Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent carcinomas and the third leading cause of cancer-related
deaths throughout the world (1).
The best curative procedures for patients with HCC are hepatectomy
and liver transplantation. However, curative therapies are not
effective in a large number of patients due to diagnosis at
advanced stage (2). Main risk
factors for the development of HCC have been defined during the
decades, nevertheless numerous aspects of the evolution of
hepatocellular carcinogenesis, progress and metastasis are still
not fully understood (3). Thus, HCC
remains an aggressive cancer with a high mortality rate. In recent
years, compelling evidence has emerged in support of the presence
of cancer stem-like features that is responsible for
chemoresistance and recurrence of HCC (4–7).
Targeting cancer stem cell stemness determinants including
self-renewal and drug-resistance has been proposed as a therapeutic
goal. The Notch pathway is one of several key pathways linked to
both stem cell biology and cancer (8). Four transmembrane Notch receptors
(Notch1-4) exist in mammals. The receptors are mainly activated by
ligands of the Delta and Jagged families, which are expressed on
the surface of adjacent cells (9,10). In
particular, dysregulated Notch2 activity has been reported to be
associated with several human tumor types (11–14).
Most importantly, Notch inhibition suppresses nasopharyngeal
carcinoma by depleting cancer stem-like side population cells
(15). Since aberrant Notch
signaling has been implicated in cancer and cancer stem cell
therapeutic strategies that effectively target Notch signaling
could have a major impact on cancer patient survival.
In the liver, Notch2 are key regulators of liver
development (16,17). The expression of Notch2 was found in
HCC patients (18). Dill et
al reported that actived Notch2 signaling in the liver leads to
upregulation of pro-proliferative genes and proliferation of
hepatocytes and biliary epithelial cells (BECs). Most importantly,
using the diethylnitrosamine (DEN) HCC carcinogenesis model, they
showed that constitutive Notch2 signaling accelerated DEN-induced
HCC formation (19). However, the
biological function of Notch2 in human HCC cells has not yet been
documented. The objective of this study was to investigate the
biological function of Notch2 in the progress of human HCC.
Materials and methods
Cell culture
The human HCC cell lines HepG2, SMMC-7721, Bel-7402
and PLC were purchased from the Shanghai Cell Collection (Chinese
Academy of Sciences). In brief, SMMC-7721 and BEL-7402 cell lines
were cultured in RPMI-1640 medium (Gibco) supplemented with 10%
fetal bovine serum (Biological Industries). HepG2 and PLC were
cultured in DMEM medium (Invitrogen) supplemented with 10% fetal
bovine serum (Biological Industries).
Small interfering RNA transfection
Validated human siRNA for Notch2 (sense,
5′-GGAGGUCUCAGUGGAUAUATT-3′, antisense,
5′-UAUAUCCACUGAGACCUCCTT-3′) and negative control siRNA (sense,
5′-UUCUUCGAACGUGUCACGUTT3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′)
were designed and purchased from Shanghai GenePharma Co., Ltd. One
day before transfection, cells were plated on 6-well plates
(2×105 cells/well) or 96-wells (5×103
cells/well), and then the cells were transiently transfected with
either Notch2 or control siRNA (100 nM) using lipofectamine 2000
(Invitrogen) according to the manufacturer's protocol.
Western blot analysis
Protein lysates from the cells in culture were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and were detected with primary
antibodies recognizing Notch2 (1:1000, ab8926, Abcam) and β-tubulin
(1: 2000, #2128, Cell Signaling Technology).
Cell proliferation analysis
Cell viability was measured using the Cell Counting
Kit-8 (CCK-8, Dojindo). Briefly, transfected cells
(5×103) on a 96-well plate with 5 replicate wells were
allowed to incubate for different time interval from day 1 to 4.
After washed in PBS, 100 µl medium containing 10% CCK-8
solution was added to each well and incubated for 2 h at 37°C.
Samples are read directly in the wells using an absorbance of the
450 nm wavelength by an enzyme linked immunosorbent assay (ELISA)
plate reader. The blank control absorbance (100 µl medium
containing 10% CCK-8 alone) was subtracted from the experimental
absorbance to adjust the background.
Cell cycle analysis
For cell cycle analysis, cells were collected 24 h
after transfection, washed with PBS and fixed in 1 ml ice-cold 70%
ethanol at 4°C overnight. Fixed cells were pelleted by
centrifugation at 3000 rpm for 5 min. Then the cells were washed by
PBS twice and stained with 1 ml staining solution containing 50
µg/ml propidium iodide (PI) and 0.5 µg/ml RNase a for
30 min and subjected to flow cytometric analysis.
Colony formation analysis
Standard colony formation assays were used to
measure cell proliferation. At 24 h after transfection, transfected
cells were seeded in 6-well tissue culture plates (50 cells per
well). After an incubation period of 10 days, the medium was
decanted and each well was washed twice with PBS. The cells were
fixed in 100% methanol for 20 min and then stained with 1% crystal
violet for 15 min, followed by detaining. Colonies (>20
cells/colony) were counted.
Mammosphere formation analysis
Cells were seeded on 6-well ultra low attachment
plates (Corning, Tewksbury, MA, USA) at a density of 3,000
cells/500 µl in MammoCult Basal Medium and MammoCult
Proliferation Supplement with 4 µg/ml heparin and 0.48
µg/ml hydrocortisone (Stem Cell Technologies, Vancouver, BC,
Canada) and were incubated for 7 days at 37°C under 5%
Co2. Mammospheres were filtered by a 70-mm cell strainer
and mammospheres >70 mm in diameter were collected and
counted.
