Introduction
Colorectal cancer (CRC) is the third most common
malignant cancer diagnosed worldwide (1), and ranks as the second leading cause
of cancer-related deaths in developed countries (2). Approximately 90% of CRC-related
mortalities are a result of metastases (3). Metastasis is a complex, multi-step
process whereby tumor cells first invade the surrounding tissues
and intravasate into the vasculature, then translocate through the
systemic circulation, and thus extravasate into the parenchyma of
distant tissues, e.g. liver and lungs, eventually, establishing
micrometastases and forming macroscopic secondary tumors (4).
Although much progress has been made in the
identification and characterization of the genetic and epigenetic
changes involved in CRC metastasis (5,6), the
underlying mechanisms remain largely unclear. The search for
predictive markers for CRC metastasis remains a priority, as it is
the major cause of the high mortality rate. Recently, microRNAs
(miRNAs) have become a hot spot due to their significant role in
the regulation of gene expression. miRNAs are ~22-nucleotide
conserved endogenous non-coding single-stranded RNA molecules
(7), which bind to the
3′-untranslated region (3′-UTR) of target mRNAs and regulate the
stability and translation of mRNAs, resulting in either inhibition
of translation or degradation of target mRNAs (8,9). They
are involved in a wide spectrum of biological processes, such as
proliferation, metabolism, cellular differentiation and apoptosis
(10,11). To date, a number of miRNAs have been
found to be associated with cancer development and progression,
including CRC. Among them, miR-375 is proven to be involved in
early invasive CRC through downregulation of its expression
(12–14); however, the functional role of
miR-375 in CRC needs further investigation (15). In the present study, we attempted to
reveal the mechanisms of miR-375 underlying the biological behavior
of CRC. We investigated the biological functions of proliferation
and invasion/migration in CRC cells and identified the target gene
in order to explore the possible molecular mechanism involving CRC
development, in hope of identifying a new predictor for prognosis
or new target for diagnosis and therapy.
Materials and methods
Cell lines and transfection
DLD1 and HCT8 cells stored in our laboratory were
maintained in RPMI-1640 medium, supplemented with 10% fetal bovine
serum (FBS) (both from Gibco, Grand Island, NY, USA) at 37°C under
5% CO2. We had entrusted Beijing Microread Genetics Co.,
Ltd., Beijing, China for the identification of the 2 cell lines.
The authentication reports (data not shown) showed no
cross-contamination of other human cell lines, and 100% matched
cell lines are found in ATCC banks with the DLD1 and HCT8 cell
lines. Precursor miRNA (pre-miR-375) and precursor negative control
(pre-neg) (Applied Biosystems, Foster City, CA, USA) were
transfected into the two cell lines using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions. Additionally, following this method, the interfering
RNA of sp1 messenger RNA (siSP1) and the negative control (si-neg)
were transfected into the HCT8 cells.
RNA extraction and quantitative RT-PCR
(qRT-PCR)
Total RNA of the cells was extracted using mirVana
isolation kit according to the manufacturer's instructions (Applied
Biosystems). For validation of miRNA and target gene mRNA
expression, quantitative reverse transcriptase-polymerase chain
reaction (qRT-PCR) using TaqMan miRNA assays and quantitative
RT-PCR using SYBR-Green PCR Master Mix kit (Applied Biosystems)
were performed according to the manufacturer's instructions. The
expression level of U47 small nuclear RNA was used for miRNA, and
GAPDH was used for mRNA as the endogenous controls. All assays were
carried out in triplicate.
Cell colony formation assay
For the colony formation assay, 1×103
cells were plated in 6-well plates for each well in medium with 20%
FBS for 72 h, and then fixed and stained with crystal violet. Then,
the colonies were photographed and counted under a microscope. All
experiments were performed in 3 replicates and were repeated 3
times, independently.
Cell Counting Kit-8 (CCK-8) assay
DLD1 (2×103) and HCT8 cells
(3×103) were suspended in RPMI-1640 medium (100
μl) containing 10% FBS and cultured into 96-well plates
overnight, and then transfected with pre-miR-375 and pre-neg. Cell
proliferation was determined using CCK-8 assay at 0, 1, 2 and 3
days after transfection, respectively, and the absorbance of the
samples was measured with a spectrophotometer reader at 450 nm.
Cell migration and invasion assays
DLD1 (2×105) and HCT8 cells
(2×105) were cultured into 6-well plates, at 24 h after
transfection of pre-miR-375 and pre-neg, as previously described.
Migration and invasion assays were performed both using 24-well
Transwell migration chambers with 8-μl pore size (Corning
Costar, Inc., Corning, NY, USA). DLD1 (1×105) and HCT8
cells (5×104) suspended in 100 μl corresponding
culture medium without FBS were loaded into the top chamber, and
the bottom chamber contained 600 μl medium with 20% FBS. For
the migration assay, the cells were allowed to migrate for 12 h. In
addition, for the invasion assay, Transwell wells were pre-coated
with Matrigel, and the cells were allowed to invade for 48 h. When
both assays were stopped, the cells that migrated or invaded into
the bottom chamber were fixed with methanol for 5 min, and then
stained with crystal violet (0.05%) for 4 min. The cells that
migrated or invaded were photographed and counted under a
microscope. In addition, the migration and invasion rates were
assessed by the formula: (motile cells transfected with
pre-miR-375/motile cells transfected with pre-neg). All experiments
were independently repeated in triplicate.
