Introduction
Endometrial carcinoma is the most frequent
malignancy in female genital tract accounting for 1–2% mortality
rate of all cancer in the western world (1). According to the statistic data
collected from 2000 to 2011 in China, the morbidity has increased
by 3.7% every year (2). Lack of an
early diagnostic marker and therapeutic targets is the main cause
of EC deaths.
In general, EC is mainly classified into two major
types (type I and type II). Type I ECs comprise 80% of new cases
(3). They are histologically well
and moderately differentiated, estrogen-dependent and less prone to
be aggressive. Type II ECs are mostly low differentiated,
estrogen-independent and with a tendency to recur, even at early
stage. The hormonal imbalance and the genetic alterations are
proved to be the main cause of EC (4,5). Most
of EC cases are diagnosed after menopause. The use of estrogen only
replacement therapy and late menopause onset can increase the risk
of EC. Genetic alteration, such as MLH1 (6), MSH2 (7), PMS2 (8), MSH6 (9), ARID1A (10), PTEN (11), KRAS2 (12), CTNNB1 (13), RB1 (11), TP53 (12), c-Kit (14) gene mutations or gene loss have been
demonstrated to be associated with the increased risk of EC, ATR
mutation in endometrioid carcinoma (EEC) is associated with
aggressiveness of the EEC (15).
Thus, we believe that new molecular-based subtype classification
can help for the early diagnosis and treatment decision.
Recently, the non-coding RNAs, including miRNA,
lncRNAs (longer than 200 bp, not encoding proteins, transcribed by
RNA polymerase II) which account for approximately 4/5 of all human
transcripts are proved to be critical regulators of cancers,
including EC. They are involved in nearly all of the metastatic
processes of cancer, such as proliferation, invasion and metastasis
through regulating the protein coding gene expression
transcriptionally, post-transcriptionally or by functioning as a
scaffold of the lncRNA, miRNA and protein interaction (16). Besides, a variety of lncRNAs have
been found to be the prognostic marker of diseases, such as breast
(17), lung (18) and ovarian cancer (19). Thus, the exploration of lncRNA
expression profile is essential for the discovery of early
diagnostic markers and the potential therapeutic candidates.
Notably, Zhai et al (20),
reported the differentially lncRNA in randomly selected in EC
samples and the adjacent non-tumor tissues. However, the mechanism
of the type I EC and type II EC is quite different, the
differential lncRNAs in type I EC and the normal endometrium (NE)
from the uterine leiomyoma patients which will give us the
important information of the potential early diagnostic marker is
poorly understood. In the present study, we aimed to find the
potential early diagnostic and therapeutic candidates of lncRNAs
for EC. We first utilized the high throughput lncRNA microarray
analysis to find the differentially expressed lncRNAs. Then, we
further analyzed the lncRNA function by Gene Ontology (GO), Kyoto
Encyclopedia of Genes and Genomes (KEGG), mRNA-lncRNA co-expression
network as well as the TCGA database to find the potential core
lncRNAs that may be involved in EC. This would be helpful for
exploring the mechanism of EC development as well as for prediction
of clinical outcomes.
Materials and methods
Preparation of samples
Samples of human endometrial carcinoma (EC) and the
NE (from the patients with hysterectomy) were collected immediately
after surgery at the Obstetrics and Gynecology Hospital, affiliated
to Nanjing Medical University. Tissue samples were collected with
the consent of the patients and approved by the Ethics Committees
in the hospital. In addition, all samples were pathologically
confirmed. All samples were placed in TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) and stored at −80°C. RNA for RT-PCR was
extracted using the RNeasy Mini kit (Qiagen, Valencia, CA, USA)
according to the manufacturers protocol. RNA samples for microarray
were extracted by Guangzhou RiboBio Co., Ltd., (Guangzhou, China).
Total RNA (1 µg) was reversely transcribed using the Thermo
Scientific ReverseAid First Strand cDNA Synthesis kit (Thermo
Fisher Scientific, Wilmington, DE, USA).
Microarray
RNA preparation and microarray hybridization were
performed by Guangzhou RiboBio Co., Ltd., according to their
manufacturers standard protocols. Briefly, RNA was purified from
total RNA after removal of rRNA. Approximately 17899 lncRNAs and
26363 mRNAs are detected using the lncRNA microarray.
Quantitative real-time (qRT) PCR
qRT-PCR was performed using SYBR-Green (Applied
Biosystems, Shanghai, China) on an Applied Biosystems ViiA™ 7 DX
machine (Life Technologies, Carlsbad, CA, USA). Assays were run in
triplicate or quadruplicate and values were normalized to
glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Primers for each
gene and lncRNA were designed according to the NCBI online
primer-blast tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and
sequences are listed in Table I.
The PCR conditions were 95°C for 30 sec, followed by 40 cycles of
95°C for 5 sec, 60°C for 31 sec.
 | Table I.Details of primer pairs used in
analysis of lncRNA and mRNA expression by qRT-PCR. |
Table I.
