Introduction
Renal cell carcinoma (RCC) is the fifth most common
cancer worldwide, accounting for 2–3% of all malignant diseases in
adults (1,2). Despite the fact that RCC is frequently
diagnosed at a small and early stage, there are approximately one
third of patients with renal cancer presenting metastatic diseases
at the time of diagnosis and 30–40% of patients with localized
renal cancer develop metastasis after surgery, which is the major
clinical challenge in RCC (3–5). RCC
tumorigenesis and tumor progression comprises diverse molecules and
mechanisms. It has been suggested that the directional trafficking
play crucial roles in driving the tumor metastasis (6,7).
CXCR4 chemokine receptor belongs to the group of
seven transmembrane G-protein coupled receptors (GPCR) which was
found in more than 20 types of tumors in human, including prostate,
ovarian and esophageal cancer, melanoma and RCC (8–13).
Growing evidence indicated that high level of CXCR4 was
significantly implicated in tumorigenesis and metastasis in many
malignancies (14,15). Previous studies have shown that
nuclear localized CXCR4 determined prognosis for colon, gastric,
lung and colorectal cancer (16–19).
While in different types of tumors, the prognostic prediction of
nuclear localized CXCR4 was different, even opposite. Nuclear CXCR4
expression was associated with a favorable prognosis in non-small
cell lung cancer (17), but a poor
prognosis in primary colon cancer (20,21).
Our previous study confirmed that high level of CXCR4 was
associated with poor overall and recurrence-free survival in RCC
patients (22), and nuclear
translocated CXCR4 may play functional roles in metastatic RCC
(23). Thus, considerable attention
has been focused on the mechanism of the nuclear translocation of
CXCR4, which may promote tumor growth and metastasis. However,
detailed mechanisms are still to be illustrated.
The surface-to-nucleus signaling pathways in a
cascade (Ras/MAP kinase) or receptor-activated proteins acting
singly (STATs) have been well-recognized. However, the ability of
cytomembranous receptors to directly translocate to the nucleus and
influence on the cellular functions is less investigated. While a
great number of nuclear translocated cytomembranous receptors have
been reported, only a few cases (Notch, APP and ErbB4) have been
shown to change nuclear function convincingly (24–26).
In addition, preliminary evidence showed that CXCR4 may be one of
the candidates.
Almost every plasm-nucleolus shuttled protein harbor
a functional nuclear localization sequence (NLS) or bind to
transport proteins which possess an NLS. Our previous study showed
that CXCR4 contained an NLS located in amino acids 90–170, which
was very long and not precise (23). Furthermore, NLS '146RPRK149', within
CXCR4 presumed by PSORTII (http://psort.nibb.ac.jp/) has been identified to
contribute to nuclear localization in prostate cancer cells
(27). Whether this putative NLS
also modulates nuclear translocation of CXCR4 in RCC remains
unknown. The present study investigated the function of nuclear
localized CXCR4 and its biological mechanisms in RCC cells.
Materials and methods
Cell lines and culture conditions
Human RCC cell lines (A498 and ACHN) were obtained
from the Chinese Academy of Sciences (Shanghai, China). The cells
were incubated in Roswell Park Memorial Institute-1640 medium
containing 10% heat-inactivated fetal bovine serum and antibiotics
(100 µg/ml of penicillin-streptomycin). Cells were grown as
a monolayer on plastic cell culture dishes at 37°C in a humidified
atmosphere containing 5% CO2.
Lentiviral vectors and infection
The lentivirus encoding EGFP, EGFP-CXCR4 or
EGFP-CXCR4MutNLS (combination of R146A, R148A and R149A point
mutations within the NLS) plasmids were packaged and purified at
HanBio Biotechnology (Shanghai, China) and infected cells following
the manufacturer's instructions.
Subcellular fractionation and western
blot analysis
RCC cells were serum-starved for 24 h. Subcellular
fractionations were performed by Nuclear/Cytosol Fractionation kit
(BioVision, San Francisco, CA, USA) following the manufacturer's
instructions. Western blotting was performed as previously
described (23) with anti-human
CXCR4 antibody (1:1,000), anti-topoisomerase I (1:1,000) (both from
Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-CD44
(1:1,000; Cell Signaling, BSN, USA) antibodies.
Point mutation
Combination of R146A, R148A and R149A point
mutations within the NLS of GFP-CXCR4 fusion protein were generated
using the gene splicing by overlap extension-PCR, SOE-PCR;
pEGFPN1-CXCR4 acted as the template (23). The forward and reverse primers
purchased from Sango Biotech (Shanghai, China), were: i) Rm689 F,
TAGA CCACCTTTTCAGCCAACAGCGCCGCTGGCGCCTGACTGTTGGTGGCGTGGACGA and R,
AAAGCTTGCTGGAGTGAAAACTTGAAGAC. The resultant plasmids were
pEGFPN1-CXCR4NLS689. Positive CXCR4 mutant clones were selected
with ampicillin and further purified by Maxiprep (Omega Bio-Tek,
Norcross, GA, USA). Accuracy of the mutations was confirmed by
Sangon Biotech (Shanghai, China).
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded into 96-well culture plates
(5×103 cells/well). At indicated time, 10 µl
CCK-8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) was added to each well and incubated for 2 h at 37°C.
Absorbance values at a wavelength of 450 nm were recorded using a
microplate reader (Varioskan Flash; Thermo Scientific, Waltham, MA,
USA). Viability (%) was calculated based on the optical density
(OD) values, as follows: (OD of time sample − blank)/(OD of control
sample − blank) × 100.
Scratch assay
When cells reached 90% confluency, a scratch was
made through each well using a sterile pipette tip. Cells were
monitored under a microscope (magnification, ×50) for indicated
time after wounding.