In vitro cytotoxicity assay
Twenty-four hours after transfection, transfected
cells on a 96-well plate with 5 replicate wells were allowed to
incubate for 48 h with the treatment of anticancer drug
5-fluorouracil (5-FU) with various concentrations (25, 50, 100, 200
and 400 µg/ml, respectively). After 48 h of incubation, cell
viability was assessed utilizing the CCK-8 assay. The rate of cell
growth inhibition (IR) was calculated according to the following
equation: IR = [1−A450 (drug) / A450 (control)] ×100%, where A450
(drug) is the absorbance of the cells exposed to 5-FU and a450
(control) is the absorbance of the cells without 5-FU
treatment.
In vivo xenograft study
All animal experimentation described in this study
was performed in accordance with protocols approved by the
Institutional Animal Care and Use Committee at Sun Yat-sen
University. Cells (3×106) were suspended in 100
µl PBS and were injected subcutaneously into 5-week-old
female nude mice 24 h after transfection. Tumor volumes were
monitored every 3 days by caliper measurement of the length and
width and calculated using the formula of (width2) ×
length/2. Mice were sacrificed 21 days after the injection and the
tumors were isolated and measured.
Immunofluorescence staining
Briefly, cells grown on coverslips were fixed in 4%
fresh paraformaldehyde for 20 min and then permeabilized with 0.3%
Triton X-100 in PBS for 10 min at room temperature. After the cells
were blocked for 15 min with 10% normal goat serum, the coverslips
were incubated overnight at 4°C with two primary antibodies, Notch2
(1:200, ab8926, Abcam) and CD90 (1:200, ab92574, Abcam). After
being extensively washed with PBS, the cells were incubated with
species-specific secondary antibodies (1:400, 711-545-152 or
715-095-150, Jackson Immuno Research) for 1 h at room temperature.
Finally, the coverslips were washed, counterstained with 4′,
6-diamidino-2-phenylindole (DAPI; 0.1 mg/ml, Sigma) for 5 min and
mounted on glass slides. Immunofluorescence images were
photographed under a fluorescence microscope (Olympus).
Statistical analysis
Each experiment was repeated 3–4 times. Statistical
analysis was conducted with the 20.0 SPSS software package. The
functional assays were compared using the paired student t-test.
P-value <0.05 was considered statistically significant.
Results
The expression of Notch2 in four HCC cell
lines
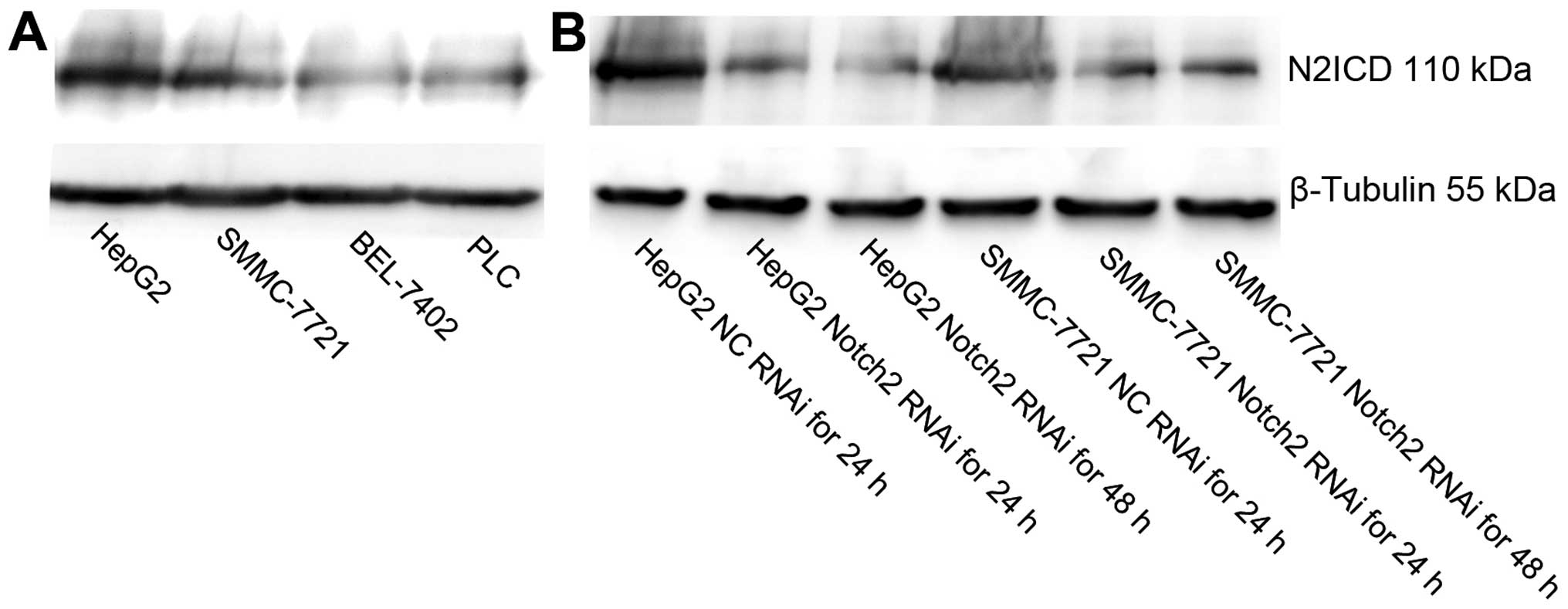
As showed in Fig. 1,
western blot analysis revealed that Notch2 was expressed in four
HCC cell lines and the expression of Notch2 was higher in HepG2 and
SMMC-7721 cells compared to BEL-7402 and PLC cell lines (Fig. 1A). These two HCC cell lines were
used in subsequent functional assays.
