Introduction
Changes in the carbohydrate structure on the surface
of tumor cells are an important feature of cancer metastasis.
Sialic acid, which is ubiquitous at terminal positions of
glycoconjugates and bears net charge, has attracted the increased
attention of researchers (1).
However, the role of sialic acids in the processes involved in
cancer metastasis has not been clarified. Previously we reported
that α2,3-sialic acid residues in breast cancer are associated with
metastatic potential. α2,3-sialyltransferase ST3Gal III transfers
sialic acid with α2,3-linkage to Gal residues located on either
Galb1-4GlcNAc or Galb1-3GlcNAc structures (2).
Changes in ST3Gal III expression have been reported
in several types of tumors. In breast cancer, high expression of
ST3Gal III was found to be positively correlated with the number of
axillary lymph nodes and reduced patient overall survival (3,4). In
extrahepatic bile duct carcinoma, ST3Gal III levels are correlated
with metastasis and poor prognosis (5). In squamous cell carcinoma of the
cervix, ST3Gal III expression levels were obviously increased in
patients with lymph node metastasis when compared to levels in
patients without metastases (6,7). In
colon cancer, ST3Gal III expression was increased in carcinoma
specimens compared with that in non-malignant colorectal mucosa
(8). In gastric cancer, high levels
of ST3Gal III were found to be correlated with tumor recurrence
(9).
Although ST3Gal III expression correlates with tumor
progression in various types of carcinomas (10), the mechanism of ST3Gal III in the
process of metastasis has not been fully clarified. ST3Gal III is
involved in the biosynthesis of sialyl-Lewis antigens, which can
bind to E-selectin expressed on activiated endothelial cells and
mediate hematogenous metastasis (11–13).
In the present study, we investigated the specific influence of
ST3Gal III on key steps in breast cancer progression, such as
adhesion, migration and metastasis. Toward this aim, we chose two
breast cancer cell lines MDA-MB-231 and Bcap37, with different
ST3Gal III expression, and we generated an ST3Gal
III-overexpressing cell line and downregulated ST3Gal III
expression in a cell line by lentiviral transfection. ST3Gal III
expression was proportional to sialyl-Lewis X (SLeX) levels and
modulated the abilities of binding to E-selectin, cell migration
and cell invasion. Furthermore, changes in expression of
invasion-related molecules, including β1 integrin,
matrix metalloproteinase (MMP)-2, MMP-9 and cyclooxygenase-2
(COX-2) were found in both cell models, which could explain the
effect of ST3Gal III on breast cancer cell adhesion and
invasion.
In conclusion, our findings in these novel models of
ST3Gal III expression revealed a critical requirement for ST3Gal
III in several steps of breast cancer metastasis. ST3Gal III
modulated breast cancer cell adhesion and invasion by altering the
expression of invasion-related molecules. This study provides new
insights into the mechanisms underlying metastasis and presents a
new target for the effective drug treatment of breast cancer
metastasis.
Materials and methods
Stable transfection
The construct human α2,3-sialyltransferase (human
ST3Gal III) and short hairpin RNAs (shRNAs) targeting ST3Gal III
and cloned into a lentivirus were synthesized by Suzhou GenePharma
(Suzhou, China). The nucleotide sequence was confirmed by DNA
sequencing. The encoding ST3Gal III lentivirus vector, four shRNA
sequences targeting the ST3Gal III lentivirus vector and the empty
lentivirus vector were transfected into human breast cancer cell
lines MDA-MB-231 and Bcap37 (purchased from the Chinese Academy of
Sciences Cell Bank, Shanghai, China) with different ST3Gal III
expression. Stable transfected cells were selected with 2 µg/ml
puromycin (InvivoGen, San Diego, CA, USA), and resistant clones
were further confirmed by real-time PCR and western blot
analysis.
Culture conditions for the transfected
cells
The stable ST3Gal III-transfected MDA-MB-231 cells
(ST3), shRNA targeting ST3Gal III-transfected MDA-MB-231 cells
(shRNA-1, shRNA-2, shRNA-3 and shRNA-4, respectively),
mock-transfected cells (M) and parental cells (P) were used in this
study. Cells were cultured in L15 medium (Gibco™ Invitrogen Corp.,
Carslbad, CA, USA) containing 10% fetal bovine serum (FBS), 100
U/ml penicillin and 100 mg/ml streptomycin. Stable transfectants
were supplemented with 2 µg/ml puromycin. Cells were fed every 3
days at 37°C in a humid atmosphere of 5% CO2 and
harvested by 0.25% trypsin. Cell growth and morphology were daily
assessed under a field microscope.
ST3Gal III expression by real-time
quantitative PCR
Total cellular RNA was extracted using TRIzol
reagent (Invitrogen). Quantitative real-time PCR was performed
using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd., Dalian,
China) according to the manufacturer's instructions. For real-time
PCR, we used ST3Gal III gene forward, 5′-AGAGAAGGACGGTGCCAAG-3′ and
reverse, 5′-CTGAATGAGGCTGAGTGCTG-3′; β-actin gene forward,
5′-GTGGACATCCGCAAAGAC-3′ and reverse, 5′-GAAAGGGTGTAACGCAACT-3′.
The following standard thermal profile was used for all PCRs: 95°C
for 10 min; 40 cycles of 95°C for 10 sec and 60°C for 31 sec. For
each sample dilution, logarithmic increase in fluorescence signal
(DRn) was obtained. DRn threshold was set at 0.02 to obtain the
corresponding Ct (threshold cycle) values. The endogenous
housekeeping gene β-actin was used to normalize the results.
Results are expressed as mean ± SD.
