Introduction
It is well known that leukemia, bone marrow or blood
cancer, is one of the most common and aggressive cancers (1), particularly in children and adults
(2). For conducting effective
treatment of this disease, accurate and sensitive diagnoses are
essential. In previous studies, DNA-silver nanocluster (3,4), mass
spectrometry (5) and aptamer-DNA
concatamer-quantum dot probes (6)
were used as a complex to detect leukemia cells. However, such
complexes have serious drawbacks, such as insensitivity and high
expense, and sophisticated instruments and meters are required.
Hence, a simpler strategy with higher sensitivity is demanded to
accurately detect leukemia cells.
Aptamers, special three-dimensional structures, can
be obtained via SELEX (systematic evolution of ligands by
exponential enrichment) in vitro process (7,8). The
simulated single-stranded oligonucleotides can selectively
recognize as well as combine with a wide range of objects, such as
drugs, proteins and even entire cells (9,10). In
comparison with conventional antibodies, aptamers show some
superiority, including the capability of being synthesized easily
with low expense, the ability to be purified to a high level, lack
of immunogenicity and good stability (11–13),
which are helpful to detect cancer cells (14,15).
The Sgc8 aptamer, selected based on the whole cell (16), has been proven to have
characteristics including high binding-affinity and specificity for
human T-acute lymphocytic leukemia (T-ALL) cell lines (17). CCRF-CEM, a T-ALL cell line, was
reported to be used as a target cell, and Romas, a human Burkitt's
lymphoma cell line was reported to be used as a negative
control.
Fluorescent quantum dots (QDs) and nanometer
semiconductors, have been used as photographic developers in the
construction of diagnostic instruments and antitumor nanodrugs
(18–21). They have attracted widespread
attention due to their superior fluorescent features. Unlike
precious metal nanoparticles and traditional organic dyes, QDs
exhibit excellent biocompatibility, low toxicity, resistance to
light bleaching, and allows stable and easy modulation (22). Several studies have confirmed that
QDs are appropriate and efficient for PDT and bio-imaging (23–25).
Furthermore, they can be applied to detect and sense biomarkers,
molecular markers, and cell surface receptors when being linked
with some guiding instruments or targeting molecules (26–28).
In the present study, we developed an innovative
complex based on the Sgc8 aptamer and QDs for the diagnosis of
leukemia. The biotin-modified Sgc8 aptamer was used to identify
CCRF-CEM cells. Then biotin-appended QDs were labeled with the
aptamer via streptavidin and biotin amplification interactions. The
sensitive detection of leukemia cells by the complex (QDs-bsb-apt)
was observed by quantifying the fluorescence signal of the QDs.
Materials and methods
Reagents
Streptavidin was purchased from Promega Corporation
(Madison, WI, USA). Biotin-QD525 was synthesized by Beijing Zhongke
Wu Yuan Biotechnology Co., Ltd. (Beijing, China) and the
biotin-Sgc8 aptamer was synthesized by Shanghai Sangon
Biotechnology (Shanghai, China). The sequence was:
biotin-T10,
5′-ATCTAACTGCTGCGCCGCCGGGAAAATACTGTACGGTTAGA-3′; and biotin-labeled
random DNA library (biotin-Lib) as control biotin-T10,
5′-NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN-3′. Sterile
phosphate-buffered saline (PBS; 10 mM, pH, 7.4) was used as the
buffer. All solutions were prepared with Milli-Q water.
Cells and animals
All cells were provided by the National Center for
International Research of Biological Targeting Diagnosis and
Therapy, Guangxi Medical University (Nanning, Guangxi, China). CEM
and Ramos cells were used as the positive and negative cells,
respectively, for the QDs-bsb-Sgc8 complex. 293T cells were used to
assess the toxicity of the complex. All of the cells were cultured
in 1640 medium, which was supplemented with 1%
penicillin-streptomycin and 10% fetal bovine serum (FBS) (both from
Gibco Co., Grand Island, NY, USA). All of the cell lines were
cultured at 37°C, in a humid environment with 5% CO2.
Nude mice were provided by the Guangxi Laboratory Animal Center
(Guangxi, China), and were raised and cared for following the
Federation of European Laboratory Animal Science Association
guidelines. All protocols were approved by the Animal Ethics
Committee of Guangxi Medical University.
Preparation of the QD-aptamer
complex
Biotin-QD525 was decorated by streptavidin, and then
conjugated with the biotin-modified Sgc8 aptamer. Firstly,
streptavidin and biotin-QDs were mingled at a ratio of 10:1 and
incubated for 40 min at 37°C to obtain streptavidin-QDs. Secondly,
QDs-bsb-apt was synthesized followed by addition of the
biotin-modified Sgc8 aptamers and incubated sequentially at a ratio
of 40:1. Then, the mixture was co-cultured for 40 min at 37°C.
Finally, unconjugated biotin-aptamers were eliminated, and
conjugated QDs-bsb-aptamer complexes were retrieved by centrifugal
filtration at 14,000 rpm for 25 min. Following washing in PBS (10
mM; pH, 7.4), an ultrasonic processor (Scientz-IID; Ningbo Scientz
Biotechnology Co., Ltd., China) was used to disperse the aggregated
conjugates by sonication (25 kHz; amplitude, 15 µm;
sonication time, 3 min) and stored at 4°C.