Transient knockdown of Notch2 by
siRNA
To investigate the possible role of Notch2 on HepG2
and SMMC-7721 cells, we employed RNAi to deplete its expression in
these cells, both of which were treated with NC-siRNA or
Notch2-siRNA. After 24 and 48 h, the cells were examined by western
blot analysis. As shown in Fig. 1B,
the expression of Notch2 intracellular domain (N2ICD) was knocked
down as determined by western blot analysis. This data indicated
that Notch2-specific siRNA could effectively and obviously suppress
the expression of Notch2 in HCC cells.
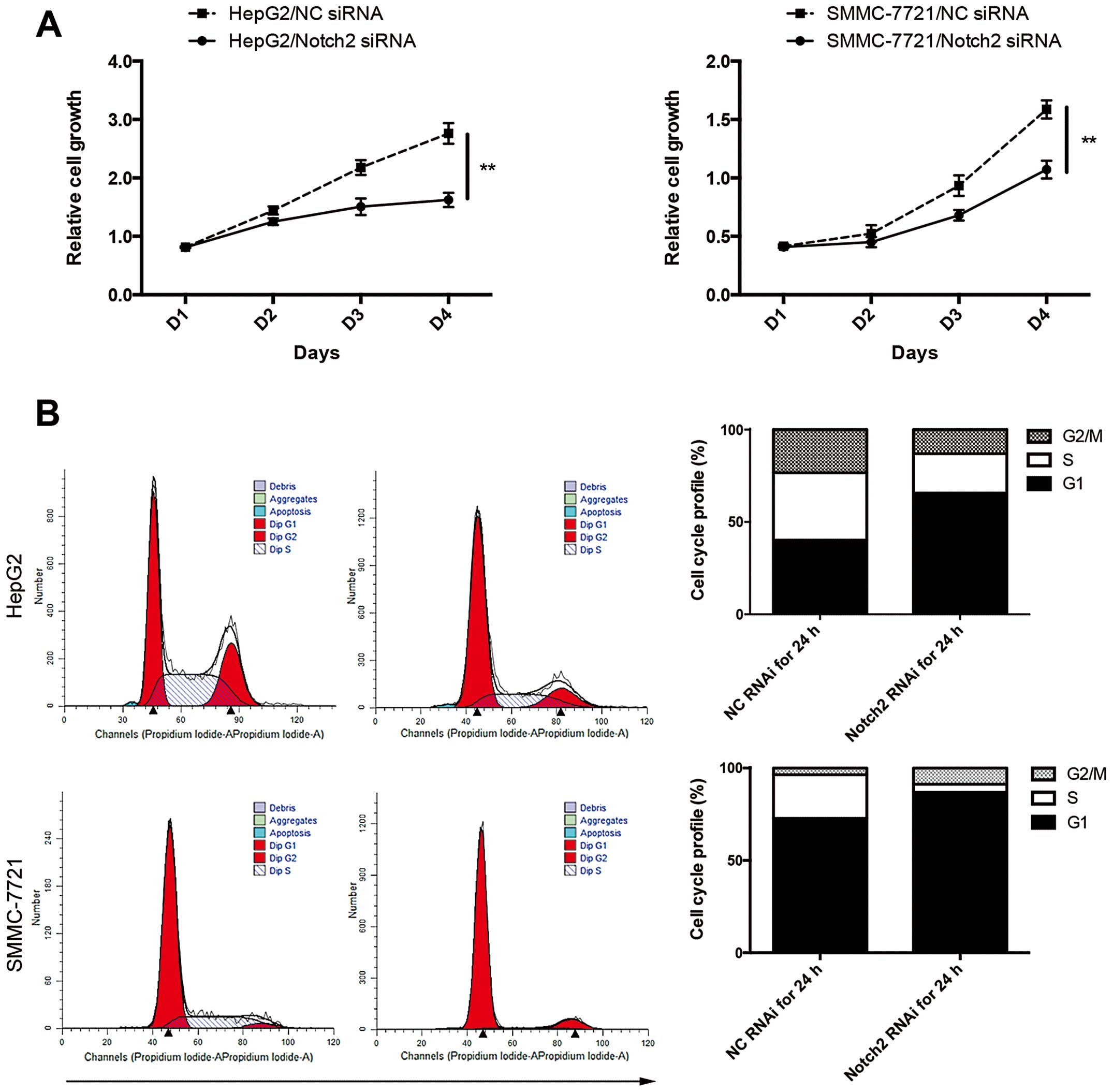
Transfection of Notch2-siRNA represses
proliferation and cell cycle progression of HCC cells in vitro
CCK-8 cell proliferation assay revealed that the
decrease in Notch2 expression caused by Notch2-siRNA significantly
inhibited the proliferation of HCC cells (Fig. 2A). Additionally, cell cycle analysis
by PI staining revealed significant increase in G0/G1 cell
population but decrease in both S and G2/M population. The
reduction of S+G2/G1 ratio after knockdown of Notch2 suggested that
depletion of Notch2 would induce G0/G1 cell cycle arrest in HCC
(Fig. 2B).