Transwell invasion and migration
assays
The invasion and migration abilities were performed
with the filters (Corning, Lowell, MA, USA) and Transwells
(Millipore, Billerica, MA, USA) following the manufacturer's
instructions. In the invasion assay cells were stained with 0.5%
crystal violet, while migrated cells were counted using
4′,6-diamidino-2-phenyl-indole (DAPI).
Immunofluorescence and confocal
microscopy
LV-EGFP-CXCR4 and LV-EGFP-CXCR4MutNLS infected RCC
cells were evaluated by immunofluorescence as previously described
(23). The coverslips were viewed
under a fluorescence microscope (Nikon C1-i; Nikon, Tokyo, Japan)
or a Leica Microsystems SP5 confocal microscope.
Immunoprecipitation
The immunoprecipitation of CXCR4 was performed by
pull-down with CXCR4 antibody from total protein lysates according
to the manufacturer's protocol (Invitrogen, Karlsruhe, Germany).
The CXCR4-pull-down products were subjected to 10% denaturing
polyacrylamide gel electrophoresis and visualized by silver
staining. The single protein band of ~230 kDa was analyzed using
MS/MS spectra and the results were confirmed by western blot
analysis.
RCC tissue
We obtained human RCC surgical resection samples at
the Department of Pathology, Changhai Hospital (Shanghai, China).
The samples originated from one patient with RCC, who provided
written informed consent to use the specimens. The study design was
approved by the Changhai Hospital Ethics Committee.
Immunofluorescent localization
Co-localization of NMMHC-IIA and CXCR4 in RCC tissue
was observed by immunofluorescence (IF). The sections were prepared
following the instructions of the manufacturer, and the slides were
double-stained with the primary antibodies, a mouse antibody
specific to CXCR4 and a rabbit antibody specific to NMMHC-IIA,
respectively.
Statistical analysis
GraphPad Prism version 6.0 (GraphPad, San Diego, CA,
USA) was used for all statistical analyses. Data are presented as
the means ± standard deviation from at least three separate
experiments. The t-test (two-tailed) was used to draw a comparison
between groups, and the significance level was set at
P<0.05.
Results
NLS mutation within CXCR4 inhibits its
nuclear localization in RCC cells
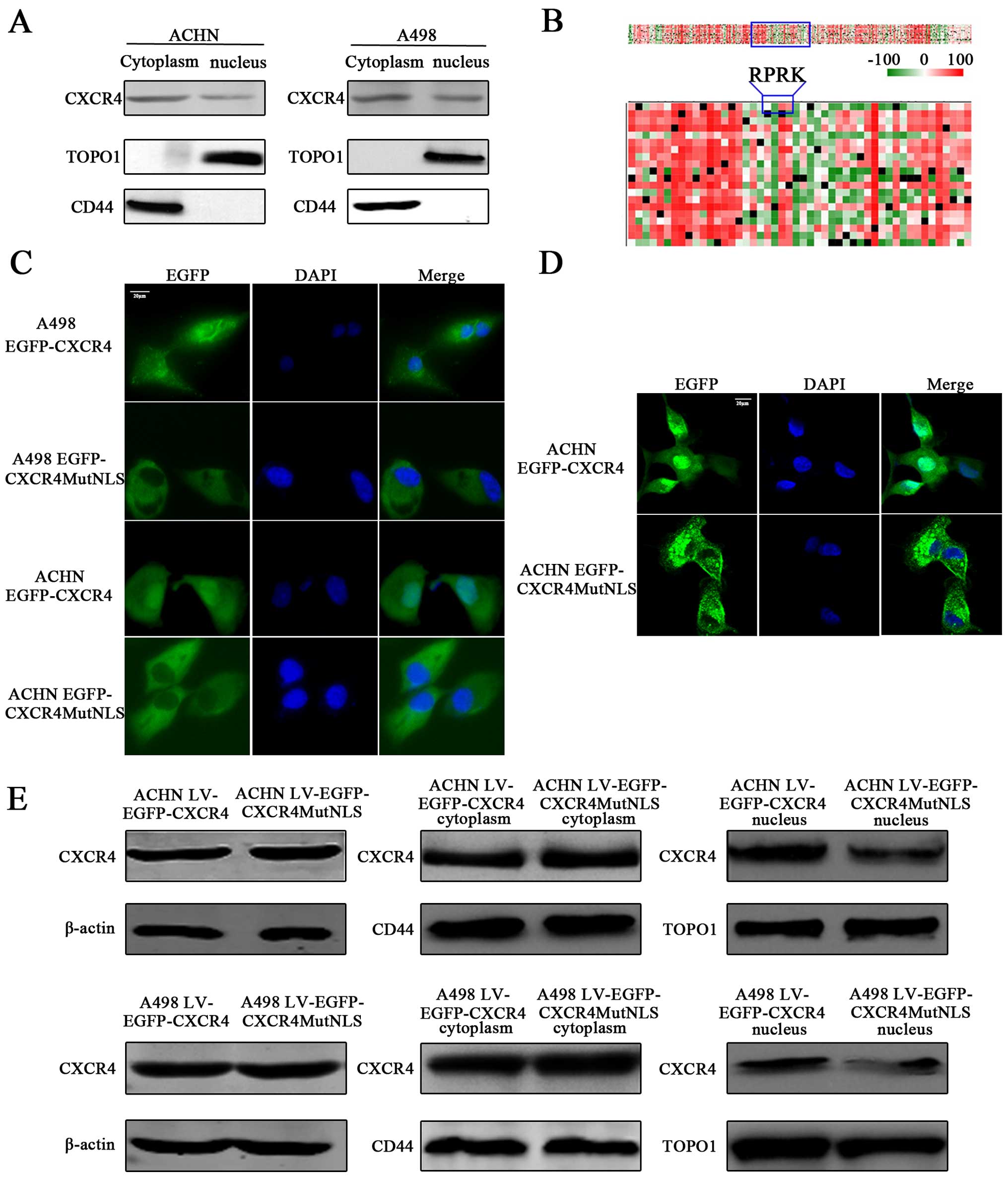
We have previously found that CXCR4 protein was
positive in A498 and ACHN cells lines (data not shown). To confirm
the subcellular localization of CXCR4, RCC cells were fractionated
into nuclear and cytoplasmic fractions, and the purity of
subcellular fractions was confirmed by expression of CD44, a
non-nuclear control (28) and
topoisomerase 1, a nuclear control (29) (Fig.