Details of primer pairs used in
analysis of lncRNA and mRNA expression by qRT-PCR.
| Gene | Forward primer
(5–3) | Reverse primer
(5–3) |
|---|
| KIAA0087 |
TCGGGGGACCGAGAAATACT |
CCAGTCCAAGAGAAGCAGCA |
| RP11-508N22 |
TTAGCAGCACATGCTCACCA |
GGTAAGTAGCTGGGCTGTGG |
| RP11-501O2 |
ACGATGAGACTTGGTGCTGAA |
TGGTCCTGTTTCTCGCTGAC |
| FAM212B-AS1 |
CCAGAGAAGAAAGGCAGCGA |
ACTCCTTCGCGTGTTCAGTT |
| LOC102723552 |
ACGAGATGCAGAAGATTACGC |
TGAAATCTTACCTGACTGCAGATT |
| RP11-140I24 |
TCAGTCGTCTTCACGCTTCC |
CAGATTAGCTGGCAGAGGGG |
| RP11-600K151 |
AGGGCTCAGTAGATTTGCCC |
AGGCAGCATCTCACCTAACG |
| ALCAM |
CTGGAGTACAAGACAACCAAGG |
GTCACCTGCTCTGTAGGATAG |
| POLR3E |
TTTCAGTACCCTGTGCGTCC |
TGGGCTTGATCTTGGCTGAG |
| TDRD10 |
GGAGATCCTGTTGGAAAGCA |
GCCAACATACACCTCTGTCTC |
Statistical analysis
The data were analyzed using the Microsoft Excel
2007 (Microsoft, Redmond, WA, USA). The values are presented as
mean ± SEM. Student's t-test was used for the comparison of two
independent groups (P<0.05, P<0.01 and P<0.001).
Results
Clinical characteristics of patient
samples
EC samples were collected from 12 patients and the
control endometrium were collected from at least 18 patients who
underwent hysterectomy after having been diagnosed with uterine
leiomyoma. According to the clinicopathological study, all the
patients are type I EC at stage I/II. Pathology analysis showed
that all EC patients are
PR+;ER+;Ki-67+ (30–80%) with
middle/high differentiated tissue morphology.
lncRNA microarray profiling
To find the potential roles of lncRNA function in
EC, we first examined the differentially expressed lncRNA by
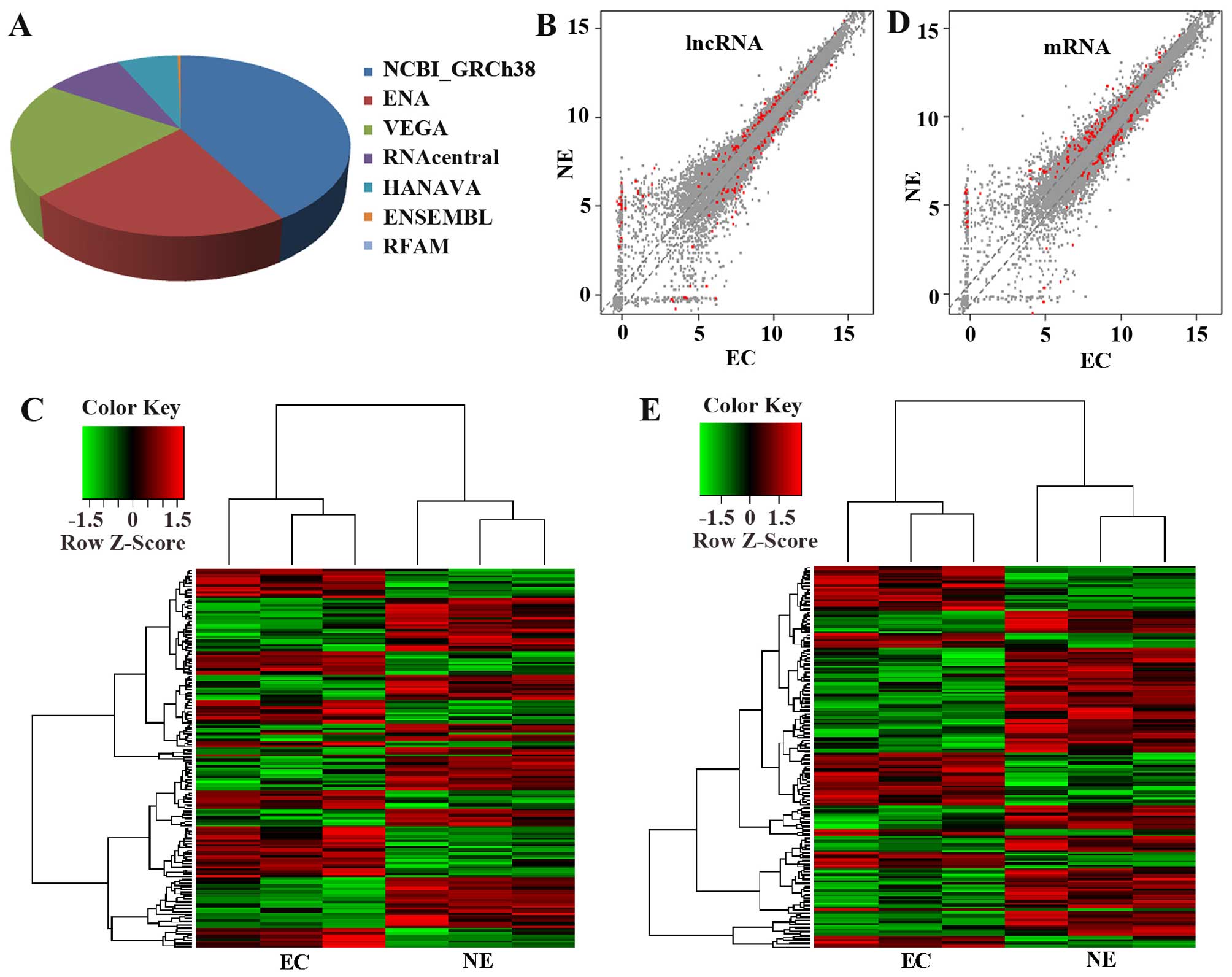
microarray profiling (Fig. 1).
lncRNAs were collected from the authoritative database NCBI-GRCh38,
ENA, VEGA, RNAcentral, HANAVA, ENSEMBL, RFAM (Fig. 1A). In the data, a total of 17899
lncRNA and 26363 mRNA were detected with 172 lncRNA and 188 mRNA
differentially expressed between type I EC (stage I/II) and their
relative control endometrium (fold >1.5 and P<0.05) as shown
in the scatter plot data (Fig. 1B and
D) (the red dots indicate the differentially expressed lncRNA
and mRNA with fold >1.5 and P<0.05) and clustering data
(Fig. 1C and E; fold >1.5 and
P<0.05). The detailed information of the differentially lncRNAs
and mRNAs are presented in Tables
II and III with the fold
change >5 and P<0.05.
 | Table II.Dysregulated lncRNAs in the EC
compared with the NE filtered by a fold-change >5.0 and
P<0.05 |
Table II.
