
expression status of LEP in Arabian female BC patients and the
possible LEP-mediated mechanism involved in BC cell
proliferation.
Materials and methods
Patients and samples
A transcriptomic study was carried out on 45 female
Saudi BC patients undergoing treatment at King Abdulaziz University
Hospital (Jeddah, Saudi Arabia) during the years 2008–2010. All
collected tissue specimens were immediately stored in RNAlater
(Invitrogen, Life Technologies, Grand Island, NY, USA), and
clinicopathological features, including, age, tumor grade and size,
hormone receptor status, lymph node involvement and pathology
reports were retrieved from the patient records. The average age of
the patients in the present cohort was 48 years, with a median of
47 years (range, 27–80 years). To estimate the effect of obesity on
BC, we also determined the mean body mass index (BMI). The present
study was approved by the Ethics Committee (no. 08-CEGMR-02-ETH) of
King Abbdulaziz University, and informed consent was provided by
all the patients included in the present study.
RNA extraction and array processing
Total RNA was extracted from fresh breast tissue
specimens according to the manufacturer's recommendations provided
with the Qiagen RNeasy Mini kit (Qiagen, Hilden, Germany). RNA
quality was verified on an Agilent 2100 Bioanalyzer (Agilent
Technologies, Palo Alto, CA, USA) with a cut-off of RNA integrity
number (RIN) >5. RNA concentrations were determined using a
NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies,
Wilmington, DE, USA) and 300 ng of each RNA sample was processed
according to the manufacturer's recommendations for Human Gene 1.0
ST GeneChip arrays (Affymetrix, Santa Clara, CA, USA).
Gene expression analysis
Affymetrix CEL files were imported to Partek
Genomics Suite (PGS version 6.6; Partek Inc., St. Louis, MO, USA)
and the robust multi-array average (RMA) was used for data
normalization. We performed principal component analysis to
visualize high dimensional data and to assess overall variance in
gene expression. Analysis of variance (ANOVA) was carried out on
the complete dataset and a differentially expressed transcript list
of genes was then conducted using an false discovery rate (FDR)
(Benjamini-Hochberg) of 0.05 with a 2-fold change cut-off. The
complete dataset and associated clinicopathological information
were submitted to the NCBI Gene Expression Omnibus (GEO) which are
accessible through accession no. GSE36295.
Real-time quantitative PCR assay
LEP expression levels were validated using the
TaqMan® quantitative real-time PCR (qPCR) assay (ID
Hs01084494_m1; life Technologies, Carlsbad, CA, USA). We used 0.3
μg of template RNA and 50 ng/μl of random hexamer in
a 5 μl mixture that was denatured at 70°C for 5 min, and ice
cooled for at least 1 min before use for cDNA conversion.
MgCl2 (1.5 mM), dNTP mix (0.5 mM), RNasin (1 U), reverse
transcriptase (2.5 U) and RT buffer (1X) were used for cDNA
conversion in the following steps: initial incubation of 10 min at
25°C, 1 h incubation at 37°C, 5 min heat inactivation at 95°C and
storage at 4°C to obtain a cDNA concentration of ~650 ng/μl.
PCR was performed using cDNA (100 ng), primer (1X) and Master Mix
(1X) for 1 cycle (2 min at 50°C), 1 cycle of denaturation (10 min
at 94°C), 40 cycles of denaturation, annealing and extension (30
sec at 97°C; 45 sec at 59°C and 1 min at 72°C), and final extension
of 10 min at 72°C. gene expression analyses were performed using
the ΔCt value model (average Ctgene − average
CtGAPDH). Statistical significances between BCs vs.
normal control were evaluated using the Student's unpaired t-test.
A p-value of <0.01 was considered to indicate a statistically
significant result.
Identification of functionally
significant proteins interacting with LEP
We used Search Tool for the Retrieval of Interacting
genes/Proteins (STRING version 9.1) to identify significant
proteins interacting with LEP. It is a biological database and web
resource of known as well as predicted protein interactions derived
from high-throughput experimental sources, text mining and
co-expression (16,17).
Functional and pathway analysis
The identified differentially expressed transcripts
with corresponding probesets ID, Entrez gene ID as clone
identifier, gene symbol, p-value and fold-change values were
uploaded into the Ingenuity Pathways Analysis (IPA) software
(Ingenuity Systems, Redwood City, CA, USA). We used IPA to identify
biological associations, and to perform interaction and functional
analysis among the differentially regulated BC genes. The impact of
the link between the expression data and canonical pathways were
calculated by ratio and/or Fisher's exact test. To develop an
assumed network of differentially expressed genes downstream of
LEP, the network was grown downstream of LEP using the
differentially regulated genes.
