Introduction
Colorectal cancer (CRC) is one of the most common
cancers worldwide and the second leading cause of cancer-related
death, despite improvements in available treatment options
(1,2). Chemotherapy is consistently used as a
palliative or adjuvant treatment, but nearly all patients
ultimately develop drug resistance, which has become one of the
major causes of CRC recurrence. Additionally, advanced CRC is often
resistant to such chemotherapy, resulting in poor outcomes. Studies
have proposed many mechanisms as being responsible for drug
resistance, including limitation of drug accumulation within cells
by reducing uptake, enhancing efflux or affecting membrane lipids;
activation of detoxification; alterations in cell cycle
checkpoints; and inhibition of cell apoptosis (3). However, only a few predictive and
prognostic biomarkers have been validated clinically despite
enormous efforts (3), and the exact
mechanisms remain unknown. Therefore, it is essential to better
understand the underlying mechanisms of chemoresistance, and to
uncover novel methods to improve the efficacy of chemotherapeutic
treatment by targeting these mechanisms.
More effective chemotherapeutic agents, such as
oxaliplatin (L-OHP), 5-fluorouracil, irinotecan and the monoclonal
antibodies cetuximab, panitumumab and bevacizumab, have been used
in clinical practice during the last two decades, but one of the
major issues of CRC treatment is the inherent or acquired
resistance to therapeutic approaches. L-OHP is a chemotherapeutic
agent that is widely used in CRC treatment; however, L-OHP
chemoresistance poses a great challenge when treating
chemorefractory patients. The treatment of increasing numbers of
metastatic CRC patients to L-OHP, as well as its introduction into
the adjuvant therapy of early-stage CRC, has led to efforts to
identify and target the mechanisms of chemoresistance to L-OHP.
Currently, although the molecular mechanisms of the chemoresistance
of CRC are not well known, several findings suggest that targeting
autophagy may be considered as a potential therapeutic strategy to
address chemoresistance in the treatment of CRC (4–6). In
the present study, we investigated the mechanisms involved in cell
autophagy underlying L-OHP-induced cell apoptosis and
chemoresistance in CRC cells, the knowledge of which may contribute
to clinical treatment.
MicroRNAs (miRNAs) are a class of small, endogenous,
non-coding, single-stranded RNAs, which have been recognized as key
regulators of gene expression at post-transcriptional levels.
miRNAs are broadly involved in tumor proliferation, invasion,
angiogenesis and drug resistance (7), which may add new dimensions to the
diagnosis and treatment of cancer. Several miRNAs are known to be
dysregulated in CRCs and have been linked to biological processes
involved in tumor progression and response to anticancer therapies
(8,9). In particular, decreased miR-218
expression has been observed in CRC tissues, and patients with low
miR-218 expression have shorter survival times; furthermore, an
increased level of serum miR-218 was noted one month after surgery
(10). Thus, we infer that miR-218
may play important roles in the development and progression of CRC,
which may serve as a potential diagnostic and prognostic biomarker
of CRC.
YEATS domain containing 4 (YEATS4) is a highly
conserved nuclear protein that was originally identified in human
glioma (11). Research has shown
that YEATS4 functions as an oncogene and is amplified in various
types of malignancy, including CRC (12). Transcriptional networks isolated
from the tissues of drug-resistant patients suggest that YEATS4
could be targeted to ameliorate drug resistance (13). In addition, several studies have
shown that therapy-induced autophagy may be a novel mechanism of
resistance to anticancer agents (14,15).
In the present study, we demonstrated that YEATS4 is a downstream
target of miR-218 and that miR-218 sensitizes CRC cells to
L-OHP-induced cell apoptosis by inhibiting cytoprotective
autophagy.
Materials and methods
Tissue samples and cell culture
Human CRC and the corresponding non-tumor colon
samples were collected at the time of surgical resection at the
Tumor Hospital of Henan Province (The Affiliated Tumor Hospital of
Zhengzhou University). The present study was conducted following
patient informed consent in accordance with the Declaration of
Helsinki and was approved by the Ethics Committee of Zhengzhou
University. The samples were immediately snap-frozen and stored in
liquid nitrogen. Samples used for molecular analysis were initially
ground into powder in liquid nitrogen and were then separately
collected for miRNA or protein analyses.
The HEK293T cell line [American Type Culture
Collection (ATCC); Manassas, VA, USA] was cultured in Dulbecco's
modified Eagles medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) (both from Gibco, Rockville, MD, USA), 100 U/ml
penicillin and 100 mg/l streptomycin (Life Technologies, Rockville,
MD, USA) in a humidified incubator with 95% air and 5%
CO2 at 37°C. The human CRC cell line HCT-116 (ATCC) and
its multidrug-resistant variant HCT-116/L-OHP (established and
maintained in our laboratory), were maintained in RPMI-1640 (Gibco)
supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/l
streptomycin in a humidified incubator with 95% air and 5%
CO2 at 37°C. To maintain the drug-resistance phenotype,
L-OHP (Sigma, St. Louis, MO, USA) was added to the culture medium
at a final concentration of 5 µg/ml for HCT-116/L-OHP cells.
Chloroquine (CQ; Sigma) was added to the cell culture at a final
concentration of 10 µM.
Quantitative real-time PCR (qRT-PCR)
for miRNA analyses
Total RNA from tissue samples or cells, including
miRNAs, was extracted using the miRNA isolation kit (Ambion,
Austin, TX, USA) according to the manufacturer's protocol. The
purity and concentration of the RNA samples were quantified using a
dual-beam ultraviolet spectrophotometer (Eppendorf, Hamburg,
Germany). The TaqMan miRNA reverse transcription kit and TaqMan
Human miRNA assay kit (both from Applied Biosystems, Foster City,
CA, USA) were used to measure miR-218 levels, which were normalized
to RNU48 expression. The relative quantification of miR-218 levels
was determined using the 2−ΔΔCt method. The above
experiment was performed in triplicate, and each assay included a
negative control that lacked cDNA.