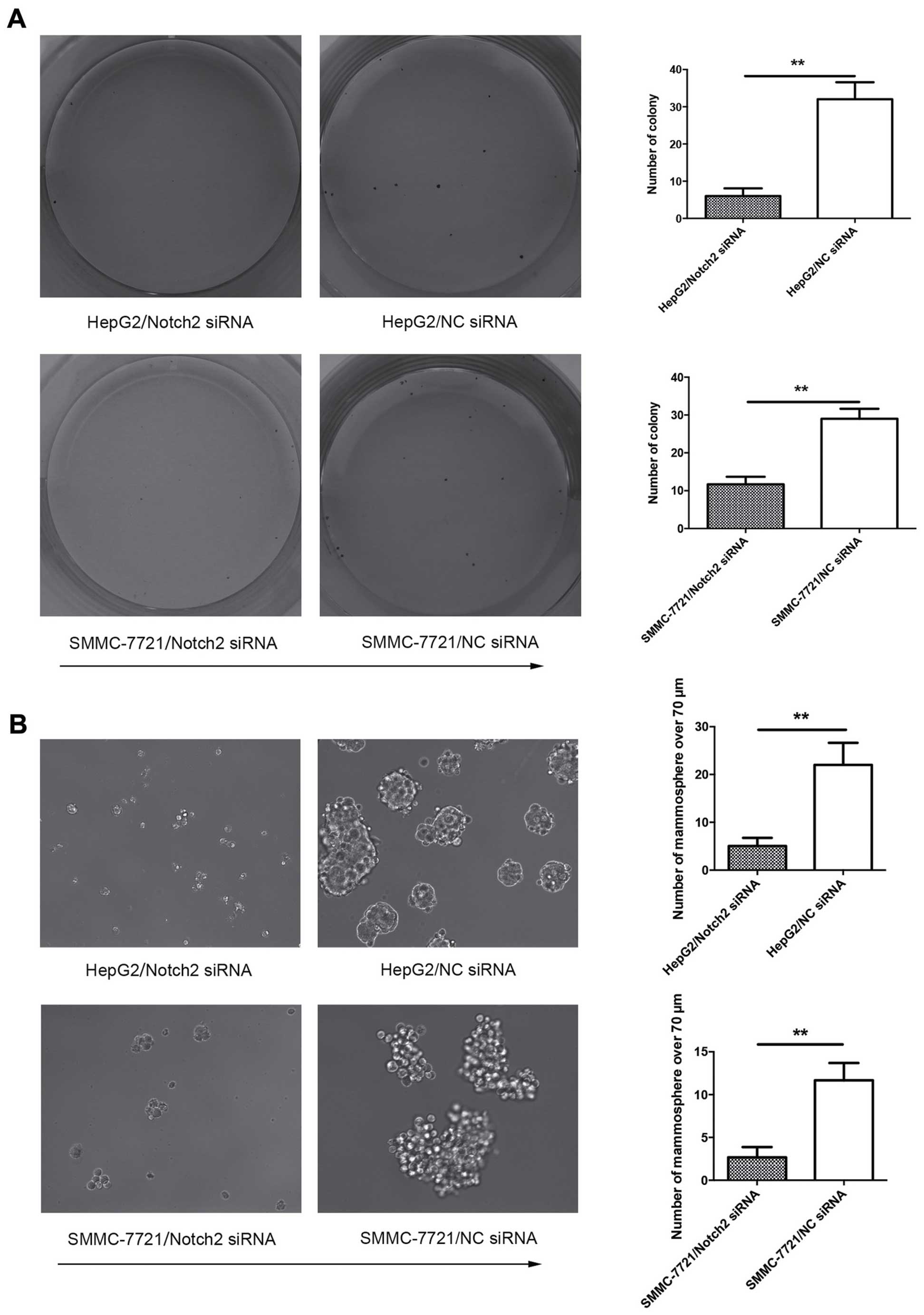
Transfection of Notch2-siRNA represses
self-renewal and increases sensitivity to 5-FU of HCC cells in
vitro
We next investigated the potential role of Notch2 on
regulation of stem-like cell characteristics in HCC. Self-renewal
and drug-resistance are two hallmarks of stemness behavior, so
colony formation assay and chemoresistance were undertaken to assay
the effect of Notch2 in HCC cells. Cell colony formation assay
revealed that suppression of Notch2 could impede the self-renewal
ability of HCC cells, which was reflected by fewer colonies and
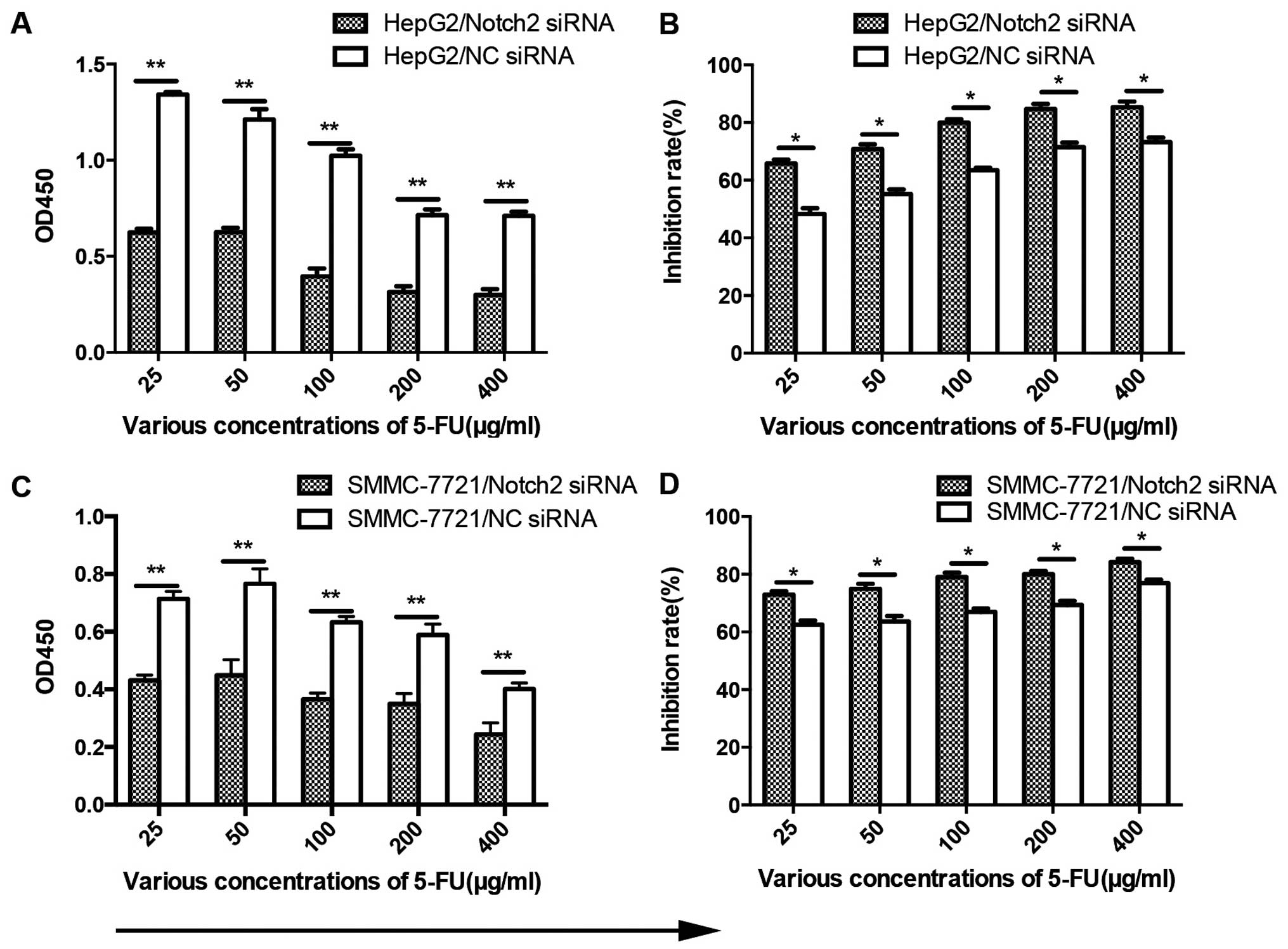
mammospheres in Notch2-depleted HCC cells (Fig. 3A and B). Moreover, knockdown of
Notch2 sensitized HCC cells to anticancer drug 5-FU (Fig. 4A and C). Effect of Notch2-siRNA or