Flow cytometric analysis
The cultured cells were collected and centrifuged
for 8 min at 1,200 rpm at 4°C, and washed two or three times in
cold PBS. To determine the cell density, a counting plate was used.
Then, 3–4×105 cells were resuspended in 200 µl
PBS. Cell samples were incubated with the QDs-bsb-apt complexes in
cell culture medium or binding buffer (200 µl) containing
FBS [10% (vol/vol)] on ice for 50 min, followed by washing using
washing buffer and suspension in serum (200 µl). The
detection sensitivity of this complex was determined by a flow
cytometer (Epics XL; Beckman Coulter, USA). EXPO32 ADC analysis
software was applied to analyze the data.
Cell imaging
CEM and Ramos cells were cultured in a 6-well cell
culture plate at a density of 3–4×105 cells/ml. After
incubation for 24 h, the cells were then placed on ice for 30 min
to avoid non-specific binding. The cells were washed with cold PBS
three times, and then fixed with 4% polyoxymethylene
(Sigma-Aldrich). After incubation with the complexes (10 nM) in
binding buffer (1 mM CaCl2, 1 mM MgCl2, 1
mg/ml bovine serum albumin and 10% FBS) on ice for 50 min, the
cells were then washed three times. Next, the cells were mounted
onto microscope slides using 4′,6-diamidino-2-phenylindole
dihydrochloride (DAPI) (Life Co., USA) to perform imaging analysis
via a fluorescence microscope (DS-Ri1; Nikon Corporation, Tokyo,
Japan).
MTT assay
Evaluation of cell toxicity of the QDs-bsb-apt was
carried out by the methylthiazol tetrazolium (MTT) assay. The
process involved in the MTT assay is briefly described as follows.
Firstly, 293T cells (2×104 in 100 µl) were seeded
into 96-well plates and cultured overnight in an incubator. Then,
the cells were treated with the complexes (10, 20, 30, 50 and 100
nM) or PBS as a control group for 24 or 48 h. After that, 10
µl of MTT (5 mg/ml) was added to each well and the cells
were incubated for a further 3 h at room temperature in a lucifugal
place. Before adding the ethanol/dimethyl sulfoxide (DMSO) into the
well, the medium in the well was discarded. After 2 min, the
absorbance was tested at λ=570 nm with an ELISA microplate reader
(Thermo Scientific, USA).
Histological analysis
Evaluation of systemic toxicity was carried out by
analysis of the histologic evidence from the heart, liver, kidney,
spleen and lung. Six-week-old nude mice were administered an
intravenous injection of QDs-bsb-apt (15 mg/kg) or PBS in the
control experiments through the tail vein. Then, changes in the
clinical behavior of the treated mice were monitored every day.
Mice were sacrificed after being treated for 20 days, for
histological examination. Tissue samples of the organs, which were
preserved in 10% formaldehyde solution, were dehydrated, following
a routine strategy, and then embedded in paraffin. The paraffin
sections were deparaffinized and immersed in distilled water,
following a routine strategy. Then, hematoxylin and eosin staining
was carried out in the following steps: i) coloration with
hematoxylin solution for 5–15 min; ii) separation of color with
acid or ammonia solution for a few seconds, respectively; iii)
washing with running water for half an hour; iv) dehydration in 70%
and 90% alcohol for 10 min sequentially; v) eosin staining for 2–3
min; and vi) dehydration, clearing and mounting with neutral gums.
The negative control group was implemented using the same steps as
described above. Hematoxylin and eosin solutions were purchased
from Beijing Solarbio Co., China.
Statistical analyses
Student's t-test was applied to make comparisons and
one-way analysis of variance (ANOVA) was applied to compare three
means or more. Data are shown as means ± SD. All statistical
analyses were two-tailed. P≤0.05 was considered to indicate a
statistically significant result. Statistical analyses were
performed using GraphPad Prism software (GraphPad Software, San
Diego, CA, USA).