Cell transfection
Cultured cells were seeded and transfected with 50
nM corresponding miRNAs, including miR-218, anti-miR-218, control
and anti-control mimics (GenePharma, Shanghai, China) using
Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in 6- or
96-well plates following the manufacturer's protocol. Transfection
efficiency was evaluated by qRT-PCR. After 48 h of transfection,
cells were harvested or processed for further analysis.
To overexpress YEATS4, the YEATS4 coding sequence
without the 3′-untranslated region (3-UTR) was selected and cloned
into pcDNA3.1 (Invitrogen), and the empty vector was used as
negative control for the detection of transfection efficiency. The
plasmids were transfected into cultured cells as indicated using
Lipofectamine 2000 according to the manufacturer's
instructions.
MTT assay
Cultured cells (5×103 cells/well) were
re-seeded in 96-well plates (Corning Inc., Corning, NY, USA) after
transfection with the indicated microRNA mimics or plasmids. After
24 h, the cells were treated with L-OHP for the indicated times,
24, 48 or 72 h. Then, 20 µl modified tetrazolium salt
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT; 5 mg/ml; Sigma Aldrich) was added to each well and samples
were incubated at 37°C for 4 h. The supernatant was then removed
and 100 µl dimethyl sulfoxide (DMSO; Sigma) was added to each well.
After the dark-blue MTT crystals were dissolved, the absorbance was
measured at 490 nm using a Bio-Rad Benchmark Microplate Reader
(Bio-Rad, Hercules, CA, USA). The data were defined as: (mean
L-OHP-treated A490 - blank)/(mean untreated control
A490 - blank) × 100, and analyzed by weighted non-linear
regression to determine the inhibitory concentration
(IC50). The IC50 value of each cell line to
L-OHP was defined as the concentration that inhibited MTT
metabolism by 50% compared with the untreated controls.
Luciferase reporter assay
The potential target of miR-218 was predicted by
TargetScan (http://www.targetscan.org) to be
YEATS4, which suggests a way to modulate drug resistance through
the miRNA level in human CRC. The 3′-UTR of human YEATS4 containing
the miR-218 targeting sequence was inserted into the pMIR-REPORT™
miRNA expression reporter vector system (Ambion). The reporter
vector plasmid containing either the YEATS4-wt 3′-UTR or the
YEATS4-mut 3′-UTR sequence was subsequently co-transfected with 50
nM of the corresponding miRNAs into HEK293T cells using
Lipofectamine 2000. After 48 h post-transfection, the cells were
harvested and lysed for luciferase assays. The luciferase assay kit
(Promega, Madison, WI, USA) was used to measure the reporter
activity according to the manufacturer's protocol.
Flow cytometric analysis of
apoptosis
HCT116/L-OHP or HCT116 cells (4×105
cells/well) were plated into 6-well plates and transfected with 50
nM of the corresponding miRNA mimics or pcDNA3.1-YEATS4 plasmids.
After 48 h, the cells were treated with 10 or 20 µg/ml L-OHP. After
24 h, the cells were collected, washed twice with ice-cold
phosphate-buffered saline (PBS), and then resuspended in 400 µl of
1X binding buffer. After addition of 5 µl of Annexin V-FITC, the
cells were gently vortexed, and then incubated for 10 min at 4°C in
the dark. Propidium iodide (PI; 10 µl) was then added, followed by
incubation for another 5 min at 4°C in the dark. Flow cytometry was
then conducted using a FACScan flow cytometer (Becton-Dickinson).
The fraction of the cell population that was present in different
quadrants was analyzed using quadrant statistics. The cells in the
lower-right quadrant represented early apoptosis and those in the
upper-right quadrant represented late apoptotic cells.
Western blot analysis
Cell lysates were gathered and protein
concentrations were determined using the Pierce BCA protein assay
kit (Thermo Scientific, Rockford, IL, USA). Equal amounts of
protein were processed for western blot analysis following the
standard protocols. The primary antibodies used were
anti-microtubule-associated protein 1 light chain 3 (LC3) antibody
(Novus Biologicals, Littleton, CO, USA); rabbit anti-YEATS4
polyclonal antibody (MyBioSource.com, San Diego, CA, USA); anti-β-actin
antibodies (Santa Cruz Biotechnology, Santa Cruz, CA, USA). After
incubation with the appropriate secondary antibody, the resultant
protein bands were visualized using enhanced chemiluminescence ECL
(Beyotime, Shanghai, China). The absorbance values of the target
proteins were performed through Gel-Pro Analyzer version 4.0
software (Media Cybernetics, Silver Spring, MD, USA), and data were
normalized to β-actin.
Statistical analysis
Data are expressed as mean ± SD of the results
derived from three independent experiments performed in triplicate.
Statistical analysis was performed using the Student's t-test and
ANOVA. P<0.05 was considered to indicate statistical
significance compared with the respective control.