Flow cytometric analysis
Detection of oligosaccharide epitopes on the cell
surface was performed by indirect fluorescence. Cells
(5×105) were incubated with anti-SLeX monoclonal
antibodies (MAbs) (Calbiochem, EMD Chemicals Inc., San Diego, CA,
USA) at 4°C for 30 min. After washing, the cells were incubated
with the secondary antibody Alexa Fluor 594 goat anti-mouse IgG
(Invitrogen Life Technologies, Frederick, MD, USA). Antibodies were
diluted in phosphate-buffered saline (PBS) containing 1% bovine
serum albumin (BSA). Mean fluorescence intensity (MFI) was
calculated using FACSCalibur (BD Biosciences, Franklin Lakes, NJ,
USA). For each sample three independent assays were performed.
E-selectin binding assay
Adhesion of breast cancer cells to recombinant human
E-selectin (rh-E-selectin) was performed. 96-well plates were
coated with rh-E-selectin (R&D Systems, Inc., Minneapolis, MN,
USA) for 30 min. The plates were blocked and 1×105
viable M, P, ST3 or shRNA-2, shRNA-4 cells, were added and
incubated at 37°C for 1 h. In the selected experiments, the cells
were previously incubated with anti-SLeX MAbs for 30 min at 4°C.
After washes, the adherent cells were fixed with methanol for 30
min. After drying, the fixed cells were stained with 0.1% crystal
violet for 30 min, and then washed and dissolved in 10% acetic
acid. The optical densities of each well were measured by an ELISA
reader at 570 nm. All the experiments were performed in
quintuplicate, and three independent assays were carried out.
Results are expressed as the mean ± SD.
Tumor cell adhesion assay to
HUVECs
Human umbilical vein endothelial cells (HUVECs)
(purchased from the Chinese Academy of Sciences Cell Bank) were
incubated for 12 h with 10 ng/ml recombinant interleukin IL-1β
(R&D Systems, Inc.). Cells were stained with calcein
(Invitrogen Life Technologies) in L15 solution. After washing, the
cells were resuspended and added to a 96-well plate and cultured
with HUVECs. The plates were incubated at 37°C for 30 min, and then
washed three times and the number of adhesive cells was counted
under a fluorescence microscope. Results are expressed as the mean
± SD. Each experiment was performed in triplicate wells and three
independents assays were carried out.
Cell migration and invasion
assays
Tumor cell migration and invasion were assessed
using Transwell chamber (Corning Costar, Inc., Corning, NY, USA).
Briefly, Matrigel was used to coat the upper chamber for the
invasion assay (not for the migration assay). Cells
(1×105) were added to the upper chamber in serum-free
medium, and the lower compartment was filled with media
supplemented with 10% FBS. After incubation at 37°C for 24 h, the
non-migrating cells on the upper surface of the filter were removed
with cotton swabs, and the migrating cells on the lower surface
were fixed and stained with 0.1% crystal violet for 30 min, then
photographed under a fluorescence microscope and incorporated dye
was dissolved in 10% acetic acid. The optical densities of each
well were measured by ELISA reader at 570 nm. Experiments were
performed in triplicate.
Western blot analysis
Total cellular proteins were extracted. Equivalent
amounts of cellular protein were electrophoresed on 10% SDS-PAGE
gel and transfered to nitrocellullose membranes (Millipore Corp.,
Billerica, MA, USA) and blocked in 5% non-fat milk in TBST for 1 h
at room temperature. The cells were incubated with primary
antibodies to human ST3Gal III (Abcam, Cambridge, MA, USA), MMP-9,
MMP-2 (Cell Signaling Technology, Inc., Beverly, MA, USA),
β1 integrin (Chemicon, Temecula, CA, USA), COX-2 and
GAPDH (both from Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) in 5% non-fat milk at 4°C overnight. The membranes were washed
with TBST and incubated with horseradish peroxidase-conjugated
secondary antibody in 5% non-fat milk for 1 h at room temperature.
Immune complexes were detected by enhanced chemoluminescence
techniques (Amersham Life Science, Piscataway, NJ, USA). Band
densities were quantified by using BandScan software.
Statistical analysis
The data are expressed as the mean ± SD and were
analyzed by SPSS 17.0 statistical software to evaluate the
statistical difference. P<0.05 was considered to indicate a
statistically significant result. One-way ANOVA analysis was used
to evaluate all quantitative data.
