Introduction
Breast cancer subtypes have been identified based on
the expression of estrogen receptor (ER), progesterone receptor
(PR) and human epidermal growth factor receptor 2 (HER2). Different
subgroups presented with distinct molecular backgrounds and
exhibited diverse clinical behavior and different treatment
response (1). Estrogen receptor α
(ERα) activation by estrogenic hormones induced breast cancer cell
proliferation of luminal molecular subtype (2). ERα has been widely accepted as a
prognostic marker and a predictor for endocrine therapy response of
breast cancer (3,4). In general, ERα-negative breast cancers
were more aggressive and unresponsive to antiestrogens (5–7). Our
previous studies demonstrated that the absence of ERα expression
associated with aberrant methylation of its CpG island in a
significant fraction of breast cancers (8–10).
Intraductal proliferative lesions of the breast have
traditionally been divided into three categories: usual ductal
hyperplasia (UDH), atypical ductal hyperplasia (ADH) and ductal
carcinoma in situ (DCIS) (11). They were associated with an
increased risk, albeit of greatly different magnitudes, for the
subsequent development of invasive carcinoma (12,13).
The relative risk of subsequent invasive ductal carcinoma of breast
was 1.5–2.0 times for UDH, 4–5 times for ADH (range, 2.4–13) and
8–10 times for DCIS (range, 2.4–13) (11,14–16).
It was suggested that the breast intraductal proliferative lesions
may be direct precursors of invasive ductal carcinoma. Many
questions remain as to the role ERα in breast intraductal
proliferative lesions. Which stage is the watershed between
ERα-positive and -negative breast carcinogenesis needs to be
established. The aim of the present study was to determine the
expression of ERα and to define the ERα methylation in breast
intraductal proliferative lesions.
Materials and methods
Patients and tissue samples
Fresh breast tissue samples were collected from
surgical resection in the Department of Breast Surgery, the First
Affiliated Hospital of China Medical University between June 2007
and December 2014, including pure UDH (N=98), ADH without DCIS
(N=160), DCIS without invasive breast cancer (N=149). None of
patients underwent chemotherapy, radiotherapy or adjuvant treatment
before operation. Patient ages ranged from 21 to 82, with an
average age of 34.5 years. Each case was reviewed independently by
two pathologists with a subspecialty focus in breast pathology, and
only those cases that both pathologists finally reached unanimous
diagnosis were used. In case of insufficient or unattainable
material, original tissue blocks were reprocessed and new slides
were created. All sections were reviewed for a comprehensive list
of pathologic features, including margins (close margins were
defined as tissue-free margins <1 mm), the presence of
concomitant UDH, ADH, DCIS and IDC. Pathology classification was
according to the WHO criteria published by Tavassoli (17–19).
The study was approved by the regional Ethics Committee of China
Medical University.
Immunohistochemical staining
Formalin-fixed, paraffin- embedded specimens were
cut into 4 µm-thick sequential sections. The sections were
dewaxed in xylene and rehydrated stepwise in ethanol. Then the
sections were boiled in citrate buffer (pH 6.0) for 90 sec in an
autoclave. Endogenous peroxidase activity and non-specific binding
were blocked with 3% H2O2 and non-immune
sera, respectively. The sections were then incubated with primary
rabbit anti-human ERα polyclonal antibody F-10 (sc-8002, dilution
1:400; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
Thereafter, the Catalyzed Signal Amplification System (Maixin
Biotechnology, Fuzhou, China) was used for ERα staining according
to the manufacturer's instructions. For the negative control,
phosphate-buffered saline (PBS) was used instead of primary
antibodies. We also adopted the German semi-quantitative scoring
system in combination of the staining intensity and area extent,
which has been widely accepted and used in previous studies
(20–22). Every lesion was given a score
according to the intensity of the nucleic staining (no staining, 0;
weak staining, 1; moderate staining, 2; strong staining, 3) and the
extent of stained cells (0%, 0; 1–10%, 1; 11–50%, 2; 51–80%, 3;
81–100%, 4; negative, 0% area staining; focally positive, 1–80%
area staining; diffusely positive, 81–100% area staining). The
final immunoreactive score was determined by multiplying the
intensity scores with the extent of positivity scores of stained
cells, with the minimum score of 0 and a maximum score of 12
(20,21). Slides were independently examined by
two pathologists as previously mentioned. If there was a
discrepancy in individual scores, both pathologists re-evaluated
them together until a consensus agreement was reached before
combining the individual scores. To obtained statistical results, a
final score ≤1 was considered as negative, while scores ≥2 were
considered as positive.
DNA extraction and bisulphate
modification
Genomic DNA was extracted by solubilization in an
SDS/proteinase K solution, followed by phenol/chloroform extraction
and ethanol precipitation. Sodium bisulphate conversion was done on
2 µg of sample DNA per sample using previously described
methods to convert all unmethylated cytosines to uracils, while
leaving methylcytosines unaltered. Alkali-denatured DNA was
incubated in 3 mol/l NaHSO3 and 0.5 mmol/hydroquinone
for 16 h at 54°C. Modified DNA was purified using the Wizard DNA
Clean-Up System (Promega, Madison, WI, USA) and eluted into 50
µl of sterile water. DNA was precipitated with 0.5 mol/l
ammonium acetate (pH 4.6), 1.5 µl of 20 mg/ml glycogen, and
ethanol and then resuspend in Tris-EDTA.