Results
The prime aim of the present study was to establish
the transcriptomic profiles of BC from a Saudi female patient
population. We profiled and compared 45 fresh BC tissue specimens
with 8 normal breast tissues. The PCA results showed a clear
difference between the BC and normal breast tissues. We also
calculated the BMI of all enrolled patients and its average was
found to be ~32.5 kg/m2.
Identification of differentially
expressed genes
Global genome-wide expression profiling of BC
identified 1,159 differentially expressed genes: 544 upregulated
and 615 downregulated (p<0.05; 2-fold change). Functional
analysis of the BC-associated genes found an overexpression of
genes associated with cell cycle progression, cell death, DNA
repair, tumor morphology and tissue developments. Specifically,
genes that are known to be linked with BC, included H3 histone
family, member A, histone 1, H3a (HIST1H3A), chemokine
(C-X-C motif) ligand 10 (CXCL10), topoisomerase 2α
(TOP2A), carcinoembryonic antigen-related cell adhesion molecule 1
(CEACAM1), cell division cycle 6 homolog (S.
cerevisiae) (CDC6), ADAM-like decysin 1
(ADAMDEC1), actin-binding protein anillin (ANLN),
centromere protein F (CENPF), mucin 1, cell
surface-associated (MUC1), protease serine 8 (PRSS8),
kinesin family member 23 (KIF23), interferon, γ-inducible
protein 6 (IFI6), chemokine (C-X-C motif) ligand 9
(CXCL9), ubiquitin-conjugating enzyme E2T (UBE2T) and
matrix metallopeptidase −9, −11 and −13 (MMP-9, −11
and −13). Genes including LEP linked to lipid
metabolism, and endocrine system development were significantly
downregulated in the BC tissues.
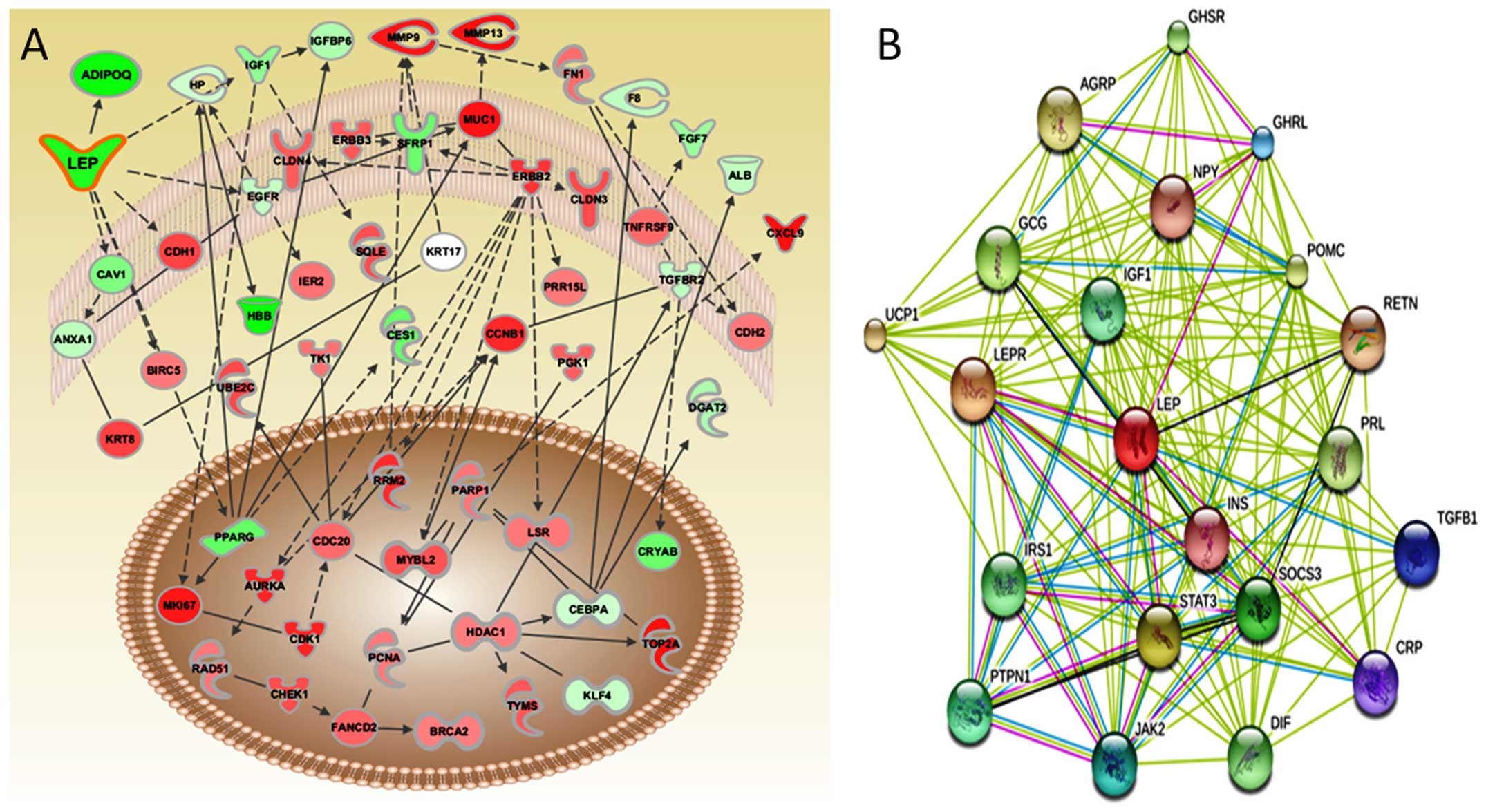
Pathways and networks underlying BC
We examined molecular networks using IPA to
comprehend the mechanisms by which the genes control a wide range
of physiological processes including cancer. Transcriptomic