100 µg/ml 5-FU alone showed an inhibition rate on cell
viability of HCC cells by 29.3 and 63.3%, respectively, however the
synthetical outcome showed a clear distinct effect of 79.9%
reduction on cell viability (Fig. 4B
and D). Collectively, these data indicate that Notch2 maintains
stem-like cell properties of HCC cells in vitro.
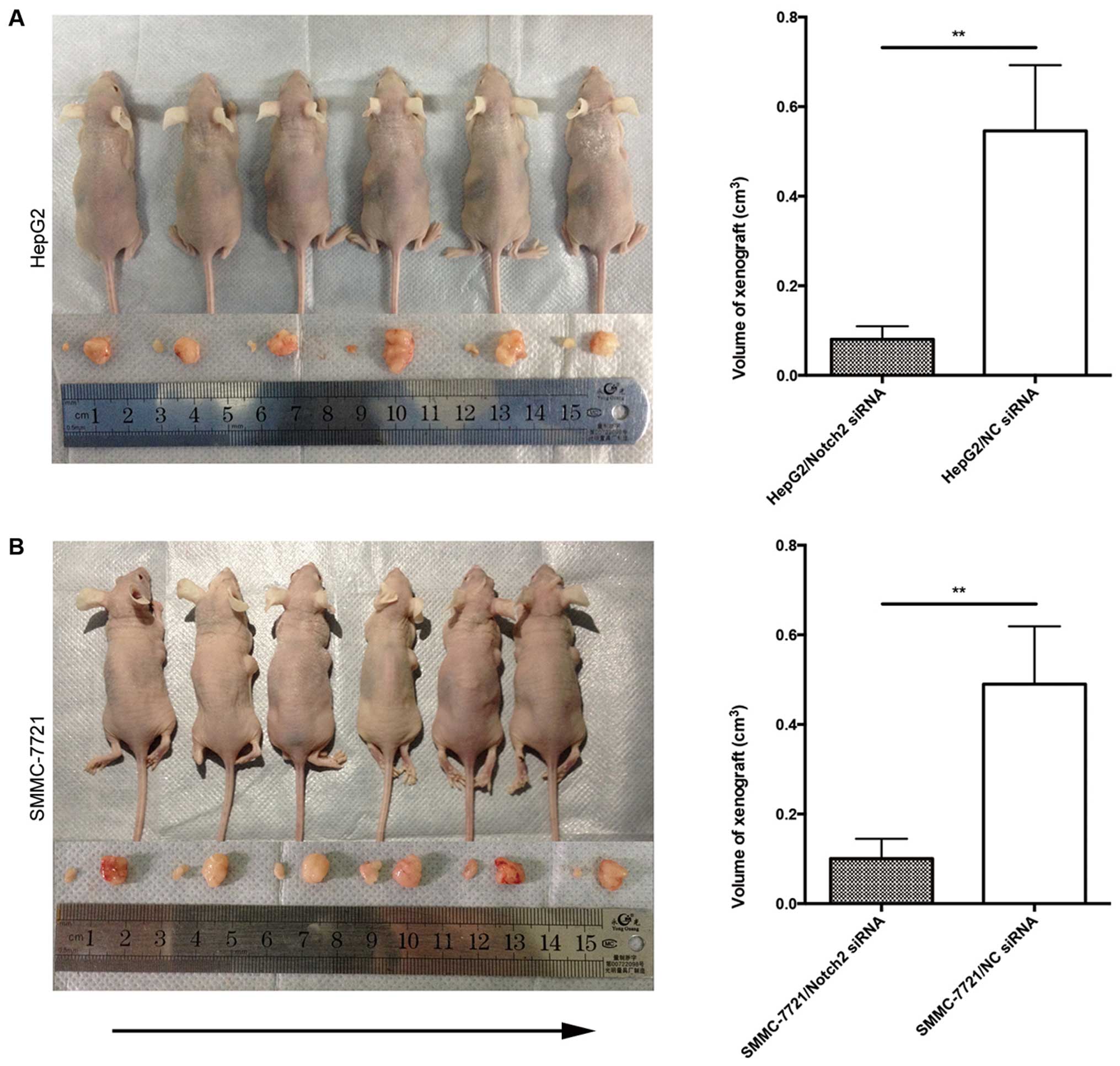
Transfection of Notch2-siRNA suppresses
tumorigenicity in vivo
Since our in vitro studies suggested that
Notch2 plays a regulatory role in proliferation and invasion of HCC
cells, the biological significance of these results was further
evaluated in an in vivo model of HCC. Both Notch2-siRNA
tumor cells and control cells were implanted subcutaneously into
nude mice and the resulting tumor formation was measured. Following