1A). We found that A498 and ACHN expressed CXCR4 in both the
nuclear and cytoplasmic fractions, which suggested that CXCR4 was
expressed in the cytoplasm, but also within the nucleus of RCC
cells.
Plasm-nucleolus shuttled proteins often contain a
functional NLS or bind to transport proteins possessing an NLS
(30). Our previous study suggested
that the NLS of CXCR4 was located in amino acids 90–170, but it was
not accurate (23). A
bioinformatics analysis using the PSORTII NLS prediction software
revealed a putative NLS, 'RPRK' (27,31)
between amino acids 146–149 within CXCR4 amino acid sequence. To
determine whether the putative NLS, '146RPRK149', was functional
and contributed to nuclear translocation of CXCR4, we evaluated the
subcellular distribution of wild-type EGFP-CXCR4, three mutational
fusion proteins in which arginine 146, 148 and 149 were separately
mutated to an alanine (CXCR4R146A, CXCR4P148A and CXCR4R149A,
respectively), as well as a fusion protein where three arginine
146, 148 and 149 in the NLS were all mutated to alanines
(CXCR4MutNLS) by PredictProtein software (Fig. 1B). Plasmids encoding EGFP-CXCR4 were
transfected into ACHN cells and examined by immunofluorescence
microscopy. Although EGFP-CXCR4 was localized in both cytoplasm and
nucleus (Fig. 1C), CXCR4R146A,
CXCR4R148A and CXCR4R149A were also detectable at the nucleus,
suggesting that R146A, R148A or R149A mutation were insufficient to
inhibit CXCR4 localization to nucleus (data not shown). However, we
detected that CXCR4 diffusely throughout the cytoplasm upon
transfection of EGFP-CXCR4 plasmid, but CXCR4 in the nucleus of
EGFP-CXCR4MutNLS transfected cells was obviously decreased
(Fig. 1C). The confocal microscopy
further precisely confirmed that, compared with EGFP-CXCR4 group,
the nuclear distribution of CXCR4 was obviously less in
EGFP-CXCR4MutNLS transfected ACHN cells (Fig. 1D). Additionally, to confirm that
CXCR4 was decreased in the nucleus, LV-EGFP-CXCR4 or
LV-EGFP-CXCR4MutNLS infected RCC cells were fractionated into
nuclear and cytoplasmic fractions for western blot analysis
(Fig. 1E). Consistent with IF
observations, we found that cells infected with wild-type
LV-EGFP-CXCR4 and LV-EGFP-CXCR4MutNLS were both presented with
cytoplasmic CXCR4 detection, while the nuclear CXCR4 was
significantly reduced in LV-EGFP-CXCR4MutNLS infected RCC cells.
Collectively, these data suggest that the '146RPRK149' may be
involved in localization of CXCR4 to the nucleus in RCC cells and
may help us further investigate the function of nuclear CXCR4.
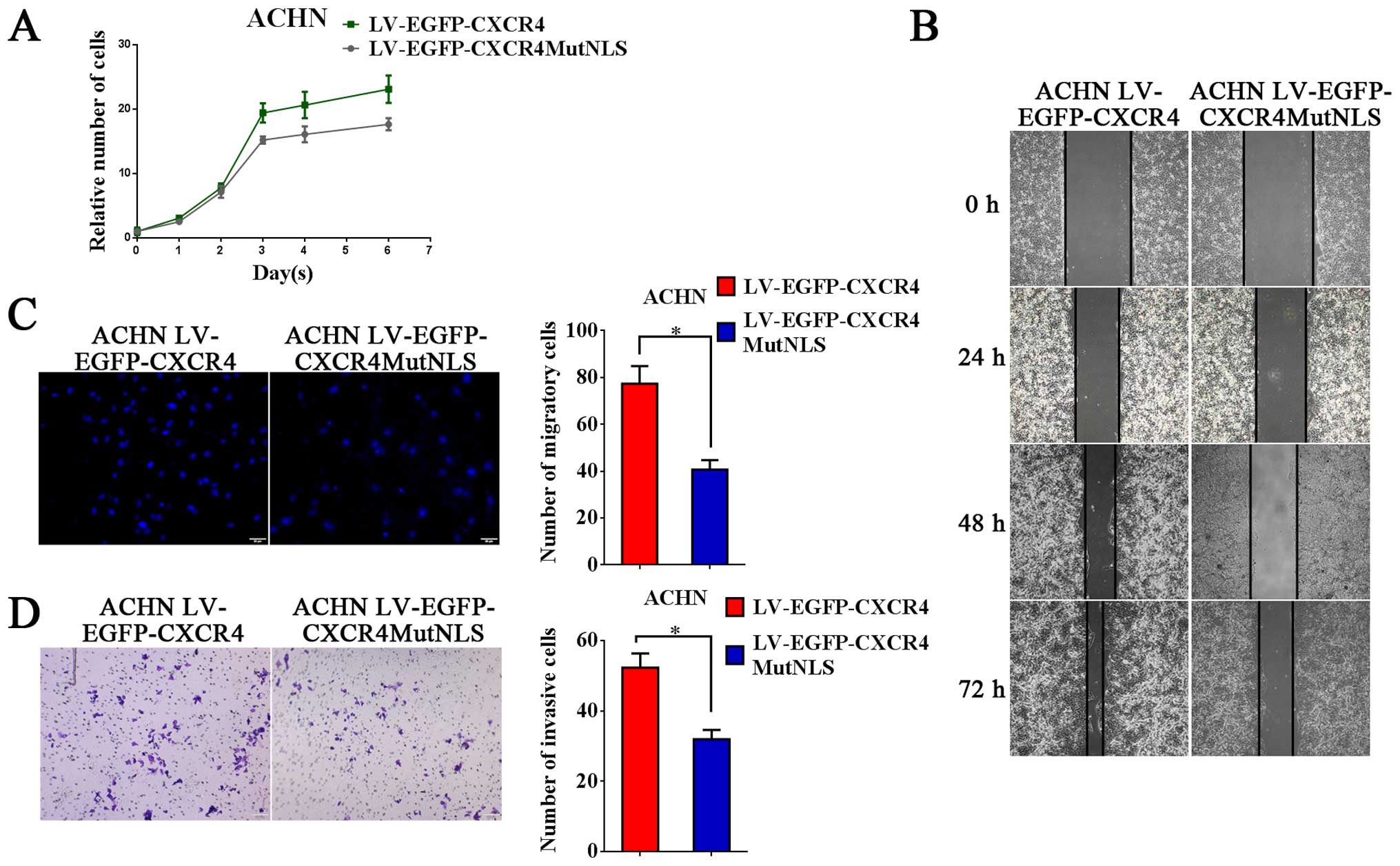
CXCR4 in the nucleus promotes
proliferation, migration and invasion of RCC cells
CCK-8 assays were performed to investigate the
effects of nuclear CXCR4 on proliferation of ACHN cell line.