signatures showed noteworthy disruption in the following signaling
pathways: glycerolipid metabolism, ATM signaling, DNA damage and
cell cycle, and ILK signaling involved in tumor initiation or
progression. We were the first to report the role of the
glycerolipid metabolism pathway in BC (18). The majority of the genes involved in
glycerolipid metabolism were downregulated. These results further
support the putative role of these pathways in rendering
susceptibility to BC. Differentially expressed genes were imported
into IPA to graphically represent all known relationships and
potential interactions among them. IPA and gene ontology based
network analysis suggested that LEP is significantly linked
with the other differentially expressed genes, leading to BC
progression (Fig. 1A).
Protein interaction study
Results of STRING showed a direct interaction and
predicted a functional association between LEP and its interacting
proteins. The following proteins showed prominent interactions with
LEP: leptin receptor (LEPR); suppressor of cytokine signaling 3
(SOCS3); insulin-like growth factor 1 (IGF1), prolactin (PRL);
ghrelin/obestatin prepropeptide (GHRL); insulin (INS); signal
transducer and activator of transcription 3 (STAT3); protein
tyrosine phosphatase, non-receptor type 1 (PTPN1); insulin receptor
substrate 1 (IRS1); growth hormone secretagogue receptor (GHSR);
resistin (RETN); glucagon (GCG); agouti-related protein homolog
(mouse) (AGRP); proopiomelanocortin (POMC); C-reactive protein,
pentraxin-related (CRP); neuropeptide Y (NPY); transforming growth
factor, β1 (TGFB1); uncoupling protein 1 (mitochondrial, proton
carrier) (UCP1); tumor necrosis factor precursor (TNF-α) (DIF); and
Janus kinase 2 (JAK2). Based on the predicted result of STRING for
LEP partners, we only used those likely candidates of protein
interactions for a limited but focused hierarchical clustering
(Fig. 1B).