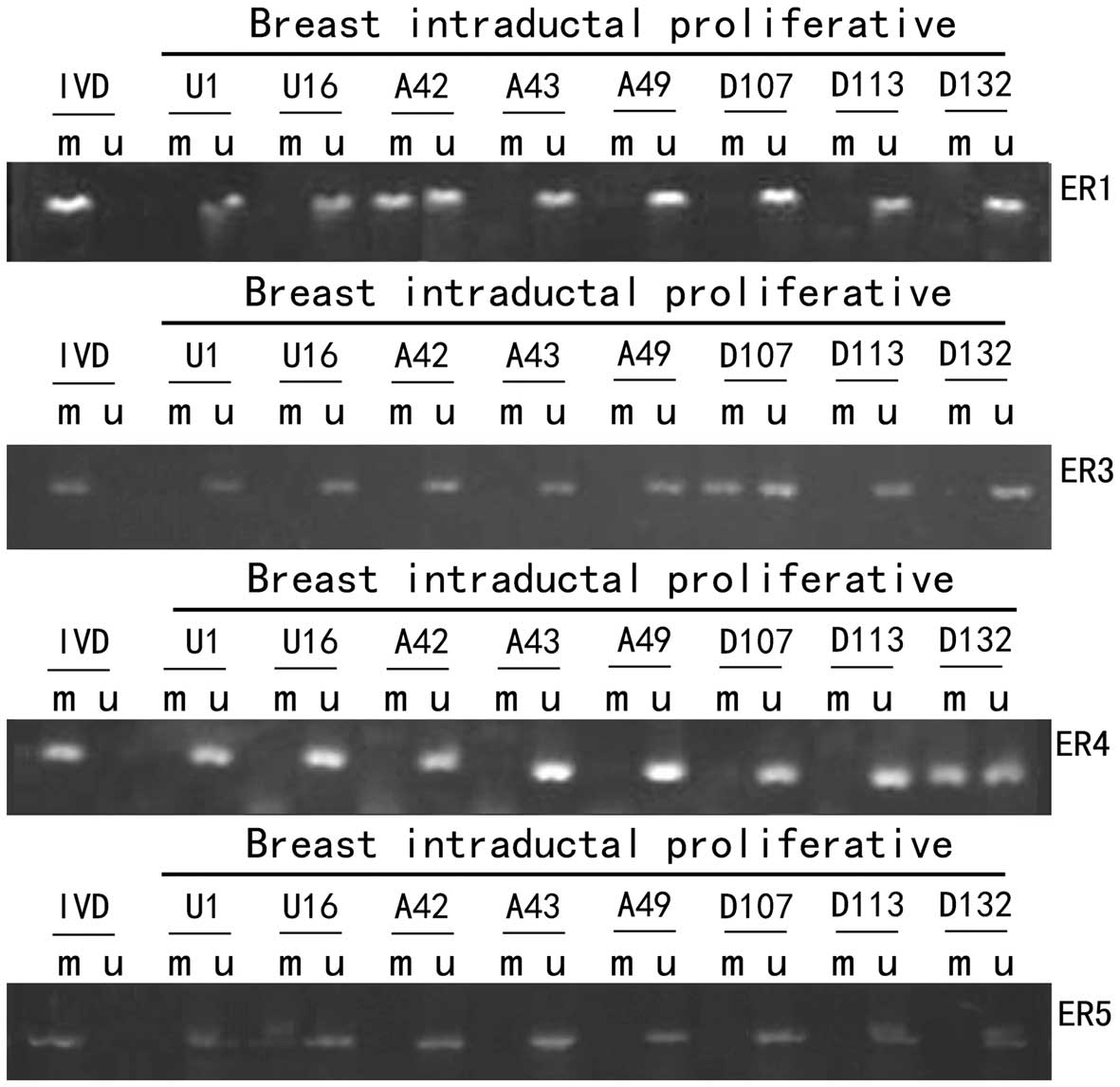
Methylation-specific PCR (MSP) of
ERα
We selected ER1, ER3, ER4 and ER5 for MSP from the
six primer pairs previously described (8,9)
because these covered the most significant methylated loci. The
oligonucleotides of primers were synthesized by Sangon Biotech Co.,
Ltd. (Shanghai, China). PCR was carried out initial denaturation at
95°C for 10 min, followed by 14 cycles of 94°C for 30 sec, 62°C
(ER1) or 59°C (ER3, ER4 and ER5) for 45 sec (−0.5°C decreased/each
cycle), 72°C for 1 min, then followed by 30 cycles of 94°C for 30
sec, 55°C (ER1) or 52°C (ER, ER4 and ER5) for 45 sec, 72°C for 45
sec, ending with a final extension of 72°C for 10 min and a quick
chill to 4°C. The products were subjected to 3% agarose gel
electrophoresis at 100 V for 60 min and visualized by UV light. DNA
from lymphocytes of healthy volunteers treated with SssI
methyltransferase (New England Biolabs, Beverly, MA, USA) and then
subjected to bisulfite modification was used as positive control
for methylated alleles and water was used as negative controls.
Statistical analysis
Statistical analysis was carried out using SPSS
version 13.0 (SPSS, Inc., Chicago, IL, USA). We compared ERα
expression or methylation in breast intraductal proliferative
lesions including UDH, ADH and DCIS using Chi-square tests of
significance. Separate analyses were carried out for percent
staining and the percent-by-intensity product term, using both the
weighted averages as well as maximal values for percent staining.
Differences were considered statistically significant at p<0.05.
The non-parametric correlations of ERα expression with methylation
were analyzed with Spearman's test.
Ethics statement
The study was approved by the regional Ethics
Committee of China Medical University. Subjects were also given
sufficient explanation of the study in writing, and provided with
written informed consent to participate. All patients providing
tissues of intraductal proliferative lesions signed a consent form
prior to breast surgery to allow for this research to be
undertaken.