Luciferase reporter assay
To elucidate the molecular mechanisms involved in
the effects of miR-375 on CRC cells, putative miR-375 target genes
were predicted through the gateway miRecords (http://mirecords.biolead.org/). To improve the
accuracy of the prediction, the genes that were predicted by at
least 4 of 11 databases (DIANA-microT, MicroInspector, miRanda,
MirTarget2, miTarget, NBmiRTar, PicTar, PITA, RNA22, RNAhybrid and
TargetScan) were selected as candidate targets. Wild and mutant
putative targets of the Sp1 transcription factor (SP1) 3′-UTR
(Fig. 3A) were cloned into
pmiReport vector (Ambion, Carlsbad, CA, USA). 293T cells
(2×104) were co-transfected with 500 ng of wild or
mutant constructs of the SP1 3′-UTR with pre-miR-375 or pre-neg.
Each sample was cotransfected with 50 ng of pRL-TK plasmid
expressing Renilla luciferase to monitor the transfection
efficiency. A luciferase activity assay was performed 48 h after
transfection with the dual-luciferase reporter assay system. The
relative luciferase activity was normalized to Renilla
luciferase activity.
Western blot analysis
DLD1 and/or HCT8 cells (1×105) were
incubated in 6-well plates for 72 h. Then, cell proteins were
harvested and homogenized with lysis buffer (Tiangen, Shanghai,
China). Proteins from the cells were resolved by 10% SDS-PAGE gel
and transferred to a nitrocellulose membrane (Millipore, Billerica,
MA, USA). The membrane was incubated with primary antibody at 4°C
overnight, then the secondary antibody for 1 h at 37°C and finally
visualized by enhanced chemiluminescence (ECL; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The antibodies used were: SP1
(1:2,000; Abcam); matrix metalloproteinase 2 (MMP2) (1:1,000),
E-cadherin (1:2,000), vimentin (1:500), snail (1:1,000), β-catenin
(1:1,000) [all from Cell Signaling Technology (CST), Beverly, MA,
USA]; β-actin (1:1,000) and GAPDH (1:2,000) (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA).
Immunofluorescence analysis
HCT8 cells (5×104) were seeded into
6-well plates containing 13-mm collagen (Sigma, St. Louis, MO,