Dysregulated lncRNAs in the EC
compared with the NE filtered by a fold-change >5.0 and
P<0.05
|
Transcript:seqname | Regulation | Ratio | Chromosome | Strand | Database |
|---|
|
URS0000781FB6:URS0000781FB6 | Downregulation | 47.28 | chr13 | − | RNAcentral |
|
ENST00000624452:RP1-41C23 | Downregulation | 61.15 | chr17 | − | HAVANA |
|
URS00004EDFF6:AC013461 | Downregulation | 89.16 | chr2 | − | VEGA |
|
XR_243449;XR_243450;XR_243448:
RP11-7O141 | Downregulation | 33.50 | chr16 | + | NCBI_GRCh38 |
|
XR_430290:LOC102723552 | Downregulation | 11.26 | chr20 | − | NCBI_GRCh38 |
|
ENST00000624279:RP11-140I24 | Downregulation | 42.61 | chr16 | − | HAVANA |
| XR_427960:TRDN | Downregulation | 19.50 | chr6 | − | NCBI_GRCh38 |
|
URS0000415D28:RP11-311B18 | Downregulation | 39.35 | chr10 | − | VEGA |
|
URS0000506C5B:AC003090 | Downregulation | 54.03 | chr7 | − | VEGA |
|
XR_426628:ANKRD13C | Downregulation | 27.04 | chr1 | − | NCBI_GRCh38 |
|
URS000034FDBD:RP11-508N22 | Downregulation | 45.24 | chr10 | + | VEGA |
|
XR_427604:LOC102724353 | Downregulation | 27.03 | chr4 | − | NCBI_GRCh38 |
|
NR_002319:PIPSL | Downregulation | 20.52 | chr10 | − | NCBI_GRCh38 |
|
URS00004A7EA2:CTD-2410N18 | Downregulation | 31.52 | chr5 | + | VEGA |
|
URS00001B388A:AC097517 | Downregulation | 15.44 | chr2 | − | VEGA |
|
ENST00000620339:KLF3-AS1 | Downregulation | 35.30 | chr4 | − | HAVANA |
|
URS00000000FD:AC113618 | Downregulation | 30.04 | chr2 | + | ENA |
|
URS0000258264:RP11-501O2 | Downregulation | 18.88 | chr3 | + | ENA |
|
NR_022006:KIAA0087 | Downregulation | 9.25 | chr7 | − | NCBI_GRCh38 |
|
NR_039986:RP11-600K151 | Downregulation | 18.06 | chr8 | − | NCBI_GRCh38 |
|
NR_038951;NR_038952:FAM212B-AS1 | Downregulation | 51.75 | chr1 | + | NCBI_GRCh38 |
|
URS000049614A:RP11-140I24 | Downregulation | 10.28 | chr16 | + | ENA |
|
URS000058EAF9:RP11-680F8 | Downregulation | 8.16 | chr15 | + | ENA |
|
URS000041DE60:RP1-35C21 | Upregulation | 12.36 | chr1 | + | ENA |
|
URS000075B1F5:ROR1-AS1 | Upregulation | 10.60 | chr1 | − | RNAcentral |
|
NR_029671:MIR125B1 | Upregulation | 21.10 | chr11 | − | NCBI_GRCh38 |
|
URS00004B2E02:RP11-573J24 | Upregulation | 18.64 | chr8 | + | ENA |
|
URS00005369A5:RP11-474P2 | Upregulation | 32.39 | chr12 | − | VEGA |
|
URS00001CE0A9:AC098828 | Upregulation | 15.07 | chr2 | − | VEGA |
|
XR_243832:RP11-856M71 | Upregulation | 78.22 | chr18 | + | NCBI_GRCh38 |
|
ENST00000623186:RP11-449M6 | Upregulation | 19.09 | chr8 | + | HAVANA |
|
URS0000610163:RP11-539G18 | Upregulation | 7.00 | chr4 | + | ENA |
|
NR_027333:GPR158-AS1 | Upregulation | 7.01 | chr10 | − | NCBI_GRCh38 |
|
URS000032778C:RP11-1143G9 | Upregulation | 9.89 | chr12 | − | VEGA |
 | Table III.Dysregulated mRNAs in the EC compared
with the NE filtered by a fold-change >5.0 and P<0.05. |
Table III.
Dysregulated mRNAs in the EC compared
with the NE filtered by a fold-change >5.0 and P<0.05.