Results
Stable expression of ST3Gal III in
MDA-MB-231 variant cells
To explore the role of ST3Gal III in breast cancer
metastasis, the human ST3Gal III gene, and shRNA targeting ST3Gal
III were cloned to lentiviral vectors. Breast cancer cells,
MDA-MB-231 and Bcap37, were transfected with the lentiviral vector
encoding the human ST3Gal III gene and MDA-MB-231 cells were
transfected with ST3Gal III shRNAs. Parental cells were
concomitantly transfected with the empty lentiviral vector. Stable
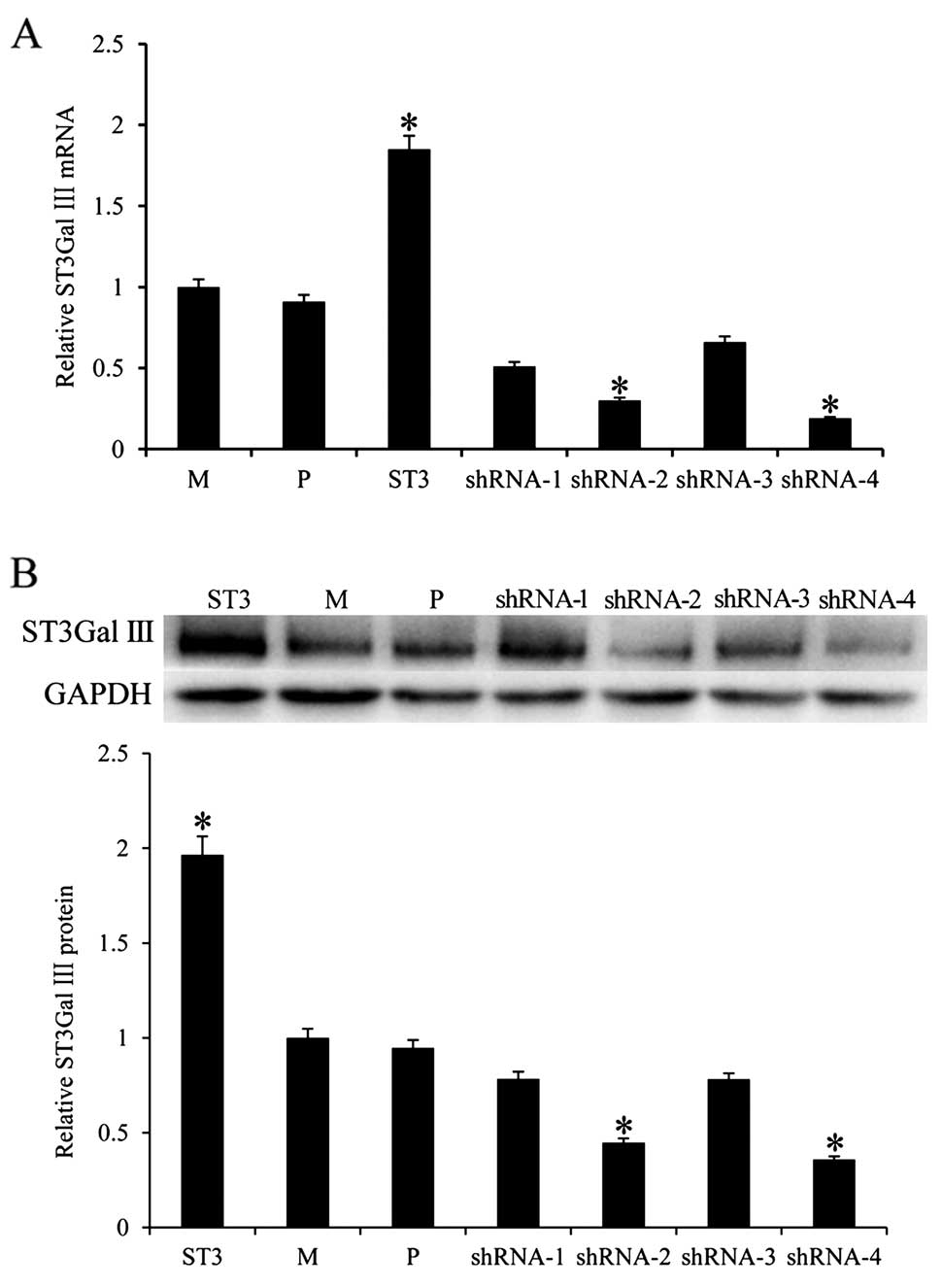
cell clones were selected by puromycin and the mRNA levels of
ST3Gal III were quantified by real-time PCR (Fig. 1A). ST3Gal III mRNA expression in the
stable ST3Gal III-transfected MDA-MB-231 cells (ST3 cells) was
1.846-fold higher (P<0.05) than that in the parental (P) cells.
For the Bcap37 cell model, there was no significant difference
between the Bcap37 ST3 and P cells (data not shown). In contrast,
ST3Gal III mRNA expression was 3.33-fold lower (P<0.05) in the
shRNA-2-transfected cells and 5.26-fold lower (P<0.05) in the
shRNA-4-transfected cells than that in the P cells. The protein
expression of ST3Gal III was examined by western blot analysis. As
expected, protein expression of ST3Gal III was increased in the ST3
cells (P<0.05) and decreased in the shRNA-2 and shRNA-4
transfected cells (P<0.05) when compared with the P cells
(Fig. 1B). For shRNA-1 and shRNA-3
transfected cells, there was no significant difference in ST3Gal
III mRNA and protein expression. Thus, ST3Gal III-overexpressing
ST3 cells (for the MDA-MB-231 model) and shRNA-2 and shRNA-4
transfected cells with downregulated ST3Gal III expression were
selected for further studies. As controls, mock-transfected clones
(MDA-MB-231), M and parental cell clones, P were used.
ST3Gal III modulates de novo
expression of SLeX
The synthesis of SLeX structures was catalyzed by
ST3Gal III. The levels of SLeX could resort to ST3Gal III enzyme
activities. Carbohydrate structure on the cell surface was studied
by flow cytometry using specific MAbs against SLeX. The results
revealed that ST3 clones displayed a large increase in SLeX
expression and a significant decrease in SLeX expression was
observed in the shRNA-2 and shRNA-4 transfected cells, compared to
the controls (P<0.05) (Fig.
2).