USA)-coated coverslips. After 12 h, the cells were transfected with
pre-miR-375 and pre-neg. Then, for another 48 h, the cells on the
coverslips were washed using cooled PBS on ice twice with PBS for 5
min. Primary antibodies were diluted in 1% (w/v) BSA/PBS and
applied to the coverslips and incubated on ice for 1 h. After 3
washes with PBS, coverslips were incubated with cy3-conjugated
secondary antibody diluted in 1% (w/v) BSA/PBS on ice for 30 min.
The cells were washed twice with PBS before coverslips were mounted
onto glass slides using Gelvatol mounting medium. Slides were
viewed using an Olympus IX71 microscope fitted with a U-RFL-T
fluorescent lamp, and images were captured and analyzed using DP
Controller software (Olympus Corporation, Tokyo, Japan).
Expression of miR-375 in metastatic CRC
tissues
Fifteen snap-frozen tumor tissues from 5 patients
who were surgically resected at Shanghai Huashan Hospital in 2008
were obtained. Each group included primary cancer (T), lymphatic
metastasis (N) and liver metastasis tissues (M) from the same
patient. The present study was approved by the Institutional Review
Board of Shanghai Medical College, Fudan University and informed
consent was obtained from all patients. The RNA extraction and
qRT-PCR protocols are referred to as above.
Statistical analysis
The data are expressed as mean ± SD. Student's
t-test was used to compare test samples with the controls. Two-way
analysis of variance was used to compare differences among three or
more experimental groups. Statistical analyses were performed using
SPSS 11.0 software. P<0.05 was considered to indicate a
statistically significant result.
Results
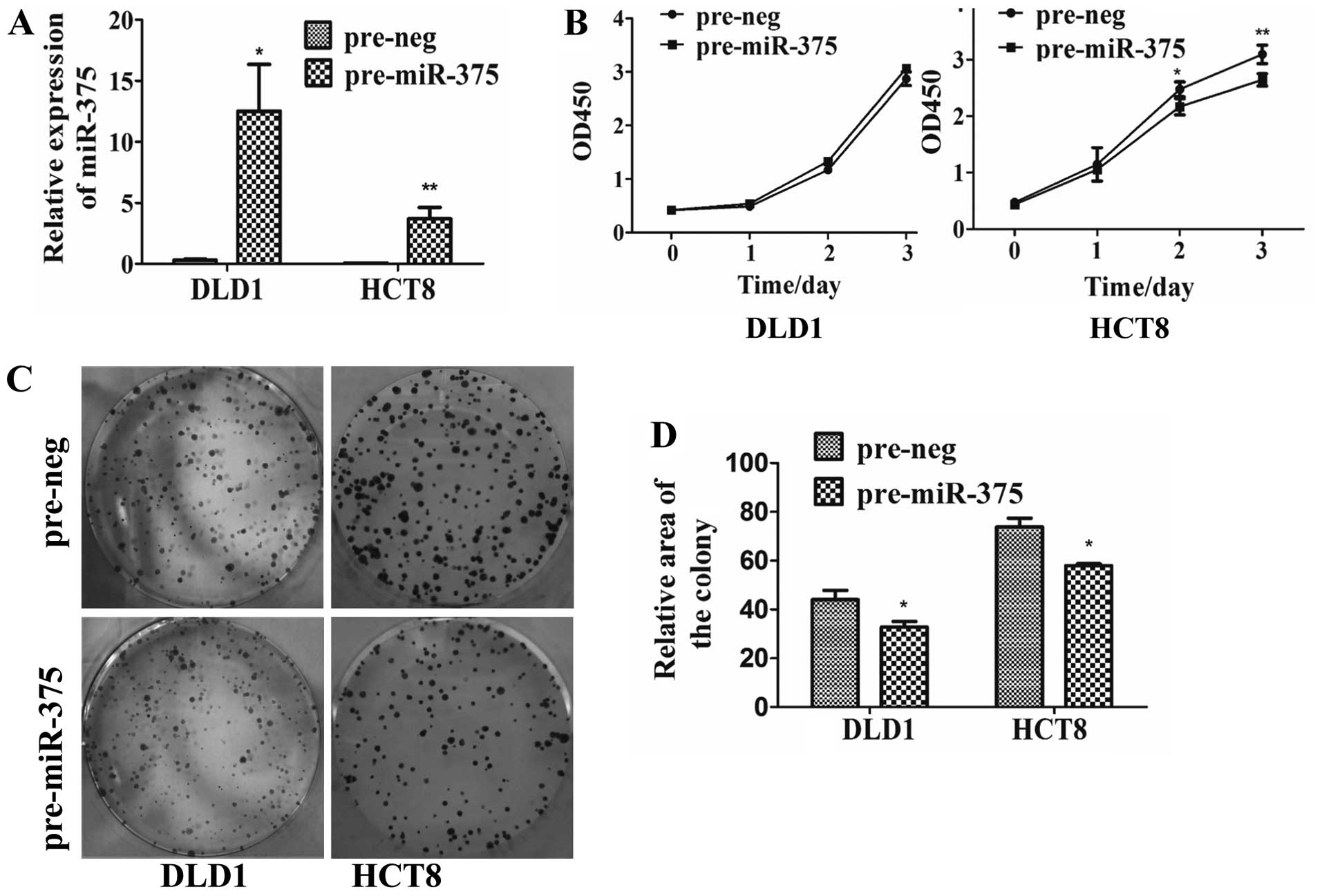
miR-375 inhibits the proliferation of CRC
cells
To further determine the role of miR-375 in CRC
cells, expression of miR-375 was upregulated in DLD1 cells by
40.9-fold (P=0.0040) and in HCT8 cells by 89.2-fold (P=0.0052)
(Fig. 1A) compared with the
negative control. Overexpression of miR-375 repressed the
proliferation of CRC cells as detected by CCK-8 and colony
