Introduction
MicroRNAs (miRNAs) are endogenously processed
non-coding RNAs that regulate gene expression by blocking
translation or decreasing mRNA stability (1,2). Since
the discovery of miRNAs in 1993 (3), they have been shown to affect multiple
cellular processes (4), and in
particular, play significant roles in cancer development and
progression (5). miRNAs can
function as tumor suppressors or oncogenes, depending on their
specific target genes (6,7). For example, miR-145, miR-335,
miR-125b-1, miR-126, miR-15a, miR-16-1, miR-31, and miR-335 are all
tumor suppressors for specific cancer types (8–11).
Recent studies have shown that miRNAs are involved in the malignant
progression of breast cancer (12–14).
In particular, miR-124 is a brain-enriched miRNA that plays a
crucial role in gastrulation and neural development (15,16),
and its deregulation is related to carcinogenesis. The expression
level of miR-124 is significantly decreased in glioma,
medulloblastoma, oral squamous cell carcinoma, hepatocellular
carcinoma, and bladder cancer, suggesting that it may function as a
tumor suppressor (17–22). However, its function in breast
cancer remains unclear. Furthermore, the molecular mechanisms by
which it modulates the malignant phenotype of breast cancer cells
are not fully understood.
In the present study, we used quantitative real-time
PCR (qPCR) to demonstrate that the expression of miR-124 in breast
cancer tissue is decreased compared to corresponding adjacent
normal tissue. In addition, miR-124 significantly inhibited breast
cancer cell proliferation and migration by targeting snail family
zinc finger 2 (SNAI2) via its 3′-untranslated region (3′-UTR). Its
expression was inversely correlated with SNAI2 mRNA expression
level in breast tissue specimens. Furthermore, knockdown of SNAI2
inhibited the proliferation and migration of breast cancer cells.
Together, these data suggest that miR-124 may function as a tumor
suppressor that targets SNAI2 to influence the proliferation and
migration of breast cancer cells.
Materials and methods
Breast tissue specimens
Documented informed consent was obtained from all
patients or guardians, and the Ethics Committee of the First
Hospital of Zibo (Zibo, Shandong, China) approved all aspects of
the study. Breast cancer and adjacent normal tissue samples were
obtained from 30 female patients with breast cancer in the
Department of Surgery, the First Hospital of Zibo from 2015 to
2016. Both tumor tissues and corresponding adjacent normal tissues
were histologically confirmed. Tissue specimens were placed in
cryovials, snap-frozen, and stored at −80°C immediately after
operation until subsequent use. The protocol for the use of patient
samples was approved by the Institutional Review Board of the
hospital.
Cell lines and culture
The human breast cancer cell lines MCF-7 and
MDA-MB-231, as well as the immortalized human embryonic kidney cell
line HEK293T, were obtained from Jiangsu University (Zhenjiang,
Jiangsu, China). The human breast cancer cell line BT-474, as well
the immortalized breast epithelial cell line MCF-10A, were obtained
from the Central Laboratory of the First Hospital of Zibo. MCF-7,
MDA-MB-231, and HEK293T cells were cultured in Dulbecco's modified
Eagles medium (DMEM; Gibco-BRL, Carlsbad, CA, USA) with low glucose
(L-DMEM) supplemented with 10% fetal bovine serum (FBS; ExCell
Biology, Inc., Shanghai, China). Another breast cancer cell line,
BT-474, was maintained in RPMI-1640 (Gibco-BRL) with 10% FBS.
HEK293T cells and the human breast epithelial cell line MCF-10A
were cultured in DMEM with 10% FBS. All of the cells were
maintained in a humidified atmosphere of 5% CO2 and 95%
air at 37°C.
Transient miRNA transfection
The MCF-7 cell line was selected for transfection of
miR-124. miR-124 mimics and negative miRNA mimic controls (NC
miR-mimics) were synthesized and purified by GenePharma (Shanghai,
China). The sequences are listed in Table I. Briefly, the cells were grown
overnight and then transfected with 100 nM miR-124 mimics or NC
miR-mimics using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Waltham, MA, USA) according to the
manufacturer's instructions.
 | Table I.Sequences of miR-124 mimics and
negative miRNA mimic controls. |
Table I.
Sequences of miR-124 mimics and
negative miRNA mimic controls.
| Name |
| Sequence
(5′-3′) |
|---|
| miR-124 | Sense: |
UAAGGCACGCGGUGAAUGCC |
| mimics | Antisense: |
CAUUCACCGCGUGCCUUAUU |
| Mimic | Sense: |
UUCUCCGAACGUGUCACGUTT |
| negative
control | Antisense: |
ACGUGACACGUUCGGAGAATT |
Short hairpin RNA transfection
Short hairpin RNA (shRNA) constructs against SNAI2
were from Origene Co. (GenePharma). MCF-7 cells were cultured until
70–80% confluence was reached. Cells were transfected with shRNA
using Lipofectamine® 2000 according to the
manufacturer's instructions. The level of SNAI2 expression was
determined by western blot analysis.