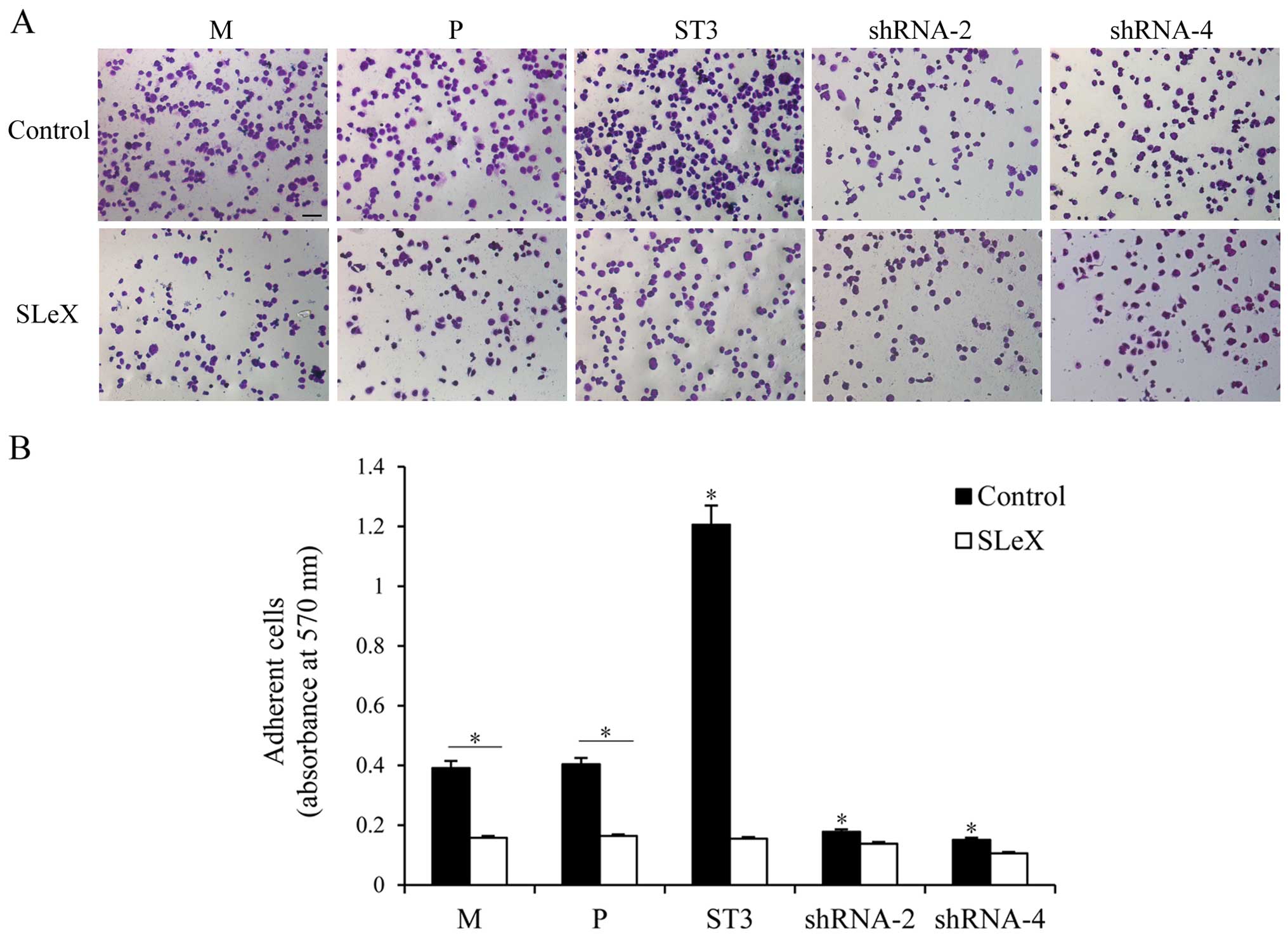
Breast cancer cell binding to
rh-E-selectin is correlated with SLeX levels
SLeX present on the tumor cell surface can mediate
tumor hematogenous spread by binding to E-selectin expressed on
activated endothelial cells. rh-E-selectin binding assays were used
to analyze whether the different levels of SLeX expression could
regulate changes in the adhesion to rh-E-selectin. As shown in
Fig. 3, both cell clones showed
different adhesion patterns. The capacity for adhesion to
rh-E-selectin was enhanced in the ST3 cells with higher SLeX
expression compared with that in the M and P cells (P<0.05).
Obviously, lower adhesion to rh-E-selectin was noted in the shRNA-2
and shRNA-4 transfected cells than that noted in the M and P cells
(P<0.05), which was consistent with the lower expression level
of SLeX. To further confirm that SLeX expression induced by ST3Gal
III caused regulation of rh-E-selectin binding, we preincubated
both cell clones with anti-SLeX MAb. Cell adhesion to E-selectin
was significantly decreased in the ST3 cells (P<0.05), whereas
shRNA cells, which had lower SLeX expression levels, did not show a
significant change in adhesion capacity to rh-E-selectin. Moreover,
there was no different adhesion ability among the groups after
incubation. These results demonstrated that the E-selectin binding
capacity of the transfectants was proportional to cell surface SLeX
levels.