Results
ERα protein expression in breast
intraductal proliferative lesions including UDH, ADH and DCIS
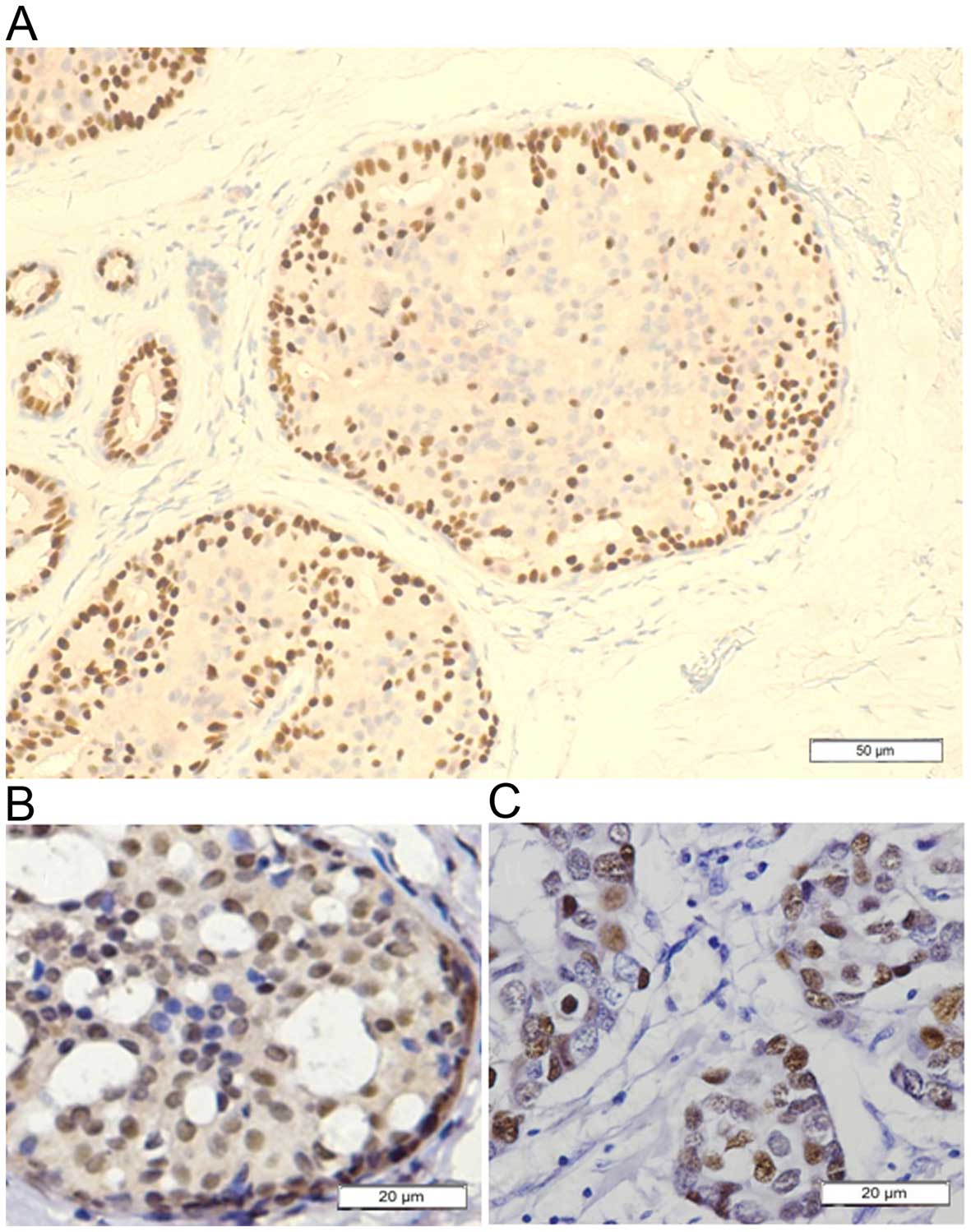
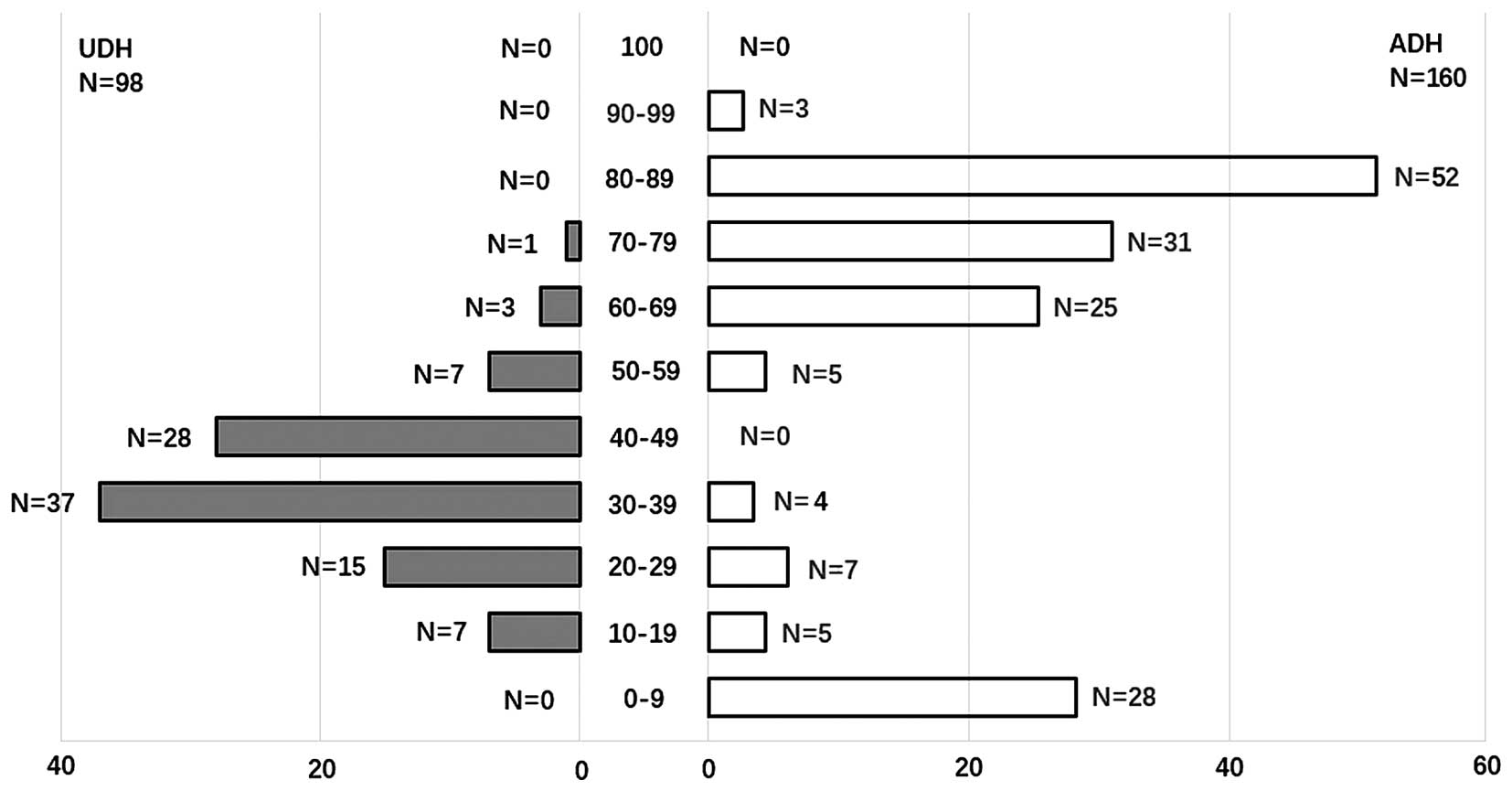
ERα has nuclear staining. UDH were positive for ERα
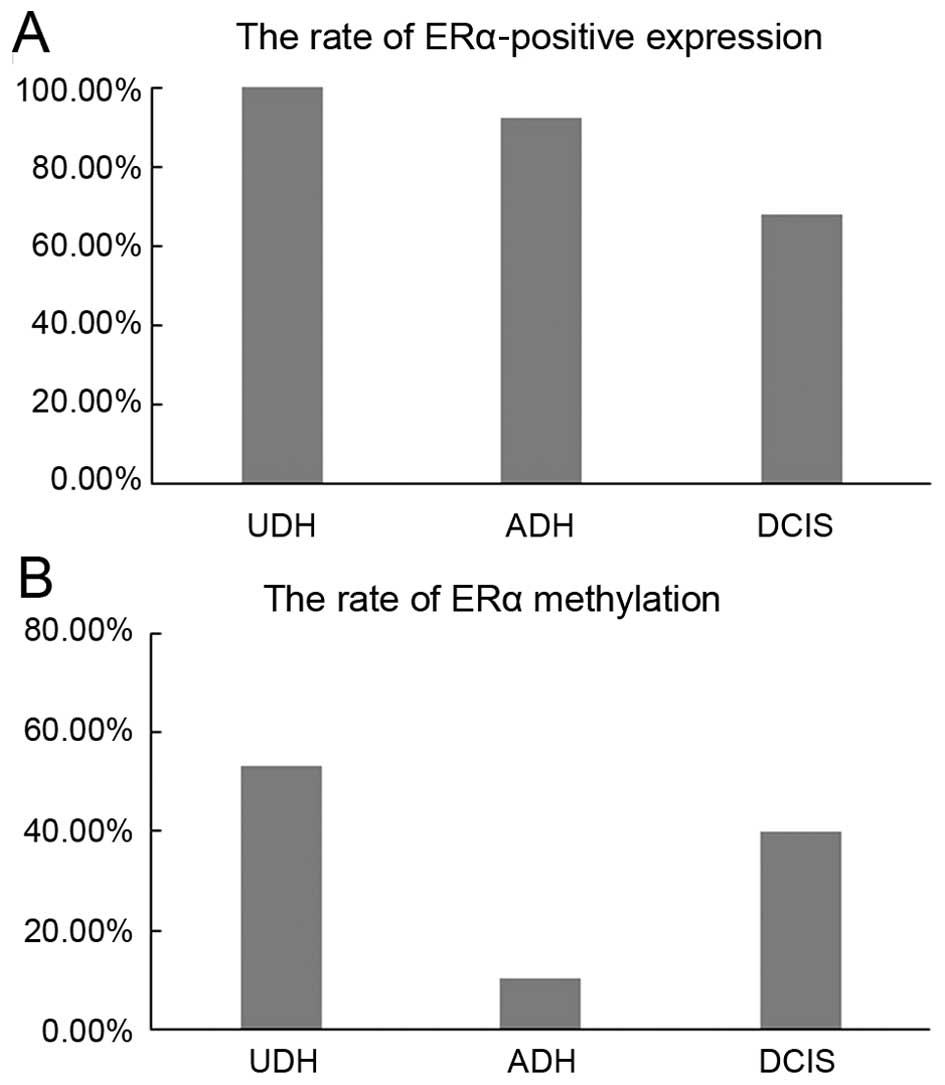
expression in 98/98 (100%) cases. ERα protein expression in ADH
(132/160) (92.5%) was higher than in DCIS (101/149) (67.8%)
(p<0.05, Fig. 1). Although the
positive rate of ERα expression in UDH was higher than in ADH. The
expression pattern of ERα was different with histological diversity
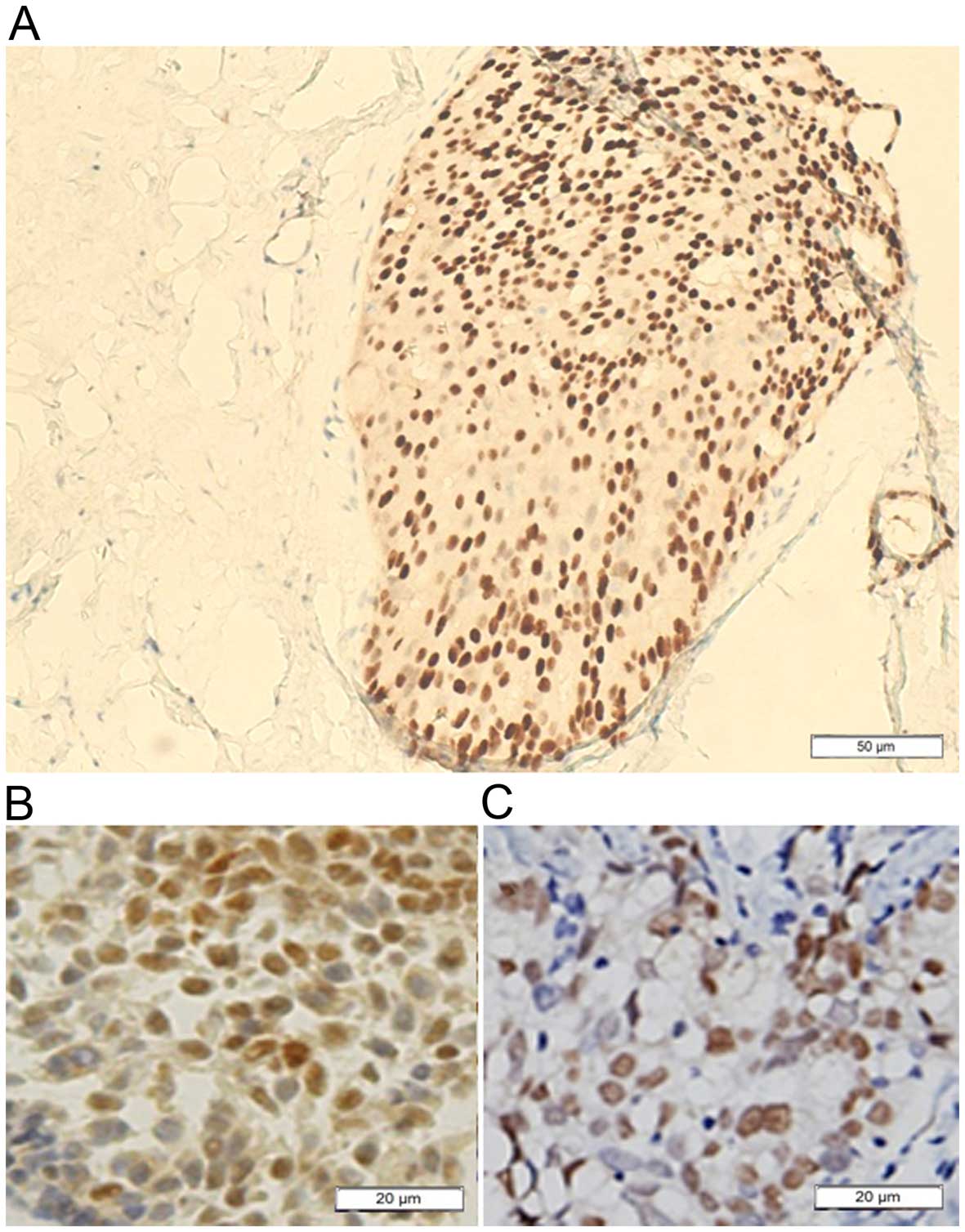
of breast intraductal proliferative lesions. Figs. 2Figure 3–4 show immunohistochemical staining of ERα