RNA isolation and qPCR
Total RNA in tissues and cell lines was isolated
with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer's instructions. RNA was also isolated with TRIzol
reagent from cell lines transfected with miR-124 mimics and NC
miR-mimics for 24, 48 and 72 h. To measure miR-124 RNA expression,
qPCR was performed using a SYBR-Green containing PCR kit
(GenePharma). For detection of SNAI2 mRNA, cDNA was synthesized
from 1 µg total RNA using the reverse transcription kit in
accordance with the manufacturer's instructions (Thermo Fisher
Scientific). Then, qPCR was performed using UltraSYBR Mixture (with
ROX) assay kits (CWBio, Beijing, China) according to the
manufacturer's instructions. The CFX-96 real-time fluorescence
thermal cycler (Bio-Rad, NJ, USA) was used for the quantitative
detection of miRNA and mRNA. The relative expression levels of
miRNA and mRNA were normalized to the expression of U6 snRNA and
β-actin mRNA, respectively. The expression of each gene was
quantified by measuring cycle threshold (Ct) values and normalized
using the 2−ΔCt or 2−ΔΔCt Ct method relative
to U6 snRNA or β-actin mRNA. All of the primer sequences are listed
in Table II.
 | Table II.Specific primers for target and
control genes. |
Table II.
Specific primers for target and
control genes.
| Name | Sequence
(5′-3′) |
|---|
| miR-124 | F:
ACGTTGTGTAGCTTATCAGACTG |
|
| R:
AATGGTTGTTCTCCACACTCTC |
| U6 snRNA | F:
ATTGGAACGATACAGAGAAGATT |
|
| F:
GGAACGCTTCACGAATTTG |
| SNAI2 | F:
ACATAAGCAGCTGCACTGCG |
|
| R:
ATGGGTCTGCAGATGAGCCC |
| β-actin | F:
TGGCACCCAGCACAATGAA |
|
| R:
CTAAGTCATAGTCCGCCTAGAAGCA |
Cell proliferation assay
Twenty-four hours after transient transfection with
miR-124 mimics or NC miR-mimics or after knockdown of SNAI2, MCF-7
breast cancer cells were harvested and sub-cultured in 96-well
plates. Cell proliferation was assessed using thiazolyl blue
tetrazolium bromide assay (MTT; Amresco, Radnor, PA, USA) according
to the manufacturer's instructions. Briefly, MTT reagent (20 µl)
was added to each well and incubated at 37°C for 4 h. Then, the
reagent was removed and dimethyl sulfoxide (150 µl) was added to
each well. Absorbance at 492 nm was measured using the FLx800
fluorescence microplate reader (BioTek, Winooski, VT, USA). The
experiment was performed in triplicate and repeated three times.
The data are summarized as means ± standard error of the mean
(SEM).
Colony formation assay
MCF-7 breast cancer cells were transfected as
described above in the presence or knockdown of SNAI2, seeded into
12-well plates (0.3×103 cells/well), and incubated for
10 days. Then, cells were fixed and stained, followed by colony
counting. The experiment was performed in triplicate, with data
summarized as means ± SEM.
Wound healing assay
MCF-7 breast cancer cells were seeded into 6-well
plates, transiently transfected as previously described to
overexpress miRNA or knockdown SNAI2, and then allowed to grow
until 100% confluent. Next, the cell layer was scratched through
the central axis using a sterile plastic tip, and loose cells were
washed away with phosphate-buffered saline. Wound healing was
observed and photographed at 0 and 48 h in three randomly selected
microscopic fields for each condition and time-point. The degree of
motility 48 h after confluent cells had been scratched was
expressed as the percentage of wound closure calculated as follows:
(Distance of cell migration at 48 h/distance of scratch at 0 h) ×
100%. The experiment was performed in triplicate and data are
summarized as means ± SEM.
Cell migration and invasion
assays
The cell migration assay was performed using
Transwell inserts (Corning, Blacksburg, VA, USA), and the cell
invasion assay was performed with the CytoSelect 24-well cell
invasion assay kit (Cell Biolabs, Inc., San Diego, CA, USA)
according to the manufacturer's instructions. The cells were
transiently transfected as previously described to overexpress
miRNA or knockdown SNAI2. Beginning 48 h after transfection, cells
were starved in L-DMEM without FBS for 2 h, and 5×104
cells were resuspended in 0.1 ml L-DMEM without FBS and seeded in
the upper chamber of a Transwell insert. Then, L-DMEM containing
20% FBS was added to the lower chamber as a chemoattractant. To
measure the effects of miR-124 or SNAI2 on MCF-7 migration and
invasion potential, the cells in the upper chamber were cultured
for 14 h at 37°C in humidified 5% CO2. Cells that had
migrated to the lower chamber were fixed and stained with 0.1%
crystal violet. Three low-magnification areas (×100) were randomly
selected, and the number of migrated or invasive cells was counted.
All of the experiments were performed in triplicate, and data are
summarized as means ± SEM.
Western blot analysis
For protein expression analyses, standard western
blotting was performed. Whole cell protein lysates were
electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels
and transferred onto polyvinylidene fluoride membranes (Beyotime
Institute of Biotechnology, Shanghai, China). The membranes were
blocked in 5% skimmed milk/Tris-buffered saline (20 mM Tris-HCl pH
7.4, 150 mM NaCl, with 0.1% Tween-20; Tris-buffered saline with
Tween-20, TBST) at room temperature for 1 h and incubated with
primary antibodies at 4°C overnight. The antibodies were anti-SNAI2
(1:1,000) and anti-glyceraldehyde 3-phosphate dehydrogenase
(1:1,000), all purchased from Cell Signaling Technology, Inc.