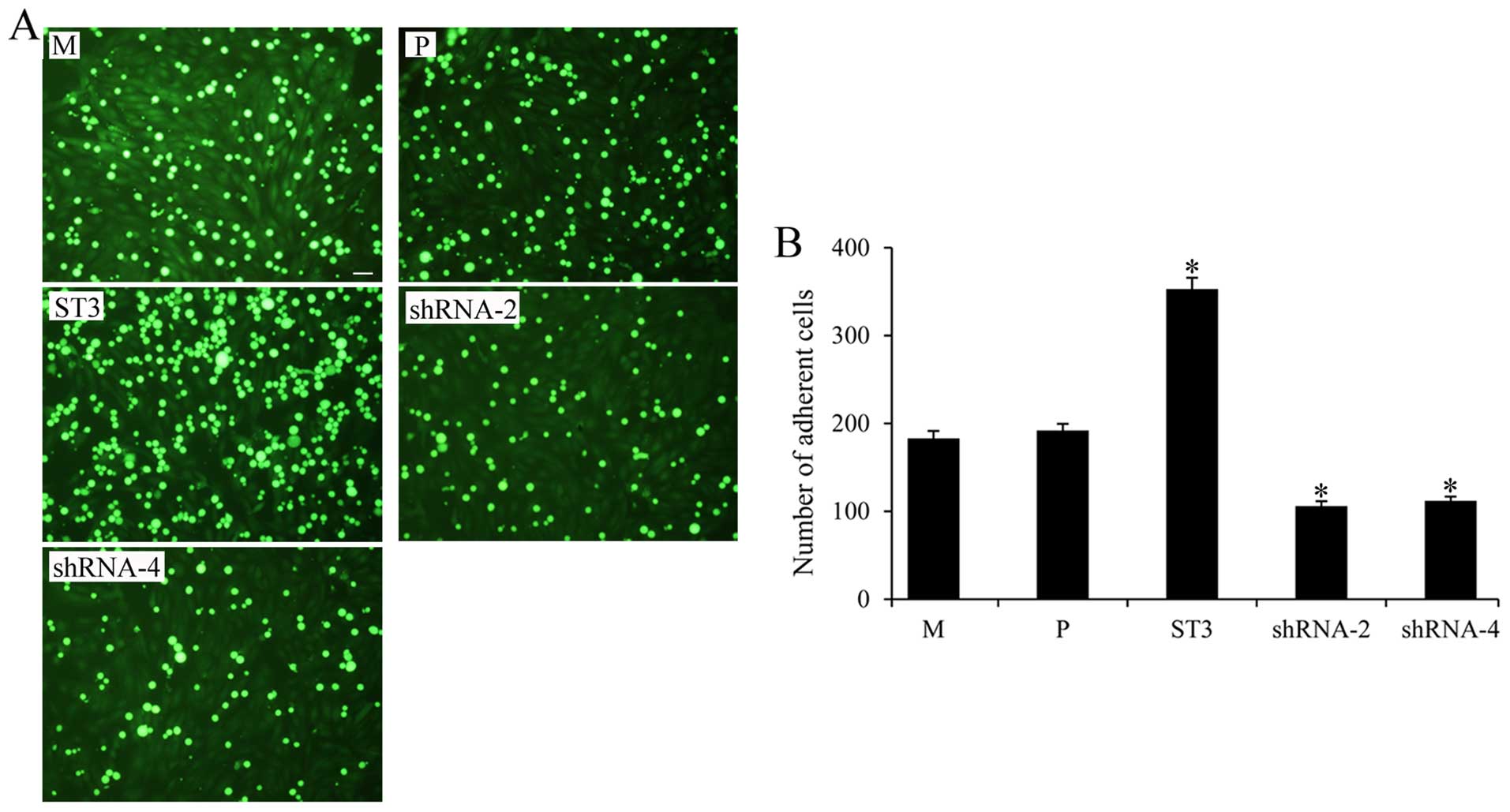
ST3Gal III expression mediates breast
cancer cell adhesion to HUVECs
We next determined the adhesion of the MDA-MB-231
cells with different ST3Gal III expression levels to HUVECs
pretreated with IL-1β. ST3 cells with high SLeX expression had a
higher adhesion to HUVECs, and shRNA-2, shRNA-4 cells showed
significantly decreased adhesion to HUVECs, compared with that
observed in the M and P cells (P<0.05) (Fig. 4). These data showed that ST3Gal III
and SLeX play an important role in mediating breast cancer adhesion
to HUVECs.
ST3Gal III expression levels modulate
cell migration and invasion
To further investigate the relationship between
ST3Gal III expression and the acquisition of a more invasive cell
phenotype, we evaluated cell migration and invasion using Transwell
chamber. As shown in Fig. 5, both
cell clones showed different migratory capabilities; the ST3Gal
III-overexpressing clone ST3 exhibited higher migratory capability
(P<0.05), and the ST3Gal III-silenced clones shRNA-2 and shRNA-4
exhibited decreased migration compared with the M and P cells
(P<0.05). The results demonstrated a positive correlation
between ST3Gal III levels and invasive capabilities.
ST3Gal III induces the expression of
COX-2, β1 integrin, MMP-2 and MMP-9
To further investigate the molecular mechanism of
ST3Gal III in modulating cell migration and invasion, we examined
the protein expression of invasion-related molecules, COX-2,
β1 integrin, MMP-2 and MMP-9 using western blot
analysis. The results showed that ST3Gal III-overexpressing ST3
cells had increased COX-2 (Fig.
6A), β1 integrin (Fig.
6C), MMP-2 and MMP-9 expression (P<0.05) (Fig. 6E). Moreover, the expression of COX-2
(Fig. 6B), β1 integrin
(Fig. 6D), MMP-2 and MMP-9
(Fig. 6F) were downregulated in the
shRNA-2 and shRNA-4 cells (P<0.05). These results showed that
ST3Gal III induced the expression of COX-2, β1 integrin,
MMP-2 and MMP-9.
 | Figure 6.ST3Gal III induces the expression of
cyclooxygenase-2 (COX-2), β1 integrin, matrix
metalloproteinase (MMP)-2 and MMP-9. The expression of
invasion-related molecules, COX-2, β1 integrin, MMP-2
and MMP-9 was determined via western blot analysis. (A, C and E)
ST3 cells displayed increased COX-2, β1 integrin, MMP-2
and MMP-9 expression, whereas (B, D and F) shRNA-2 and shRNA-4
cells showed a significant reduction in expression levels. The
density of the bands (normalized to GAPDH) are shown as mean ± SD
(n=3). *P<0.05, **P<0.01 compared to P. M, MDA-MB-231 mock
cells; P, MDA-MB-231 parental cells; ST3, MDA-MB-231 cells
transfected with the ST3Gal III gene; shRNA-2 and shRNA-4, ST3Gal
III-silenced cells. |
Discussion
Our previous study showed that α2,3-sialic acid
residues in breast cancer are associated with metastatic potential
(14). We used
α2,3-sialyltransferase ST3Gal III-overexpressing clone ST3 and
shRNA-targeted ST3Gal III clone shRNA cells, and extended our study
to investigate the mechanistic role of ST3Gal III in the process of
metastasis, such as adhesion, migration and invasion in MDA-MB-231
breast cancer cell lines. The results showed that ST3Gal III
expression increased sialylation structure SLeX expression. ST3Gal
III regulated the abilities of breast cancer cell binding to
rh-E-selectin and IL1-β-pretreated HUVECs, via SLeX-E-selectin
interaction. In addition, a positive correlation between invasive
capacity and ST3Gal III expression was found. Our finding showed
that ST3Gal III expression plays a critical role in several steps
of the process of breast cancer metastasis.