Results
miR-218 downregulation and YEATS4
upregulation in CRC tissues and HCT-116/L-OHP cells are associated
with L-OHP resistance
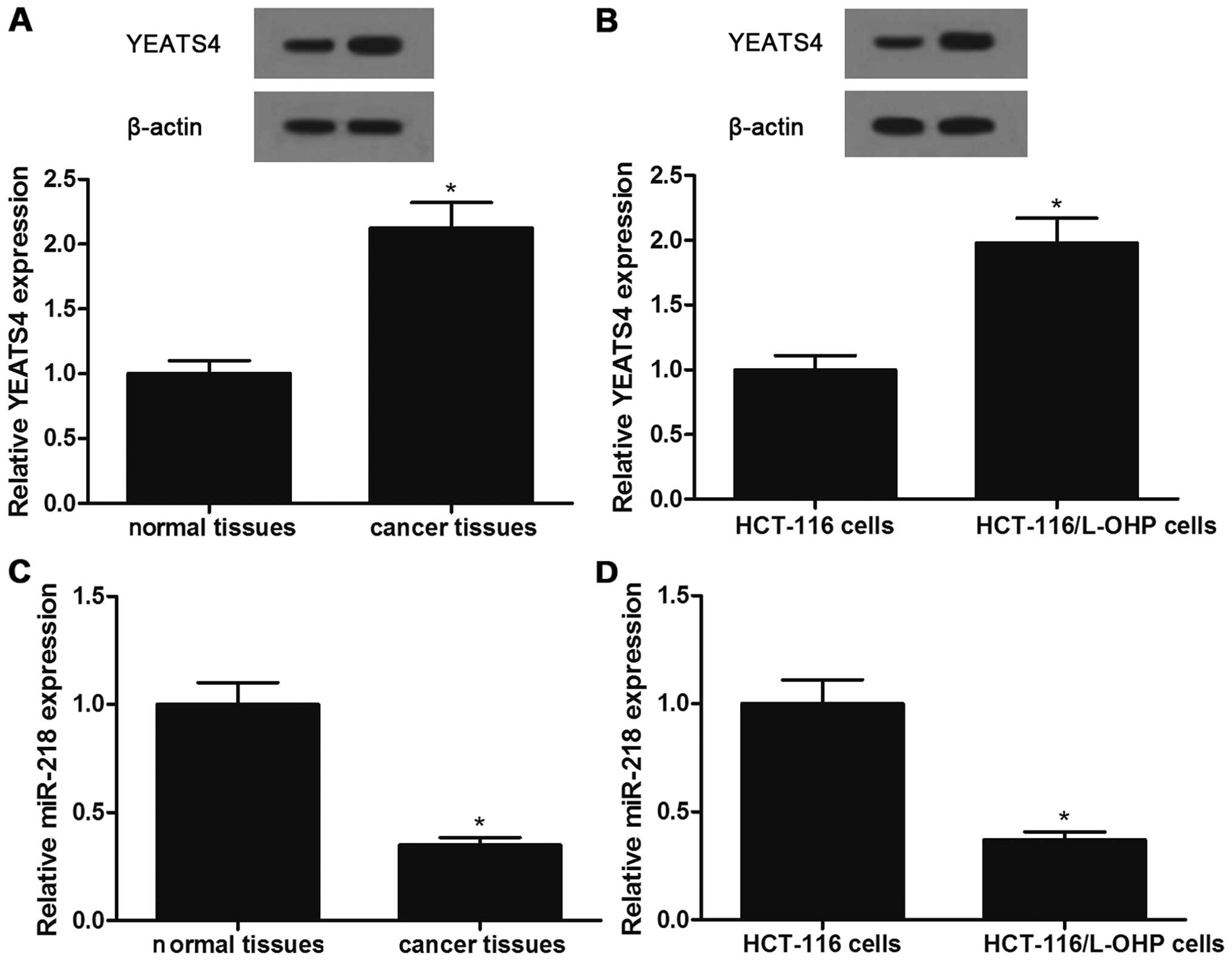
Western blot analysis showed that the protein level
of YEATS4 was significantly upregulated in both CRC tissues and
HCT-116/L-OHP cells compared with this level in the corresponding
non-tumor colon tissues and parental HCT-116 cells, respectively
(Fig. 1A and B). qRT-PCR showed
that the level of miR-218 was lower in both the CRC tissues and
HCT-116/L-OHP cells compared with the level in the respective
control group (Fig. 1C and D).
These data suggest that miR-218 expression was downregulated and
YEATS4 expression was upregulated in the CRC tissues and
HCT116/L-OHP cells, in association with L-OHP resistance.
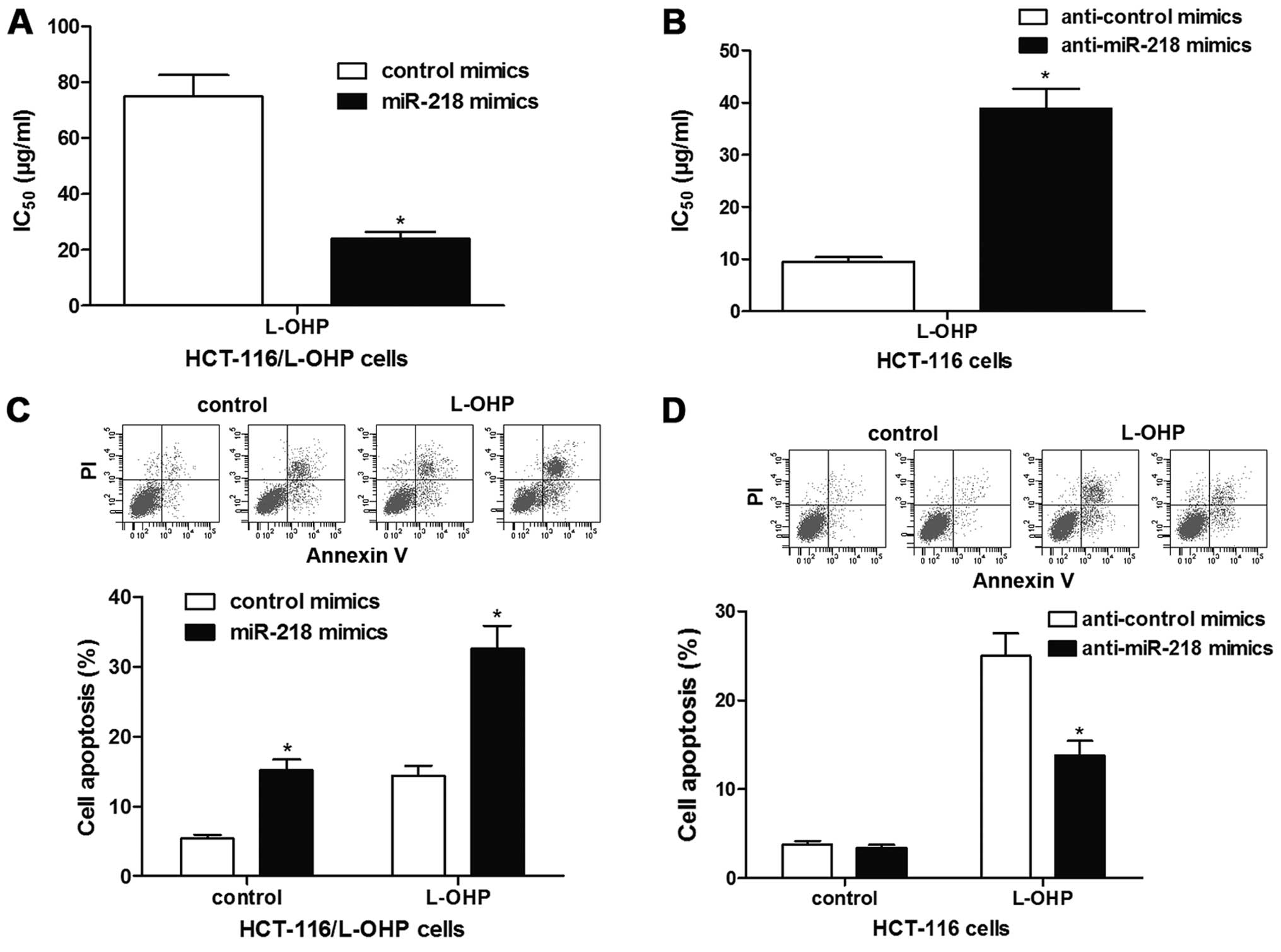
miR-218 overexpression sensitizes
HCT-116 cells to L-OHP-induced cell apoptosis
To investigate whether miR-218 expression influences
L-OHP resistance in HCT-116 and HCT-116/L-OHP cells, we transfected
the cells with corresponding mimics followed by L-OHP treatment,
and then the MTT assay was performed. The results showed that
miR-218 overexpression significantly decreased the IC50
value of L-OHP in the HCT-116/L-OHP cells, and miR-218 suppression
significantly enhanced the IC50 value of L-OHP in the
HCT-116 cells (Fig. 2A and B).