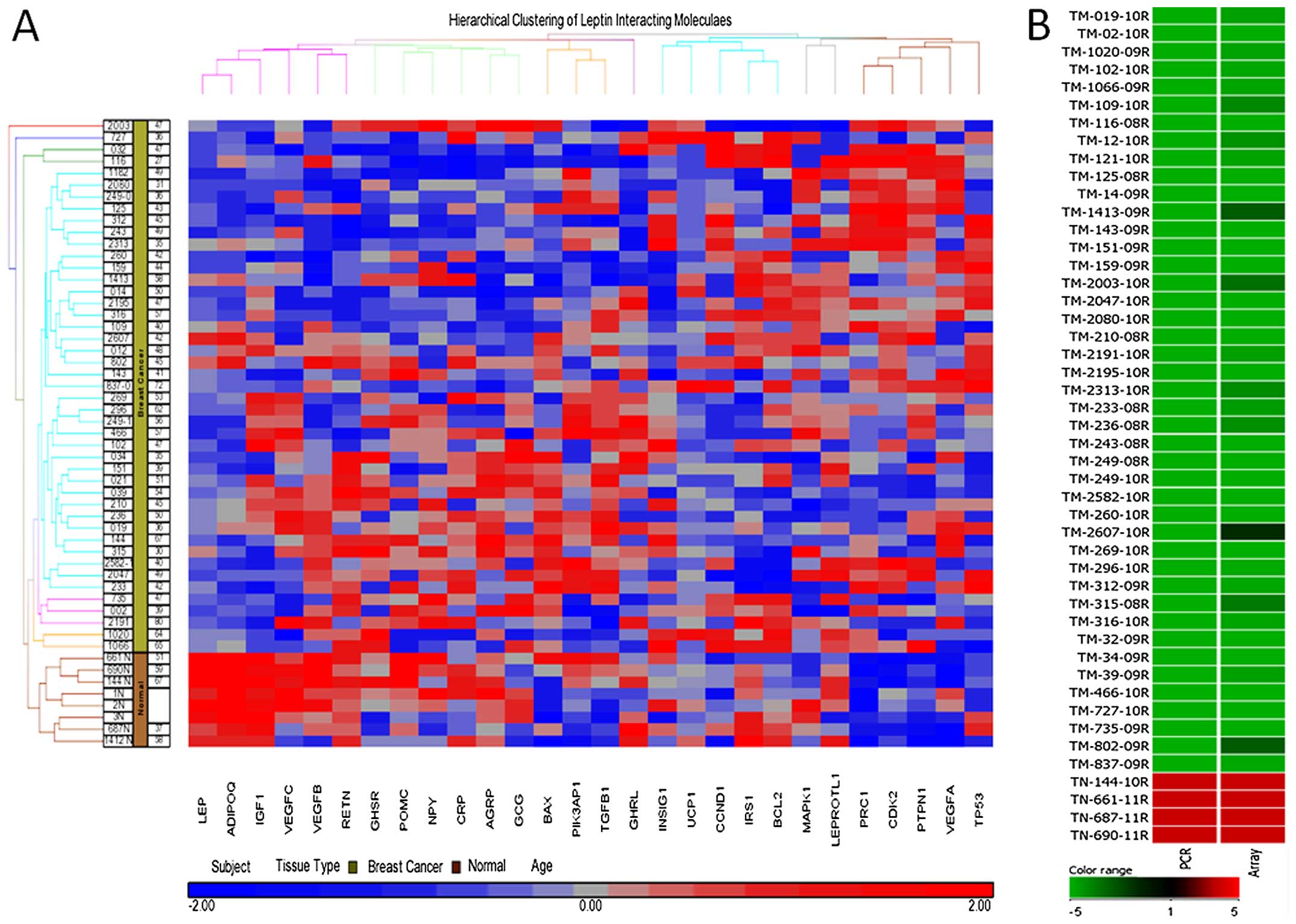
Real-time qPCR validation of LEP
The microarray results showed downregulation of
LEP and most of its interacting molecules in the BC tissues
(Fig. 2A). To evaluate the
expression level of LEP in the BC tissues, we validated it
using qPCR in 43 BC and 4 normal tissues (Fig. 2B). Our results further confirmed LEP
downregulation in all the 43 BC samples with significant
differences between the BC and the control groups
(p<0.0001).
Discussion
The therapeutic strategies currently employed for
breast cancer (BC) are generally based on histopathological
characterization, tumor size, grade and axillary lymph node and
receptor status (1). However,
patients when diagnosed with similar conditions and when treated
with similar agents can present with extensive differences in the
development and relapse of BC. Studies have demonstrated that women
who have had a full-term pregnancy at an age younger than 30; and
who have had three full-term pregnancies, and three or more years
of breast feeding, were significantly protected against BC
(12). However, the incidence of BC
is high among women from Saudi Arabia. BC risk factors, including
nulliparity or low parity, first full-term pregnancy at a late age,
no history of breast feeding, are not common in the Saudi female
population (11). These factors
combined with the early onset of BC among this ethnic population
prompted us to study the molecular mechanisms underlying this
malignancy.
In the present study, we characterized the role of
the specific gene LEP from identified transcriptomic
signature in BC tissues from Saudi Arabian patients that were
associated with clinical and histological parameters. Further
pathway analysis of the differentially regulated transcripts
provided new hypotheses underlying metastatic BC progression as
reported in our previous publications (18,19).
The prevalence of obesity is high in Saudia Arabia
with an incidence of 33.9% (20).