The molecular mechanisms regulating breast cancer
metastasis are still poorly understood. Sialic acids overexpressed
in some tumors have the potential to modulate interactions between
molecules and cells. One crucial molecule is SLeX with α2,3-sialic
acid residues, which involves the attachment of tumor cells to
activated endothelial cells. SLeX has been reported to participate
in the processes of extravasation by interacting with E-selectin
(15,16). Several studies have reported that
SLeX expression is directly correlated with the binding to
endothelial cells via E-selectin-SLeX interaction in colon cancer
(17), H7721 hepatocarcinoma
(18) and lung adenocarcinoma cells
(19). In agreement with these
data, our results showed that the ST3Gal III-overexpressing ST3
cells exhibited enhanced adherence to rh-E-selectin and the ST3Gal
III-silenced shRNA cells had reduced adhesion ability. Moreover,
when studying the adhesion of breast cancer cells to
IL-1β-stimulated HUVECs, similar results further demonstrated that
ST3Gal III and SLeX levels were correlated with E-selectin
expressed on HUVECs. When we pretreated ST3 cells with anti-SLeX,
the enhanced adhesion was obviously decreased, whereas shRNA cells
with low SLeX levels had no significant change. After incubation,
there was no difference among the groups, which could account for
breast cancer cells binding to stimulated HUVECs mediated by
SLeX.
Cell migration and invasion are multistep processes
that play pivotal role in cancer metastasis. To our knowledge,
α2,3-sialic acid residues are associated with the migration
processes in cancer. When α2,3-sialic acid levels on the tumor cell
surface were increased, cell migration was significantly enhanced
(20). We aimed to ascertain
whether ST3Gal III expression and the subsequent changes in
α2,3-sialic acid levels have a role in invasion in MDA-MB-231
breast cancer cells. ST3Gal III-overexpressing ST3 cells exhibited
high migration and invasion abilities, whereas ST3Gal III-silenced
shRNA cells demonstrated lower invasion capabilities when compared
with the mock and parental cells, which was in agreement that
α2,3-sialic acid expression resulted in a more invasive phenotype
in vitro (21). In addition,
both cell clones ST3 and shRNA displayed different amounts of SLeX
expression, which consequently show a different invasive potential.
These results reinforce the importance of α2,3-sialic acids in
potentiating cell invasion and metastasis. In accordance, decreased
α2,3-sialic acid levels in a cancer model resulted in suppression
of invasion and metastasis in lung cancer (22) and hepatocarcinoma (23). Moreover, migration was inhibited
after sialic acid residue was elimination (24). To the best of our knowledge, this is
the first report concerning a positive correlation between cell
migration and ST3Gal III expression levels in breast cancer
cells.
Integrins as extracellular matrix (ECM) adhesion
molecules, are involved in metastatic progression, such as the
formation of invadopodia (25), the
interaction of metastatic tumor cells with their surrounding ECM
(26), and acquisition of invasive
behavior (27). The levels of
β1 integrin are correlated with metastatic potential.
The results described here suggest that the ST3Gal III gene
regulates the levels of β1 integrin expression. We found
that β1 integrin expression was constitutively increased
in the ST3Gal III-overexpressing ST3 cells, while low expression
was noted in the ST3Gal III-silenced shRNA cells, which could be
the machanism involved in the influence on cell invasion abilities.
Several studies have demonstrated that sialylation influences
migration capability by modulating the integrin function (28,29).
Thus, we infer that ST3Gal III could alter the sialylation levels
of β1 integrin, modulating their metastatic potential.
Nevertheless, further investigations are required to address this
issue.
COX-2 is undetectable in most normal tissues but can
be induced in many cancers. High COX-2 expression has been reported
in colon (30), bladder (31), and breast (32) cancers, and is associated with cell
motility and invasion (33),
angiogenesis (34), and lymph node
metastasis (35). Interestingly,
all these cancers show increased expression of ST3Gal III. The
relationship between ST3Gal III and COX-2 protein expression was
investigated. The results found that the ST3Gal III gene could
induce the expression of COX-2. COX-2 expression was increased in
the ST3 cells with ST3Gal III overexpression, and downregulated in
the ST3Gal III-silenced shRNA cells compared with the mock and
parental cells, which could be one of the mechanism of the
influence on cell invasion abilities mediated by ST3Gal III.
MMP-9 and MMP-2, are important isoforms in the MMP
family, and have been reported to be overexpressed and degrade
collagen and gelatin, which can aid cell migration and angiogenesis
(36). Recently, it was shown that
expression of MMP-9 has been associated with shorter survival rates
in breast cancer patients (37).
Additionally, the pulmonary metastasis ability of cancer cells was
found to be reduced in MMP-2- or MMP-9-deficient mice compared to
wild-type mice (38) and cancer
cell proliferation was suppressed in tumors obtained from
MMP-9-knockout mice (39). Our
results found that ST3 cells with ST3Gal III overexpression had
increased expression of MMP-2 and MMP-9, which was decreased in the
ST3Gal III-silenced shRNA cells, which may have resulted in the
breast cancer cell invasion abilities mediated by ST3Gal III.