in the breast intraductal proliferative lesions. The average
percent of cells staining positive for ERα was 35.33% in UDH,
87.75% in ADH and 71.45% in DCIS. ADH was more likely to show
increased ER percent of cell staining, with mean percent of cells
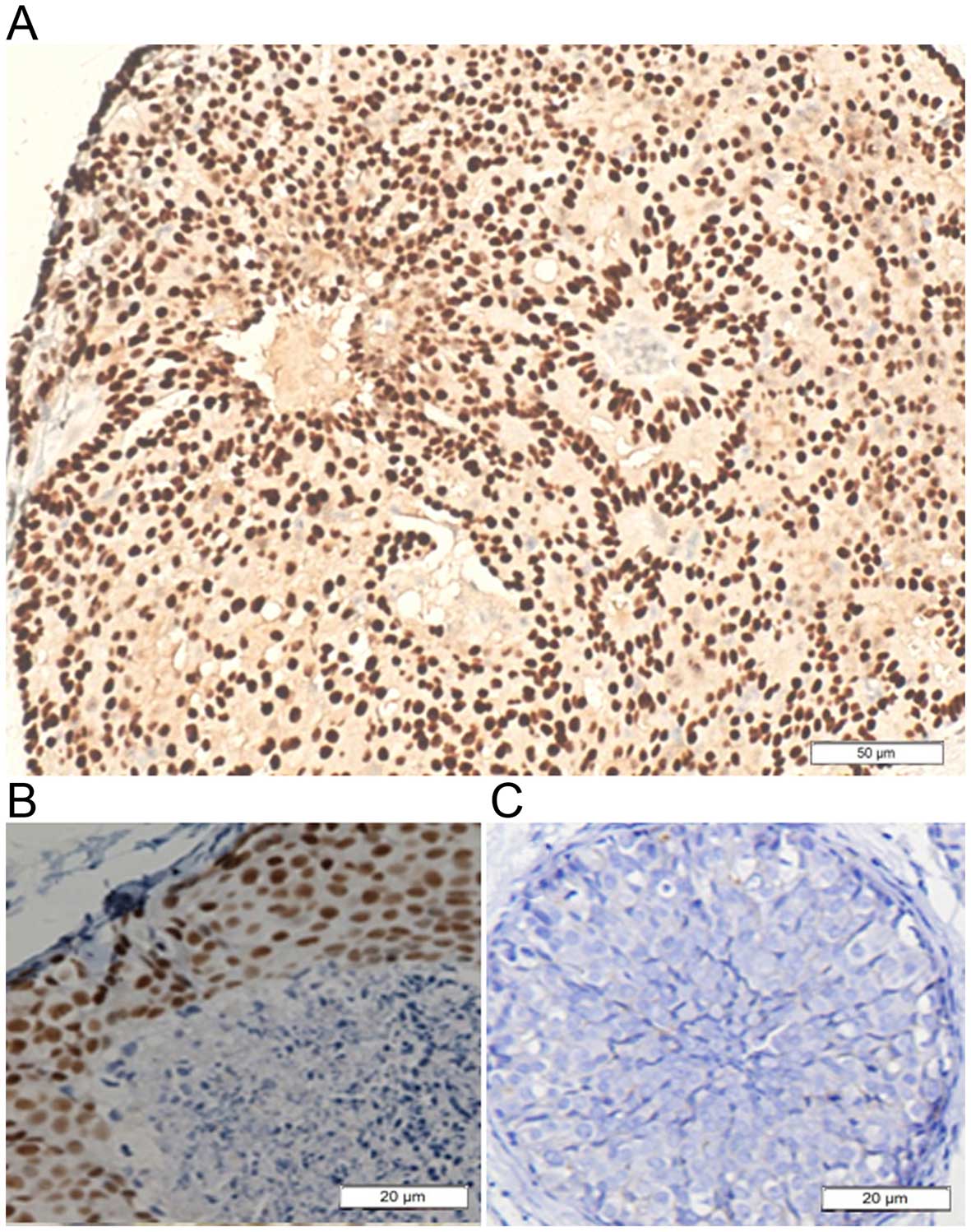
staining of 87.75%. As can be seen in Figs. 5 and 6, 116/160 (72.5%) of the ADH lesions had
ERα staining in over half of the atypical cells. Conversely, only
11/98 (11.2%) of the UDH lesions demonstrated ERα expression in
>50% of the hyperplasia cells. DCIS had ERα staining in over
half of the tumor cells (76/149) (51.0%). In contrast, UDH showed
less ERα staining (Fig. 5). As can
be seen in Fig. 2, the ERα-positive
cells uniformly scattered in the benign proliferative ductal
epithelial cells of UDH. In ADH, the ERα-positive cells clustered
in ductal hyperplasia with thick cell layers which exhibited
positivity of contiguous cells accounting for the majority in the
lesions (Fig. 3). In some cases of
DCIS, all or none ERα-positive cells constructed the morphological
variants (Fig. 4). On the whole,
the stain pattern was variable in DCIS. It exhibits considerable
tumor heterogeneity, and in a given patient can have more than a
single microscopic structural, cytologic, or immunocytochemical
phenotype of ERα.