Compared with the control mimic group, miR-218 overexpression alone
in the HCT-116/L-OHP cells elevated the cell apoptosis rate, which
was further enhanced in response to L-OHP treatment (Fig. 2C). miR-218 inhibition decreased the
HCT-116 cell apoptosis rate following treated with L-OHP (Fig. 2D). These results indicate that
miR-218 is capable of sensitizing HCT-116 cells to L-OHP
treatment.
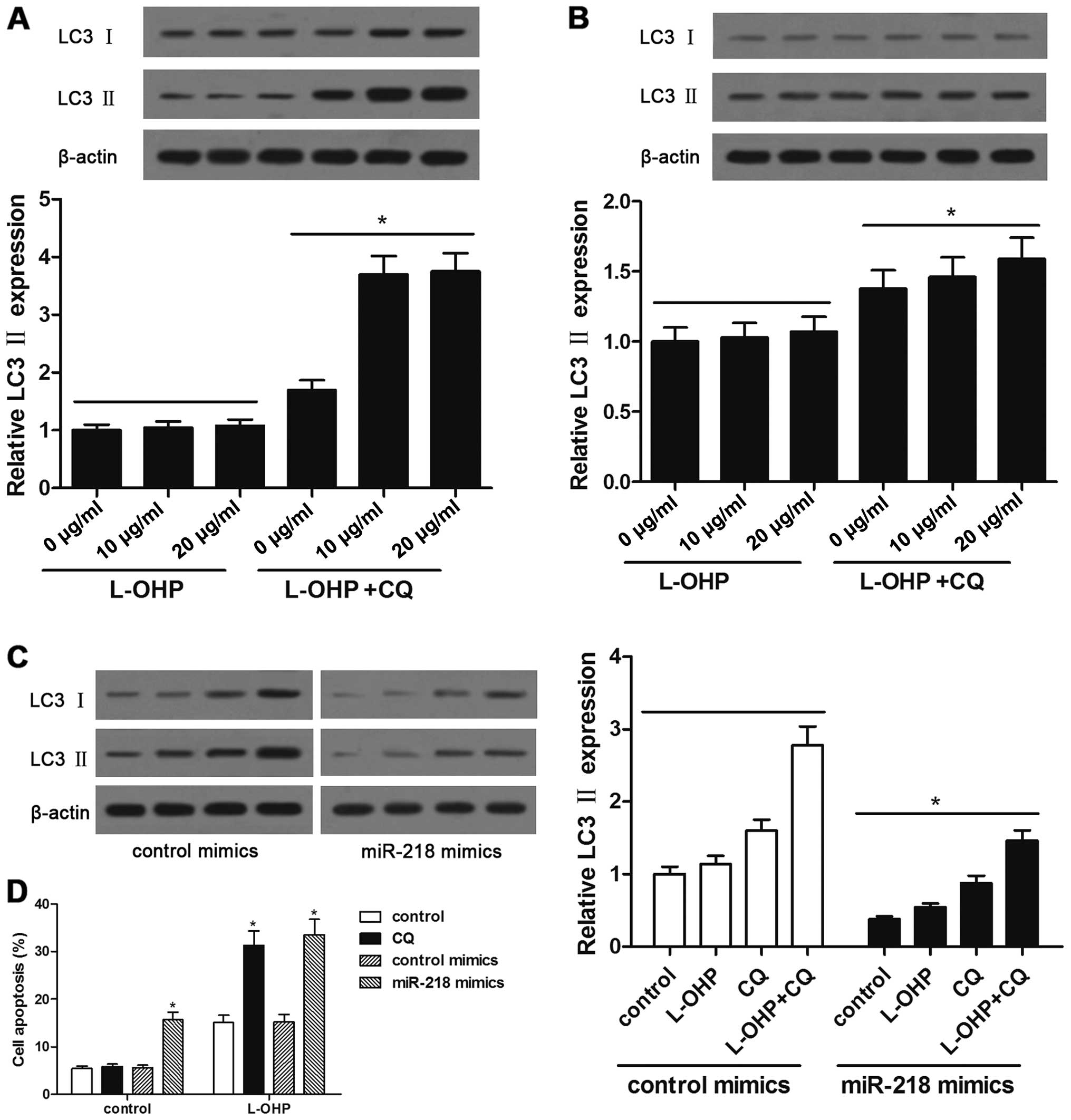
miR-218 overexpression increases the
sensitivity of HCT-116/L-OHP cells to L-OHP treatment via
inhibition of autophagy
Detection of LC3 is regarded as a common way to
monitor autophagy. As LC3 II is rapidly degraded by lysosomal
turnover during autophagy, we treated cells with L-OHP in the
presence or absence of the lysosome inhibitor CQ. Autophagic flux
can be reflected by the fold-change in LC3 II levels with or
without CQ (16,17). We found that, compared with the
small increase observed in parental L-OHP-sensitive HCT-116 cells,
the relative LC3 II levels in L-OHP-resistant HCT-116/L-OHP cells
after lysosome inhibition via CQ was markedly upregulated in
response to L-OHP treatment, suggesting induction of autophagy
(Fig. 3A and B). In addition,
exposure of HCT-116/L-OHP cells to L-OHP after control mimic
transfection increased autophagic flux, as reflected by increased
LC3 II levels. However, miR-218 overexpression partly reversed
L-OHP-mediated LC3 II accumulation in the HCT-116/L-OHP cells
(Fig. 3C). To test the hypothesis
that L-OHP resistance is due to autophagy protection, we treated
cells with CQ to determine whether inhibition of autophagy enhances
L-OHP-induced apoptosis. Indeed, CQ treatment promoted
L-OHP-induced cell apoptosis in the HCT-116/L-OHP cells, and
miR-218 overexpression obtained the same results. These results
indicate that miR-218 overexpression increased the sensitivity of
HCT-116/L-OHP cells to L-OHP treatment via inhibition of
autophagy.