The mean body mass index (BMI) of the BC subjects in the present
study was 32.5 kg/m2. One study demonstrated that native
Hawaiian women with a BMI 30 kg/m2 or greater, or obese
women, had an 82% higher risk of BC, compared to those who had a
BMI of 20–24.9 kg/m2 (21). The strong association between
increased BMI and postmenopausal BC was also found specifically in
Asia-Pacific populations from 34 studies of a total sample size of
2,559,829 (22).
LEP, a product of the obesity gene and an
adipose derived hormone is mainly secreted by adipose tissue that
regulates energy intake and energy expenditure, including
metabolism (23,24). Disruption of lipid metabolism has
been implicated in breast tumorigenesis in several studies
(25–29). Studies have reported that increased
adiposity (body fat mass) is associated with higher circulating
levels of LEP (30,31). Studies have also shown that
increased expression of LEP in epithelial mammary cells may
promote tumorigenesis via cell proliferation, angiogenesis,
apoptosis, cell cycle regulation and cell survival mechanisms
(32). Furthermore, clinical
reports have also shown a strong correlation between BC risk and
blood LEP levels (33).
Similarly, Saxena et al showed the existence of
bidirectional crosstalk between LEP and insulin-like growth
factor 1 (IGF1) upregulation in triple-negative breast
cancer (TNBC) cell lines (34).
Additionally, increased LEP and decreased ADIPOQ
levels interrupt cellular signaling networks that are linked to
cell survival, angiogenesis, proliferation and cell cycle
regulation (35), and the
LEP-ADIPOQ axis has been well implicated in BC tumorigenesis
(35). However, most of the lipid
metabolism genes including LEP, ADIPOQ and
IGF-1 were found to be downregulated in our analysis, even
though the mean BMI of the BC subjects in the present study was
32.5 kg/m2. The effect of the downregulation of
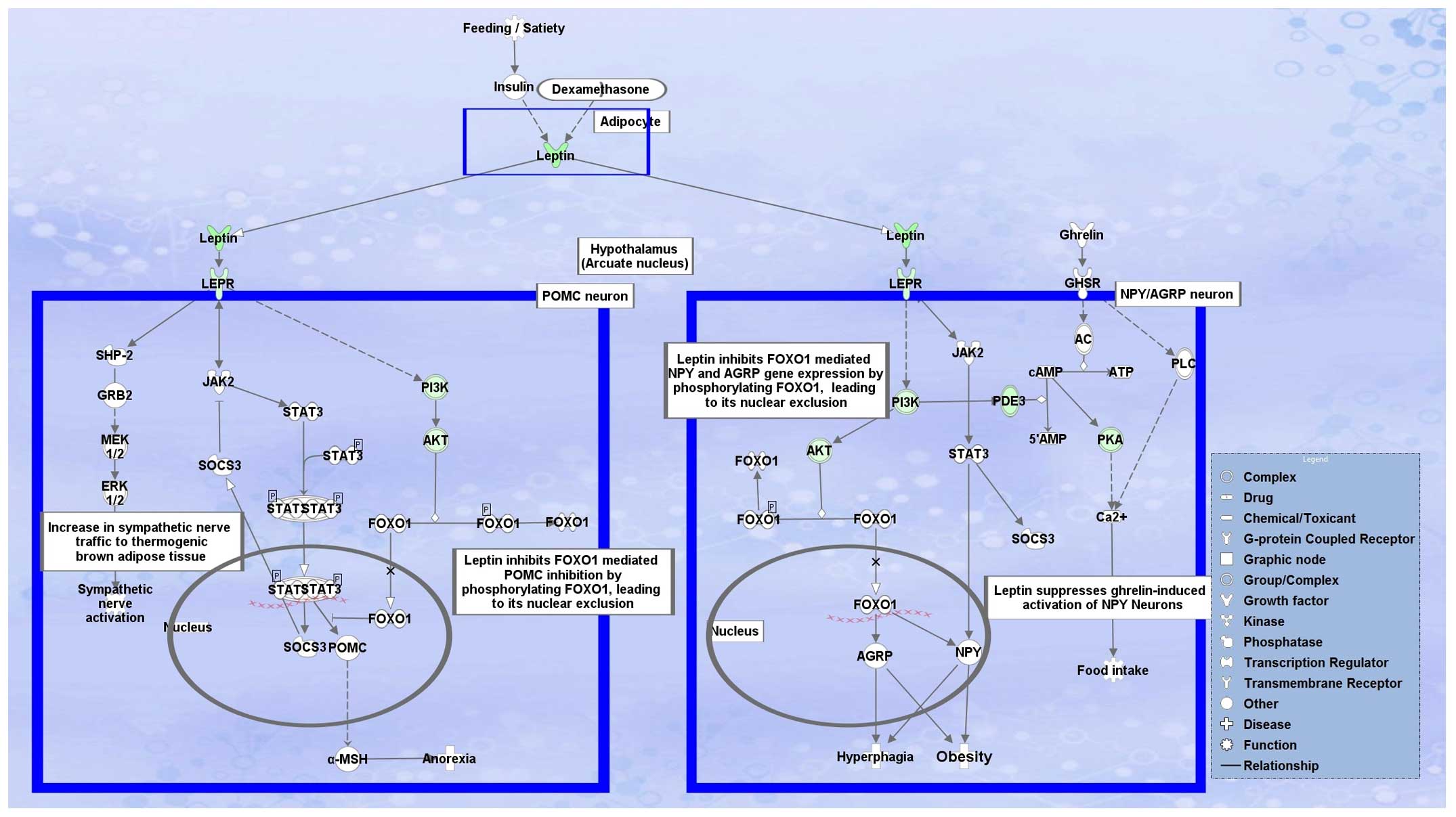
LEP is somewhat unclear. The LEP signaling pathway clearly
shows that LEP is at the top of the pathway and almost all
downstream genes were found to be downregulated, suggesting the
downregulation of LEP; however, regulation of LEP is still
unclear. Studies have shown that decreased levels of insulin
(36), glucosamine (37), glucocorticoids (38), cyclic AMP (39) and stimulation of adipose tissue
β-andrenergic receptor (40)
inhibit LEP expression (41). Thus,
we overlaid the differentially regulated genes onto the canonical
pathway for LEP signaling in obesity and our analysis showed that
most of the proteins present in the pathway to the downstream of