| Name | Regulation | Ratio |
|---|
| CLDN23 | Downregulation | 64.81788 |
| NEIL3 | Downregulation | 60.71055 |
| ALCAM | Downregulation | 27.00711 |
| TMEM68 | Downregulation | 68.43251 |
| POLR3E | Downregulation | 18.53199 |
| TDRD10 | Downregulation | 18.66308 |
| SELP | Downregulation | 40.2663 |
| FGFR2 | Downregulation | 15.83808 |
| DISP1 | Downregulation | 32.15958 |
| TBC1D22A | Downregulation | 27.66319 |
| GALK2 | Downregulation | 19.62079 |
| RTKN2 | Upregulation | 39.20362 |
| CADPS | Upregulation | 40.45117 |
| NLRP10 | Upregulation | 16.82022 |
| UPP1 | Upregulation | 5.53434 |
| EPC2 | Upregulation | 23.74738 |
| NXNL2 | Upregulation | 37.67642 |
| TXNL4B | Downregulation | 5.196587 |
| PCDH17 | Downregulation | 8.172227 |
| HSD17B2 | Downregulation | 7.949501 |
| LRRC1 | Downregulation | 5.796955 |
| STK25 | Downregulation | 5.488636 |
Expression signature of dysregulated
lncRNAs in EC compared to NE
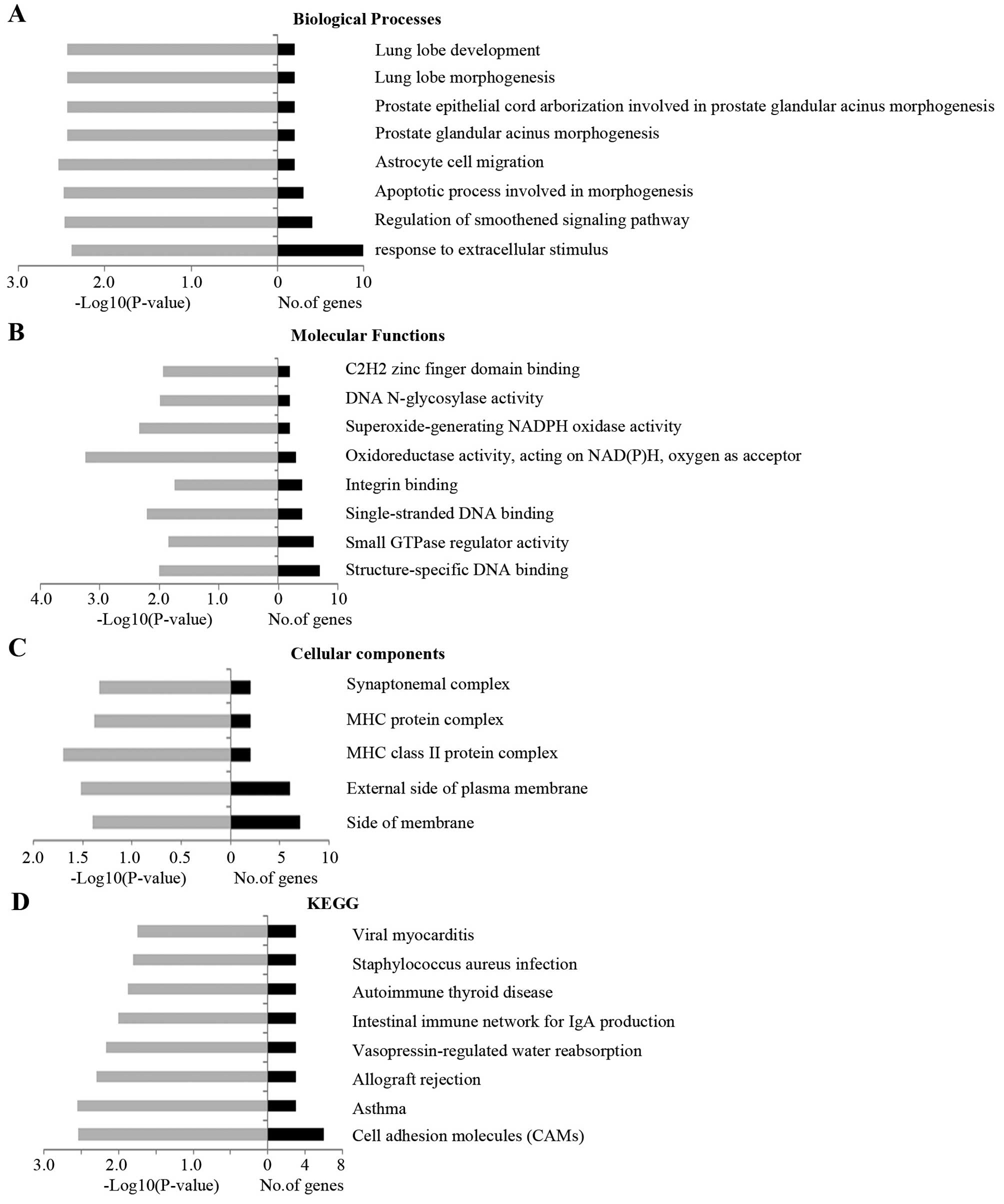
To investigate the signature of the dysregulated
lncRNAs in EC, we studied the dysregulated lncRNA classification,
lengths and chromosome distribution (Fig. 2). In the present study, the majority
of the lncRNAs that were differentially expressed in EC were
intergenic lncRNAs, antisense lncRNAs as well as the TEC (to be
experimentally confirmed) lncRNAs (Fig.
2A). The lncRNA lengths are mainly between 400 and 64000 bp
(Fig. 2B). The upregulated lncRNAs
and the downregulated lncRNAs are distributed in different
chromosomes (Fig. 2C and D).