formation assays. CRC HCT8 cells transfected with pre-miR-375 were
observed to grow more slowly by 18.8% (P=0.0096) on day 3 (Fig. 1B). The cell colony average area in
the DLD1 cells transfected with pre-miR-375 was reduced by 25%
(P=0.0201), and by 21.5% in the HCT8 cells (P=0.0136) (Fig. 1C and D).
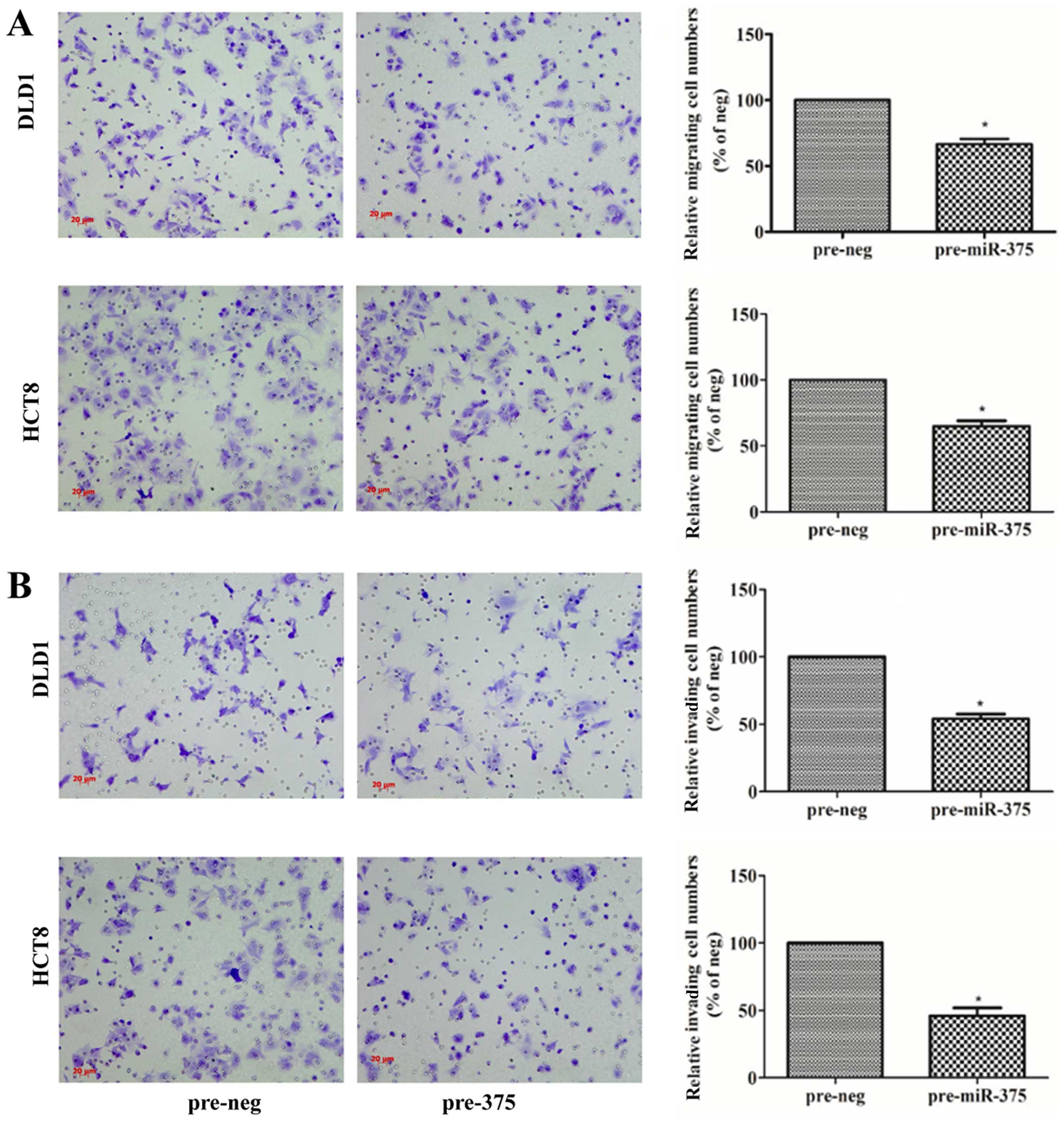
miR-375 inhibits the migration and
invasion of CRC cells
To validate the involvement of miR-375 in
metastasis, migration and invasion assays were performed in the
DLD1 and HCT8 cells. In the migration assay, the numbers of
migrating cells transfected with pre-miR-375 were significantly
reduced, and the migration rate decreased by 33% in the DLD1 cells
(P=0.0138), and 36% in the HCT8 cells (P=0.0140) compared with the
pre-neg groups (Fig. 2A). In the
invasion assay, the numbers of invading cells transfected with
pre-miR-375 were significantly reduced, and the invasion rate
decreased by 46% in the DLD1 cells (P=0.0479), and 52.3% in the
HCT8 cells (P=0.0422) compared with the pre-neg groups (Fig. 2B).
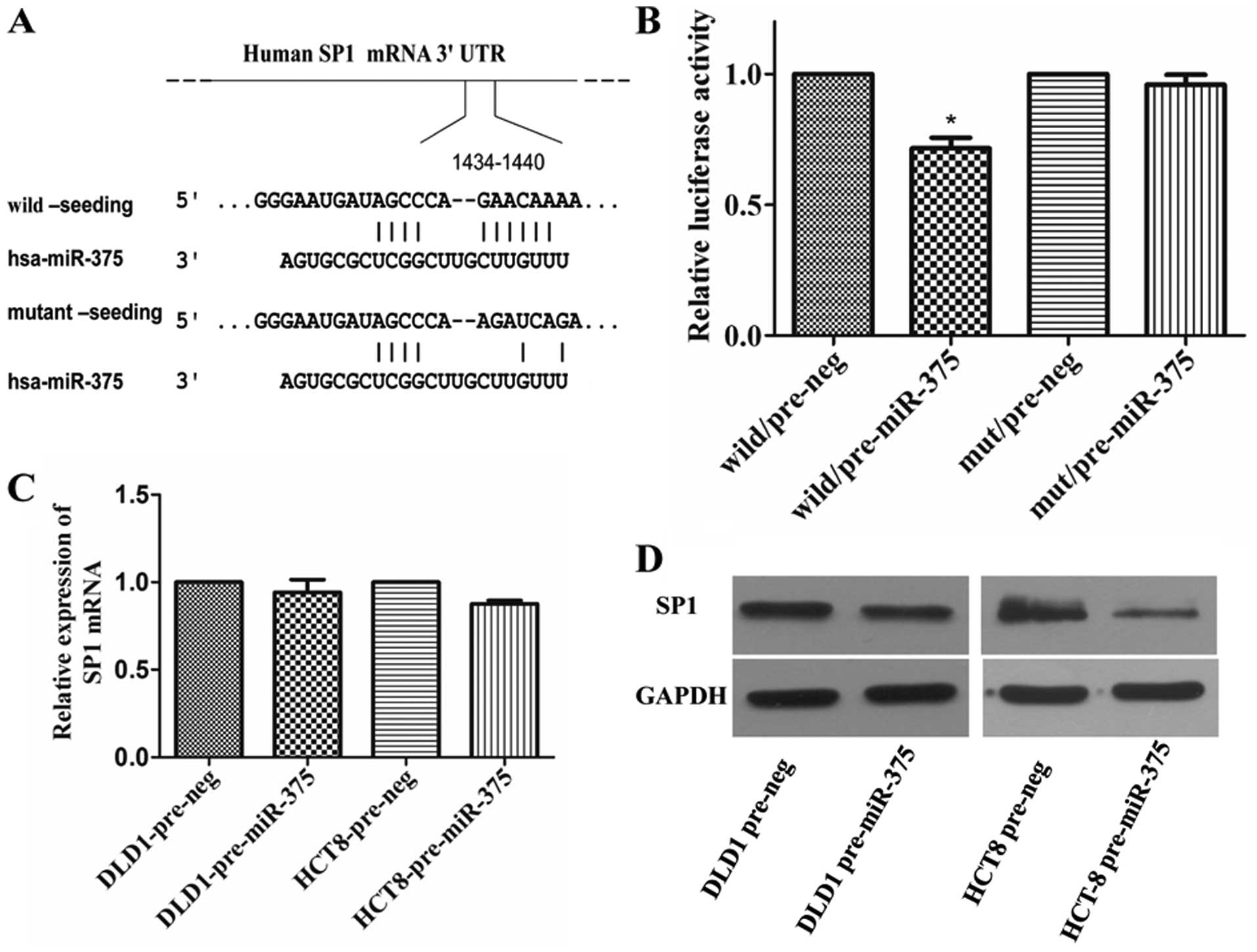
SP1 is identified as a target of
miR-375
SP1 was found to be one of the candidate target
genes, which bears a miR-375 binding sites in its 3′-UTR. To
determine whether miR-375 can regulate the expression of SP1,
luciferase reporter assay was performed with a vector containing
the wild-type (WT) or mutant (MUT) (Fig. 3A) putative SP1 3′-UTR target site
downstream of the luciferase reporter pMIR-REPORT vector. When
pre-miR-375 was co-transfected, the relative luciferase activity of
the reporter containing WT-3′-UTR was significantly suppressed by
28.5% (P=0.0203) compared with that of the reporter containing
NC-3′ UTR. In contrast, the luciferase activity of the reporter
containing MUT-3′-UTR was unaffected by simultaneous transfection
with NC-3′-UTR (Fig. 3B). Then,
western blotting and qRT-PCR results showed that overexpression of
miR-375 decreased the SP1 expression at the protein (Fig. 3D) but not the messenger RNA level
(Fig. 3C).