downregulation of Notch2 expression, HCC cells showed significantly
diminish in vivo tumor growth compared to control cells
(Fig. 5A and B). These data
corroborate our in vitro observations and support the notion
that Notch2-siRNA significantly inhibits tumor growth of HCC.
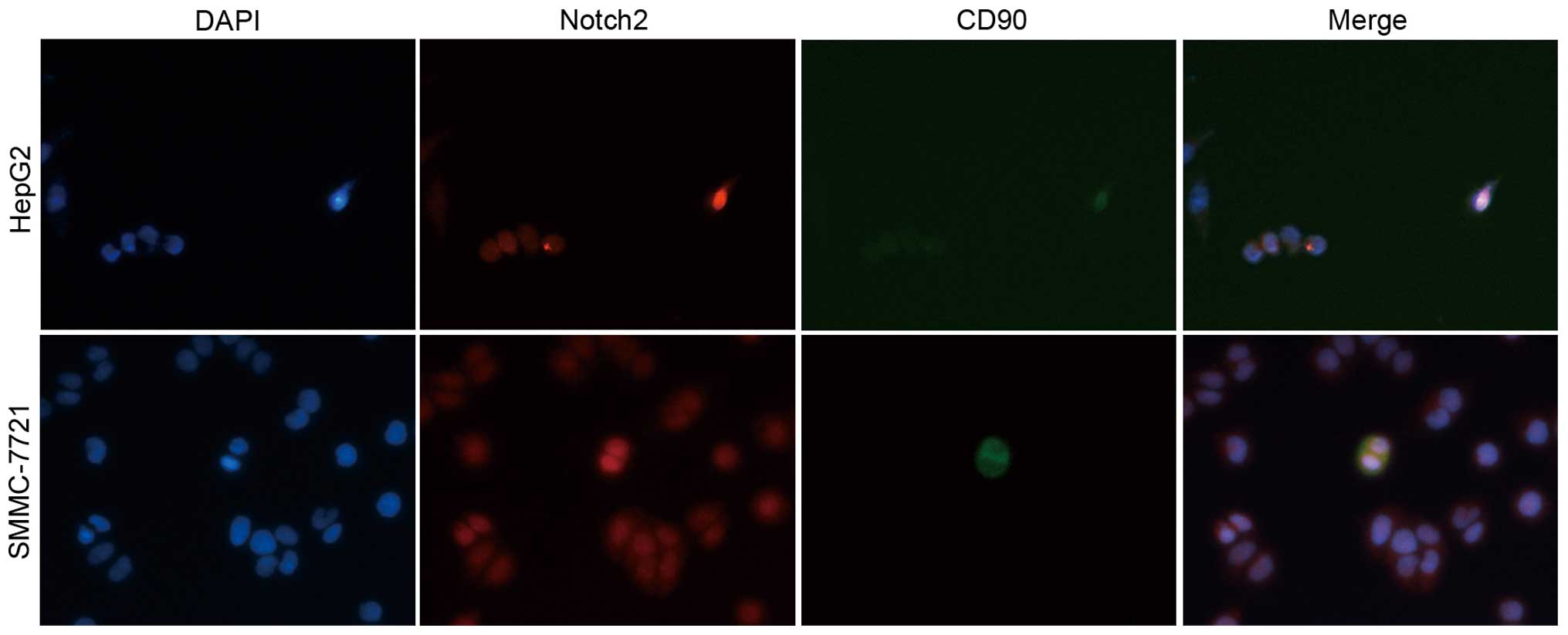
Notch2 was highly expressed in CD90
positive HCC cells
To further determine the expression of Notch2 in HCC
cells, expression of Notch2 and hepatic stem cell marker CD90 were
examined by immunofluorescence staining. Of note, Notch2 was
markedly upregulated in CD90-positive HCC cells (Fig. 6). Based on this finding, we
speculated that the extensive anticancer effects in HCC by
silencing Notch2 might be through targeting cancer stem cells.