Compared with LV-EGFP-CXCR4MutNLS infected group, LV-EGFP-CXCR4
infected ACHN cells had a significant growth promoting capacity
(Fig. 2A) indicating that high
level of CXCR4 in the nucleus promoted RCC cell growth. To further
investigate the function of CXCR4 in the nucleus, the migration
ability of ACHN cell was assessed by scratch-wound and Transwell
assays. The scratch-wound assay showed that the migration ability
of the LV-EGFP-CXCR4MutNLS infected ACHN cells was lower than the
LV-EGFP-CXCR4 infected ACHN cells. We found that the cell-free area
of the LV-EGFP-CXCR4 group was obviously narrower than the
LV-EGFP-CXCR4MutNLS group at 24, 48 and 72 h after drawing the
'scratch' line on the monolayer cells (Fig. 2B) and confirmed the results by
Transwell migration assay (Fig.
2C). In Transwell invasion assays, the number of invaded cells
stained with crystal violet was significantly higher in the
LV-EGFP-CXCR4 group (Fig. 2E).
These results showed that CXCR4 in the nucleus can promote the
proliferation, migration and invasion ability of RCC cells.
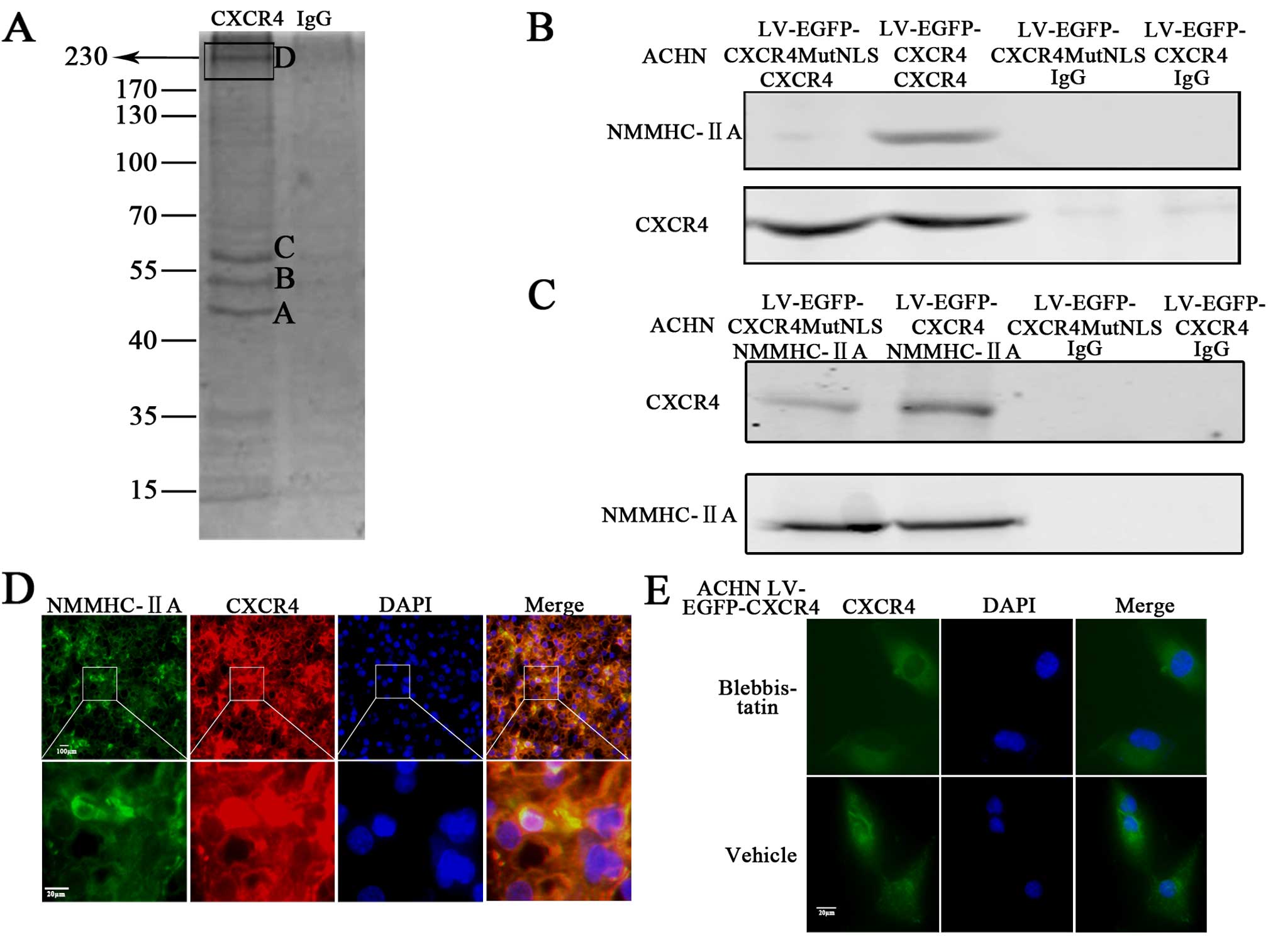
Nuclear localization of CXCR4 partly
depends on NMMHC-IIA
Next, we investigated whether CXCR4 specifically
bound intracellular partners that affected its subcellular
localization. Immunoprecipitation with cell lysates of ACHN cells
(Fig. 3A) revealed the presence of
protein bands of spots (Fig.