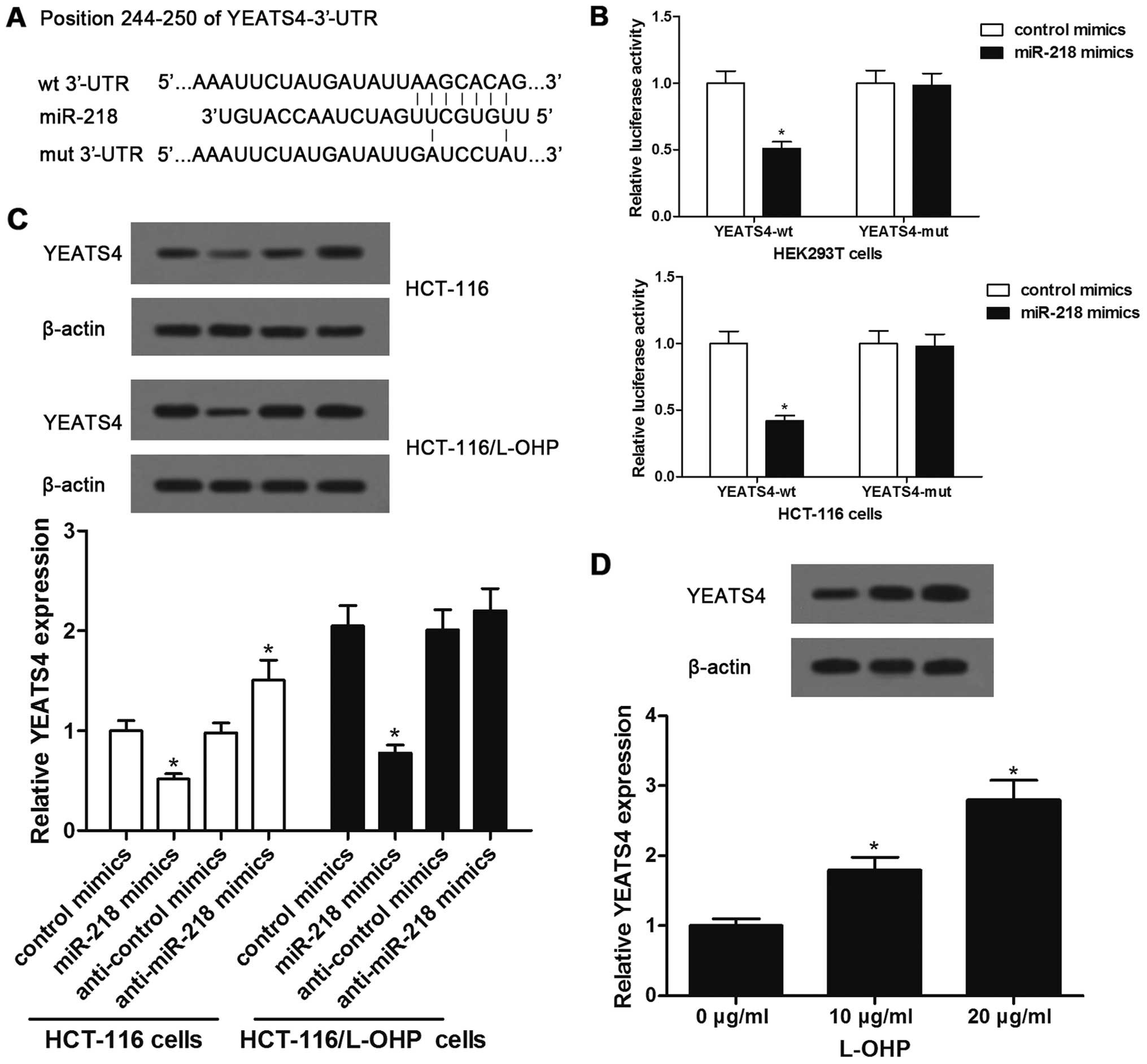
YEATS4 is a direct target of
miR-218
To elucidate the underlying mechanisms involving
miR-218 and YEATS4, we performed a bioinformatic prediction and the
results showed that there was one putative binding site between
miR-218 and the 3′-UTR of YEATS4 (Fig.
4A). To confirm the binding, a luciferase reporter assay was
performed by evaluating the luciferase activity of HEK293T and
HCT-116 cells transfected with pMIR-YEATS4 3′-UTR plasmids and
comparing this activity with that of cells transfected with control
plasmids. The results showed that miR-218 suppressed the luciferase
expression of YEATS4-wt, whereas YEATS4-mut induced no suppressive
effect in either HEK293T or HCT-116 cells (Fig. 4B). Western blot analysis indicated
that the basal level of YEATS4 in the HCT-116/L-OHP cells was
higher than that in the HCT-116 cells, and miR-218 overexpression
resulted in a reduction in YEATS4 protein levels in both cells.