LEP were also downregulated (Fig.
3).
Obesity is perhaps related to LEP resistance not
deficiency, as an elevated level of LEP is found in many obese
persons. Whether elevated LEP concentrations are responsible for an
increased cancer risk is not yet certain. The variable reports on
circulating LEP and its level in cancer may be a consequence of
different analytic methods, or difference in sample preparation
where a number of influencing aspects, such as food intake and
circadian rhythm, were inappropriately controlled. Understanding
the underlying genetics and complex interactions of lifestyle, diet
and other known risk factors will remain a key area of
research.
In conclusion, the present study described
genome-wide profiling of BC from Saudi ethnic females. We
identified differential expression signatures, biological functions
and pathways that were significantly altered in BC and may serve as
targets for novel therapeutics. In our analysis, genes associated
with lipid metabolism and small-molecule biochemistry were
significantly downregulated in the BC tissues. LEP was one of the
most downregulated genes, and biological network analysis revealed
a strong connection between LEP and other molecules of the lipid
metabolism pathway. Further studies are needed to determine the
relationship between dysregulation of the lipid metabolism and the
mechanisms underlying BC.
Acknowledgments
The present study was funded by the NSTIP Strategic
Technologies Program in the Kingdom of Saudi Arabia, KACST
strategic project award no. (10-BIO1073-03, 10-BIO1258-03 and
08-MED120-03), the Center of Excellence in Genomic Medicine
Research (CEGMR-N08-14), and the Deanship of Scientific Research
(434/019-T and HiCi-1434-117-2), King Abdulaziz University, Jeddah,
Saudi Arabia. The authors also, acknowledge with thanks the
laboratory staff for the technical support. The authors would like
to thank Nuha AlAnsari, Alaa Albomgi, Manal Shabaat, Manar Ata and
Maha Al-Quaiti for performing RNA extraction, bioanalyzer assays
and microarray experiments. We thank the patients, and the
contributions of the physicians, nurses and the pathologists of the
King Abdulaziz University Hospital, King Faisal Specialist Hospital
and Research Center, and Bakhsh Hospital, Jeddah, the Kingdom of
Saudi Arabia.
Abbreviations:
|
BC
|
breast cancer
|
|
LEP
|
leptin
|
References
|
1
|
American Cancer Society: Cancer Facts and
Figures 2012. http://www.cancer.org/research/cancerfactsfigures/cancerfactsfigures/cancer-facts-figures-2012.