3A-A-D) that were specifically associated with CXCR4. We
focused on the spots D (230 kDa), the bands were excised, digested
with trypsin, and the peptides were analyzed by mass spectrometry.
The 230 kDa band was unequivocally identified as non-muscle myosin
IIA by MS/MS analysis (Table I). To
confirm the identity of the 230 kDa protein, western blot analysis
with an antibody against NMMHC-IIA and CXCR4 were performed in
CXCR4 or NMMHC-IIA binding protein lysates of LV-EGFP-CXCR4 and
LV-EGFP-CXCR4MutNLS infected ACHN cells (Fig. 3B and C), which showed that CXCR4 and
NMMHC-IIA could bind with each other mutually. Additionally,
colocalization pattern of NMMHC-IIA and CXCR4 was presented by
immunofluorescence in RCC tissue (Fig.
3E). The result showed that the distribution of CXCR4 and
NMMHC-IIA were almost overlapping, which further revealed that
NMMHC-IIA specifically bound with CXCR4.
 | Table IMass spectrometry analysis of the 230
kDa band. |
Table I
Mass spectrometry analysis of the 230
kDa band.
| Groups | Protein mass
(Da) | PepCount | Unique
PepCount | Sequence
header | Relative abundance
(%) |
|---|
| 1 | 226,532.68 | 34 | 23 | Non-muscle myosin
IIA | 13.62 |
| 2 | 85,015.53 | 6 | 4 | Raichu-404X | 5.54 |
| 3 | 39,745.56 | 3 | 2 | Cysteine (C)-X-C
receptor 4 | 5.68 |
| 4 | 14,728.26 | 1 | 1 | Ubiquitin-60S
ribosomal protein L40 | 7.03 |
| 5 | 83,673.87 | 1 | 1 | Protein kinase
Cε | 0.81 |
| 6 | 16,387.7 | 1 | 1 | Cystatin-SN | 12.06 |
To assess the involvement of NMMHC-IIA in the
subcellular localization of CXCR4, blebbistatin, a specific
inhibitor of NMMHC-IIA, was used to inhibit NMMHC-IIA activation,
and CXCR4 nucleus localization was observed by IF. Treatment with
50 µM blebbistatin, CXCR4 in the nucleus was significantly
decreased in LV-EGFP-CXCR4 infected ACHN cells (Fig. 3F). These results suggested that
targeting NMMHC-IIA interfered with CXCR4 subcellular
localization.
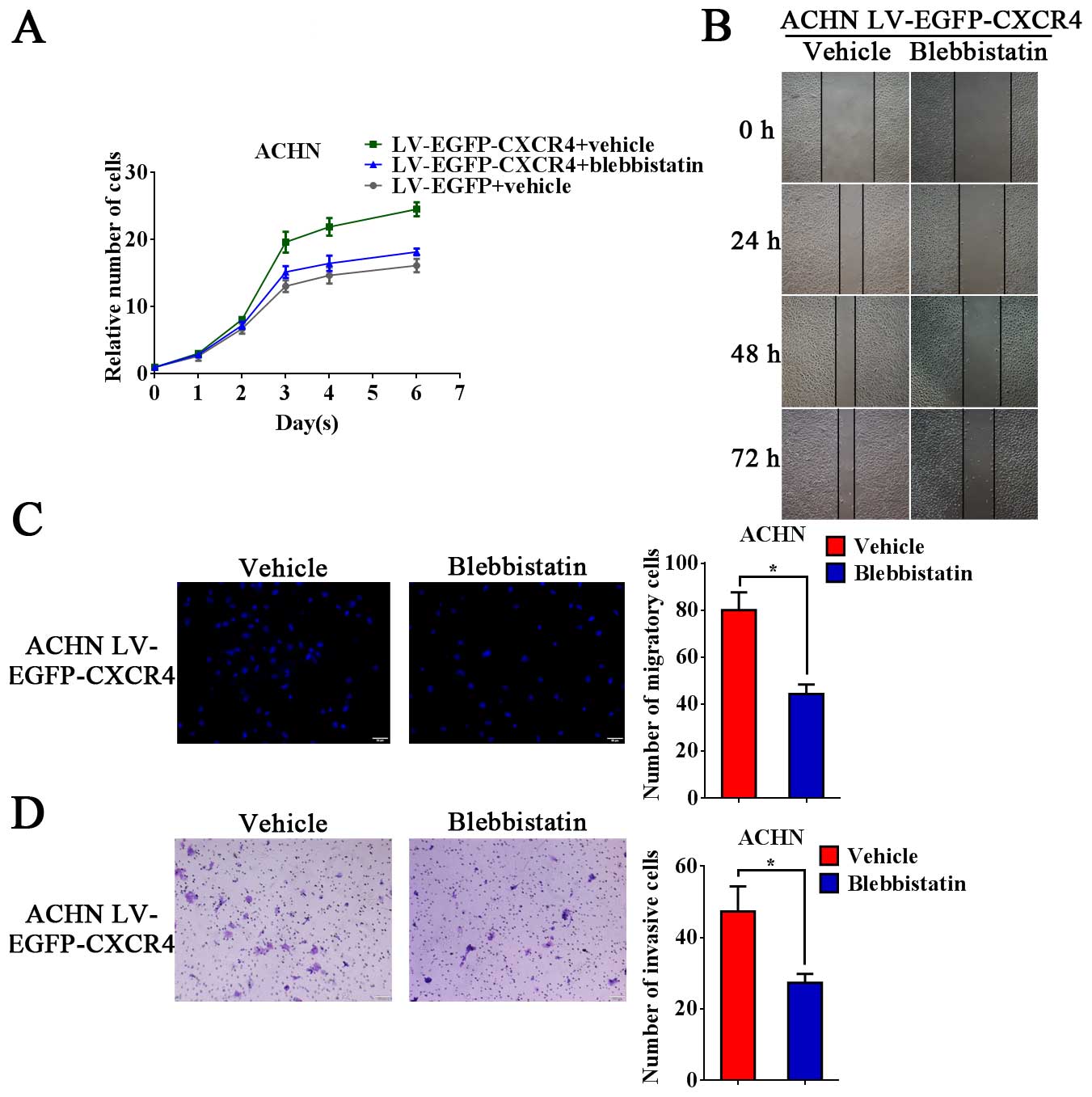
Inhibition of NMMHC-IIA suppresses the
metastatic potential of RCC cells
Then, the functional relationship between NMMHC-IIA
and CXCR4 was analyzed. Blebbistatin suppressed the growth ability
of LV-EGFP-CXCR4 infected ACHN cells (Fig. 4A). We also found that blebbistatin
decreased the migratory and invasive ability in LV-EGFP-CXCR4
infected ACHN cells (Fig. 4B–D).
Collectively, these results indicated that inhibition of NMMHC-IIA
suppressed the growth, migration and invasion capacity of RCC cells
by inhibiting the nuclear location of CXCR4.
Discussion
CXCR4 has been identified as a key factor involving
in organ-specific metastasis of RCC (32). Our previous study observed that
nuclear localization of CXCR4 was found in metastatic but not
primary RCC lesions (33). It is
known that within thousands of cancer cells, just a few cells
escaped the primary tumor by migration and invasion to target
organs, so we hypothesized CXCR4 played important roles in
metastasis when it translocated into the nucleus.
In the present study, we demonstrated that mutation
in NLS '146RPRK149' within CXCR4 prevented its translocation to the
nucleus and inhibited proliferation, migration and invasion of RCC
cells, suggesting that nuclear localization of CXCR4 may be a
mechanism by which RCC cells survive and spread. It indicated to us