Furthermore, suppression of miR-218 in HCT-116 cells enhanced the
YEATS4 protein level in the HCT-116 cells (Fig. 4C). In addition, L-OHP treatment
dose-dependently elevated YEATS4 expression in the HCT-116/L-OHP
cells (Fig. 4D). Taken together,
these data strongly suggest that miR-218 directly targets YEATS4
expression in both HCT-116 and HCT-116/L-OHP cells.
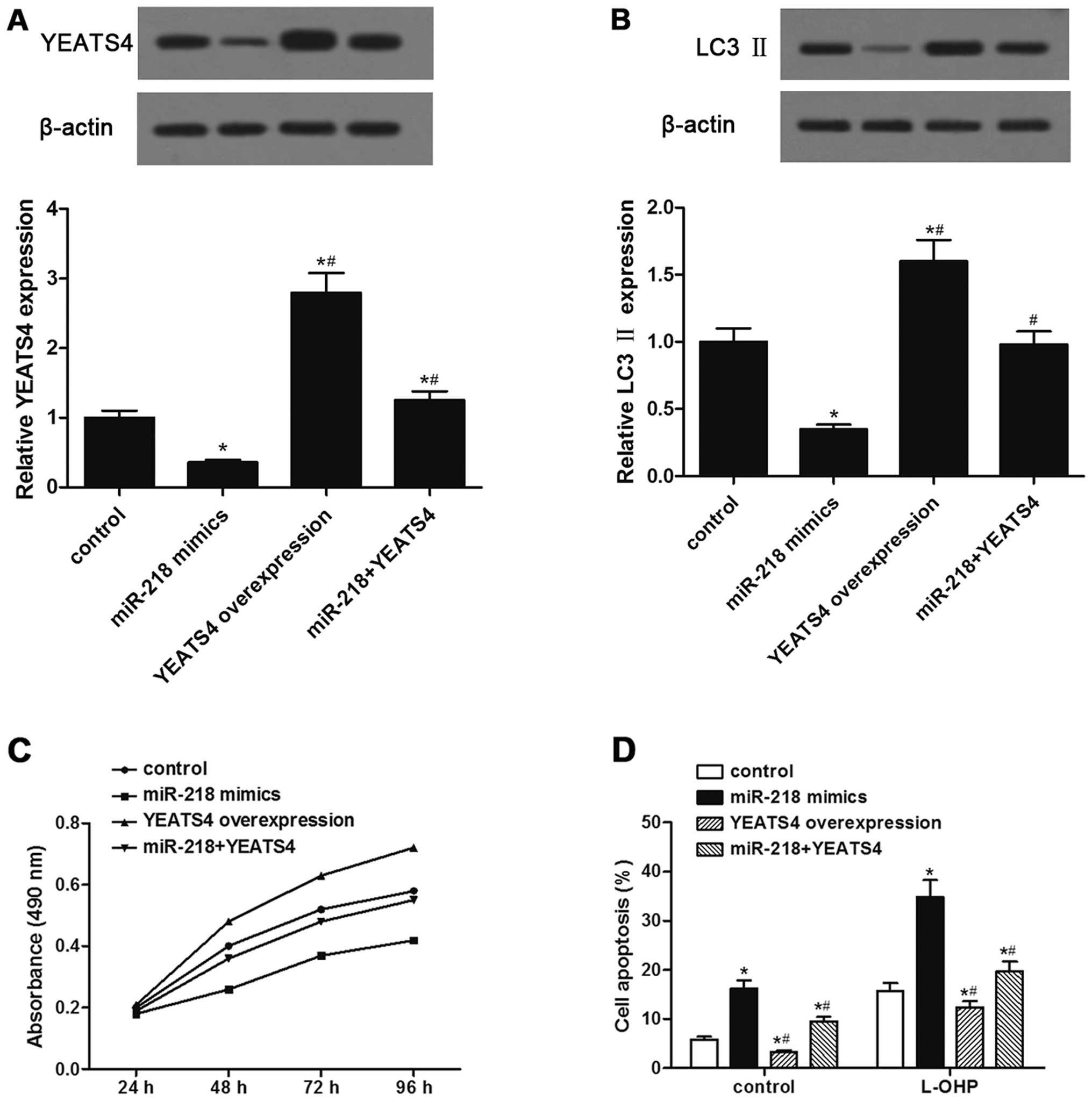
YEATS4 overexpression reverses the
stimulatory effect of miR-218 on cell apoptosis in response to
L-OHP via promotion of basal autophagy in HCT-116/L-OHP cells
Western blot analysis indicated that YEATS4
overexpression without the 3′-UTR robustly enhanced the expression
of YEATS4, which was significantly reduced by miR-218
overexpression (Fig. 5A). YEATS4
overexpression restored LC3 II expression following reduction by
miR-218 in the presence of CQ (Fig.
5B). In addition, YEATS4 overexpression abolished the loss of
cell viability and increase in cell apoptosis promoted by miR-218
in response to L-OHP (Fig. 5C and
D). Thus, the data confirmed that YEATS4 overexpression
reversed the stimulatory effect of miR-218 on cell apoptosis in
response to L-OHP by promoting basal autophagy in HCT-116/L-OHP
cells.
Discussion
Colorectal cancer (CRC) is one of the leading causes
of cancer-related mortality worldwide. Resistance to chemotherapy
remains the primary reason for treatment failure in advanced CRC
patients (3). YEATS4 overexpression
has been linked to the aggressive phenotypes of certain types of
cancers, including CRC. Several important miRNAs have been
identified as regulators of L-OHP treatment in CRC, such as miR-378
(18), miR-21 (19) and miR-222 (20). Therefore, the identification of
aberrantly expressed YEATS4 and miRNAs is an important first step
towards elucidating YEATS4-mediated or miRNA-mediated oncogenic
pathways.
Previous studies have demonstrated that miRNAs are
important regulators of carcinogenesis, progression, invasion,
angiogenesis and metastases in CRC, which may serve as potential
predictive and prognostic factors and even as therapeutic targets
themselves. A large body of research has demonstrated that miR-218
is downregulated in multiple malignancies, including glioblastoma
(21), nasopharyngeal (22) and lung cancer (23), and CRC (24), and it functions as a
tumor-suppressor gene in various cancers targeting many genes to
globally regulate biological processes (25–27).