GO and pathway analysis
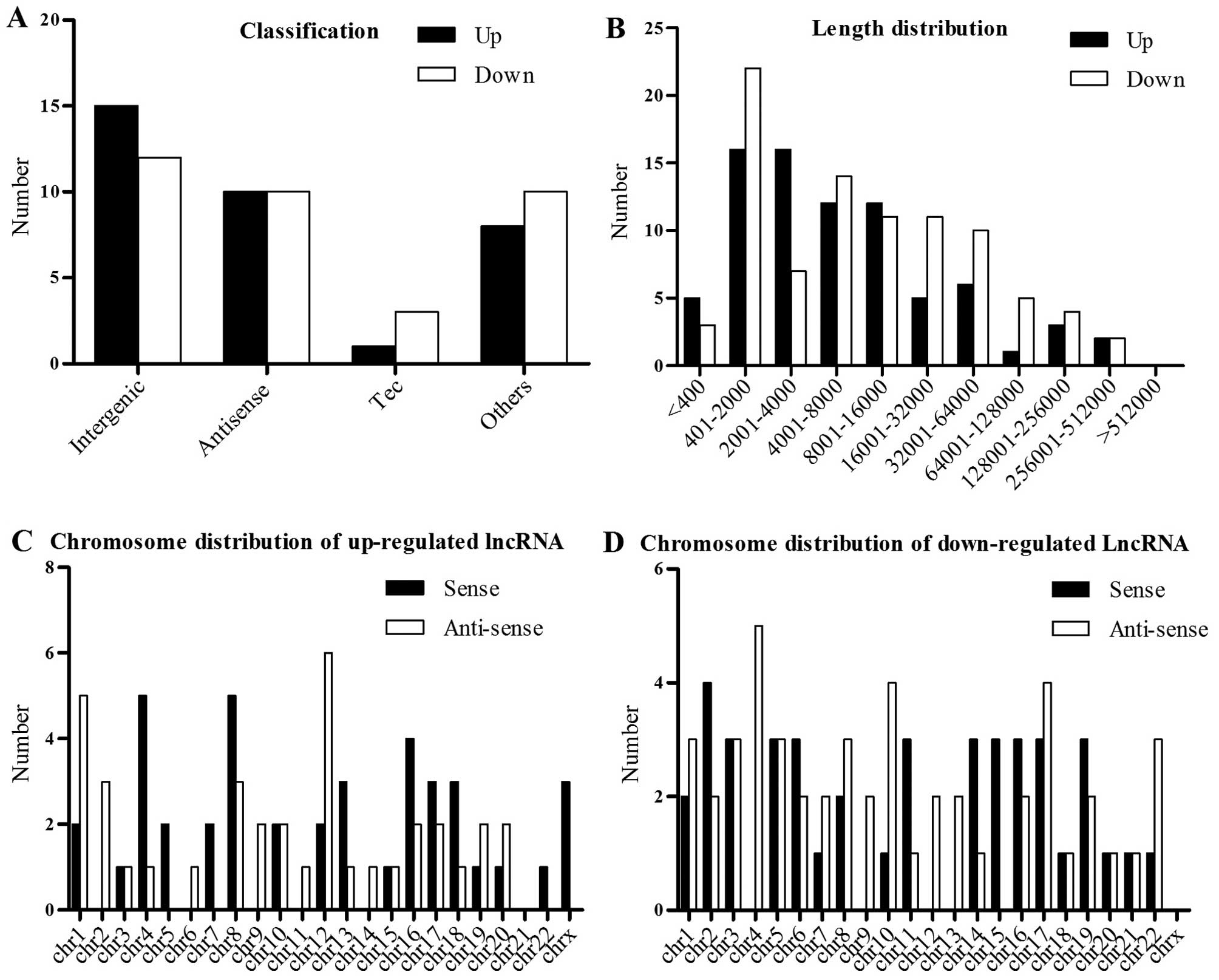
To explore the potential function of the
dysregulated lncRNAs in EC, we predicted the lncRNA target genes
according to their chromosome location. In addition, we carried out
GO and pathway analysis of these lncRNAs. The Gene Ontology
consortium which incorporate many databases and developed three
ontologies (biological processes, cellular components and molecular
functions) (http://www.geneontology.org) revealed that numerous
biological processes, cellular components and molecular function
are altered in EC compared to control (Fig. 3A-C). To find which pathways are
altered, we also mapped the genes to KEGG pathways. It is
noteworthy that the plasma components which are the main components
during cell response to the intracellular or extracellular signals,
such as the cell adhesion molecules are the most dysregulated
molecules in EC as shown in the result of biological pathway
analysis, the cellular component and KEGG pathway analyses
(Fig. 3A, C and D). Besides, the
structure specific DNA binding components which may function during
transcription and the oxidoreductase component which may be
involved in cell metabolism and other processes are also
dysregulated in EC (Fig. 3B).
Cell adhesion molecules are important for tissue
integrity and also cell migration in tumor cells or other
developmental cells (16,21). E-cadherin, N-cadherin and integrins
which are the most famous adhesion molecules have been demonstrated
to be associated with cell invasion of various cancer types
(22–24), the transmembrane glycoprotein ALCAM
(also known as CD166) which belongs to the immunoglobulin
superfamily has been considered as the marker of breast cancer
(25). In the present study, the
cell adhesion molecules, such as ALCAM are also the most
dysregulated mRNA.
mRNA-lncRNA co-expression network
lncRNA function has been implicated in several steps
of carcinogenesis by acting as structural, catalytic or regulatory
RNA or interacting with DNA, RNA and protein (16). However, only a few lncRNA functions
have been elucidated. mRNA and lncRNA co-expression network, which
can provide evidence for the involvement of new genes or lncRNAs in
core biological functions, is very important for the prediction of
the lncRNA function preliminarily (19,26).