(Boston, MA, USA). The next day, the membranes were washed with
TBST, and then incubated with horseradish peroxidase-linked
secondary antibody (anti-rabbit IgG, 1:1,000; Cell Signaling
Technology, Inc.). The protein bands were developed with enhanced
chemiluminescence reagents (Beyotime Institute of
Biotechnology).
Dual-luciferase reporter assay
Putative miR-124 binding sites in the 3′-UTR of the
SNAI2 mRNA was synthesized and inserted into the XbaI and
FseI sites of the pGL3-control vector (Promega, Madison, WI,
USA). These constructs were named pGL3-SNAI2-wt and pGL3-SNAI2-mut.
For the reporter assay, 293T cells were plated onto 24-well plates
and transfected into pGL3-SNAI2-wt or pGL3-SNAI2-mut and
pLL3.7-miR-124 or pLL3.7-miR-control vector using
Lipofectamine® 2000 according to the manufacturer's
instructions. The Renilla luciferase vector pRL-SV50
(Promega) was also co-transfected to normalize the differences in
transfection efficiency. After transfection for 48 h, cells were
harvested and assayed with the Dual-Luciferase Reporter assay
system (Promega) according to the manufacturer's instructions. The
experiment was performed in triplicate and data are summarized as
means ± SEM.
Statistical analysis
For statistical analyses, mean values ± SEM were
generated from several repeats of each experiment. The P-values
were obtained from t-tests with paired or unpaired samples and
P<0.05 was considered statistically significant. The correlation
between miR-124 and SNAI2 expression was analyzed using Spearman
correlation and P<0.05 was considered statistically
significant.
Results
miR-124 is decreased in breast cancer
tissues and cells
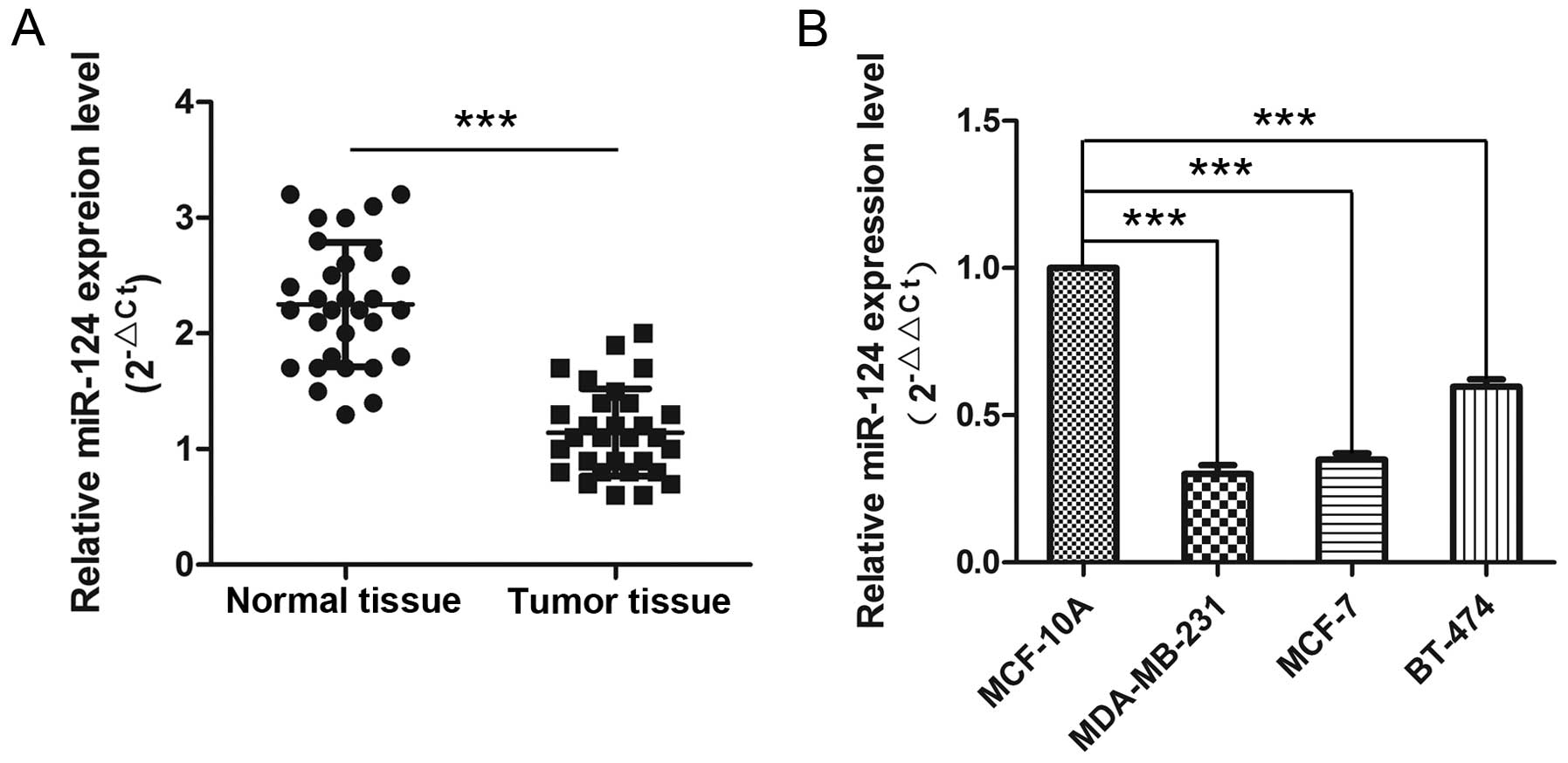
To investigate the role of miR-124 in the initiation
and progression of human breast cancer, we used qPCR to determine
miR-124 levels in 30 breast cancer tissues and adjacent normal
breast tissues. As shown in Fig.