Results
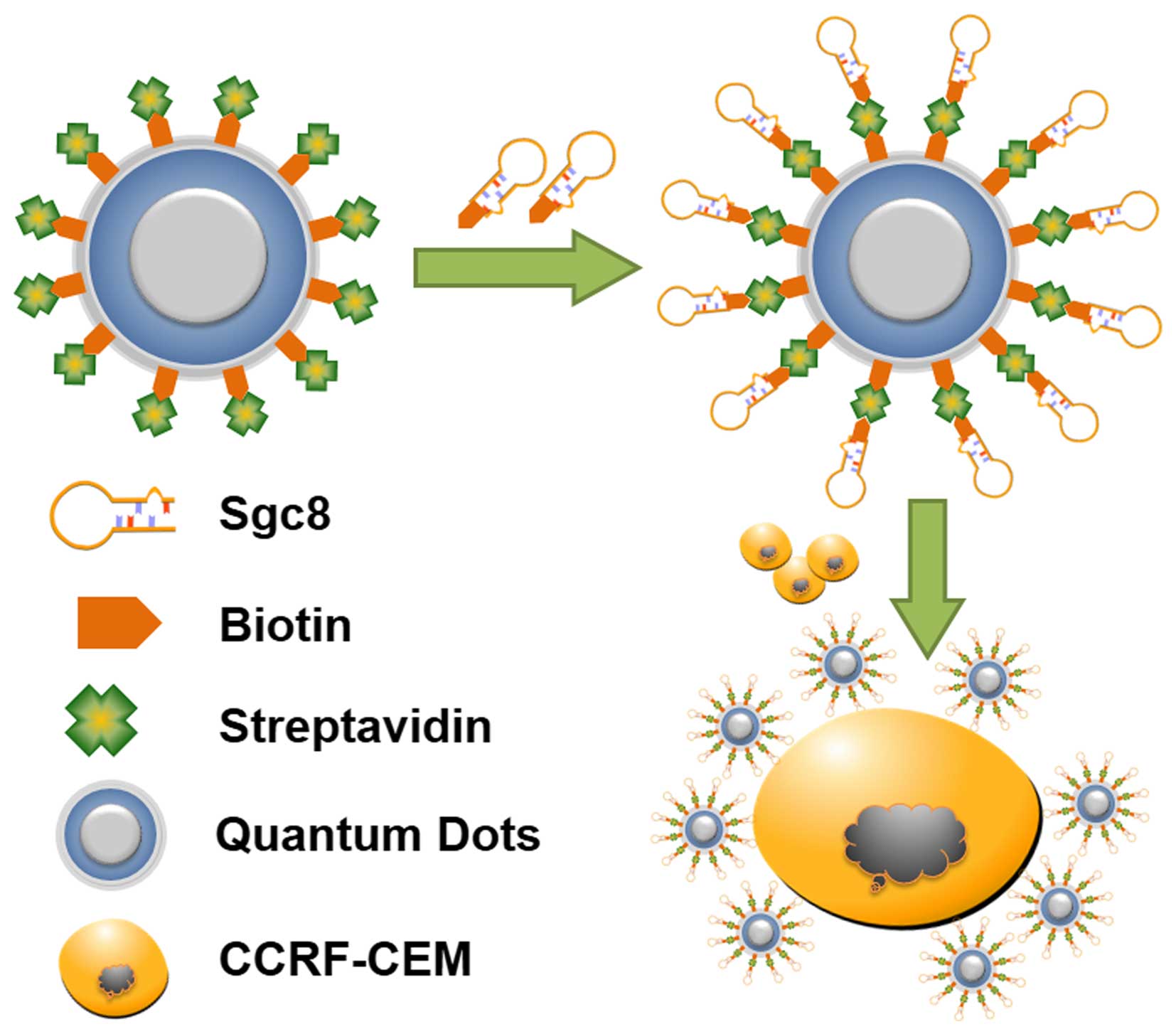
Experimental principle
Recently, QDs and aptamers have been combined and
are widely used for cell imaging and biomedical application. In
most cases, QD-aptamer conjugates are connected directly for cell
recognition, such as biotin-QDs conjugated with a
streptavidin-aptamer (29). Using
such an approach, the aptamers in recognizing target cells may be
baffled by steric hindrance caused by QDs, and the limited
amplification may cause insensitive examination in detection
processing. To avoid these issues, a strategy of linking biotin-QDs
and the biotin-aptamer was proposed in which biotin-QDs were
coupled to streptavidin, and the streptavidin-appended QDs were
then bound to the biotin-aptamer to allow the target cancer cells
to be detected by fluorescence microscopy and flow cytometry
(Fig. 1). CEM cells, a common
leukemia cancer cell line, were treated as target cells. Sgc8, an
aptamer of CEM cells, that has high affinity and specificity, was
selected by cell-SELEX. This method may provide an efficient
approach for tumor cell detection.
Characterization of the QDs-bsb-apt
complex
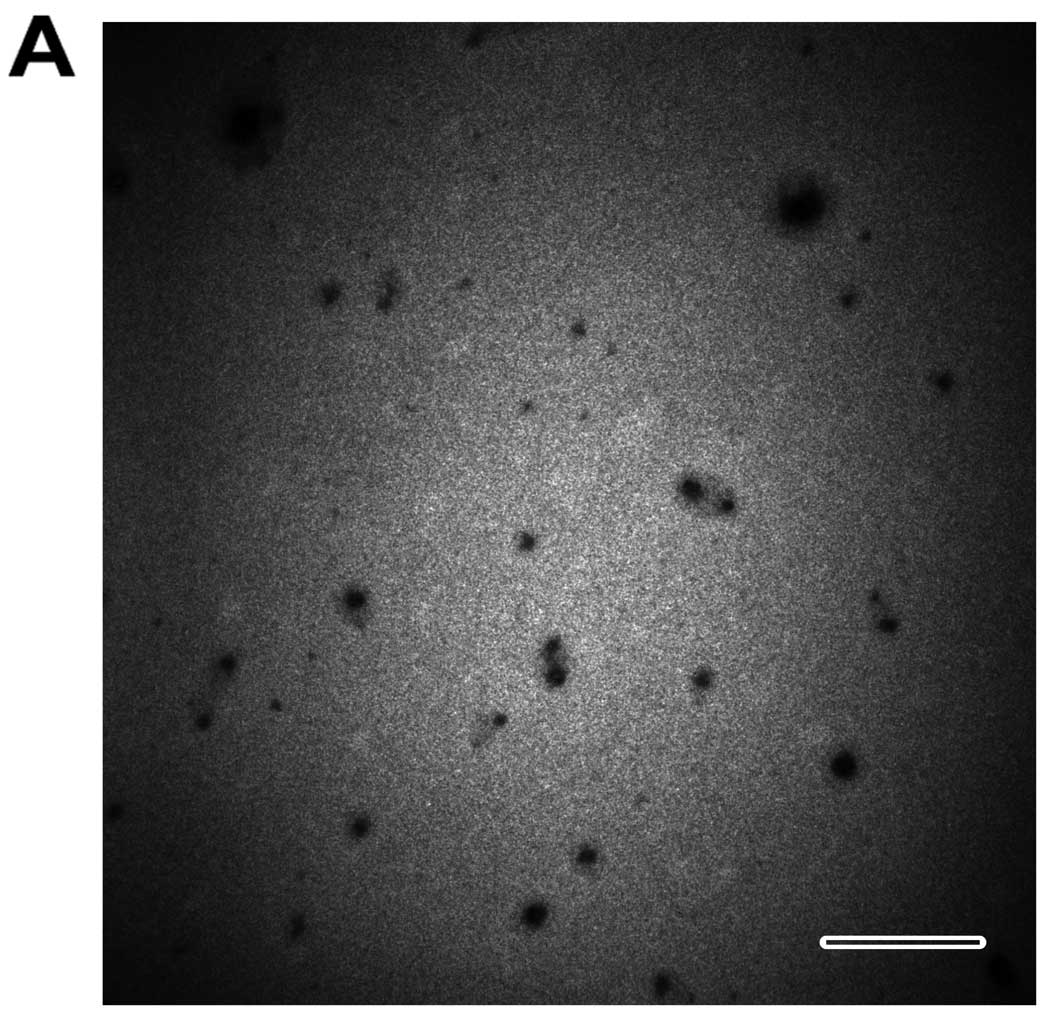