|
|
2
|
Billoski TV: Introduction to Paleontology
2. Institutional Press; New York: 1992
|
|
3
|
Morris SR and Carey LA: Molecular
profiling in breast cancer. Rev Endocr Metab Disord. 8:185–198.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hartmann A, Blaszyk H, Saitoh S, Tsushima
K, Tamura Y, Cunningham JM, McGovern RM, Schroeder JJ, Sommer SS
and Kovach JS: High frequency of p53 gene mutations in primary
breast cancers in Japanese women, a low-incidence population. Br J
Cancer. 73:896–901. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hartmann A, Rosanelli G, Blaszyk H,
Cunningham JM, McGovern RM, Schroeder JJ, Schaid DJ, Kovach JS and
Sommer SS: Novel pattern of P53 mutation in breast cancers from
Austrian women. J Clin Invest. 95:686–689. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hedenfalk I, Duggan D, Chen Y, Radmacher
M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M,
Kallioniemi OP, et al: gene-expression profiles in hereditary
breast cancer. N Engl J Med. 344:539–548. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang JC, Wooten EC, Tsimelzon A,
Hilsenbeck Sg, Gutierrez MC, Elledge R, Mohsin S, Osborne CK,
Chamness GC, Allred DC, et al: Gene expression profiling for the
prediction of therapeutic response to docetaxel in patients with
breast cancer. Lancet. 362:362–369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sabatier R, Finetti P, Adelaide J, Guille
A, Borg JP, Chaffanet M, Lane L, Birnbaum D and Bertucci F:
Down-regulation of ECRG4, a candidate tumor suppressor gene, in
human breast cancer. PLoS One. 6:e276562011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ezzat AA, Ibrahim EM, Raja MA, Al-Sobhi S,
Rostom A and Stuart RK: Locally advanced breast cancer in Saudi
Arabia: High frequency of stage III in a young population. Med
Oncol. 16:95–103. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai FM, Chen P, Ku HC, Lee MS, Chang SC,
Chang TM and Liou SH: A case-control study of parity, age at first
full-term pregnancy, breast feeding and breast cancer in Taiwanese
women. Proc Natl Sci Counc Repub China B. 20:71–77. 1996.PubMed/NCBI
|
|
13
|
Renehan AG, Soerjomataram I and Leitzmann
MF: Interpreting the epidemiological evidence linking obesity and
cancer: A framework for population-attributable risk estimations in
Europe. Eur J Cancer. 46:2581–2592. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ishikawa M, Kitayama J and Nagawa H:
Enhanced expression of leptin and leptin receptor (OB-R) in human
breast cancer. Clin Cancer Res. 10:4325–4331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Surmacz E: Obesity hormone leptin: A new
target in breast cancer? Breast Cancer Res. 9:3012007.PubMed/NCBI
|
|
16
|
Jensen LJ, Kuhn M, Stark M, Chaffron S,
Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et
al: STRING 8 - a global view on proteins and their functional
interactions in 630 organisms. Nucleic Acids Res. 37:D412–D416.
2009. View Article : Google Scholar
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering
C, et al: STRING v9.1: Protein-protein interaction networks, with
increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar :
|
|
18
|
Merdad A, Karim S, Schulten HJ, Jayapal M,
Dallol A, Buhmeida A, Al-Thubaity F, GariI MA, Chaudhary AG,
Abuzenadah AM, et al: Transcriptomics profiling study of breast
cancer from Kingdom of Saudi Arabia revealed altered expression of
Adiponectin and Fatty Acid Binding Protein4: Is lipid metabolism
associated with breast cancer? BMC Genomics. 16(Suppl 1): S112015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Merdad A, Karim S, Schulten HJ, Dallol A,
Buhmeida A, Al-Thubaity F, Gari MA, Chaudhary AG, Abuzenadah AM and
Al-Qahtani MH: Expression of matrix metalloproteinases (MMPs) in
primary human breast cancer: MMP-9 as a potential biomarker for
cancer invasion and metastasis. Anticancer Res. 34:1355–1366.
2014.PubMed/NCBI
|
|
20
|
Al-Othaimeen AI, Al-Nozha M and Osman AK:
Obesity: An emerging problem in Saudi Arabia. Analysis of data from
the National Nutrition Survey. East Mediterr Health. 13:441–448.
2007.
|
|
21
|
White KK, Park SY, Kolonel LN, Henderson
BE and Wilkens LR: Body size and breast cancer risk: The
Multiethnic Cohort. Int J Cancer. 131:E705–E716. 2012. View Article : Google Scholar
|
|
22
|
Renehan AG, Tyson M, Egger M, Heller RF
and Zwahlen M: Body-mass index and incidence of cancer: A
systematic review and meta-analysis of prospective observational
studies. Lancet. 371:569–578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Blüher S and Mantzoros CS: Leptin in
humans: Lessons from translational research. Am J Clin Nutr.