1A, the expression of miR-124 was significantly reduced in
cancer tissues compared to adjacent normal tissues (P<0.001). We
further evaluated the expression levels of miR-124 in breast cancer
cell lines, and found that its expression was decreased to varying
degrees in the three tested breast cancer cell lines compared to
MCF-10A (P<0.001), an immortalized breast epithelial cell line
(Fig. 1B). These results indicate
that the reduced miR-124 expression is a frequent event in human
breast cancer tissues and cells, and may play a role in breast
carcinoma progression. We selected the MCF-7 breast cancer cell
line for subsequent experiments.
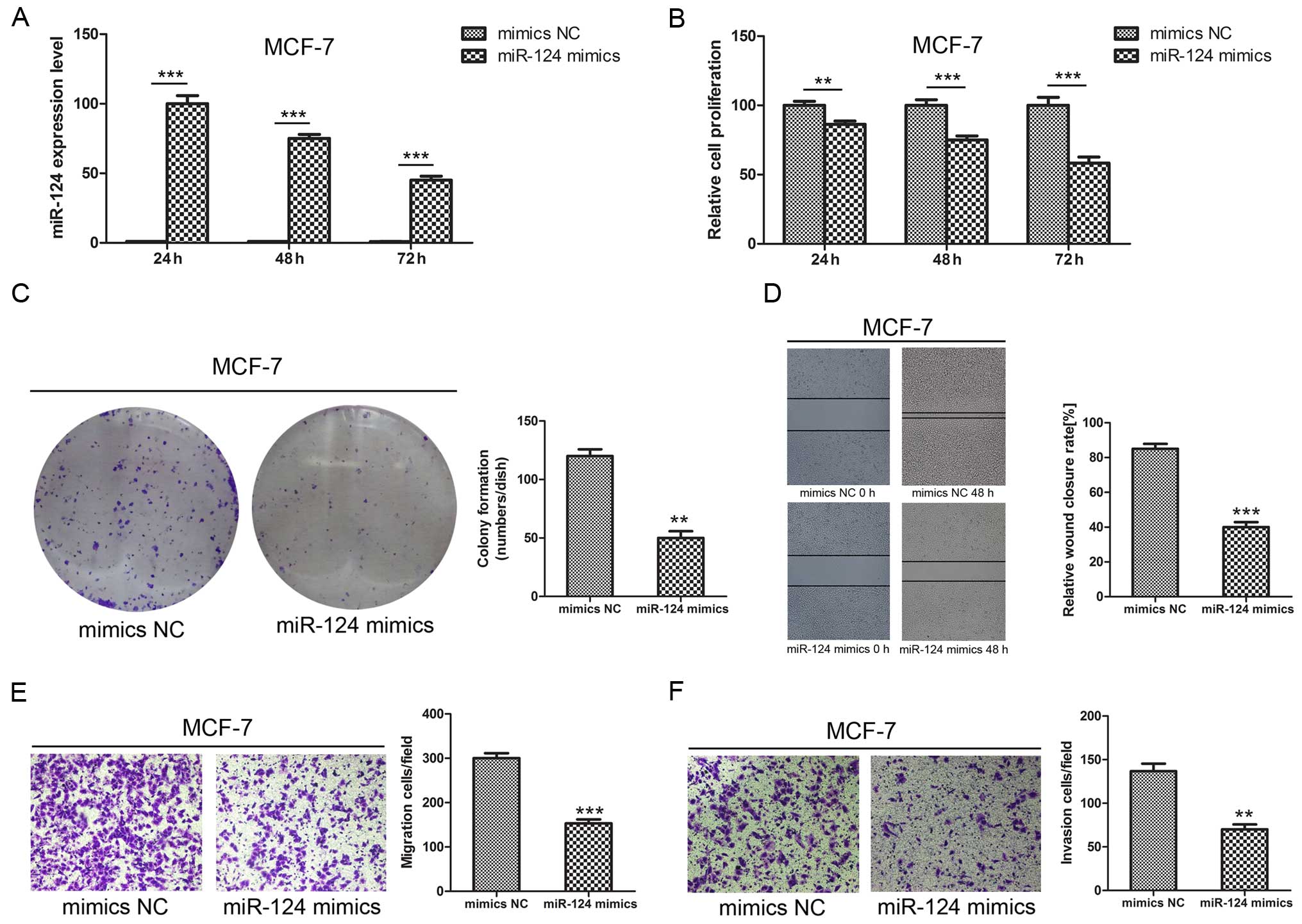
miR-124 inhibits breast cancer cell
proliferation, colony formation, migration, and invasion
To confirm the function of miR-124, we transfected
miR-124 mimics or negative control sequences into MCF-7 breast
cancer cells. Transfection efficiency was estimated by fluorescence
microscopy 6 h after transfection, and miR-124 expression was
determined by qPCR at 24, 48 and 72 h. The results showed that the
miR-124 mimics significantly increased miR-124 RNA expression in
these cells (Fig. 2A). The
functional analyses showed a significant decrease in cell
proliferation (Fig. 2B), colony
formation (Fig. 2C), migration
(Fig. 2D and E), and invasion
(Fig. 2F) in cells transfected with
miRNA-124 mimics compared to those transfected with NC-miR mimics
(P<0.001).