that antagonizing nuclear transport pathways or the function of
nuclear CXCR4 is a rational approach for RCC therapy.
To date, increasing evidence supports that CXCR4 is
highly expressed in tumor tissues and correlated with poor
progression in RCC (34). However,
few studies have evaluated the biological function of subcellular
localization of CXCR4 in RCC cells. The present study showed that
CXCR4 presented in both the nucleus and cytoplasm in RCC cells and
determined the accurate location of the NLS and the impact that
nuclear CXCR4 may have on RCC growth and metastasis. Our previous
study suggested that 90–170 amino acid sequence within CXCR4 served
as NLS (23). A prior study by
Ayesha et al presumed a non-traditional NLS, '146RPRK149',
within CXCR4 by PSORTII and demonstrated that removal of the
putative NLS attenuated its nuclear localization in prostate cancer
cells (30). We observed that
nuclear localization of CXCR4 decreased obviously when arginine
146, 148 and 149 were simultaneously mutated to alanine, which
indicated that the 'RPRK' motif may be involved in localization of
CXCR4 to the nucleus in RCC.
Previous studies found a correlation of CXCR4
expression levels with metastatic occurrence of RCC cells, but had
not shown an association of nuclear localization of CXCR4 and
biological behavior of RCC cells, we then examined whether 'RPRK'
motif mutation and subsequently CXCR4 subcellular localization,
could alter the growth and metastatic potential of RCC cells. Our
results suggested that nuclear CXCR4 was a positive regulator in
the growth of RCC cells. We also found that migration, and invasion
of RCC cells bearing CXCR4MutNLS were significantly repressed.
Prevention of CXCR4 translocation to nucleus effectively inhibited
proliferation, migration and invasion in RCC cells. This may partly
explain the fact that nuclear CXCR4 was detected in many metastatic
RCC patients, and the subcellular distribution of CXCR4 may play a
significant role in RCC progression.
Since CXCR4 nuclear localization determined RCC
progression, blockage of this process may be a novel promising
therapeutic approach for RCC patients. In the present study, we
found that NMMHC-IIA could particularly bind with CXCR4 and promote
its nuclear translocation. NMMHC-IIA belongs to the myosin II
subfamily and comprise a complex of two non-muscle myosin II heavy
chains, two essential light chains and two regulatory light chains
(35). NMMHC-IIA has been reported
to participate in many steps of cancer metastasis (36). A previous study found that CXCR4
could bind with NMMHC-IIA in T lymphocytes(37), but its association with CXCR4
subcellular distribution was almost unknown. The present study
confirmed that NMMHC-IIA could bind with CXCR4 and also promoted
its nuclear translocation. Furthermore, pretreatment with
blebbistatin inhibited nuclear CXCR4-induced proliferation,
migration and invasion of RCC cells. This finding is consistent
with previous studies demonstrating impaired migration in
NMMHC-IIA-depleted MDA-MB-231 breast cancer cells (38) and blebbistatin-treated pancreatic
adenocarcinoma cells (39). The
present study suggested that CXCR4 translocated to the nucleus and
subsequently promoted RCC cell growth and metastasis partly
dependent on NMMHC-IIA.