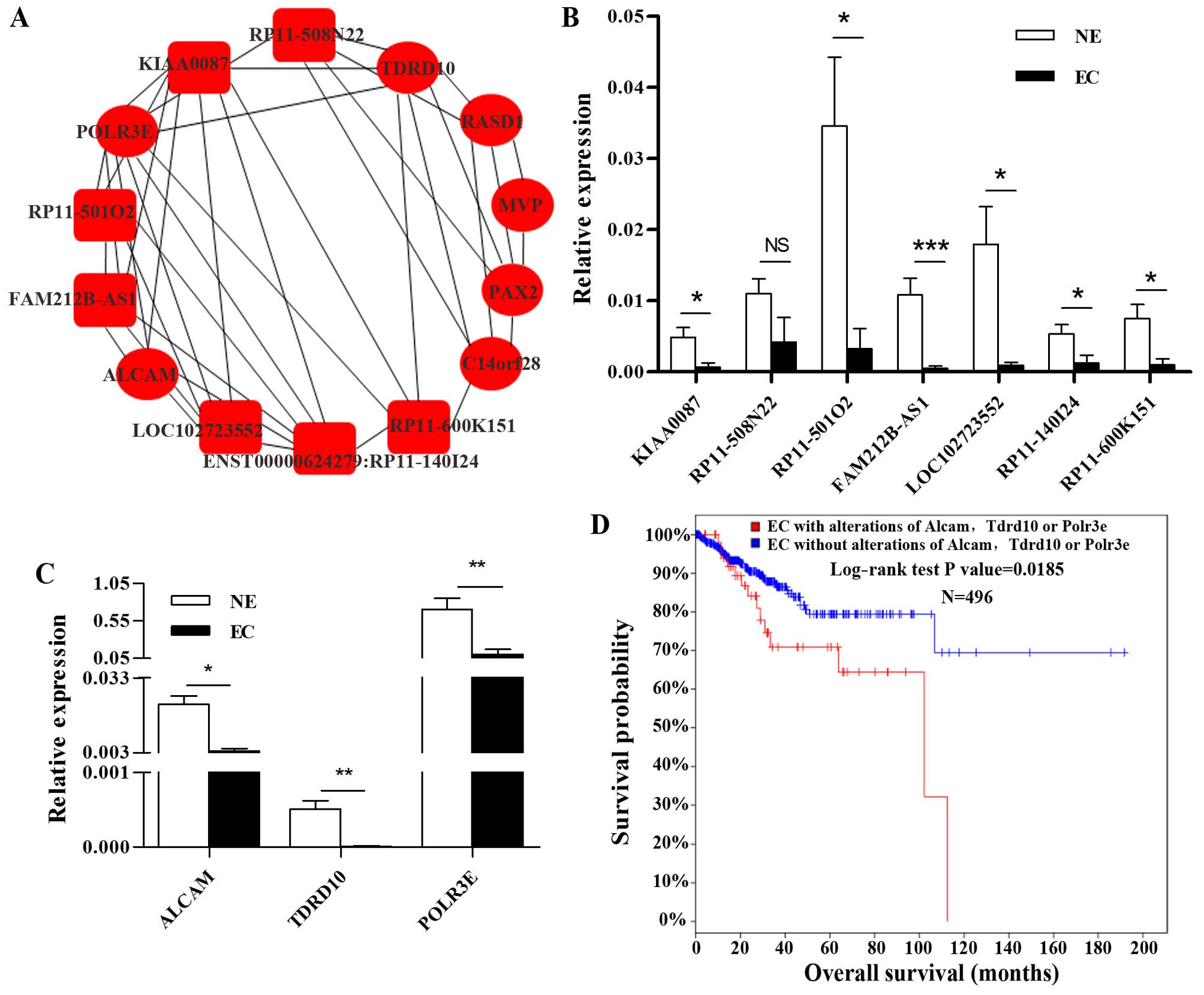
Thus, we carried out the lncRNA and mRNA co-expression network in
Guangzhou RiboBio Co. The co-expression network in Fig. 4A shows that ALCAM is co-expressed
with 5 lncRNAs (ENST00000624279:RP11-140I24, LOC102723552,
KIAA0087, RP11-600K151 and FAM212B-AS1) and 1 mRNA (POLR3E), POLR3E
is co-expressed with 6 lncRNAs (FAM212B-AS1, RP11-140I24,
LOC102723552, RP11-508N22, RP11-501O2 and KIAA0087) and 2 mRNAs
(ALCAM and TDRD10), TDRD10 is co-expressed with 3 lncRNAs
(RP11-600K151, RP11-508N22 and KIAA0087) and 1 mRNA (POLR3E) which
are differentially expressed in EC (>5-fold, P<0.05).
Real-time PCR verified that 6 of the 7 lncRNAs and all of the 3
mRNAs are dramatically downregulated in EC, which is consistent
with the microarray data (Fig. 4B and
C). Interestingly, most of these lncRNAs are relatively
conserved between human, rhesus, rat and mouse as analyzed using
UCSC website (http://genome.ucsc.edu/) (data not
shown).
The Cancer Genome Atlas (TCGA) database includes
most of the tumor types and information on each case for community
research use. cBioPortal for Cancer Genomics provides visualization
and analysis of the TCGA datasets. Notably, the results presented
in the cBioPortal of cancer genomics (http://www.cbioportal.org/) showed that EC cases with
ALCAM, TDRD10 and/or POLR3E alterations showed poorer overall
survival rate as compared with cases without alterations of these
genes (Fig. 4D).
Discussion
According to the ENCODE Project Consortium, the
protein coding genes only account for 1% of the human genome
(27). Most of the regions are not
transcribed, such as a large class of the non-coding RNA. lncRNA
functions as structural, catalytic, regulatory RNAs have been found
to be involved in a variety of processes, such as development
(18), tumorigenesis (16), cardiovascular disease (28) and neurodegenerative diseases
(29). Similarly to the protein
coding genes, lncRNAs which are distributed in the cytoplasm,
nucleus and also transported to the extracellular matrix can be
identified as the biomarkers of various diseases. For example,
studies found that lncRNA-DANCR can be used as a prognostic
biomarker of hepatocellular carcinoma (30). The circulating lncRNA-LIPCAR can be
used to predict survival of patients with heart failure (31). lncRNAs are also known as biomarkers
or therapeutic target of cancers, such as breast (32), lung (33) and ovarian cancer (19). Since most of the lncRNAs function
through the regulation of the mRNAs the lncRNA-mRNA co-expression
network provides us with a powerful platform for the prediction of
the lncRNA function (19). For
example, RP11-284N8.3.1 and AC104699.1.1 which were identified
using the lncRNA-mRNA co-expression network are found to be the
independent predictive biomarkers of ovarian cancer (19). In this study, we first used
microarray analysis to find the differentially expressed lncRNAs in
EC, and then by using GO and KEGG as well as the lncRNA-mRNA
co-expression network, we predicted that the 6 lncRNAs and 3 mRNAs
are probably the early biomarkers of EC.