Tumor invasion and metastasis are known to be the
major causes of mortality in breast cancer patients. In the present
study, we engineered stable transfectants of ST3Gal
III-overexpressing clone and shRNA-targeted ST3Gal III clones to
globally evaluate the role of ST3Gal III in key steps of the
process of breast cancer metastasis. In the present study, we
demonstrated that ST3Gal III and its downstream product SLeX,
conferred to breast cancer cells high adhesion to E-selectin, high
migration and invasion capacities, and regulated expression of
invasion-related molecules β1 integrin, COX-2, MMP-9 and
MMP-2. In addition to the role of ST3Gal III in these key steps,
this enzyme has been reported to produce resistance to Taxol
therapy in ovarian cancer cells (40). This study highlights the importance
of ST3Gal III in cancer processes. A next step would be to further
design inhibitors targeted ST3Gal III that may inhibit breast
cancer metastasis.
Acknowledgements
This study was supported by the National Youth
Science Foundation of China (no. 81302308), the Natural Science
Foundation of Heilongjiang Province, China (H201353) and the Youth
Leading Scholar Supporting Program in General Colleges and
Universities of Heilongjiang, China (no. 1253G067).
References
|
1
|
Varki A, Cummings RD, Esko JD, Freeze HH,
Stanley P, Bertozzi CR, Hart GW and Etzler ME: Essentials of
Glycobiology. 2nd. Cold Spring Harbor Laboratory Press; New York,
NY: 2009, View Article : Google Scholar
|
|
2
|
Harduin-Lepers A, Vallejo-Ruiz V,
Krzewinski-Recchi MA, Samyn-Petit B, Julien S and Delannoy P: The
human sialyltransferase family. Biochimie. 83:727–737. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Recchi MA, Hebbar M, Hornez L,
Harduin-Lepers A, Peyrat JP and Delannoy P: Multiplex reverse
transcription polymerase chain reaction assessment of
sialyltransferase expression in human breast cancer. Cancer Res.
58:4066–4070. 1998.PubMed/NCBI
|
|
4
|
Hebbar M, Krzewinski-Recchi MA, Hornez L,
Verdière A, Harduin-Lepers A, Bonneterre J, Delannoy P and Peyrat
JP: Prognostic value of tumoral sialyltransferase expression and
circulating E-selectin concentrations in node-negative breast
cancer patients. Int J Biol Markers. 18:116–122. 2003.PubMed/NCBI
|
|
5
|
Jin XL, Zheng SS, Wang BS and Chen HL:
Correlation of glycosyltransferases mRNA expression in extrahepatic
bile duct carcinoma with clinical pathological characteristics.
Hepatobiliary Pancreat Dis Int. 3:292–295. 2004.PubMed/NCBI
|
|
6
|
Wang PH, Li YF, Juang CM, Lee YR, Chao HT,
Ng HT, Tsai YC and Yuan CC: Expression of sialyltransferase family
members in cervix squamous cell carcinoma correlates with lymph
node metastasis. Gynecol Oncol. 86:45–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang PH, Lee WL, Lee YR, Juang CM, Chen
YJ, Chao HT, Tsai YC and Yuan CC: Enhanced expression of alpha
2,6-sialyltransferase ST6Gal I in cervical squamous cell carcinoma.
Gynecol Oncol. 89:395–401. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Petretti T, Kemmner W, Schulze B and
Schlag PM: Altered mRNA expression of glycosyltransferases in human
colorectal carcinomas and liver metastases. Gut. 46:359–366. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gretschel S, Haensch W, Schlag PM and
Kemmner W: Clinical relevance of sialyltransferases ST6GAL-I and
ST3GAL-III in gastric cancer. Oncology. 65:139–145. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Varki A: Glycan-based interactions
involving vertebrate sialic-acid-recognizing proteins. Nature.
446:1023–1029. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hosono J, Narita T, Kimura N, Sato M,
Nakashio T, Kasai Y, Nonami T, Nakao A, Takagi H and Kannagi R:
Involvement of adhesion molecules in metastasis of SW1990, human
pancreatic cancer cells. J Surg Oncol. 67:77–84. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mas E, Pasqualini E, Caillol N, El Battari
A, Crotte C, Lombardo D and Sadoulet MO: Fucosyltransferase
activities in human pancreatic tissue: Comparative study between
cancer tissues and established tumoral cell lines. Glycobiology.
8:605–613. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Peracaula R, Tabarés G, López-Ferrer A,
Brossmer R, de Bolós C and de Llorens R: Role of sialyltransferases
involved in the biosynthesis of Lewis antigens in human pancreatic
tumour cells. Glycoconj J. 22:135–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui H, Lin Y, Yue L, Zhao X and Liu J:
Differential expression of the α2,3-sialic acid residues in breast
cancer is associated with metastatic potential. Oncol Rep.
25:1365–1371. 2011.PubMed/NCBI
|
|
15
|
Kannagi R, Izawa M, Koike T, Miyazaki K
and Kimura N: Carbohydrate-mediated cell adhesion in cancer
metastasis and angiogenesis. Cancer Sci. 95:377–384. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi H, Boelte KC and Lin PC:
Endothelial cell adhesion molecules and cancer progression. Curr
Med Chem. 14:377–386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matsushita Y, Kitajima S, Goto M, Tezuka
Y, Sagara M, Imamura H, Tanabe G, Tanaka S, Aikou T and Sato E:
Selectins induced by interleukin-1beta on the human liver
endothelial cells act as ligands for sialyl Lewis X-expressing
human colon cancer cell metastasis. Cancer Lett. 133:151–160. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu SL, Ma J, Qi HL, Zhang Y, Zhang XY and
Chen HL: Forskolin up-regulates metastasis-related phenotypes and
molecules via protein kinase B, but not PI-3K, in H7721 human
hepato-carcinoma cell line. Mol Cell Biochem. 254:193–202. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martín-Satué M, de Castellarnau C and
Blanco J: Overexpression of alpha(1,3)-fucosyltransferase VII is
sufficient for the acquisition of lung colonization phenotype in
human lung adenocarcinoma HAL-24Luc cells. Br J Cancer.