Various characteristics of the prepared QDs-bsb-apt
complex were analyzed via the following methods. Transmission
electron microscopy (TEM; H-7650; Hitachi, Ltd., Tokyo, Japan) was
employed to image the dissemination and the size. It was observed,
via the image, that the QDs-bsb-apt complexes were well
disseminated and the size was increased (Fig. 2A). The hydrodynamic diameter of the
QDs-bsb-apt complexes was tested by dynamic light scattering. It
was 18.92±3.05 nm (Fig. 2C). The
fluorescence emission peak of the QDs with or without the aptamer
was 525 nm and the absorbance peak of the aptamers with or without
QDs was 260 nm (Fig. 2D).
Electrophoretic mobility shift assay was performed using 3% agarose
gel electrophoresis. As shown in Fig.
2B, the band in lane 2 indicates the QDs-bsb-apt complex. Its
size is >500 bp. The band in lane 1 represents the aptamer
alone. Its size is ~50 bp).
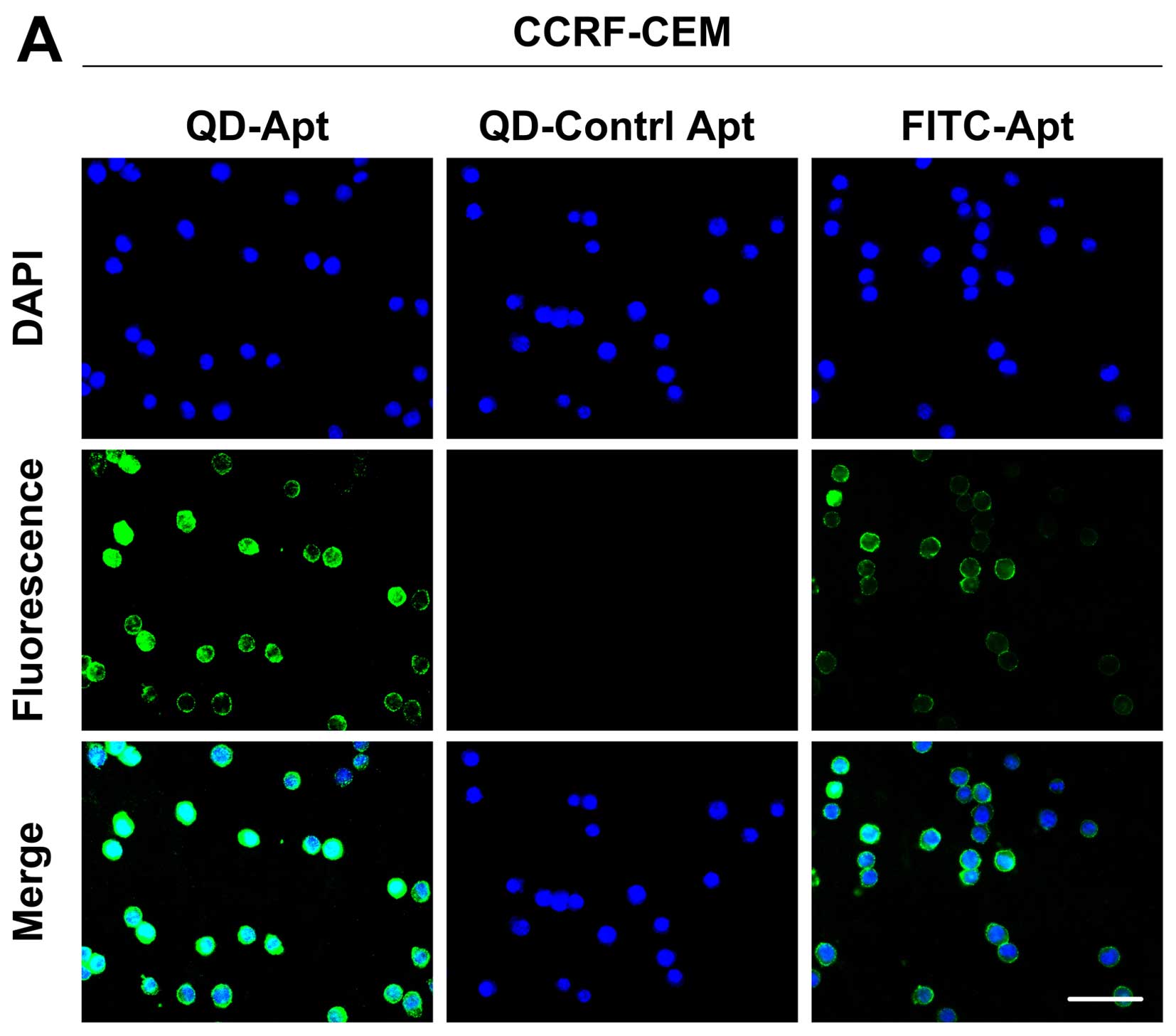
Fluorescence microscopic analysis
demonstrates the high sensitivity of the detection of leukemia
cells using QDs-bsb-apt in buffer
At first, feasibility analysis of the QDs-bsb-apt
strategy was carried out using fluorescence microscopy. Human
Burkitt's lymphoma Romas cells were used as negative control and