The glycolysis which produces ATP more quickly is
strongly enhanced in tumor cells (34,35).
Here, we used the GO analysis to prove that the main molecular
function involved is the oxidoreductase activity, emphasizing the
difference between the metabolism between the tumor and normal
tissue (Fig. 3B).
The adhesion molecules have been demonstrated to be
the important regulators of endometrial epithelium integrity, its
decreased expression will disrupt the integrity of the NE and may
be involved in the pathology of the endometrium (22). Asthma which affects 5–10% of the
population may increase the patients risk of cancer is also altered
in EC as compared with control (36). The chronic inflammatory disease
associated gene has been found to help cancers metastasize and may
be the predictor of brain cancer in children (37,38).
Notably, the KEGG analysis showed that the adhesion molecules and
asthma associated genes are the top two differentially altered
pathways in EC.
Endometrial carcinoma which is a common malignancy
of female reproductive tract accounts for ~1–2% of the tumor deaths
in the western world (1). It is
also becoming more prevalent in China each year. Thus, it is very
urgent for us to find the predictive biomarkers of EC. Here, in
order to find whether lncRNAs can be used as EC biomarkers, we
performed the lncRNA microarray. By using the microarray data and
the lncRNA-mRNA co-expression network, we found 6 lncRNAs
(KIAA0087, RP11-501O2, FAM212B-AS1, LOC102723552, RP11-140I24 and
RP11-600K151) and 3 mRNAs (ALCAM, TDRD10 and POLR3E) that are
functionally related in EC. Interestingly, ALCAM is highly
expressed in the endometrium glandular cells and its expression was
decreased in ~50% of the endometrial cancer tissues while POLR3E is
moderately expressed in the endometrium glandular cells and its
expression was decreased in ~50% of EC according to the human
protein atlas website which is consistent with our data (http://www.proteinatlas.org/). The TCGA data also
confirmed that these three gene alterations indicated poorer
overall survival as compared with the cases without these gene
alterations. We also validated that the lncRNAs which are
co-expressed with these three mRNAs are downregulated in EC.
Since the mechanism of the type I EC and type II EC
is quite different, distinguish the different type of EC is quite
important. Different from the differentially expressed lncRNAs
between the randomly selected EC samples and the adjacent non-tumor
tissue reported before (20), we
reported the differentially expressed lncRNAs in type I EC as
compared with NE using the microarray and the lncRNAs and mRNA
co-expression network for the first time. The differentially
expressed lncRNAs may be indicators of the type I EC and may
provide the potential information for the evaluation of EC.
Acknowledgements
The present study was financially supported by the
National Natural Science Foundation of China (81302304 and
81572556) and the Key Program of Science and Technology Development
Fund of Nanjing Medical University (2013NJMU145, 2014NJMU103,
2014NJMU098 and 2015NJMUZD063).
Glossary
Abbreviations
Abbreviations:
|
EC
|
endometrial carcinoma
|
|
EEC
|
endometrioid carcinoma
|
|
lncRNA
|
long non-coding RNA
|
|
mRNA
|
messenger RNA
|
|
GAPDH
|
glyceradehyde 3-phosphate
dehydrogenase
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
UCSC
|
University of California Santa
Cruz
|
References
|
1
|
Plataniotis G and Castiglione M: ESMO
Guidelines Working Group: Endometrial cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21:(Suppl 5). v41–v45. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bokhman JV: Two pathogenetic types of
endometrial carcinoma. Gynecol Oncol. 15:10–17. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weiderpass E, Persson I, Adami HO,
Magnusson C, Lindgren A and Baron JA: Body size in different
periods of life, diabetes mellitus, hypertension, and risk of
postmenopausal endometrial cancer (Sweden). Cancer Causes Control.
11:185–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yasa C, Takmaz O, Dural O and Akhan SE:
The value of tumor markers in endometrial carcinoma: Review of
literature. J Cancer Ther. 4:966–970. 2013. View Article : Google Scholar
|
|
6
|
Dudley B, Brand RE, Thull D, Bahary N,
Nikiforova MN and Pai RK: Germline MLH1 mutations are frequently
Identified in Lynch Syndrome patients withcolorectal and
endometrial carcinoma demonstrating isolated loss of PMS2
immunohistochemical expression. Am J Surg Pathol. 39:1114–1120.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vasen HF, Stormorken A, Menko FH,
Nagengast FM, Kleibeuker JH, Griffioen G, Taal BG, Moller P and
Wijnen JT: MSH2 mutation carriers are at higher risk of cancer than
MLH1 mutation carriers: A study of hereditary nonpolyposis
colorectal cancer families. J Clin Oncol. 19:4074–4080.
2001.PubMed/NCBI
|
|
8
|
Basil JB, Swisher EM, Herzog TJ, Rader JS,
Elbendary A, Mutch DG and Goodfellow PJ: Mutational analysis of the
PMS2 gene in sporadic endometrial cancers with microsatellite
instability. Gynecol Oncol. 74:395–399. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Devlin LA, Graham CA, Price JH and
Morrison PJ: Germline MSH6 mutations are more prevalent in
endometrial cancer patient cohorts than hereditary non polyposis
colorectal cancer cohorts. Ulster Med J. 77:25–30. 2008.PubMed/NCBI
|
|
10
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Albitar L, Carter MB, Davies S and Leslie
KK: Consequences of the loss of p53, RB1, and PTEN: Relationship to
gefitinib resistance in endometrial cancer. Gynecol Oncol.