In conclusion, the data presented above provided
clear evidence of CXCR4 translocated into the nucleus and acted as
a functional, ligand-responsive receptor in RCC cells. Inhibition
of CXCR4 nuclear localization could effectively suppress the
proliferation, migration and invasion of RCC cells. Given that
nuclear CXCR4 promotes metastatic ability in RCC, this
investigation provided a novel mechanism to illuminate the nuclear
distribution of CXCR4 as previously reported. We also demonstrated
that NMMHC-IIA was functionally involved in the process of CXCR4
nuclear translocation and then affected its biology functions,
possibly providing promises for new therapeutic interventions for
metastatic RCC. Further investigation is still required to explore
the accurate mechanism of nuclear CXCR4 promoting RCC progression
and its association with clinical prognosis in RCC patients.
Abbreviations:
|
CXCR4
|
cysteine (C)-X-C receptor 4
|
|
RCC
|
renal cell carcinoma
|
|
NMMHC-IIA
|
non-muscle myosin heavy chain-IIA
|
|
GPCR
|
G-protein coupled receptor
|
Acknowledgments
The present study was supported by the National
Nature Science Foundation of China (nos. 81272817 and
81172447).
References
|
1
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
An H, Xu L, Zhu Y, Lv T, Liu W, Liu Y, Liu
H, Chen L, Xu J and Lin Z: High CXC chemokine receptor 4 expression
is an adverse prognostic factor in patients with clear-cell renal
cell carcinoma. Br J Cancer. 110:2261–2268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stewart GD, O'Mahony FC, Powles T, Riddick
AC, Harrison DJ and Faratian D: What can molecular pathology
contribute to the management of renal cell carcinoma? Nat Rev Urol.
8:255–265. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Karakiewicz PI, Briganti A, Chun FK, Trinh
QD, Perrotte P, Ficarra V, Cindolo L, De la Taille A, Tostain J,
Mulders PF, et al: Multi-institutional validation of a new renal
cancer-specific survival nomogram. J Clin Oncol. 25:1316–1322.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lam JS, Leppert JT, Figlin RA and
Belldegrun AS: Role of molecular markers in the diagnosis and
therapy of renal cell carcinoma. Urology. 66(Suppl 5): S1–S9. 2005.
View Article : Google Scholar
|
|
6
|
Libura J, Drukala J, Majka M, Tomescu O,
Navenot JM, Kucia M, Marquez L, Peiper SC, Barr FG,
Janowska-Wieczorek A, et al: CXCR4-SDF-1 signaling is active in
rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and
adhesion. Blood. 100:2597–2606. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Phillips RJ, Burdick MD, Lutz M, Belperio
JA, Keane MP and Strieter RM: The stromal derived
factor-1/CXCL12-CXC chemokine receptor 4 biological axis in
non-small cell lung cancer metastases. Am J Respir Crit Care Med.
167:1676–1686. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taichman RS, Cooper C, Keller ET, Pienta
KJ, Taichman NS and McCauley LK: Use of the stromal cell-derived
factor-1/CXCR4 pathway in prostate cancer metastasis to bone.
Cancer Res. 62:1832–1837. 2002.PubMed/NCBI
|
|
9
|
Hall JM and Korach KS: Stromal
cell-derived factor 1, a novel target of estrogen receptor action,
mediates the mitogenic effects of estradiol in ovarian and breast
cancer cells. Mol Endocrinol. 17:792–803. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kaifi JT, Yekebas EF, Schurr P, Obonyo D,
Wachowiak R, Busch P, Heinecke A, Pantel K and Izbicki JR:
Tumor-cell homing to lymph nodes and bone marrow and CXCR4
expression in esophageal cancer. J Natl Cancer Inst. 97:1840–1847.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SY, Lee CH, Midura BV, Yeung C,
Mendoza A, Hong SH, Ren L, Wong D, Korz W, Merzouk A, et al:
Inhibition of the CXCR4/CXCL12 chemokine pathway reduces the
development of murine pulmonary metastases. Clin Exp Metastasis.
25:201–211. 2008. View Article : Google Scholar
|
|
12
|
Zagzag D, Krishnamachary B, Yee H, Okuyama
H, Chiriboga L, Ali MA, Melamed J and Semenza GL: Stromal
cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma
and clear cell-renal cell carcinoma: Von Hippel-Lindau
loss-of-function induces expression of a ligand and its receptor.
Cancer Res. 65:6178–6188. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
D'Alterio C, Cindolo L, Portella L,
Polimeno M, Consales C, Riccio A, Cioffi M, Franco R, Chiodini P,
Cartenì G, et al: Differential role of CD133 and CXCR4 in renal
cell carcinoma. Cell Cycle. 9:4492–4500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cho KS, Yoon SJ, Lee JY, Cho NH, Choi YD,
Song YS and Hong SJ: Inhibition of tumor growth and
histopathological changes following treatment with a chemokine
receptor CXCR4 antagonist in a prostate cancer xenograft model.