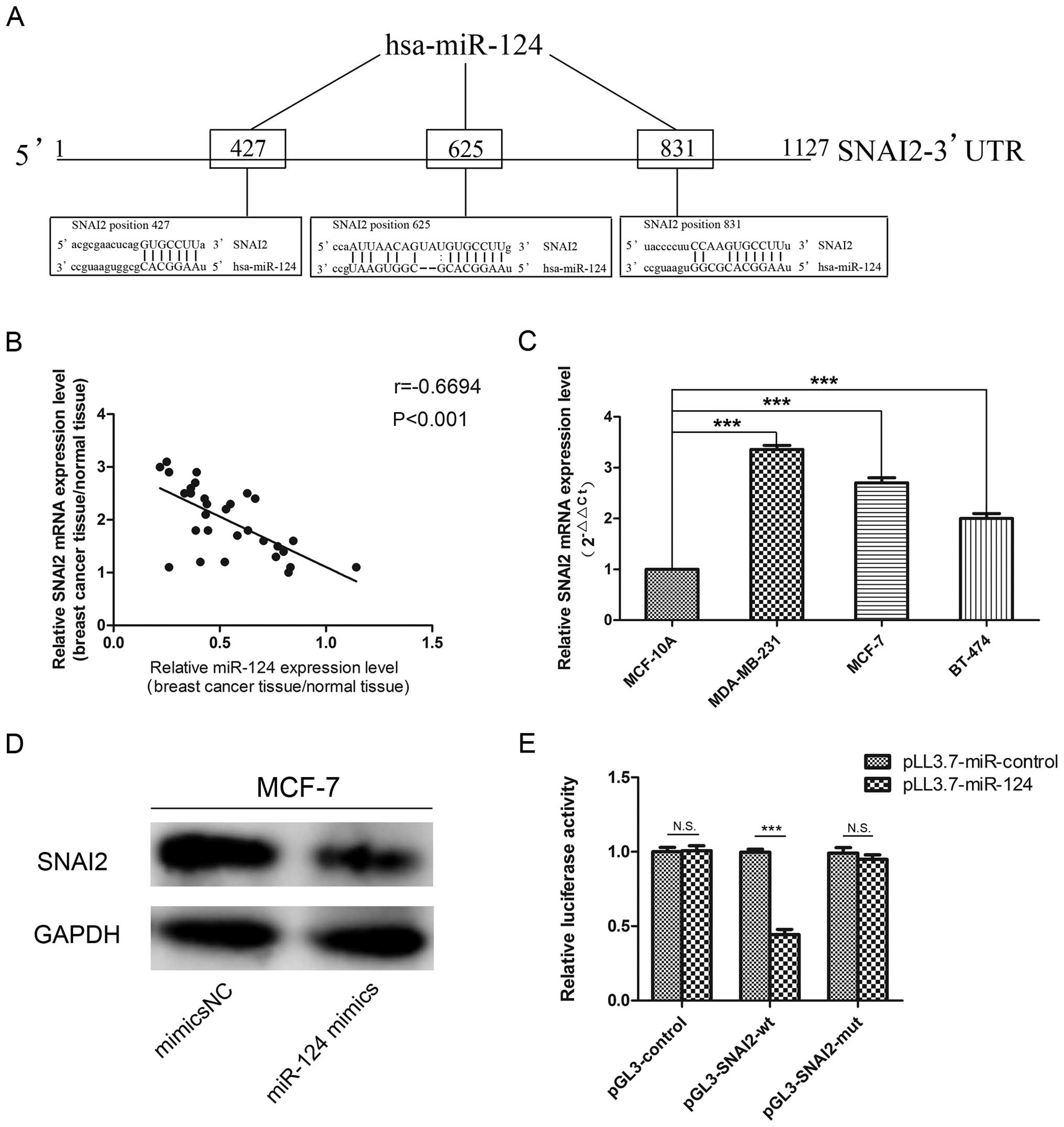
SNAI2 is a target gene of miR-124
We performed bioinformatic analyses to identify the
target genes of miR-124 using the miRanda (www.microrna.org) database, and found that SNAI2 is
targeted by miR-124. SNAI2 has three potential complimentary
binding sites for miR-124 within its 3′-UTR (Fig. 3A). Based on these results, we used
qPCR to examine the expression level of SNAI2 mRNA in breast cancer
tissue specimens and cell lines. We observed that decreased miR-124
expression is associated with increased SNAI2 mRNA levels
(r=−0.6694, P<0.001) in patients with breast cancer (Fig. 3B and C). We performed western blot
analysis to assess the effects of miR-124 on SNAI2 expression, and
found that SNAI2 protein expression was decreased in MCF-7 cells
after treatment with miR-124 mimics compared to the control
(Fig. 3D). We used a luciferase
assay to determine if miR-124 directly regulates SNAI2, and found
that the relative luciferase activities were significantly
decreased in the HEK293T cells (P<0.001) (Fig. 3E). These data show that SNAI2 is a
target gene of miR-124.