Methylation status of the ERα promoter in
breast intraductal proliferative lesions including UDH, ADH and
DCIS
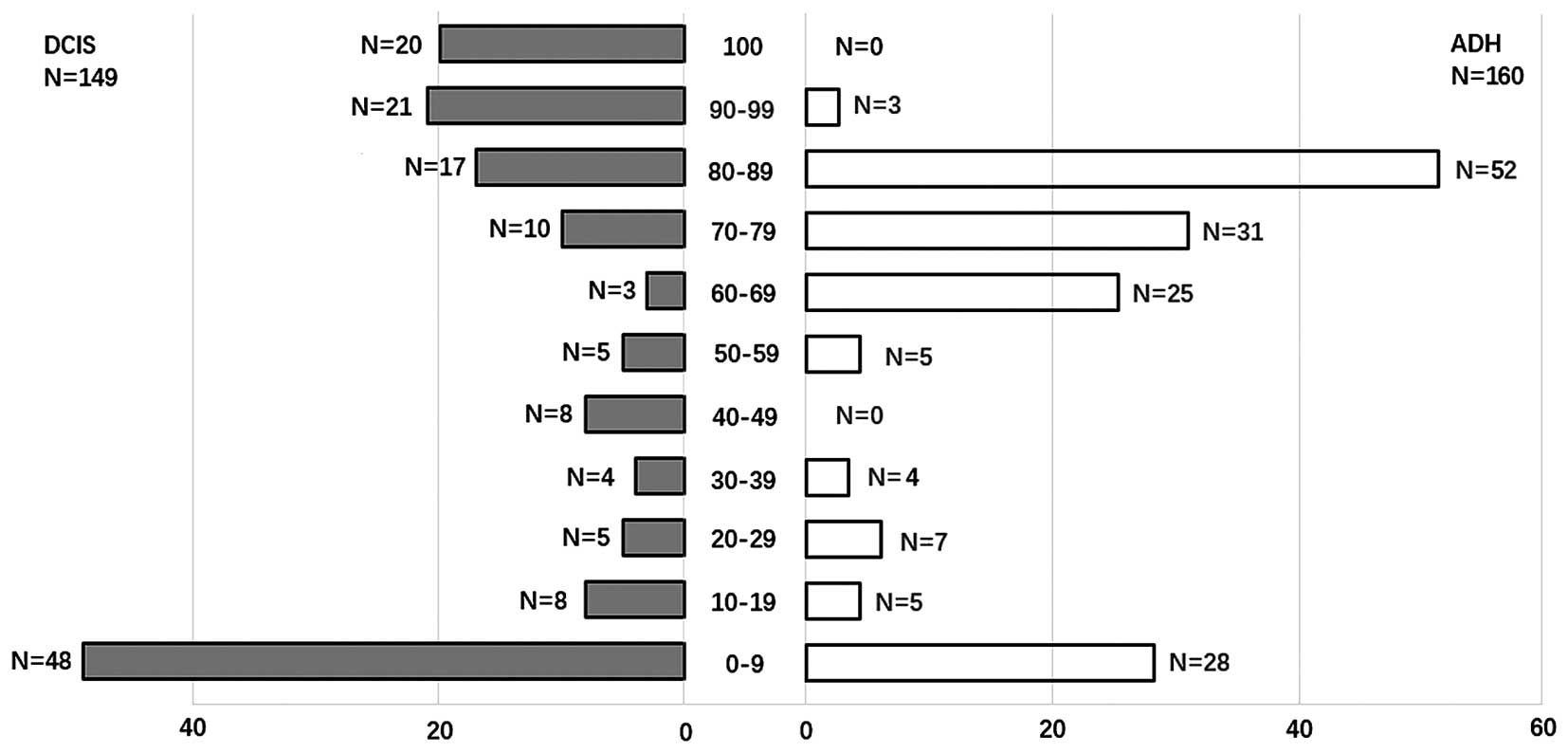
Adequate tissue of UDH, ADH and DCIS for DNA
extraction was available for 248/407 cases. Samples were classified
as methylated if one or more regions were positive for MSP.
Bisulphite-treated DNA was amplified with primers for ER1, ER3, ER4
and ER5 as shown in Fig. 7. We
detected ERα methylation in 32/60 (53.3%) UDH, 11/77 (10.2%) ADH
and 32/80 (40.0%) DCIS (Table I and
Fig. 1). There was significant
difference between the methylation in ADH and in DCIS, in UDH and
in ADH. There was no significant difference between the methylation
in UDH and in DCIS, p>0.05. There was no significant difference
in the overall average percent methylation between the four primers
in the non-invasive lesions.
 | Table IThe methylation and expression of ERα
in UDH, ADH and DCIS. |
Table I
The methylation and expression of ERα
in UDH, ADH and DCIS.
| Groups | ERα methylation
| ERα expression
|
|---|
| Tn | N (%) | Tn | N (%) |
|---|
| UDH | 60 | 32 (53.3) | 98 | 98 (100.0) |
| ADH | 108 | 11 (10.2)a | 160 | 132 (92.5)a |
| DCIS | 80 | 32 (40.0)b | 149 | 101 (67.8)a,b |
Correlation analysis of ERα methylation
and expression
In Spearman's correlation test, ERα methylation and
expression had inverse patterns of alterations in breast
intraductal proliferative lesions including UDH, ADH and DCIS. As
shown in Table II, there is a
strong negative correlation (r=−0.831, p<0.001) between the
percent cells staining positive for ERα expression and ERα
methylation, and a weaker but statistically significant negative
correlation between ERα methylation and expression (r=−0.401,
p<0.001).
 | Table IICorrelation of ERα expression with
ERα methylation in breast intraductal proliferative lesions. |
Table II
Correlation of ERα expression with
ERα methylation in breast intraductal proliferative lesions.
| ERα methylation
| Statistical
values |
|---|
| Methylation | Unmethylation |
|---|
| ERα expression
positive | 42 | 157 | r=−0.401 |
| ERα expression
negative | 33 | 16 | p<0.001 |
Discussion
A history of proliferative breast disease is a
significant risk factor for development of invasive breast cancer.
UDH is considered to represent a benign proliferation of ductal
epithelial cells, and patients with UDH carried only a small
increased risk of developing subsequent breast cancer compared with
patients without proliferative breast disease (23). ADH indicates that there are more
cells lining the duct than would normally be there, and some of
these cells are not typical - they are irregular in shape and size.
Usually, a milk duct is lined with one even layer of uniformly
shaped cells, but in ductal hyperplasia there may be many layers of
cells (24). Patients diagnosed
with ADH, have a risk of developing breast cancer 4–5 times the
average lifetime risk. DCIS is a proliferation of malignant
epithelial cells confined to the ductolobular system of the breast.
It is considered a precursor lesion for invasive breast cancer.
Estrogen plays a major role in promoting normal growth of breast
epithelium and is thought to be important in the pathogenesis of
breast cancer via the uptake into the cell through the mechanism of
the ER (25). The role of ERα in
breast intraductal proliferative lesions is a major dilemma. Which
stage is the watershed between ERα-positive and -negative breast
carcinogenesis needs to be established.
In this study we confirmed that 98/98 (100%) of the
UDH cases were positive for ERα expression. ERα protein expression
in ADH (132/160) (92.5%) was higher than in DCIS (101/149) (67.8%).