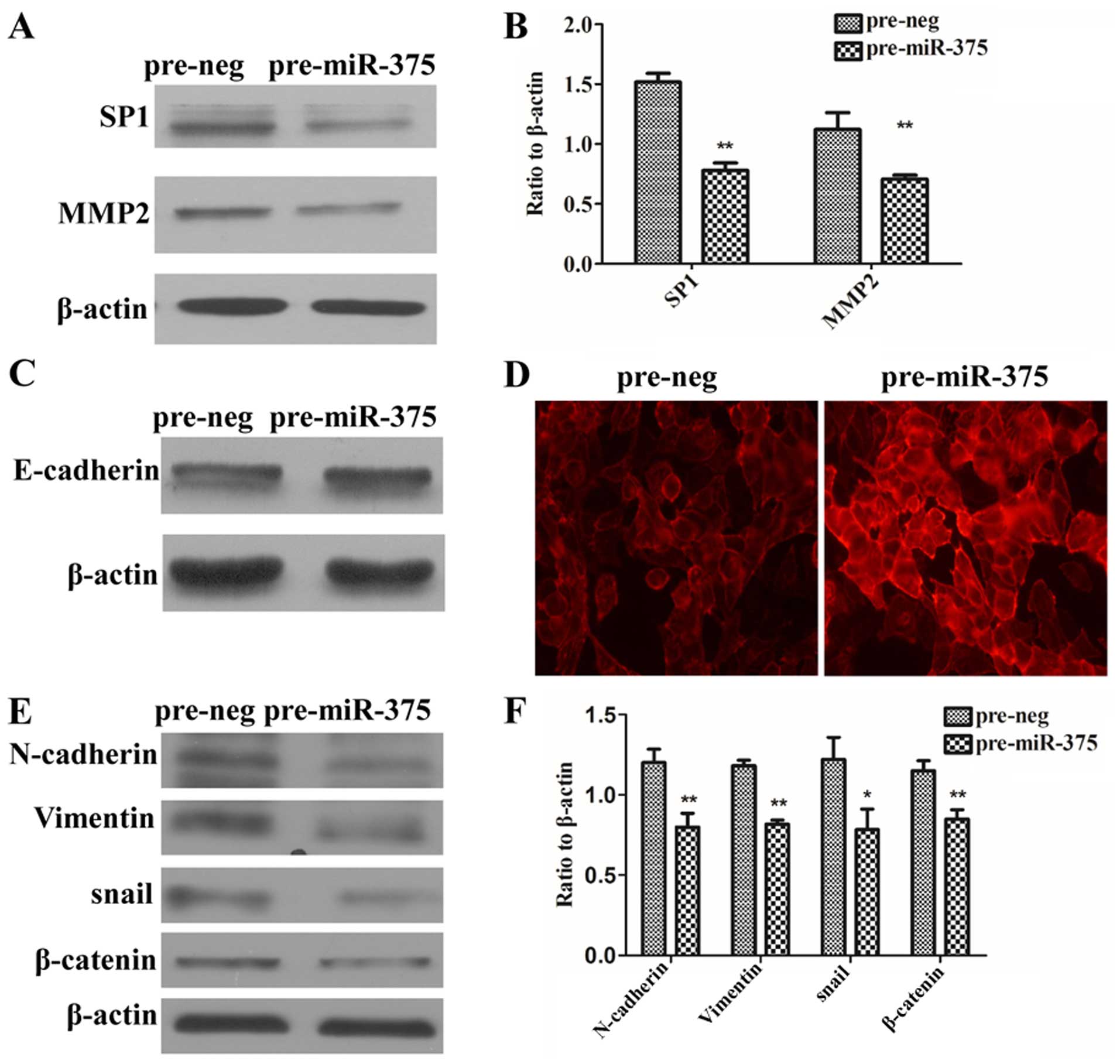
miR-375 is involved in the
epithelial-mesenchymal transition (EMT) of CRC cells via regulation
of SP1
To further explore the molecular mechanisms of how
miR-375 regulates migration and invasion development, we tested
whether miR-375 regulates the expression of MMP-2 and EMT-related
genes through SP1. The data showed that overexpression of miR-375
in the HCT8 cells reduced SP1 and MMP2 protein (Fig. 4A and B). In addition,
immunofluorescence assay showed that upregulation of miR-375
predominantly increased E-cadherin protein on the HCT8 cell
membrane (Fig. 4D), even though the
whole protein of E-cadherin did not increase obviously (Fig. 4C). Upregulation of miR-375 reduced
N-cadherin, vimentin, snail and β-catenin proteins in the HCT8
cells (P<0.05, P<0.01) (Fig. 4E
and F).