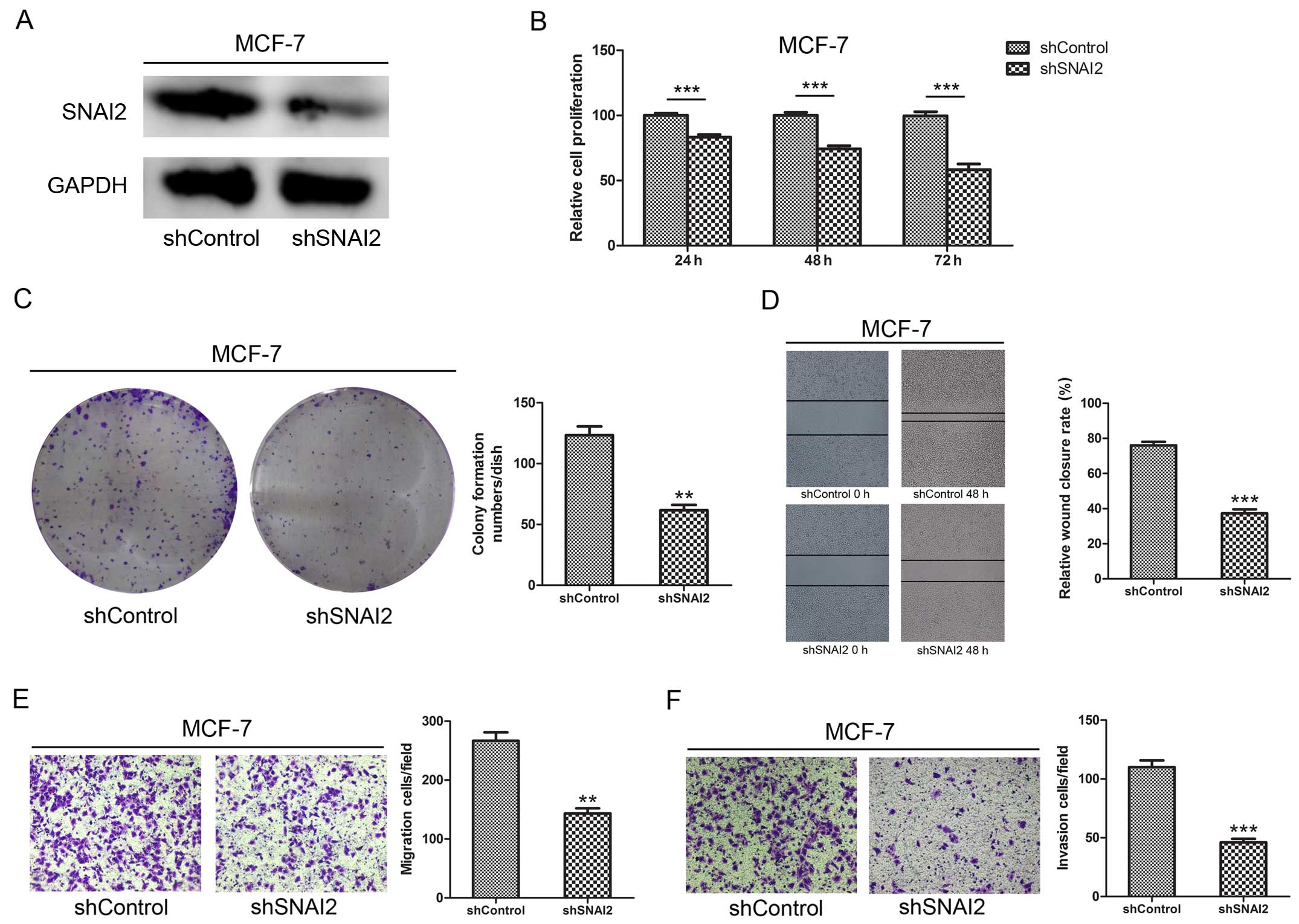
SNAI2 promotes breast cancer cell
proliferation, colony formation, migration, and invasion
To evaluate the effects of SNAI2 on breast cancer
cells, we suppressed SNAI2 by shRNA (Fig. 4A). Compared to the negative
controls, cell proliferation, colony formation, migration, and
invasion were significantly suppressed when cells were treated with
shRNA (P<0.01, P<0.001) (Fig.
4B-E). These data demonstrate that SNAI2 promotes breast cancer
cell proliferation, colony formation, migration, and invasion.
Discussion
Breast cancer is a common and highly lethal
malignancy (23). As many as 1.2
million women worldwide are diagnosed each year, and approximately
500,000 women die annually from this disease (24). The incidence of breast cancer
accounts for 7–8% of the total number of malignant tumors (25). Metastatic progression of breast
cancer is a complex and clinically daunting process (26–29).