the random DNA library (lib) was used as a control sequence. In the
fluorescence images, CEM cells exerted a distinctly stronger
fluorescence intensity on their surface with the presence of
QDs-bsb-Sgc8 than that of FITC-Sgc8, while no fluorescence signal
was observed for the random library (Fig. 3A). When CCRF-CEM cells were replaced
by Ramos cells, no obvious fluorescence signal was observed
(Fig. 3B). This result indicated
that due to the the specific properties of the Sgc8 aptamer,
QDs-bsb-Sgc8 complexes were able to be applied in the detection of
the CEM cells with high sensitivity.
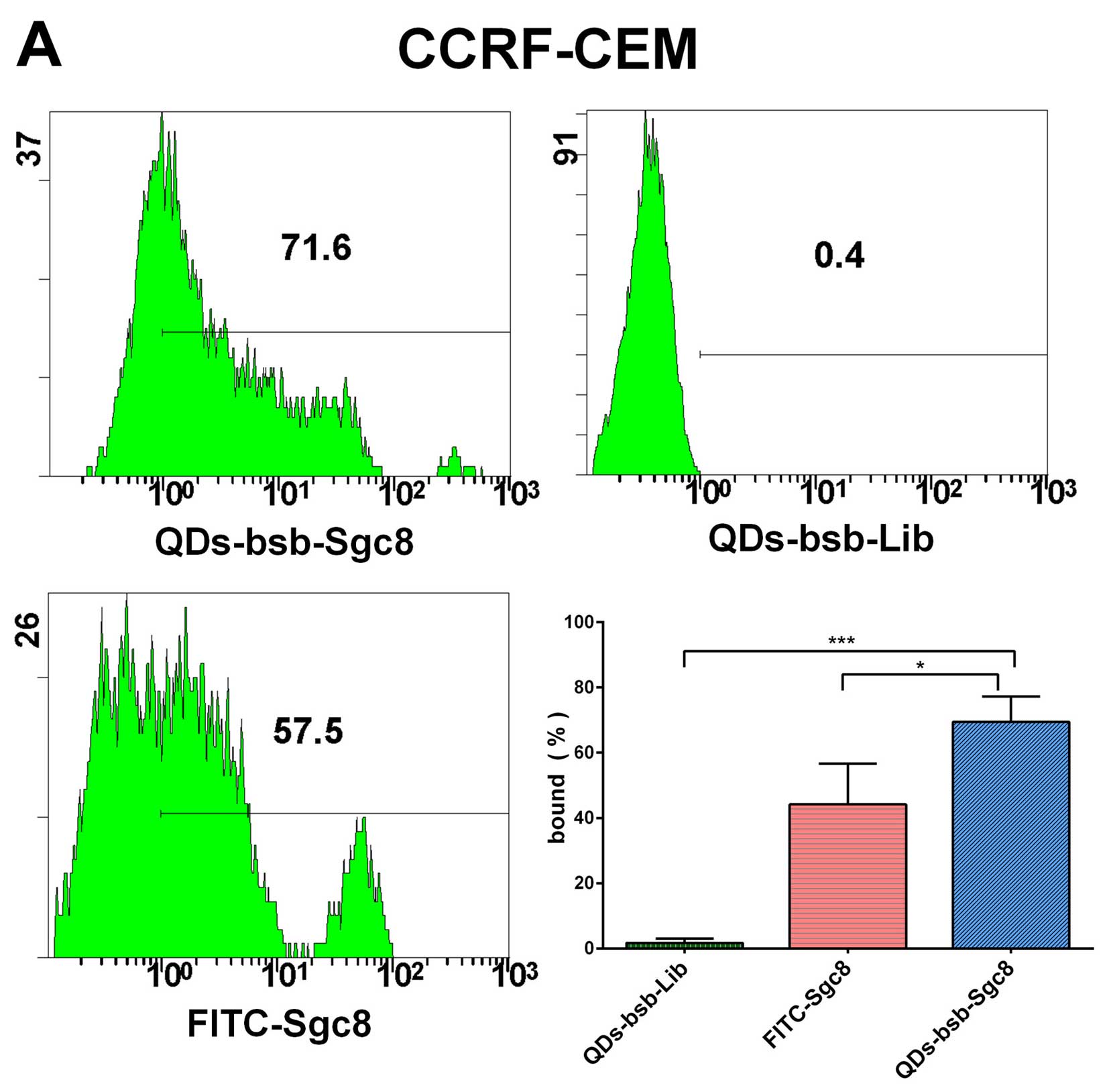
Flow cytometric analysis demonstrates the
high sensitivity of detection of leukemia cells using QDs-bsb-apt
in serum
Flow cytometric analysis confirmed that the positive
CEM cells shown a stronger fluorescent signal property in serum
than the QDs-bsb-Lib, Ramos cells and the FITC-Sgc8 (Fig. 4). CEM cells (71.6%) could be
detected by employing the QDs-bsb-Sgc8 complex, while 57.5% of
cells were detected by FITC-Sgc8. The detection efficiency of
QDs-bsb-Sgc8 was 1.2-fold higher than the traditional organic dye
modified aptamer FITC (Fig. 4).