Discussion
Recent studies on Notch signaling pathway showed
that it plays a crucial role in cell fate decision, tissue
patterning and morphogenesis (20–23).
The Notch pathway is one of several key pathways linked to both
stem cell biology and cancer (8).
Targeting Notch2 inhibits tumor growth and decreases
tumor-initiating cells (24). In
particular, dysregulated Notch2 activity has been associated with
several human tumor types, including ovarian, breast, pancreas and
colon cancers (11–14).
In the liver, Giovannini et al previously
demonstrated that Notch1, Notch3 and Notch4 were over expressed in
human HCC and Notch3 contributed to the doxorubicin resistance in
HCC lines (25). In a recent study,
Litten et al suggested that Notch2 expression and
activation, independent of Jagged1 expression, might contribute to
the pathogenesis of hepatoblastoma (26). Furthermore, Dill et al
reported that constitutive Notch2 signaling accelerated DEN-induced
HCC formation in the DEN HCC carcinogenesis model (19). These studies disclosed a key
oncogenic role of Notch2 in HCC development. However, a previous
report showed that Notch2 was downregulated in human HCC compared
with adjacent non-tumor liver tissue (18). Of great interest, no existing data
of functional assays elucidate the effect of Notch2 in human HCC
cells.
In the present study, the expression of Notch2 was
found in four human HCC cells. Functional studies disclosed that
knockdown of Notch2 results in decreased cell proliferation, cell
cycle progression, colony and mammosphere formation in HepG2 and
SMMC-7721 cells, suggesting that Notch2 is essential for
proliferation and self-renewal of HCC. Moreover, depletion of
Notch2 leads to sensitized HCC cells to chemotherapeutic agent
5-FU, indicating that Notch2 plays an essential role in the
drug-resistance of HCC cells. In addition, Notch2-knockdown
inhibited the growth of HepG2 tumors in a xenograft model, which
would in turn suggest that Notch2 plays an essential role in the
progression of HCC.
One significant breakthrough in cancer research is
the discovery of cancer stem cells (CSCs) (27), and it has received much attention.
CSCs, characterized as a subset of tumor cells with stem cell
properties, have been identified in mounting types of cancers
helping to explain the cellular heterogeneity of tumor tissue,
drug-resistance and tumor recurrence (28,29).
In HCC, it is generally accepted that genes controlling
self-renewal may sustain stem-like properties and chemoresistance
ability. In line with this point of view, our present study deduced
that Notch2 fortified stemness features can be seen in some types
of HCC as depletion of Notch2 in HpeG2 and SMMC-7721 cell lines
lead to an extensive reduction in growth and self-renewal ability
both in vitro and in vivo. Moreover, high expression
of Notch2 was found in CD90-positive HCC cells. CD90 is the hepatic
cancer stem cell maker in HCC (30). The results demonstrated that
extensive anticancer effects by downregulation of Notch2 in HCC
cells might be through targeting cancer stem cells.
In summary, Notch2 may confer stemness properties in
HCC. Downregulation of Notch2 gene by RNAi can inhibit the
proliferation and carcinogenesis of HCC cells and increase their
sensitivity to 5-FU, which could provide a novel strategy for
treatment of this fatal disease.
Acknowledgments
This work was supported by the National Natural
Science Foundation of China (81301865 and 81172068); the Science
Foundation of Guangdong Province (2015a030313033).
References
|
1
|
Singh S, Singh PP, Roberts LR and Sanchez
W: Chemopreventive strategies in hepatocellular carcinoma. Nat Rev
Gastroenterol Hepatol. 11:45–54. 2014. View Article : Google Scholar
|
|
2
|
Motola-Kuba D, Zamora-Valdés D, Uribe M
and Méndez-Sánchez N: Hepatocellular carcinoma. An overview. Ann
Hepatol. 5:16–24. 2006.PubMed/NCBI
|
|
3
|
Li Y, Tang ZY and Hou JX: Hepatocellular
carcinoma: Insight from animal models. Nat Rev Gastroenterol
Hepatol. 9:32–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woo HG, Wang XW, Budhu A, Kim YH, Kwon SM,
Tang ZY, Sun Z, Harris CC and Thorgeirsson SS: Association of TP53
mutations with stem cell-like gene expression and survival of
patients with hepatocellular carcinoma. Gastroenterology.
140:1063–1070. 2011. View Article : Google Scholar :
|
|
5
|
Oishi N and Wang XW: Novel therapeutic
strategies for targeting liver cancer stem cells. Int J Biol Sci.
7:517–535. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma S, Tang KH, Chan YP, Lee TK, Kwan PS,
Castilho A, Ng I, Man K, Wong N, To KF, et al: miR-130b Promotes
CD133(+) liver tumor-initiating cell growth and self-renewal via
tumor protein 53-induced nuclear protein 1. Cell Stem Cell.