89:991S–997S. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Janeckova R: The role of Leptin in human
physiology and pathophysiology. Physiol Res. 50:443–459.
2001.PubMed/NCBI
|
|
25
|
Körner A, Pazaitou-Panayiotou K, Kelesidis
T, Kelesidis I, Williams CJ, Kaprara A, Bullen J, Neuwirth A,
Tseleni S, Mitsiades N, et al: Total and high-molecular-weight
adiponectin in breast cancer: In vitro and in vivo studies. J Clin
Endocrinol Metab. 92:1041–1048. 2007. View Article : Google Scholar
|
|
26
|
Hammamieh R, Chakraborty N, Barmada M, Das
R and Jett M: Expression patterns of fatty acid binding proteins in
breast cancer cells. J Exp Ther Oncol. 5:133–143. 2005.
|
|
27
|
Boneberg EM, Legler DF, Hoefer MM,
Ohlschlegel C, Steininger H, Füzesi L, Beer GM, Dupont-Lampert V,
Otto F, Senn HJ, et al: Angiogenesis and lymphangiogenesis are
downregulated in primary breast cancer. Br J Cancer. 101:605–614.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pauwels EK and Kairemo K: Fatty acid
facts, part II: Role in the prevention of carcinogenesis, or, more
fish on the dish? Drug News Perspect. 21:504–510. 2008. View Article : Google Scholar
|
|
29
|
Escrich E, Solanas M, Moral R and Escrich
R: Modulatory effects and molecular mechanisms of olive oil and
other dietary lipids in breast cancer. Curr Pharm Des. 17:813–830.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moschos S, Chan JL and Mantzoros CS:
Leptin and reproduction: A review. Fertil Steril. 77:433–444. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Considine RV, Sinha MK, Heiman ML,
Kriauciunas A, Stephens TW, Nyce MR, Ohannesian JP, Marco CC, McKee
LJ, Bauer TL, et al: Serum immunoreactive-leptin concentrations in
normal-weight and obese humans. N Engl J Med. 334:292–295. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jardé T, Perrier S, Vasson MP and
Caldefie-Chézet F: Molecular mechanisms of leptin and adiponectin
in breast cancer. Eur J Cancer. 47:33–43. 2011. View Article : Google Scholar
|
|
33
|
Tessitore L, Vizio B, Jenkins O, De
Stefano I, Ritossa C, Argiles JM, Benedetto C and Mussa A: Leptin
expression in colorectal and breast cancer patients. Int J Mol Med.
5:421–426. 2000.PubMed/NCBI
|
|
34
|
Saxena NK, Taliaferro-Smith L, Knight BB,
Merlin D, Anania FA, O'Regan RM and Sharma D: Bidirectional
crosstalk between leptin and insulin-like growth factor-I signaling
promotes invasion and migration of breast cancer cells via
transactivation of epidermal growth factor receptor. Cancer Res.
68:9712–9722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen DC, Chung YF, Yeh YT, Chaung HC, Kuo
FC, Fu OY, Chen HY, Hou MF and Yuan SS: Serum adiponectin and
leptin levels in Taiwanese breast cancer patients. Cancer Lett.
237:109–114. 2006. View Article : Google Scholar
|
|
36
|
Russell CD, Petersen RN, Rao SP, Ricci MR,
Prasad A, Zhang Y, Brolin RE and Fried SK: Leptin expression in
adipose tissue from obese humans: Depot-specific regulation by
insulin and dexamethasone. Am J Physiol. 275:E507–E515.
1998.PubMed/NCBI
|
|
37
|
Wang J, Liu R, Hawkins M, Barzilai N and
Rossetti L: A nutrient-sensing pathway regulates leptin gene
expression in muscle and fat. Nature. 393:684–688. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rosmond R, Dallman MF and Björntorp P:
Stress-related cortisol secretion in men: Relationships with
abdominal obesity and endocrine, metabolic and hemodynamic
abnormalities. J Clin Endocrinol Metab. 83:1853–1859.
1998.PubMed/NCBI
|
|
39
|
Gong DW, Bi S, Pratley RE and Weintraub
BD: genomic structure and promoter analysis of the human obese
gene. J Biol Chem. 271:3971–3974. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li H, Matheny M and Scarpace PJ: beta
3-Adrenergic-mediated suppression of leptin gene expression in
rats. Am J Physiol. 272:E1031–E1036. 1997.PubMed/NCBI
|
|
41
|
Fried SK, Ricci MR, Russell CD and
Laferrère B: Regulation of leptin production in humans. J Nutr.
130:3127S–3131S. 2000.
|