Recent progress in cancer biology has also shown that YEATS4
overexpression is observed in some cancers such as non-small cell
lung cancer (28), astrocytomas
(11,29), glioblastomas (29), liposarcoma (30) and CRC (12). Furthermore, miR-218 enhances
5-fluorouracil-induced apoptosis in CRC (31), and increases the sensitivity of
gastric cancer cells to anticancer drugs, including L-OHP (25). Moreover, knockdown of YEATS4
inhibits CRC cell proliferation and induces apoptosis (12). Notably, YEATS4 is implicated as the
upstream transcription factor that most often regulates genes
implicated in drug resistance (32,33),
and 70% knockdown of YEATS4 siRNA was found to result in ~20–30%
recovery of drug sensitivity in ovarian cancer (13). In the present study, we reported
that both miR-218 downregulation and YEATS4 upregulation were
observed in CRC tissues and HCT-116/L-OHP cells, which were
associated with L-OHP resistance. Moreover, miR-218 overexpression
sensitized HCT-116 cells to L-OHP-induced cell apoptosis by
targeting YEATS4 expression.
Presently, autophagy has been reported to play a
complex role at multiple levels of tumor development, but it is
generally accepted that autophagy promotes cancer cell survival in
the process of tumor development. Autophagy has been considered a
mechanism through which cancer cells can survive the stress of
damage caused by anticancer therapy and subsequently become
resistant to treatment. Our data also showed that L-OHP induced
autophagy in HCT-116/L-OHP cells, which was inhibited by miR-218
overexpression. More importantly, autophagy partially results in
drug resistance (34), and
inhibition of autophagy may enhance the effectiveness of cancer
treatment. Furthermore, it is well documented that miRNAs do not
only regulate tumor survival and sensitivity to chemotherapy and
radiotherapy, but also affect autophagy (35). miR-218 was found to inhibit
HMGB1-mediated autophagy in endometrial carcinoma cells during
chemotherapy (36). Similarly, the
present study showed that overexpression of miR-218 increased the
sensitivity of HCT-116/L-OHP cells to L-OHP treatment via
inhibition of autophagy. Further research revealed that YEATS4 is a
novel oncogene amplified in non-small cell lung cancer, which
negatively regulates the p53 pathway (28). Inhibition of autophagy exerts
anti-CRC effects via apoptosis induced by p53 activation and ER
stress (37). We speculated that
activation of autophagy was involved in YEATS4
overexpression-mediated cell apoptosis in the HCT-116/L-OHP cells.
Indeed, YEATS4 overexpression reversed miR-218-stimulated cell
apoptosis in response to L-OHP via promoting basal autophagy in
HCT-116/L-OHP cells.
Taken together, the results of the present study
highlight the role of miR-218 in the mechanism of chemotherapy
sensitivity in CRC, and revealed that miR-218 sensitized
HCT-116/L-OHP cells to L-OHP-induced cell apoptosis via inhibition
of cytoprotective autophagy by targeting YEATS4 expression. Thus,
along with further research into its targets and upstream
regulators, miR-218 could be considered as a predictor of L-OHP
treatment for personalized treatment, and restoration of miR-218
levels could be a potential novel strategy by which to enhance
chemosensitivity to L-OHP-based treatment in the clinic.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
YEATS4
|
YEATS domain containing 4
|
|
L-OHP
|
oxaliplatin
|
|
CQ
|
chloroquine
|
|
miRNAs
|
microRNAs
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
ATCC
|
American Type Culture Collection
|
|
FBS
|
fetal bovine serum
|
|
qRT-PCR
|
quantitative real-time PCR
|
|
UTR
|
untranslated region
|
|
DMSO
|
dimethyl sulfoxide
|
|
LC3
|
microtubule-associated protein 1 light
chain 3
|
References
|
1
|
Stiegelbauer V, Perakis S, Deutsch A, Ling
H, Gerger A and Pichler M: MicroRNAs as novel predictive biomarkers
and therapeutic targets in colorectal cancer. World J
Gastroenterol. 20:11727–11735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang HZ, Ma Y, Zhou Y, Xu LM, Chen XJ,
Ding WB and Zou HB: Autophagy contributes to the enrichment and
survival of colorectal cancer stem cells under oxaliplatin
treatment. Cancer Lett. 361:128–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Y, Han Y, Xie F, Wang A, Feng X, Li N,
Guo H and Chen D: ASPP2 enhances oxaliplatin (L-OHP)-induced
colorectal cancer cell apoptosis in a p53-independent manner by
inhibiting cell autophagy. J Cell Mol Med. 19:535–543. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu W, Zhang Z, Zhang Y, Chen X, Guo S,
Lei Y, Xu Y, Ji C, Bi Z and Wang K: HMGB1-mediated autophagy
modulates sensitivity of colorectal cancer cells to oxaliplatin via
MEK/ERK signaling pathway. Cancer Biol Ther. 16:511–517. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie L, Jing R, Qi J, Lin Z and Ju S: Drug
resistance-related microRNAs in hematological malignancies:
Translating basic evidence into therapeutic strategies. Blood Rev.
29:33–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Taniguchi K, Sugito N, Kumazaki M,
Shinohara H, Yamada N, Nakagawa Y, Ito Y, Otsuki Y, Uno B, Uchiyama
K, et al: MicroRNA-124 inhibits cancer cell growth through
PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett.
363:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhong X, Xiao Y, Chen C, Wei X, Hu C, Ling
X and Liu X: MicroRNA-203-mediated posttranscriptional deregulation
of CPEB4 contributes to colorectal cancer progression. Biochem
Biophys Res Commun. 466:206–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao
X, Jia W and Huang J: Decreased expression of miR-218 is associated
with poor prognosis in patients with colorectal cancer. Int J Clin
Exp Pathol. 6:2904–2911. 2013.PubMed/NCBI
|
|
11
|
Fischer U, Heckel D, Michel A, Janka M,
Hulsebos T and Meese E: Cloning of a novel transcription
factor-like gene amplified in human glioma including astrocytoma
grade I. Hum Mol Genet. 6:1817–1822. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tao K, Yang J, Hu Y and Deng A: Knockdown
of YEATS4 inhibits colorectal cancer cell proliferation and induces
apoptosis. Am J Transl Res. 7:616–623. 2015.PubMed/NCBI
|
|
13
|
Kim YR, Park MS, Eum KH, Kim J, Lee JW,
Bae T, Lee DH and Choi JW: Transcriptome analysis indicates TFEB1
and YEATS4 as regulatory transcription factors for drug resistance
of ovarian cancer. Oncotarget. 6:31030–31038. 2015.PubMed/NCBI
|
|
14
|
Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY,
Hung SC, Hsiao M, Yao CJ and Shieh MJ: Autophagy promotes
resistance to photodynamic therapy-induced apoptosis selectively in
colorectal cancer stem-like cells. Autophagy. 10:1179–1192. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: Therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stanton MJ, Dutta S, Zhang H, Polavaram
NS, Leontovich AA, Hönscheid P, Sinicrope FA, Tindall DJ, Muders MH
and Datta K: Autophagy control by the VEGF-C/NRP-2 axis in cancer
and its implication for treatment resistance. Cancer Res.
73:160–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mizushima N, Yoshimori T and Levine B:
Methods in mammalian autophagy research. Cell. 140:313–326. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang KY, Ma J, Zhang FX, Yu MJ, Xue JS and
Zhao JS: MicroRNA-378 inhibits cell growth and enhances
L-OHP-induced apoptosis in human colorectal cancer. IUBMB Life.
66:645–654. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Faltejskova P, Besse A, Sevcikova S,
Kubiczkova L, Svoboda M, Smarda J, Kiss I, Vyzula R and Slaby O:
Clinical correlations of miR-21 expression in colorectal cancer
patients and effects of its inhibition on DLD1 colon cancer cells.
Int J Colorectal Dis. 27:1401–1408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu K, Liang X, Shen K, Sun L, Cui D, Zhao
Y, Tian J, Ni L and Liu J: MiR-222 modulates multidrug resistance
in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell
Res. 318:2168–2177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao X and Jin W: The emerging role of
tumor-suppressive microRNA-218 in targeting glioblastoma stemness.
Cancer Lett. 353:25–31. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alajez NM, Lenarduzzi M, Ito E, Hui AB,
Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, et al: MiR-218
suppresses nasopharyngeal cancer progression through downregulation
of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 71:2381–2391.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zarogoulidis P, Petanidis S, Kioseoglou E,
Domvri K, Anestakis D and Zarogoulidis K: MiR-205 and miR-218
expression is associated with carboplatin chemoresistance and
regulation of apoptosis via Mcl-1 and Survivin in lung cancer
cells. Cell Signal. 27:1576–1588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan
FK, Sung JJ and Yu J: MicroRNA-218 inhibits cell cycle progression
and promotes apoptosis in colon cancer by downregulating BMI1
polycomb ring finger oncogene. Mol Med. 18:1491–1498.
2013.PubMed/NCBI
|
|
25
|
Zhang XL, Shi HJ, Wang JP, Tang HS and Cui
SZ: MiR-218 inhibits multidrug resistance (MDR) of gastric cancer
cells by targeting Hedgehog/smoothened. Int J Clin Exp Pathol.
8:6397–6406. 2015.PubMed/NCBI
|
|
26
|
Yamamoto N, Kinoshita T, Nohata N, Itesako
T, Yoshino H, Enokida H, Nakagawa M, Shozu M and Seki N: Tumor
suppressive microRNA-218 inhibits cancer cell migration and
invasion by targeting focal adhesion pathways in cervical squamous
cell carcinoma. Int J Oncol. 42:1523–1532. 2013.PubMed/NCBI
|
|
27
|
Hu Y, Xu K and Yagüe E: miR-218 targets
survivin and regulates resistance to chemotherapeutics in breast
cancer. Breast Cancer Res Treat. 151:269–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pikor LA, Lockwood WW, Thu KL, Vucic EA,
Chari R, Gazdar AF, Lam S and Lam WL: YEATS4 is a novel oncogene
amplified in non-small cell lung cancer that regulates the p53
pathway. Cancer Res. 73:7301–7312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fischer U, Meltzer P and Meese E: Twelve
amplified and expressed genes localized in a single domain in
glioma. Hum Genet. 98:625–628. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barretina J, Taylor BS, Banerji S, Ramos
AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho
A, et al: Subtype-specific genomic alterations define new targets
for soft-tissue sarcoma therapy. Nat Genet. 42:715–721. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li PL, Zhang X, Wang LL, Du LT, Yang YM,
Li J and Wang CX: MicroRNA-218 is a prognostic indicator in
colorectal cancer and enhances 5-fluorouracil-induced apoptosis by
targeting BIRC5. Carcinogenesis. 36:1484–1493. 2015.PubMed/NCBI
|
|
32
|
Park JH and Roeder RG: GAS41 is required
for repression of the p53 tumor suppressor pathway during normal
cellular proliferation. Mol Cell Biol. 26:4006–4016. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carro MS, Lim WK, Alvarez MJ, Bollo RJ,
Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al:
The transcriptional network for mesenchymal transformation of brain
tumours. Nature. 463:318–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sui X, Chen R, Wang Z, Huang Z, Kong N,
Zhang M, Han W, Lou F, Yang J, Zhang Q, et al: Autophagy and
chemotherapy resistance: A promising therapeutic target for cancer
treatment. Cell Death Dis. 4:e8382013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et
al: VHL-regulated miR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ran X, Yang J, Liu C, Zhou P, Xiao L and
Zhang K: MiR-218 inhibits HMGB1-mediated autophagy in endometrial
carcinoma cells during chemotherapy. Int J Clin Exp Pathol.
8:6617–6626. 2015.PubMed/NCBI
|
|
37
|
Sakitani K, Hirata Y, Hikiba Y, Hayakawa
Y, Ihara S, Suzuki H, Suzuki N, Serizawa T, Kinoshita H, Sakamoto
K, et al: Inhibition of autophagy exerts anti-colon cancer effects
via apoptosis induced by p53 activation and ER stress. BMC Cancer.
15:7952015. View Article : Google Scholar : PubMed/NCBI
|