The QDs-bsb-apt complex exhibits
non-toxicity
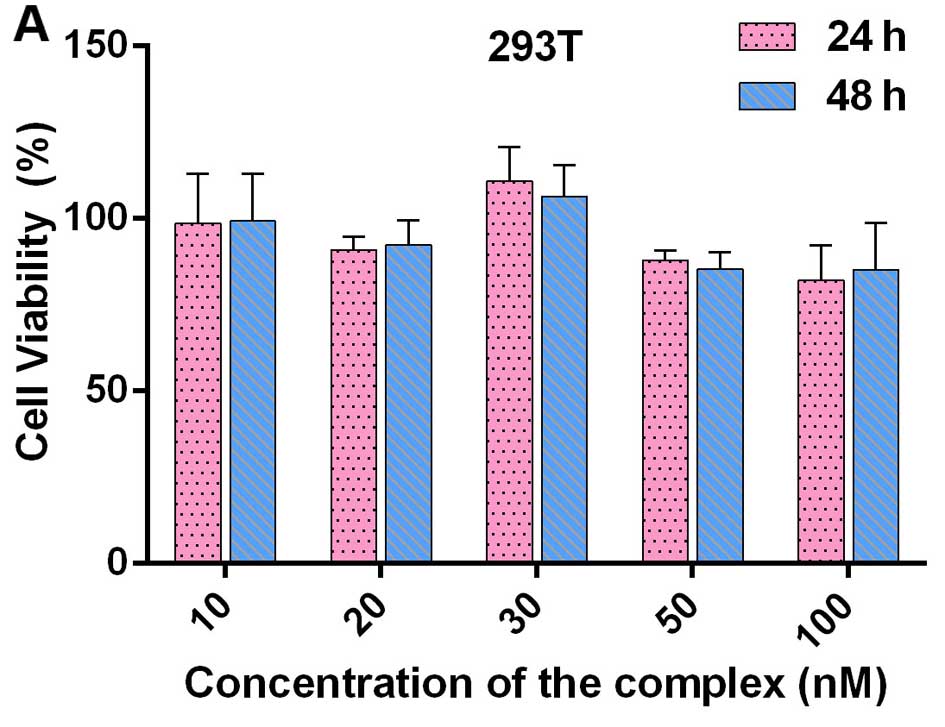
MTT assay demonstrated that various concentrations
of the QDs-bsb-apt complexes were non-toxic to normal cells such as
293T when co-incubated with the QDs-bsb-apt complexes (Fig. 5A). Additionally, after injecting the
QDs-bsb-apt complexes into nude mice, hematoxylin and eosin
staining was conducted. The staining results demonstrated that the
complexes were non-toxic to the entire organism (Fig. 5B).
Discussion
The synthesis of an aptamer with quantum dots (QDs)
in this strategy is a simple and efficient method for generating a
complex to detect tumor cells. Such a complex possesses the
characteristics of high and specific affinity, low immunogenicity,
high photoluminescence quantum yield and a remarkable high
extinction ratio. The synthesis of the Sgc8 aptamer with QDs
creates a hybrid complex possessing properties from both the
aptamer and the QDs. The Sgc8 aptamer provides specific binding to
CEM cells, while the QDs supply a sensitive signal for accurate
detection.
To demonstrate that the QDs-bsb-apt complex was
synthesized successfully, various properties were assessed. TEM
imaging and hydrodynamic diameter detection showed that the
complexes were satisfactory. The increased size and its narrow
distribution suggested that the complexes were uniform and
well-proportioned. Fluorescence emission spectra and absorbance of
the nucleic acid showed that biotin-QDs modified by the
biotin-aptamer preserved the characteristics of the aptamers and
the superior spectral properties of the QDs after modification. To
further determine the formation of the QDs-bsb-apt complex,
electrophoretic mobility shift assay was performed using agarose
gel electrophoresis. QDs-bsb-apt was hardly run on the gel, which
indicated that the complex was synthesized successfully and
unconjugated aptamers were removed.