But the ERα expression pattern was different with histological
diversity of breast intraductal proliferative lesions. The average
percent of cells staining positive for ERα was 35.33% in UDH,
87.75% in ADH and 71.45% in DCIS. ERα methylation in 32/60 (53.3%)
UDH, 11/77 (10.2%) ADH and 32/80 (40.0%) DCIS. Our results
demonstrated a strong negative correlation between the percent of
cells staining positive for ERα and ERα methylation (r=−0.831,
p<0.001); and weak to moderate negative correlations between ERα
expression and methylation (r=−0.401, p<0.001). Could that mean
that methylation alone is not enough to result in negative ERα
expression and that in ERα-negative cases there are additional
genetic or epigenetic events that result in loss of staining? The
ERα expression detected by immumohistochemistry is to understand
the distribution and localization of ERα within the tissue
examined. The MSP requires only small quantities of DNA, in
analysis of ERα methylation patterns in CpG islands in a small
fraction of cells. So we found strong negative correlation between
the percent of cells staining positive for ERα and ERα methylation.
The downregulation of ERα through promoter methylation was highly
prevalent in breast intraductal proliferative lesions, which is
consistent with previously published reports (8–10). Our
data revealed that the levels of ERα protein expression diminished
with the methylation of ERα. It is possible that the percent of
cells staining positive for ERα can reflect the status of ERα more
accurately.
A few recent studies have examined ERα expression in
ductal neoplasia. In our cohort, the ERα-positive cells uniformly
scattered in the terminal duct lobular units which contains benign
proliferative ductal epithelial cells of UDH. ADH may represent the
first clonal neoplastic expansion of these cells (24). Immunohistochemical study showed that
ERα-positive cells clustered in ductal hyperplasia with thick cell
layers which exhibited positivity of contiguous cells accounting
for the majority in the lesions. It has been reported that ERα is
the primary ER for mammary epithelial cell proliferation and
differentiation (26). Mammary
glands of adult female ERα knock-out mice fail to respond to
ovarian hormones resulting at rudimentary duct stage without
further development (27). In this
study we found the average percent of cells staining positive for
ERα was higher in ADH than in UDH. The upregulation of ERα
expression involved in the conversion of UDH to atypia hyperplasia.
DCIS represented a heterogeneous group of lesions with diverse
malignant potential (28). The
majority of available evidence suggests that ER expression range
from very high to very low levels characteristic (29,30).
The staining pattern was erratic in DCIS in our results which was
consistent with previous published research (31). We thought that the DCIS is a
heterogeneous group of lesions with diverse pattern of ERα
expression resulted from the chaos methylation of ERα. The breast
cancer risk is not consistent with the ERα expression or
methylation. Possibly the ERα expression or methylation were not
the initial factors in breast carcinogenesis. The ERα methylation
is a common epigenetic signaling tool that is used to control ER
expression. One of the most provocative recent observations in
human solid tumors is the discovery of large hypomethylated blocks
in cancer epigenetics (32). DNA
methylation is a reversible signal, similar to other physiological
biochemical modifications (33,34).
The altered DNA methylation in CpG islands in solid tumors
associated with loss of both epigenetic and gene expression
regulation, resulting in hyper-variability of gene expression
(35,36). These changes could even have
interaction with the change of biological behavior of cells which
resulted from the loss of structural integrity of
heterochromatin.
DNA methylation is a reversible biological signal
(34,37). Our new working hypothesis is that
the methylation and expression of ERα shows dynamic variation in
breast carcinogenesis. Our previous study indicated that breast
cancer can convert into other molecular subtypes with the treatment
of neoadjuvant chemotherapy (38).
ERα expression or methylation may be involved in the breast
carcinogenesis and advancement, thus it is not parallel to breast
cancer risk in breast intraductal proliferative lesions. No obvious
watershed exists between ERα-positive and -negative breast
carcinogenesis. ER methylation or expression is a reversible signal
in breast carcinogenesis affecting biological behavior of
cells.
Acknowledgments
This study was supported by the Project of
Scientific Technology and Social Development, Liaoning (no.
2009225008-3) and the National Natural Science Foundation of China
(nos. 81201886 and 81341063). The funders had no role in study
design, data collection and analysis, decision to publish, or
preparation of the manuscript.
References
|
1
|
Liu CY, Hung MH, Wang DS, Chu PY, Su JC,
Teng TH, Huang CT, Chao TT, Wang CY, Shiau CW, et al: Tamoxifen
induces apoptosis through cancerous inhibitor of protein
phosphatase 2A-dependent phospho-Akt inactivation in estrogen
receptor-negative human breast cancer cells. Breast Cancer Res.
16:4312014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miano V, Ferrero G, Reineri S, Caizzi L,
Annaratone L, Ricci L, Cutrupi S, Castellano I, Cordero F and De
Bortoli M: Luminal long non-coding RNAs regulated by estrogen
receptor alpha in a ligand-independent manner show functional roles
in breast cancer. Oncotarget. 7:3201–3216. 2016.
|
|
3
|
Goldhirsch A, Gelber RD and Coates AS:
What are the long-term effects of chemotherapy and hormonal therapy
for early breast cancer? Nat Clin Pract Oncol. 2:440–441. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen JQ and Russo J: ERalpha-negative and
triple negative breast cancer: Molecular features and potential
therapeutic approaches. Biochim Biophys Acta. 1796:162–175.
2009.PubMed/NCBI
|
|
5
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MC, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Giacinti L, Claudio PP, Lopez M and
Giordano A: Epigenetic information and estrogen receptor alpha
expression in breast cancer. Oncologist. 11:1–8. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ottaviano YL, Issa JP, Parl FF, Smith HS,
Baylin SB and Davidson NE: Methylation of the estrogen receptor
gene CpG island marks loss of estrogen receptor expression in human
breast cancer cells. Cancer Res. 54:2552–2555. 1994.PubMed/NCBI
|
|
8
|
Wei J, Han B, Mao XY, Wei MJ, Yao F and
Jin F: Promoter methylation status and expression of estrogen
receptor alpha in familial breast cancer patients. Tumour Biol.
33:413–420. 2012. View Article : Google Scholar
|
|
9
|
Jing MX, Mao XY, Li C, Wei J, Liu C and
Jin F: Estrogen receptor-alpha promoter methylation in sporadic
basal-like breast cancer of Chinese women. Tumour Biol. 32:713–719.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao L, Wang L, Jin F, Ma W, Ren J, Wen X,
He M, Sun M, Tang H and Wei M: Silencing of estrogen receptor alpha
(ERalpha) gene by promoter hypermethylation is a frequent event in
Chinese women with sporadic breast cancer. Breast Cancer Res Treat.