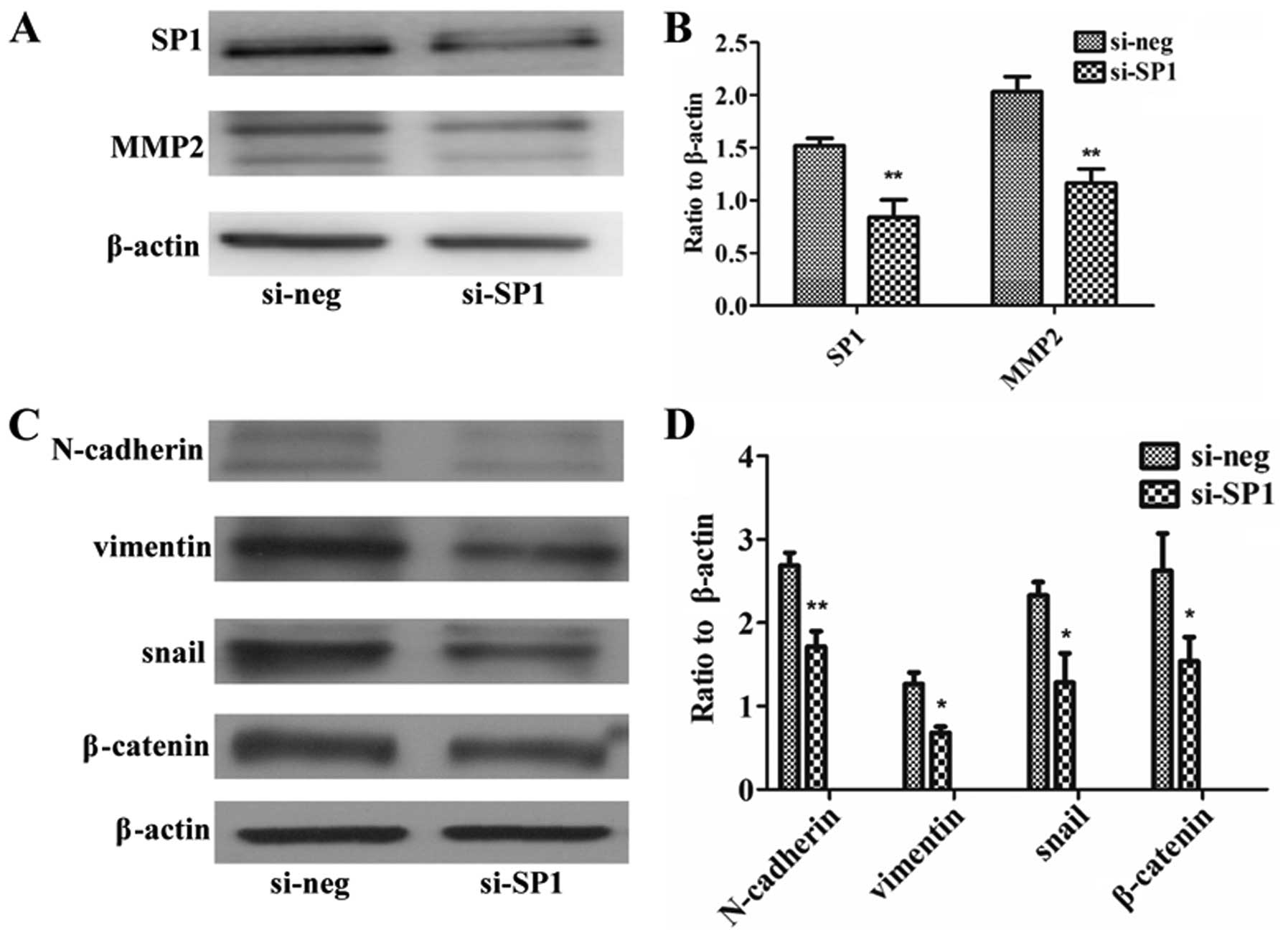
Downregulation of SP1 by siSP1 regulates
MMP2 and EMT-associated genes
To further explore whether SP1 regulates MMP2 and
EMT-associated genes, SP1 was interfered by siSP1 in the HCT8
cells. In addition, the results showed that interference with siSP1
reduced SP1 protein, and at the same time reduced expression of
MMP2 (Fig. 5A and B) and
EMT-associated genes (Fig. 5C and
D) (P<0.05, P<0.01).