80:1169–1174. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bassagañas S, Pérez-Garay M and Peracaula
R: Cell surface sialic acid modulates extracellular matrix adhesion
and migration in pancreatic adenocarcinoma cells. Pancreas.
43:109–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pérez-Garay M, Arteta B, Pagès L, de
Llorens R, de Bolòs C, Vidal-Vanaclocha F and Peracaula R:
alpha2,3-sialyltransferase ST3Gal III modulates pancreatic cancer
cell motility and adhesion in vitro and enhances its metastatic
potential in vivo. PLoS One. 5:e125242010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lalor PF, Edwards S, McNab G, Salmi M,
Jalkanen S and Adams DH: Vascular adhesion protein-1 mediates
adhesion and transmigration of lymphocytes on human hepatic
endothelial cells. J Immunol. 169:983–992. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu F, Qi HL, Zhang Y, Zhang XY and Chen
HL: Transfection of the c-erbB2/neu gene upregulates the expression
of sialyl Lewis X, alpha1,3-fucosyltransferase VII, and metastatic
potential in a human hepatocarcinoma cell line. Eur J Biochem.
268:3501–3512. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Zhang XY, Liu F, Qi HL and Chen
HL: The roles of terminal sugar residues of surface glycans in the
metastatic potential of human hepatocarcinoma. J Cancer Res Clin
Oncol. 128:617–620. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beaty BT, Sharma VP, Bravo-Cordero JJ,
Simpson MA, Eddy RJ, Koleske AJ and Condeelis J: β1 integrin
regulates Arg to promote invadopodial maturation and matrix
degradation. Mol Biol Cell. 24:1661–1675, S1-S11. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shibue T, Brooks MW and Weinberg RA: An
integrin-linked machinery of cytoskeletal regulation that enables
experimental tumor initiation and metastatic colonization. Cancer
Cell. 24:481–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nieto MA and Cano A: The
epithelial-mesenchymal transition under control: Global programs to
regulate epithelial plasticity. Semin Cancer Biol. 22:361–368.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shaikh FM, Seales EC, Clem WC, Hennessy
KM, Zhuo Y and Bellis SL: Tumor cell migration and invasion are
regulated by expression of variant integrin glycoforms. Exp Cell
Res. 314:2941–2950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Seales EC, Jurado GA, Brunson BA,
Wakefield JK, Frost AR and Bellis SL: Hypersialylation of beta1
integrins, observed in colon adenocarcinoma, may contribute to
cancer progression by up-regulating cell motility. Cancer Res.
65:4645–4652. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eberhart CE, Coffey RJ, Radhika A,
Giardiello FM, Ferrenbach S and DuBois RN: Up-regulation of
cyclooxygenase 2 gene expression in human colorectal adenomas and
adenocarcinomas. Gastroenterology. 107:1183–1188. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eltze E, Wülfing C, Von Struensee D,
Piechota H, Buerger H and Hertle L: Cox-2 and Her2/neu
co-expression in invasive bladder cancer. Int J Oncol.
26:1525–1531. 2005.PubMed/NCBI
|
|
32
|
Ristimäki A, Sivula A, Lundin J, Lundin M,
Salminen T, Haglund C, Joensuu H and Isola J: Prognostic
significance of elevated cyclooxygenase-2 expression in breast
cancer. Cancer Res. 62:632–635. 2002.PubMed/NCBI
|
|
33
|
Larkins TL, Nowell M, Singh S and Sanford
GL: Inhibition of cyclooxygenase-2 decreases breast cancer cell
motility, invasion and matrix metalloproteinase expression. BMC
Cancer. 6:181–192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Davies G, Salter J, Hills M, Martin LA,
Sacks N and Dowsett M: Correlation between cyclooxygenase-2
expression and angiogenesis in human breast cancer. Clin Cancer
Res. 9:2651–2656. 2003.PubMed/NCBI
|
|
35
|
Costa C, Soares R, Reis-Filho JS, Leitão
D, Amendoeira I and Schmitt FC: Cyclo-oxygenase 2 expression is
associated with angiogenesis and lymph node metastasis in human
breast cancer. J Clin Pathol. 55:429–434. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mehner C, Hockla A, Miller E, Ran S,
Radisky DC and Radisky ES: Tumor cell-produced matrix
metalloproteinase 9 (MMP-9) drives malignant progression and
metastasis of basal-like triple negative breast cancer. Oncotarget.
5:2736–2749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Scorilas A, Karameris A, Arnogiannaki N,
Ardavanis A, Bassilopoulos P, Trangas T and Talieri M:
Overexpression of matrix-metalloproteinase-9 in human breast
cancer: A potential favourable indicator in node-negative patients.
Br J Cancer. 84:1488–1496. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Itoh T, Tanioka M, Yoshida H, Yoshioka T,
Nishimoto H and Itohara S: Reduced angiogenesis and tumor
progression in gelatinase A-deficient mice. Cancer Res.
58:1048–1051. 1998.PubMed/NCBI
|
|
39
|
Coussens LM, Tinkle CL, Hanahan D and Werb
Z: MMP-9 supplied by bone marrow-derived cells contributes to skin
carcinogenesis. Cell. 103:481–490. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huang S, Day TW, Choi MR and Safa AR:
Human beta-galactoside alpha-2,3-sialyltransferase (ST3Gal III)
attenuated Taxol-induced apoptosis in ovarian cancer cells by
downregulating caspase-8 activity. Mol Cell Biochem. 331:81–88.
2009. View Article : Google Scholar : PubMed/NCBI
|