117:253–259. 2009. View Article : Google Scholar
|
|
11
|
Ellis IO: Intraductal proliferative
lesions of the breast: Morphology, associated risk and molecular
biology. Mod Pathol. 23(Suppl 2): S1–S7. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hilton HN, Kantimm S, Graham JD and Clarke
CL: Changed lineage composition is an early event in breast
carcinogenesis. Histol Histopathol. 28:1197–1204. 2013.PubMed/NCBI
|
|
13
|
Beer N, Ali AS, de Savigny D, Al-Mafazy
AW, Ramsan M, Abass AK, Omari RS, Björkman A and Källander K:
System effectiveness of a targeted free mass distribution of long
lasting insecticidal nets in Zanzibar, Tanzania. Malar J.
9:1732010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bane A: Ductal carcinoma in situ: What the
pathologist needs to know and why. Int J Breast Cancer.
2013:9140532013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goodwin A, Parker S, Ghersi D and Wilcken
N: Post-operative radiotherapy for ductal carcinoma in situ of the
breast. Cochrane Database Syst Rev. 11:CD0005632013.PubMed/NCBI
|
|
16
|
Costarelli L, Campagna D, Mauri M and
Fortunato L: Intraductal proliferative lesions of the
breast-terminology and biology matter: Premalignant lesions or
preinvasive cancer. Int J Surg Oncol. 2012:5019042012.
|
|
17
|
Tavassoli FA: Correlation between gene
expression profiling-based molecular and morphologic classification
of breast cancer. Int J Surg Pathol. 18(Suppl): 167S–169S. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tavassoli FA: Lobular and ductal
intraepithelial neoplasia. Pathologe. 29(Suppl 2): 107–111. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tavassoli FA: Breast pathology: Rationale
for adopting the ductal intraepithelial neoplasia (DIN)
classification. Nat Clin Pract Oncol. 2:116–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kok LF, Lee MY, Tyan YS, Wu TS, Cheng YW,
Kung MF, Wang PH and Han CP: Comparing the scoring mechanisms of
p16INK4a immunohistochemistry based on independent nucleic stains
and independent cytoplasmic stains in distinguishing between
endocervical and endometrial adenocarcinomas in a tissue microarray
study. Arch Gynecol Obstet. 281:293–300. 2010. View Article : Google Scholar
|
|
21
|
Koo CL, Kok LF, Lee MY, Wu TS, Cheng YW,
Hsu JD, Ruan A, Chao KC and Han CP: Scoring mechanisms of p16INK4a
immunohistochemistry based on either independent nucleic stain or
mixed cytoplasmic with nucleic expression can significantly signal
to distinguish between endocervical and endometrial adenocarcinomas
in a tissue microarray study. J Transl Med. 7:252009. View Article : Google Scholar
|
|
22
|
Manne U, Myers RB, Moron C, Poczatek RB,
Dillard S, Weiss H, Brown D, Srivastava S and Grizzle WE:
Prognostic significance of Bcl-2 expression and p53 nuclear
accumulation in colorectal adenocarcinoma. Int J Cancer.
74:346–358. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong G, DeVries S, Chew KL, Cha I, Ljung
BM and Waldman FM: Genetic changes in paired atypical and usual
ductal hyperplasia of the breast by comparative genomic
hybridization. Clin Cancer Res. 7:2410–2414. 2001.PubMed/NCBI
|
|
24
|
Di Bonito M, Cantile M, De Cecio R,
Liguori G and Botti G: Prognostic value of molecular markers and
cytogenetic alterations that characterize breast cancer precursor
lesions (Review). Oncol Lett. 6:1181–1183. 2013.PubMed/NCBI
|
|
25
|
Barr FE, Degnim AC, Hartmann LC, Radisky
DC, Boughey JC, Anderson SS, Vierkant RA, Frost MH, Visscher DW and
Reynolds C: Estrogen receptor expression in atypical hyperplasia:
Lack of association with breast cancer. Cancer Prev Res (Phila).
4:435–444. 2011. View Article : Google Scholar
|
|
26
|
Tan H, Zhong Y and Pan Z: Autocrine
regulation of cell proliferation by estrogen receptor-alpha in
estrogen receptor- alpha-positive breast cancer cell lines. BMC
Cancer. 9:312009. View Article : Google Scholar
|
|
27
|
Okolowsky N, Furth PA and Hamel PA:
Oestrogen receptor-alpha regulates non-canonical
Hedgehog-signalling in the mammary gland. Dev Biol. 391:219–229.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sprague BL, Trentham-Dietz A and Burnside
ES: Socioeconomic disparities in the decline in invasive breast
cancer incidence. Breast Cancer Res Treat. 122:873–878. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Allred DC, Wu Y, Mao S, Nagtegaal ID, Lee
S, Perou CM, Mohsin SK, O'Connell P, Tsimelzon A and Medina D:
Ductal carcinoma in situ and the emergence of diversity during
breast cancer evolution. Clin Cancer Res. 14:370–378. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leonard GD and Swain SM: Ductal carcinoma
in situ, complexities and challenges. J Natl Cancer Inst.
96:906–920. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Allred DC, Brown P and Medina D: The
origins of estrogen receptor alpha-positive and estrogen receptor
alpha-negative human breast cancer. Breast Cancer Res. 6:240–245.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hovestadt V, Jones DT, Picelli S, Wang W,
Kool M, PA, Sultan M, Stachurski K, Ryzhova M, Warnatz HJ, et al:
Decoding the regulatory landscape of medulloblastoma using DNA
methylation sequencing. Nature. 510:537–541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ballestar E and Esteller M: Epigenetic
gene regulation in cancer. Adv Genet. 61:247–267. 2008.PubMed/NCBI
|
|
34
|
Ramchandani S, Bhattacharya SK, Cervoni N
and Szyf M: DNA methylation is a reversible biological signal. Proc
Natl Acad Sci USA. 96:6107–6112. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Timp W, Bravo HC, McDonald OG, Goggins M,
Umbricht C, Zeiger M, Feinberg AP and Irizarry RA: Large
hypomethylated blocks as a universal defining epigenetic alteration
in human solid tumors. Genome Med. 6:612014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hansen KD, Timp W, Bravo HC, Sabunciyan S,
Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al:
Increased methylation variation in epigenetic domains across cancer
types. Nat Genet. 43:768–775. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Reik W, Dean W and Walter J: Epigenetic
reprogramming in mammalian development. Science. 293:1089–1093.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lv M, Li B, Li Y, Mao X, Yao F and Jin F:
Predictive role of molecular subtypes in response to neoadjuvant
chemotherapy in breast cancer patients in Northeast China. Asian
Pac J Cancer Prev. 12:2411–2417. 2011.
|