To illustrate the sensitive detection of leukemia
cells using the QDs-bsb-Sgc8 complex, fluorescence microscopy and
flow cytometric analysis were carried out. The fluorescent images
demonstrated that the CEM cells incubated with QDs-bsb-Sgc8
exhibited stronger fluorescent intensity than that with FITC-Sgc8,
which indicated that the QDs-bsb-apt complex had the ability to
target cancer cells specifically. Furthermore, the efficiency of
detection of the complex was higher than that of the FITC-apt. In
the flow cytometric analysis, we observed some shift in the
fluorescence intensity among the positive and control samples,
suggesting that the sensitivity of this complex was higher. We
presumed that the reason for this was that our method of synthesis
of the QDs-bsb-apt complex facilitated the binding of more aptamers
with QDs to increase the specificity and fluorescence intensity. On
the basis of the above results of the fluorescence imaging and the
flow cytometric analysis, we conclude that the sensitivity of the
QDs-bsb-apt complex was better than that of the FITC-apt
complex.
To confirm the non-toxicity of the QDs-bsb-apt
complex, MTT assay and hematoxylin and eosin staining of
histological sections were executed. The results showed that the
complex was non-toxic to normal cells and to the whole organism,
which suggests that the complex could not only be safely used as a
detector in vitro, but also has the potential to be used
safely in vivo for tumor assessment or treatment. After
referring to the reported literature, it was quite meaningful that
the present study described the toxicity of the QDs-bsb-apt complex
for the first time, which could lay a foundation for further
research on the aptamer-conjugated QD complex.
In the previous study, the Sgc8 aptamer selected by
Shangguan et al (16) was
combined with organic dyes or metal elements. For instance,
aptamer-silver conjugates were used as theragnostic agents for
specific cancer cell therapy and fluorescence-enhanced cell imaging
by Li et al (3). In
addition, aptamer-DNA concatamer-quantum dots probes were used for
selective and ultrasensitive detection of cancer cells by Liu et
al (6), and a DNA-silver
nanocluster fluorescence was applied in cancer cell detection by
Yin et al (4). These
conjugates were applied to capture leukemia cells for detection or
cure. The present study extends the previous research: i) by
demonstrating that QDs conjugated with the Sgc8 aptamer can detect
CEM cells efficiently; ii) by demonstrating that the method of
conjugating biotin-QDs with biotin-Sgc8 aptamer through coupling of
biotin-QDs to streptavidin is more sensitive than the FITC-labeled
aptamer directly; and iii) by proving the non-toxicity of the
QDs-bsb-apt complex at the cell and systemic level.
In summary, an innovative structure based on aptamer
and QDs for leukemia diagnosis was provided. We showed that
QDs-bsb-Sgc8 can recognize CEM cells specifically and sensitively.
This complex exhibits properties of both the aptamer and QDs,
including specificity of targeting and sensitivity to fluoresce.
The connections to chemistry also enable it to be used for tumor
cell imaging in vitro or in vivo, for clinical
application to realize the diagnosis of disease at the early stage
of the development of tumors. Furthermore, future investigations
may focus on constructing a drug transporter combined with such a
complex, which could be used for simultaneously treating and
detecting tumors without toxicity and with high specificity. In
this way, clinicians may be provided with significant and timely
guidance through the detection signal to implement individualized
patient therapy. Our research group will conduct further
research.
Acknowledgments
The present study was supported, in part, by grants
from the National Natural Scientific Foundation of China (nos.
81430055 and 81372452), Programs for Changjiang Scholars and
Innovative Research Team in University (No.IRT_15R13), the
International Cooperation Project of the Ministry of Science and
Technology of China (no. 20 15DFA31320), the Project for Innovative
Research Team in Guangxi Natural Science Foundation
(2015GXNSFFA139001), and the Project of Science and Technology of
Guangxi (nos. 14125008-2-12 and 1599005-2-10).
References
|
1
|
Nelson BP, Treaba D, Goolsby C, Williams
S, Dewald G, Gordon L and Peterson LC: Surface immunoglobulin
positive lymphoblastic leukemia in adults; A genetic spectrum. Leuk
Lymphoma. 47:1352–1359. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karanu FN, Murdoch B, Miyabayashi T, Ohno
M, Koremoto M, Gallacher L, Wu D, Itoh A, Sakano S and Bhatia M:
Human homologues of Delta-1 and Delta-4 function as mitogenic
regulators of primitive human hematopoietic cells. Blood.
97:1960–1967. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Hu H, Zhao Y, Chen X, Li W, Qiang W
and Xu D: Multifunctional aptamer-silver conjugates as theragnostic
agents for specific cancer cell therapy and fluorescence-enhanced
cell imaging. Anal Chem. 87:3736–3745. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin J, He X, Wang K, Xu F, Shangguan J, He
D and Shi H: Label-free and turn-on aptamer strategy for cancer
cells detection based on a DNA-silver nanocluster fluorescence upon
recognition-induced hybridization. Anal Chem. 85:12011–12019. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiong L, Zhang J, Yuan B, Dong X, Jiang X
and Wang Y: Global proteome quantification for discovering
imatinib-induced perturbation of multiple biological pathways in
K562 human chronic myeloid leukemia cells. J Proteome Res.
9:6007–6015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu H, Xu S, He Z, Deng A and Zhu JJ:
Supersandwich cytosensor for selective and ultrasensitive detection
of cancer cells using aptamer-DNA concatamer-quantum dots probes.