Breast cancer is both genetically and histopathologically
heterogeneous. Although many molecular triggers play a vital role
in breast cancer development, the mechanisms underlying this
process remain largely unknown. However, an understanding of these
mechanisms is crucial for developing effective treatments for this
disease.
miRNAs are small noncoding RNAs that usually
negatively regulate gene expression by degrading target mRNAs,
inhibiting the translation of these mRNAs, or both (30). Many studies have demonstrated that
miRNAs can function as oncogenes or tumor suppressors, and are
often dysregulated in tumors (31).
miRNAs play a vital role in many crucial cellular processes,
including apoptosis, differentiation, invasion, and proliferation
(32–34). The abnormal expression of miRNA has
been reported in many cancers, including breast (35), gastric (36), colorectal (37), and liver cancers (38), as well as leukemia (39) and lymphoma (40). miR-124 has been described as a
brain-specific miRNA in mammals, and may play a role in defining
and maintaining neuron-specific characteristics (41,42).
It has also been reported to function as an important regulator of
many human cancers (43,44). In particular, recent studies have
indicated that miR-124 targets specific genes to regulate
proliferation and migration in breast cancer (45–47).
In this study, we showed that the expression level
of miR-124 was significantly decreased in human breast cancer
tissues compared to adjacent normal tissues. To determine the role
of miR-124 in breast cancer, we transfected miR-124 mimics into
breast cancer cells to induce its overexpression. The exogenous
overexpression of miR-124 significantly inhibited the cell growth,
as indicated by MTT and colony formation assays. Moreover, cell
migration and invasion were also significantly decreased by the
overexpression of miR-124 in breast cancer cells, as shown by wound
healing and Transwell migration assays.
Next, we used online biological software to predict
that SNAI2 is a potential target gene of miR-124. SNAI2, a member
of the snail family of C2H2-type zinc-finger
transcription factors, promotes epithelial-mesenchymal transition
and has antiapoptotic activity (48). These data demonstrate that SNAI2 is
a direct target of miR-124, and that miR-124-mediated inhibition of
SNAI2 is dependent on a conversed motif in the 3′-UTR of SNAI2.
Recent independent studies have shown that the overexpression of
SNAI2 alters cell invasion, motility, chemoresistance, metastasis,
and poor prognosis in several human cancers (49–52).
Furthermore, we showed that the proliferation and migration of
breast cancer cells were reduced by inhibiting SNAI2 with shRNA,
consistent with the aforementioned conclusion that SNAI2 promotes
tumor development.
In conclusion, our observations suggest that a low
level of miR-124 expression may result in the elevated expression
of SNAI2. Increased SNAI2 levels would allow breast cancer cells to
proliferate and migrate, and would favor tumor progression.
Additionally, our results showed that miR-124 inhibited breast
cancer cell proliferation and migration by regulating SNAI2
expression. Our findings may advance our understanding of the
complex molecular mechanisms underlying the development and
maintenance of miR-124-SNAI2 levels that are associated with breast
cancer. Based on these data, we suggest that the restoration of
miR-124 activity may represent an attractive strategy for breast
cancer therapy, and future studies should be focused on this
area.
Acknowledgements
This study was supported by the Natural Scientific
Foundation of Shandong Province (grant no. ZR2015PH056).
References
|
1
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sonoki T, Iwanaga E, Mitsuya H and Asou N:
Insertion of microRNA-125b-1, a human homologue of lin-4, into a
rearranged immunoglobulin heavy chain gene locus in a patient with
precursor B-cell acute lymphoblastic leukemia. Leukemia.
19:2009–2010. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michael MZ, O'Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
11
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Negrini M and Calin GA: Breast cancer
metastasis: a microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee MR, Kim JS and Kim KS: miR-124a is
important for migratory cell fate transition during gastrulation of
human embryonic stem cells. Stem Cells. 28:1550–1559. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng LC, Pastrana E, Tavazoie M and
Doetsch F: miR-124 regulates adult neurogenesis in the
subventricular zone stem cell niche. Nat Neurosci. 12:399–408.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia H, Cheung WK, Ng SS, Jiang X, Jiang S,
Sze J, Leung GK, Lu G, Chan DT, Bian XW, et al: Loss of
brain-enriched miR-124 microRNA enhances stem-like traits and
invasiveness of glioma cells. J Biol Chem. 287:9962–9971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hunt S, Jones AV, Hinsley EE, Whawell SA
and Lambert DW: MicroRNA-124 suppresses oral squamous cell
carcinoma motility by targeting ITGB1. FEBS Lett. 585:187–192.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y
and Liu R: miR-124 radiosensitizes human glioma cells by targeting
CDK4. J Neurooncol. 114:263–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang T, Wang J, Zhai X, Li H, Li C and
Chang J: MiR-124 retards bladder cancer growth by directly
targeting CDK4. Acta Biochim Biophys Sin (Shanghai). 46:1072–1079.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Justo N, Wilking N, Jönsson B, Luciani S
and Cazap E: A review of breast cancer care and outcomes in Latin
America. Oncologist. 18:248–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Key TJ, Verkasalo PK and Banks E:
Epidemiology of breast cancer. Lancet Oncol. 2:133–140. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Talmadge JE and Fidler IJ: AACR centennial
series: the biology of cancer metastasis: historical perspective.