Oncol Lett. 6:933–938. 2013.PubMed/NCBI
|
|
15
|
Teicher BA and Fricker SP: CXCL12
(SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 16:2927–2931.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Na IK, Scheibenbogen C, Adam C, Stroux A,
Ghadjar P, Thiel E, Keilholz U and Coupland SE: Nuclear expression
of CXCR4 in tumor cells of non-small cell lung cancer is correlated
with lymph node metastasis. Hum Pathol. 39:1751–1755. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Spano JP, Andre F, Morat L, Sabatier L,
Besse B, Combadiere C, Deterre P, Martin A, Azorin J, Valeyre D, et
al: Chemokine receptor CXCR4 and early-stage non-small cell lung
cancer: Pattern of expression and correlation with outcome. Ann
Oncol. 15:613–617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Masuda T, Nakashima Y, Ando K, Yoshinaga
K, Saeki H, Oki E, Morita M, Oda Y and Maehara Y: Nuclear
expression of chemokine receptor CXCR4 indicates poorer prognosis
in gastric cancer. Anticancer Res. 34:6397–6403. 2014.PubMed/NCBI
|
|
19
|
Wagner PL, Hyjek E, Vazquez MF, Meherally
D, Liu YF, Chadwick PA, Rengifo T, Sica GL, Port JL, Lee PC, et al:
CXCL12 and CXCR4 in adenocarcinoma of the lung: Association with
metastasis and survival. J Thorac Cardiovasc Surg. 137:615–621.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yoshitake N, Fukui H, Yamagishi H,
Sekikawa A, Fujii S, Tomita S, Ichikawa K, Imura J, Hiraishi H and
Fujimori T: Expression of SDF-1 alpha and nuclear CXCR4 predicts
lymph node metastasis in colorectal cancer. Br J Cancer.
98:1682–1689. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Speetjens FM, Liefers GJ, Korbee CJ,
Mesker WE, van de Velde CJ, van Vlierberghe RL, Morreau H,
Tollenaar RA and Kuppen PJ: Nuclear localization of CXCR4
determines prognosis for colorectal cancer patients. Cancer
Microenviron. 2:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Chen W, Gao L, Yang Q, Liu B, Wu
Z, Wang Y and Sun Y: High expression of CXCR4, CXCR7 and SDF-1
predicts poor survival in renal cell carcinoma. World J Surg Oncol.
10:2122012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang LH, Liu Q, Xu B, Chen W, Yang Q, Wang
ZX and Sun YH: Identification of nuclear localization sequence of
CXCR4 in renal cell carcinoma by constructing expression plasmids
of different deletants. Plasmid. 63:68–72. 2010. View Article : Google Scholar
|
|
24
|
Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng
L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, et al: A
TAG1-APP signalling pathway through Fe65 negatively modulates
neurogenesis. Nat Cell Biol. 10:283–294. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lyu J, Yamamoto V and Lu W: Cleavage of
the Wnt receptor Ryk regulates neuronal differentiation during
cortical neurogenesis. Dev Cell. 15:773–780. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carpenter G and Liao HJ: Trafficking of
receptor tyrosine kinases to the nucleus. Exp Cell Res.
315:1556–1566. 2009. View Article : Google Scholar :
|
|
27
|
Don-Salu-Hewage AS, Chan SY, McAndrews KM,
Chetram MA, Dawson MR, Bethea DA and Hinton CV: Cysteine (C)-x-C
receptor 4 undergoes transportin 1-dependent nuclear localization
and remains functional at the nucleus of metastatic prostate cancer
cells. PLoS One. 8:e571942013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan L, Cai Q and Xu Y: The ubiquitin-CXCR4
axis plays an important role in acute lung infection-enhanced lung
tumor metastasis. Clin Cancer Res. 19:4706–4716. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tripathi A, Davis JD, Staren DM, Volkman
BF and Majetschak M: CXC chemokine receptor 4 signaling upon
co-activation with stromal cell-derived factor-1α and ubiquitin.
Cytokine. 65:121–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Burgess A, Buck M, Krauer K and Sculley T:
Nuclear localization of the Epstein-Barr virus EBNA3B protein. J
Gen Virol. 87:789–793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ren M, Qiu S, Venglat P, Xiang D, Feng L,
Selvaraj G and Datla R: Target of rapamycin regulates development
and ribosomal RNA expression through kinase domain in Arabidopsis.
Plant Physiol. 155:1367–1382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao FL and Guo W: Expression of stromal
derived factor-1 (SDF-1) and chemokine receptor (CXCR4) in bone
metastasis of renal carcinoma. Mol Biol Rep. 38:1039–1045. 2011.
View Article : Google Scholar
|
|
33
|
Wang L, Wang Z, Yang B, Yang Q, Wang L and
Sun Y: CXCR4 nuclear localization follows binding of its ligand
SDF-1 and occurs in metastatic but not primary renal cell
carcinoma. Oncol Rep. 22:1333–1339. 2009.PubMed/NCBI
|
|
34
|
Almofti A, Uchida D, Begum NM, Tomizuka Y,
Iga H, Yoshida H and Sato M: The clinicopathological significance
of the expression of CXCR4 protein in oral squamous cell carcinoma.
Int J Oncol. 25:65–71. 2004.PubMed/NCBI
|
|
35
|
Vicente-Manzanares M, Ma X, Adelstein RS
and Horwitz AR: Non-muscle myosin II takes centre stage in cell
adhesion and migration. Nat Rev Mol Cell Biol. 10:778–790. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Aguilar-Cuenca R, Juanes-García A and
Vicente-Manzanares M: Myosin II in mechanotransduction: Master and
commander of cell migration, morphogenesis, and cancer. Cell Mol
Life Sci. 71:479–492. 2014. View Article : Google Scholar
|
|
37
|
Carpenter G and Red Brewer M: EpCAM:
Another surface-to-nucleus missile. Cancer Cell. 15:165–166. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hindman B, Goeckeler Z, Sierros K and
Wysolmerski R: Non-muscle myosin II isoforms have different
functions in matrix rearrangement by MDA-MB-231 cells. PLoS One.
10:e01319202015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Duxbury MS, Ashley SW and Whang EE:
Inhibition of pancreatic adenocarcinoma cellular invasiveness by
blebbistatin: A novel myosin II inhibitor. Biochem Biophys Res
Commun. 313:992–997. 2004. View Article : Google Scholar : PubMed/NCBI
|