Anal Chem. 85:3385–3392. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ellington AD and Szostak JW: In vitro
selection of RNA molecules that bind specific ligands. Nature.
346:818–822. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tuerk C and Gold L: Systematic evolution
of ligands by exponential enrichment: RNA ligands to bacteriophage
T4 DNA polymerase. Science. 249:505–510. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan W, Donovan MJ and Jiang J: Aptamers
from cell-based selection for bioanalytical applications. Chem Rev.
113:2842–2862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen H, Yuan F, Wang S, Xu J, Zhang Y and
Wang L: Aptamer-based sensing for thrombin in red region via
fluorescence resonant energy transfer between
NaYF4:Yb,Er upconversion nanoparticles and gold
nanorods. Biosens Bioelectron. 48:19–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vinkenborg JL, Mayer G and Famulok M:
Aptamer-based affinity labeling of proteins. Angew Chem Int Ed
Engl. 51:9176–9180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao Z and Farokhzad OC:
Aptamer-functionalized nanoparticles for medical applications:
Challenges and opportunities. ACS Nano. 6:3670–3676. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong Q, Wang J, Ahmad KM, Csordas AT, Zhou
J, Nie J, Stewart R, Thomson JA, Rossi JJ and Soh HT: Selection
strategy to generate aptamer pairs that bind to distinct sites on
protein targets. Anal Chem. 84:5365–5371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Famulok M, Hartig JS and Mayer G:
Functional aptamers and aptazymes in biotechnology, diagnostics,
and therapy. Chem Rev. 107:3715–3743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang H, Zhang XB, Lv Y, Gong L, Wang R,
Zhu X, Yang R and Tan W: Functional DNA-containing nanomaterials:
Cellular applications in biosensing, imaging, and targeted therapy.
Acc Chem Res. 47:1891–1901. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shangguan D, Li Y, Tang Z, Cao ZC, Chen
HW, Mallikaratchy P, Sefah K, Yang CJ and Tan W: Aptamers evolved
from live cells as effective molecular probes for cancer study.
Proc Natl Acad Sci USA. 103:11838–11843. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang X and Tan W: Aptamers generated from
cell-SELEX for molecular medicine: A chemical biology approach. Acc
Chem Res. 43:48–57. 2010. View Article : Google Scholar :
|
|
18
|
Zhang M, Liu H, Chen L, Yan M, Ge L, Ge S
and Yu J: A disposable electrochemiluminescence device for
ultrasensitive monitoring of K562 leukemia cells based on aptamers
and ZnO@carbon quantum dots. Biosens Bioelectron. 49:79–85. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang FB, Rong Y, Fang M, Yuan JP, Peng CW,
Liu SP and Li Y: Recognition and capture of metastatic
hepatocellular carcinoma cells using aptamer-conjugated quantum
dots and magnetic particles. Biomaterials. 34:3816–3827. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sheng Z, Hu D, Zhang P, Gong P, Gao D, Liu
S and Cai L: Cation exchange in aptamer-conjugated CdSe
nanoclusters: A novel fluorescence signal amplification for cancer
cell detection. Chem Commun. 48:4202–4204. 2012. View Article : Google Scholar
|
|
21
|
Hu D, Zhang P, Gong P, Lian S, Lu Y, Gao D
and Cai L: A fast synthesis of near-infrared emitting CdTe/CdSe
quantum dots with small hydrodynamic diameter for in vivo imaging
probes. Nanoscale. 3:4724–4732. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Medintz IL, Uyeda HT, Goldman ER and
Mattoussi H: Quantum dot bioconjugates for imaging, labelling and
sensing. Nat Mater. 4:435–446. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee J, Kang HJ, Jang H, Lee YJ, Lee YS,
Ali BA, Al-Khedhairy AA and Kim S: Simultaneous imaging of two
different cancer biomarkers using aptamer-conjugated quantum dots.
Sens Basel. 15:8595–8604. 2015. View Article : Google Scholar
|
|
24
|
Alibolandi M, Abnous K, Ramezani M,
Hosseinkhani H and Hadizadeh F: Synthesis of
AS1411-aptamer-conjugated CdTe quantum dots with high fluorescence
strength for probe labeling tumor cells. J Fluoresc. 24:1519–1529.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mashinchian O, Johari-Ahar M, Ghaemi B,
Rashidi M, Barar J and Omidi Y: Impacts of quantum dots in
molecular detection and bioimaging of cancer. Bioimpacts.
4:149–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xing Y and Rao J: Quantum dot
bioconjugates for in vitro diagnostics & in vivo imaging.
Cancer biomarkers: Section. Dis Markers. 4:307–319. 2008.
|
|
27
|
Savla R, Taratula O, Garbuzenko O and
Minko T: Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin
conjugate for imaging and treatment of cancer. J Control Release.
153:16–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tada H, Higuchi H, Wanatabe TM and Ohuchi
N: In vivo real-time tracking of single quantum dots conjugated
with monoclonal anti-HER2 antibody in tumors of mice. Cancer Res.
67:1138–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu C, Liu J, Zhang P, Li J, Ji H, Yang X
and Wang K: A recognition-before-labeling strategy for sensitive
detection of lung cancer cells with a quantum dot-aptamer complex.
Analyst. 140:6100–6107. 2015. View Article : Google Scholar : PubMed/NCBI
|