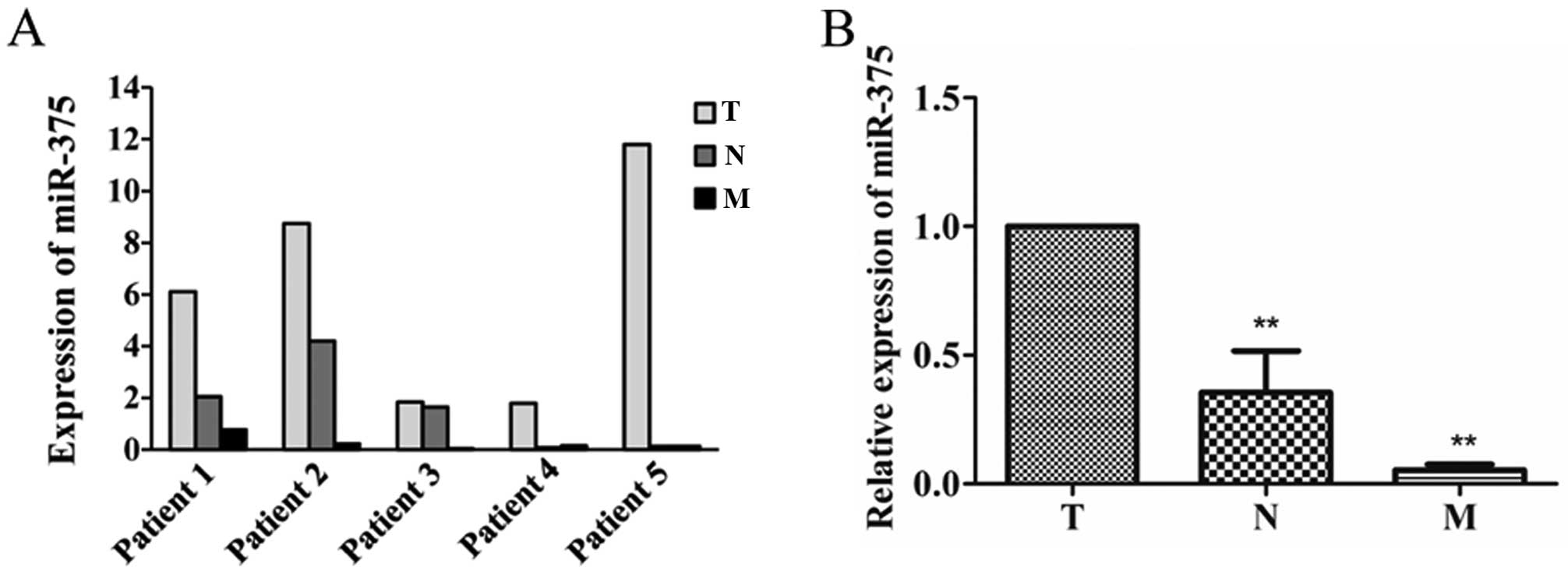
miR-375 is downregulated in metastatic
CRC
Expression of miR-375 in primary cancer (T),
lymphatic metastasis (N) and liver metastasis tissues (M) were
examined. In addition, miR-375 was downregulated in all the 5
lymphatic metastasis and liver metastasis tissues compared with
primary cancer tissues (Fig. 6A).
In addition, miR-375 was significantly downregulated in lymphatic
metastasis by 65% (P=0.0163) and liver metastasis tissues by 94.7%
(P=0.0000) compared with primary cancer (Fig. 6B).
Discussion
MicroRNAs (miRNAs) are considered as potential
specific biomarkers and play important roles in the diagnosis,
progression and prognosis of many types of cancers (16–18).
miR-375 was first identified as a pancreatic islet-specific miRNA
regulating insulin secretion (19,20).
Current studies show that miR-375 is decreased in liver, lung and
gastric cancer, and could participate in the regulation of cancer
development as a tumor-suppressor gene by targeting YAP1 and JAK2
(21,22), but its association with colorectal
cancer (CRC) development was not carefully investigated. Our
previous study showed that miR-375 was weakly expressed in CRC, and
coupled with another two miRNAs could distinguish early invasive
CRC from high-grade intraepithelial neoplasms (13). In the present study, the functions
and underlying mechanism of miR-375 in the regulation of CRC
development were explored. miR-375 was found to inhibit CRC cell
growth to some extent, and particularly suppress CRC cell migration
and invasion ability. One study found that miR-375 could inhibit
CRC cell growth by targeting PIK3CA (15), but there were no data documented
concerning whether miR-375 could regulate CRC metastasis. The
present study firstly found that miR-375 expression was negatively
associated with metastasis, and also could suppress CRC cell
migration and invasion ability. The functional research found that
miR-375 expression in the metastatic CRC samples was less than that
in the primary ones.
The molecular mechanisms underlying the function was
further explored. SP1 was predicted and validated as a direct
target of miR-375. SP1 is a ubiquitous nuclear transcription factor
that regulates gene expression via multiple mechanisms (23), for example, by regulating cell cycle
(24,25), activating MMP2 (26) and promoting EMT process (27), to participate in the development of
cell proliferation and invasion. In the present study, SP1 was
verified as a direct target gene of miR-375. Thus, we predicted
that miR-375 inhibited CRC development via targeting SP1.
In the present study, we found that miR-375 was
significantly downregulated in CRC tissues from lymphoma and liver
metastasis compared with the primary tumors. In addition, MMPs and
the EMT process are important molecular events in cancer metastasis
(28–31). In addition, we found that HCT8 cells
had weak expression of miR-375, but higher invasion ability
compared the DLD1, HCT116, HT29, LS174T cell lines stored in our
laboratory, thus HCT8 was selected for further study of EMT (data
not shown). EMT is a complicated process, with the most obvious
feature: loss of E-cadherin and increased vimentin (32). In addition, genes such as snail,
N-cadherin, β-catenin, are involved in the process (33–36).
Further study was carried out to ascertain whether miR-375 could
regulate MMP2 and EMT-associated genes. The results showed that
overexpression of miR-375 inhibited the expression of SP1, MMP2,
and also vimentin, snail, β-catenin, N-cadherin, but increased
E-cadherin, particularly on the cell membrane. In addition, the
similar results were found when SP1 mRNA was interfered in CRC
cells. Thus, we recognized that miR-375 is a key factor to inhibit
CRC migration and invasion, by targeting SP1 through downregulating
MMP2 and inhibiting the EMT process.
In conclusion, miR-375 inhibited proliferation,
invasion and migration in CRC via directly targeting SP1 through
EMT. In addition, miR-375 could be metastasis predictor and a
treatment target for clinical application.
References
|
1
|
Gryfe R, Swallow C, Bapat B, Redston M,
Gallinger S and Couture J: Molecular biology of colorectal cancer.