106:94–104. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Swisher EM, Peiffer-Schneider S, Mutch DG,
Herzog TJ, Rader JS, Elbendary A and Goodfellow PJ: Differences in
patterns of TP53 and KRAS2 mutations in a large series of
endometrial carcinomas with or without microsatellite instability.
Cancer. 85:119–126. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Senol S, Sayar I, Ceyran AB, Ibiloglu I,
Akalin I, Firat U, Kosemetin D, Engin ZP and Aydin A: Stromal clues
in endometrial carcinoma: Loss of expression of beta-catenin,
epithelial-mesenchymal transition regulators, and
estrogen-progesterone receptor. Int J Gynecol Pathol. 35:238–248.
2015. View Article : Google Scholar
|
|
14
|
Kafshdooz T, Ardabili SM Mohaddes,
Kafshdooz L, Tabrizi AD, Ghojazadeh M, Gharesouran J and Akbarzadeh
A: C-kit mutations in endometrial cancer: Correlation with tumor
histologic type. Asian Pac J Cancer Prev. 16:7449–7452. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zighelboim I, Schmidt AP, Gao F, Thaker
PH, Powell MA, Rader JS, Gibb RK, Mutch DG and Goodfellow PJ: ATR
mutation in endometrioid endometrial cancer is associated with poor
clinical outcomes. J Clin Oncol. 27:3091–3096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv M, Xu P, Wu Y, Huang L, Li W, Lv S, Wu
X, Zeng X, Shen R, Jia X, et al: LncRNAs as new biomarkers to
differentiate triple negative breast cancer from non-triple
negative breast cancer. Oncotarget. 7:13047–13059. 2016.PubMed/NCBI
|
|
18
|
Herriges MJ, Swarr DT, Morley MP, Rathi
KS, Peng T, Stewart KM and Morrisey EE: Long noncoding RNAs are
spatially correlated with transcription factors and regulate lung
development. Genes Dev. 28:1363–1379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo Q, Cheng Y, Liang T, He Y, Ren C, Sun
L and Zhang G: Comprehensive analysis of lncRNA-mRNA co-expression
patterns identifies immune-associated lncRNA biomarkers in ovarian
cancer malignant progression. Sci Rep. 5:176832015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhai W, Li X, Wu S, Zhang Y, Pang H and
Chen W: Microarray expression profile of lncRNAs and the
upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int J
Oncol. 46:2125–2137. 2015.PubMed/NCBI
|
|
21
|
Singh H and Aplin JD: Adhesion molecules
in endometrial epithelium: Tissue integrity and embryo
implantation. J Anat. 215:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Avizienyte E, Wyke AW, Jones RJ, McLean
GW, Westhoff MA, Brunton VG and Frame MC: Src-induced de-regulation
of E-cadherin in colon cancer cells requires integrin signalling.
Nat Cell Biol. 4:632–638. 2002.PubMed/NCBI
|
|
23
|
Park JH, Lee BI, Song ES, Whang SO, Lee WY
and Cho SJ: Hypermethylation of E-cadherin in endometrial
carcinoma. J Gynecol Oncol. 19:241–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hecht JL, Dolinski BM, Gardner HA,
Violette SM and Weinreb PH: Overexpression of the alphavbeta6
integrin in endometrial cancer. Appl Immunohistochem Mol Morphol.
16:543–547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kulasingam V, Zheng Y, Soosaipillai A,
Leon AE, Gion M and Diamandis EP: Activated leukocyte cell adhesion
molecule: A novel biomarker for breast cancer. Int J Cancer.
125:9–14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al: Childrens Hospital Oakland Research
Institute: Identification and analysis of functional elements in 1%
of the human genome by the ENCODE pilot project. Nature.
447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Congrains A, Kamide K, Oguro R, Yasuda O,
Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, et
al: Genetic variants at the 9p21 locus contribute to
atherosclerosis through modulation of ANRIL and CDKN2A/B.
Atherosclerosis. 220:449–455. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johnson R: Long non-coding RNAs in
Huntingtons disease neurodegeneration. Neurobiol Dis. 46:245–254.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan SX, Wang J, Yang F, Tao QF, Zhang J,
Wang LL, Yang Y, Liu H, Wang ZG, et al: Long noncoding RNA DANCR
increases stemness features of hepatocellular carcinoma by
derepression of CTNNB1. Hepatology. 63:499–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumarswamy R, Bauters C, Volkmann I, Maury
F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F and Thum
T: Circulating long noncoding RNA, LIPCAR, predicts survival in
patients with heart failure. Circ Res. 114:1569–1575. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lei B, Xu SP, Liang XS, Li YW, Zhang JF,
Zhang GQ and Pang D: Long non-coding RNA MVIH is associated with
poor prognosis and malignant biological behavior in breast cancer.
Tumour Biol. 37:5257–5264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang CH, Qiu J, OSullivan D, Buck MD,
Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ,
et al: Metabolic competition in the tumor microenvironment is a
driver of cancer progression. Cell. 162:1229–1241. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Raccosta L, Fontana R, Corna G, Maggioni
D, Moresco M and Russo V: Cholesterol metabolites and tumor
microenvironment: The road towards clinical translation. Cancer
Immunol Immunother. 65:111–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ji J, Shu X, Li X, Sundquist K, Sundquist
J and Hemminki K: Cancer risk in hospitalised asthma patients. Br J
Cancer. 100:829–833. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roncarolo F and Infante-Rivard C: Asthma
and risk of brain cancer in children. Cancer Causes Control.
23:617–623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin
C and Flavell RA: Inflammation-induced cancer: Crosstalk between
tumours, immune cells and microorganisms. Nat Rev Cancer.
13:759–771. 2013. View Article : Google Scholar : PubMed/NCBI
|