Cancer Res. 70:5649–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Valastyan S and Weinberg RA: MicroRNAs:
Crucial multi-tasking components in the complex circuitry of tumor
metastasis. Cell Cycle. 8:3506–3512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Leskelä S, Leandro-García LJ, Mendiola M,
Barriuso J, Inglada-Pérez L, Muñoz I, Martínez-Delgado B, Redondo
A, de Santiago J, Robledo M, et al: The miR-200 family controls
beta-tubulin III expression and is associated with paclitaxel-based
treatment response and progression-free survival in ovarian cancer
patients. Endocr Relat Cancer. 18:85–95. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frank D, Gantenberg J, Boomgaarden I, Kuhn
C, Will R, Jarr KU, Eden M, Kramer K, Luedde M, Mairbäurl H, et al:
MicroRNA-20a inhibits stress-induced cardiomyocyte apoptosis
involving its novel target Egln3/PHD3. J Mol Cell Cardiol.
52:711–717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dill H, Linder B, Fehr A and Fischer U:
Intronic miR-26b controls neuronal differentiation by repressing
its host transcript, ctdsp2. Genes Dev. 26:25–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang H, Liu P, Yang L and Xie X, Ye F, Wu
M, Liu X, Chen B, Zhang L and Xie X: miR-185 suppresses tumor
proliferation by directly targeting E2F6 and DNMT1 and indirectly
upregulating BRCA1 in triple-negative breast cancer. Mol Cancer
Ther. 13:3185–3197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tang H, Deng M, Tang Y and Xie X, Guo J,
Kong Y, Ye F, Su Q and Xie X: miR-200b and miR-200c as prognostic
factors and mediators of gastric cancer cell progression. Clin
Cancer Res. 19:5602–5612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun Y, Shen S, Tang H, Xiang J, Peng Y,
Tang A, Li N, Zhou W, Wang Z, Zhang D, et al: miR-429 identified by
dynamic transcriptome analysis is a new candidate biomarker for
colorectal cancer prognosis. OMICS. 18:54–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giordano S and Columbano A: MicroRNAs: new
tools for diagnosis, prognosis, and therapy in hepatocellular
carcinoma? Hepatology. 57:840–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang X, Huang H, Li Z, He C, Li Y, Chen
P, Gurbuxani S, Arnovitz S, Hong GM, Price C, et al: MiR-495 is a
tumor-suppressor microRNA down-regulated in MLL-rearranged
leukemia. Proc Natl Acad Sci USA. 109:19397–19402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ralfkiaer U, Hagedorn PH, Bangsgaard N,
Løvendorf MB, Ahler CB, Svensson L, Kopp KL, Vennegaard MT,
Lauenborg B, Zibert JR, et al: Diagnostic microRNA profiling in
cutaneous T-cell lymphoma (CTCL). Blood. 118:5891–5900. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao X, Pfaff SL and Gage FH: A functional
study of miR-124 in the developing neural tube. Genes Dev.
21:531–536. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Taniguchi K, Sugito N, Kumazaki M,
Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama
K, et al: MicroRNA-124 inhibits cancer cell growth through
PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett.
363:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun Y, Ai X, Shen S and Lu S:
NF-κB-mediated miR-124 suppresses metastasis of non-small-cell lung
cancer by targeting MYO10. Oncotarget. 6:8244–8254. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Han ZB, Yang Z, Chi Y, Zhang L, Wang Y, Ji
Y, Wang J, Zhao H and Han ZC: MicroRNA-124 suppresses breast cancer
cell growth and motility by targeting CD151. Cell Physiol Biochem.
31:823–832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li L, Luo J, Wang B, Wang D, Xie X, Yuan
L, Guo J, Xi S, Gao J, Lin X, et al: Microrna-124 targets
flotillin-1 to regulate proliferation and migration in breast
cancer. Mol Cancer. 12:1632013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li W, Zang W, Liu P, Wang Y, Du Y, Chen X,
Deng M, Sun W, Wang L, Zhao G, et al: MicroRNA-124 inhibits
cellular proliferation and invasion by targeting Ets-1 in breast
cancer. Tumour Biol. 35:10897–10904. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Casas E, Kim J, Bendesky A, Ohno-Machado
L, Wolfe CJ and Yang J: Snail2 is an essential mediator of
Twist1-induced epithelial mesenchymal transition and metastasis.
Cancer Res. 71:245–254. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Katafiasz D, Smith LM and Wahl JK III:
Slug (SNAI2) expression in oral SCC cells results in altered
cell-cell adhesion and increased motility. Cell Adhes Migr.
5:315–322. 2011. View Article : Google Scholar
|
|
50
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tang P, Yu Z, Zhang K, Wang Y, Ma Z, Zhang
S, Chen D and Zhou Y: Slug down-regulation by RNA interference
inhibits invasion growth in human esophageal squamous cell
carcinoma. BMC Gastroenterol. 11:602011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang C, Liu X, Huang H, Ma H, Cai W, Hou
J, Huang L, Dai Y, Yu T and Zhou X: Deregulation of Snai2 is
associated with metastasis and poor prognosis in tongue squamous
cell carcinoma. Int J Cancer. 130:2249–2258. 2012. View Article : Google Scholar : PubMed/NCBI
|