Curr Probl Cancer. 21:233–300. 1997. View Article : Google Scholar
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wittekind C and Neid M: Cancer invasion
and metastasis. Oncology. 69(Suppl 1): S14–S16. 2005. View Article : Google Scholar
|
|
4
|
Fidler IJ, Yano S, Zhang RD, Fujimaki T
and Bucana CD: The seed and soil hypothesis: Vascularisation and
brain metastases. Lancet Oncol. 3:53–57. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rudmik LR and Magliocco AM: Molecular
mechanisms of hepatic metastasis in colorectal cancer. J Surg
Oncol. 92:347–359. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takayama T, Miyanishi K, Hayashi T, Sato Y
and Niitsu Y: Colorectal cancer: Genetics of development and
metastasis. J Gastroenterol. 41:185–192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: The potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar
|
|
12
|
Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao
P and Xu H: Expression levels of microRNA-375 in colorectal
carcinoma. Mol Med Rep. 5:1299–1304. 2012.PubMed/NCBI
|
|
13
|
Wang S, Wang L, Bayaxi N, Li J, Verhaegh
W, Janevski A, Varadan V, Ren Y, Merkle D, Meng X, et al: A
microRNA panel to discriminate carcinomas from high-grade
intraepithelial neoplasms in colonoscopy biopsy tissue. Gut.
62:280–289. 2013. View Article : Google Scholar
|
|
14
|
Xu L, Li M, Wang M, Yan D, Feng G and An
G: The expression of microRNA-375 in plasma and tissue is matched
in human colorectal cancer. BMC Cancer. 14:7142014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Tang Q, Li M, Jiang S and Wang X:
MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA.
Biochem Biophys Res Commun. 444:199–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
17
|
Le XF, Merchant O, Bast RC and Calin GA:
The roles of microRNAs in the cancer invasion-metastasis cascade.
Cancer Microenviron. 3:137–147. 2010. View Article : Google Scholar
|
|
18
|
Liu M and Chen H: The role of microRNAs in
colorectal cancer. J Genet Genomics. 37:347–358. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3′-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Poy MN, Hausser J, Trajkovski M, Braun M,
Collins S, Rorsman P, Zavolan M and Stoffel M: miR-375 maintains
normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci
USA. 106:5813–5818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y,
Yao H, Liu X, Ke Y, Si J, et al: MiR-375 frequently downregulated
in gastric cancer inhibits cell proliferation by targeting JAK2.
Cell Res. 20:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishikawa E, Osada H, Okazaki Y, Arima C,
Tomida S, Tatematsu Y, Taguchi A, Shimada Y, Yanagisawa K, Yatabe
Y, et al: miR-375 is activated by ASH1 and inhibits YAP1 in a
lineage-dependent manner in lung cancer. Cancer Res. 71:6165–6173.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Willoughby JA Sr, Sundar SN, Cheung M, Tin
AS, Modiano J and Firestone GL: Artemisinin blocks prostate cancer
growth and cell cycle progression by disrupting Sp1 interactions
with the cyclin-dependent kinase-4 (CDK4) promoter and inhibiting
CDK4 gene expression. J Biol Chem. 284:2203–2213. 2009. View Article : Google Scholar :
|
|
25
|
Wei M, Liu B, Gu Q, Su L, Yu Y and Zhu Z:
Stat6 cooperates with Sp1 in controlling breast cancer cell
proliferation by modulating the expression of
p21Cip1/WAF1 and p27Kip1. Cell Oncol.
36:79–93. 2013. View Article : Google Scholar
|
|
26
|
Chen Y, Huang Y, Huang Y, Xia X, Zhang J,
Zhou Y, Tan Y, He S, Qiang F, Li A, et al: JWA suppresses tumor
angiogenesis via Sp1-activated matrix metalloproteinase-2 and its
prognostic significance in human gastric cancer. Carcinogenesis.
35:442–451. 2014. View Article : Google Scholar
|
|
27
|
Nam EH, Lee Y, Park YK, Lee JW and Kim S:
ZEB2 upregulates integrin α5 expression through cooperation with
Sp1 to induce invasion during epithelial-mesenchymal transition of
human cancer cells. Carcinogenesis. 33:563–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat. 154:8–20. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu QC, Gao RY, Wu W and Qin HL:
Epithelial-mesenchymal transition and its role in the pathogenesis
of colorectal cancer. Asian Pac J Cancer Prev. 14:2689–2698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Said AH, Raufman JP and Xie G: The role of
matrix metalloproteinases in colorectal cancer. Cancers. 6:366–375.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim K, Lu Z and Hay ED: Direct evidence
for a role of beta-catenin/LEF-1 signaling pathway in induction of
EMT. Cell Biol Int. 26:463–476. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakajima S, Doi R, Toyoda E, Tsuji S, Wada
M, Koizumi M, Tulachan SS, Ito D, Kami K, Mori T, et al: N-cadherin
expression and epithelial-mesenchymal transition in pancreatic
carcinoma. Clin Cancer Res. 10:4125–4133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zavadil J, Cermak L, Soto-Nieves N and
Böttinger EP: Integration of TGF-beta/Smad and Jagged1/Notch
signalling in epithelial-to-mesenchymal transition. EMBO J.
23:1155–1165. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Medici D, Hay ED and Olsen BR: Snail and
Slug promote epithelial-mesenchymal transition through
beta-catenin-T-cell factor-4-dependent expression of transforming
growth factor-beta3. Mol Biol Cell. 19:4875–4887. 2008. View Article : Google Scholar : PubMed/NCBI
|