7:694–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tong CM, Ma S and Guan XY: Biology of
hepatic cancer stem cells. J Gastroenterol Hepatol. 26:1229–1237.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Penton AL, Leonard LD and Spinner NB:
Notch signaling in human development and disease. Semin Cell Dev
Biol. 23:450–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gray GE, Mann RS, Mitsiadis E, Henrique D,
Carcangiu ML, Banks A, Leiman J, Ward D, Ish-Horowitz D and
Artavanis-Tsakonas S: Human ligands of the Notch receptor. Am J
Pathol. 154:785–794. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoneya T, Tahara T, Nagao K, Yamada Y,
Yamamoto T, Osawa M, Miyatani S and Nishikawa M: Molecular cloning
of delta-4, a new mouse and human Notch ligand. J Biochem.
129:27–34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Galic V, Shawber CJ, Reeves C, Shah M,
Murtomaki A, Wright J, Herzog T, Tong GX and Kitajewski J: NOTCH2
expression is decreased in epithelial ovarian cancer and is related
to the tumor histological subtype. Pathol Discov. 1:42013.
View Article : Google Scholar
|
|
12
|
Sehrawat A, Sakao K and Singh SV: Notch2
activation is protective against anticancer effects of zerumbone in
human breast cancer cells. Breast Cancer Res Treat. 146:543–555.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazur PK, Einwächter H, Lee M, Sipos B,
Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Klöppel G,
et al: Notch2 is required for progression of pancreatic
intraepithelial neoplasia and development of pancreatic ductal
adenocarcinoma. Proc Natl Acad Sci USA. 107:13438–13443. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang WJ, Yao Y, Jiang LL, Hu TH, Ma JQ,
Ruan ZP, Tian T, Guo H, Wang SH and Nan KJ: Increased LEF1
expression and decreased Notch2 expression are strong predictors of
poor outcomes in colorectal cancer patients. Dis Markers.
35:395–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu S, Zhang R, Liu F, Wang H, Wu J and
Wang Y: Notch inhibition suppresses nasopharyngeal carcinoma by
depleting cancer stem-like side population cells. Oncol Rep.
28:561–566. 2012.PubMed/NCBI
|
|
16
|
Sparks EE, Huppert KA, Brown MA,
Washington MK and Huppert SS: Notch signaling regulates formation
of the three-dimensional architecture of intrahepatic bile ducts in
mice. Hepatology. 51:1391–1400. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geisler F, Nagl F, Mazur PK, Lee M,
Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM and Siveke JT:
Liver-specific inactivation of Notch2, but not Notch1, compromises
intrahepatic bile duct development in mice. Hepatology. 48:607–616.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao J, Song Z, Chen Y, Xia L, Wang J, Fan
R, Du R, Zhang F, Hong L, Song J, et al: Deregulated expression of
Notch receptors in human hepatocellular carcinoma. Dig Liver Dis.
40:114–121. 2008. View Article : Google Scholar
|
|
19
|
Dill MT, Tornillo L, Fritzius T,
Terracciano L, Semela D, Bettler B, Heim MH and Tchorz JS:
Constitutive Notch2 signaling induces hepatic tumors in mice.
Hepatology. 57:1607–1619. 2013. View Article : Google Scholar
|
|
20
|
Jeliazkova P, Jörs S, Lee M, Zimber-Strobl
U, Ferrer J, Schmid RM, Siveke JT and Geisler F: Canonical Notch2
signaling determines biliary cell fates of embryonic hepatoblasts
and adult hepatocytes independent of Hes1. Hepatology.
57:2469–2479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kamath BM, Bauer RC, Loomes KM, Chao G,
Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz
ID, et al: NOTCH2 mutations in Alagille syndrome. J Med Genet.
49:138–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McCright B, Lozier J and Gridley T: A
mouse model of Alagille syndrome: Notch2 as a genetic modifier of
Jag1 haploinsufficiency. Development. 129:1075–1082.
2002.PubMed/NCBI
|
|
23
|
Tchorz JS, Kinter J, Müller M, Tornillo L,
Heim MH and Bettler B: Notch2 signaling promotes biliary epithelial
cell fate specification and tubulogenesis during bile duct
development in mice. Hepatology. 50:871–879. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yen WC, Fischer MM, Axelrod F, Bond C,
Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, et al:
Targeting notch signaling with a notch2/notch3 antagonist
(tarextumab) inhibits tumor growth and decreases tumor-initiating
cell frequency. Clin Cancer Res. 21:2084–2095. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Giovannini C, Gramantieri L, Chieco P,
Minguzzi M, Lago F, Pianetti S, Ramazzotti E, Marcu KB and Bolondi
L: Selective ablation of Notch3 in HCC enhances doxorubicin's death
promoting effect by a p53 dependent mechanism. J Hepatol.
50:969–979. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Litten JB, Chen TT, Schultz R, Herman K,
Comstock J, Schiffman J, Tomlinson GE and Rakheja D: Activated
NOTCH2 is overexpressed in hepatoblastomas: an immunohistochemical
study. Pediatr Dev Pathol. 14:378–383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature. 367:645–648. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou BB, Zhang H, Damelin M, Geles KG,
Grindley JC and Dirks PB: Tumour-initiating cells: Challenges and
opportunities for anticancer drug discovery. Nat Rev Drug Discov.
8:806–823. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sukowati CH, Anfuso B, Torre G,
Francalanci P, Crocè LS and Tiribelli C: The expression of
CD90/Thy-1 in hepatocellular carcinoma: An in vivo and in vitro
study. PLoS One. 8:e768302013. View Article : Google Scholar : PubMed/NCBI
|