Introduction
Lung cancer is the most common cause of
cancer-related deaths worldwide, accounting for 1.8 million new
cases and 1.6 million deaths in 2012 (1). During the past decade, major progress
has been achieved in the treatment and management of non-small cell
lung cancer (NSCLC), especially for adenocarcinomas (ADC), with the
discovery of its biological and therapeutic importance of acquired
genomic alterations, such as mutations in the epidermal growth
factor receptor gene (EGFR) (2,3) or
inversions involving the anaplastic lymphoma kinase gene
(ALK) (4,5). EGFR encodes pharmacologically
targetable tyrosine kinases and it has been recognized that NSCLC
patients with EGFR mutations such as deletions in exon 19
and L858R in exon 21 show dramatic prolonged response to
EGFR tyrosine kinase inhibitors (EGFR-TKIs), such as
gefitinib or erlotinib, in clinical trials of advanced-stage lung
cancer (6–8). The reliable detection of EGFR
mutations is necessary for the personalized treatment of lung
cancer patients.
A number of methods have been used to detect
EGFR mutations in NSCLC patients (9). While the sensitivity of EGFR
mutation assays has improved, the majority of these methods,
however, need at least a few hours to detect EGFR mutations.
In the case of obtaining EGFR test results from outside
laboratories, it generally requires 7–14 days after tumor sampling.
The turnaround time (TAT) for EGFR mutation testing has
become increasingly important, especially for advanced-stage
patients, in order to start EGFR-TKI therapy earlier.
Therefore, a more rapid and simple EGFR mutation assay is
needed.
Recently, we developed novel rapid assays using a
real-time droplet-polymerase chain reaction (PCR) machine (Seiko
Epson Corp. Head Office, Suwa, Japan) for the detection of the
human influenza virus (10,11) and the promyelocytic leukemia
(PML)-retinoic acid receptor α (RARA) fusion gene
(12,13), for the genotyping of single
nucleotide polymorphisms (14,15)
and for the detection of bovine respiratory syncytial virus
(16) and breakpoint cluster region
(BCR)-Abelson murine leukemia viral oncogene (ABL)
fusion mRNA (17). All of these
assays provide a much shorter TAT than the conventional assays
currently used in clinical settings while demonstrating almost the
same reactivity.
The purpose of this study was to develop a novel
rapid EGFR mutation assay in lung cancer patients by using a
real-time droplet-PCR machine (EGFR d-PCR assay). In this
study, we used fresh liquid cytology specimens, such as bronchial
lavage fluid (BLF), pleural effusion (PE) and cardiac effusion (CE)
from the patients, which are processed in more simple steps and a
shorter time for DNA extraction than formalin-fixed
paraffin-embedded (FFPE) specimens. To validate the performance of
the EGFR d-PCR assay, we compared its reactivity and
reaction time to a conventional real-time PCR assay. In addition,
we validated the sensitivity of the EGFR d-PCR assay by
using fresh liquid samples containing variable percentages of cell
lines with EGFR mutations.
Materials and methods
Patients and samples
This study was reviewed and approved by the Medical
Ethics Committee of Shinshu University School of Medicine (Nagano,
Japan). BLF, PE and CE specimens were collected from 245 patients
who were clinically diagnosed with lung cancer from January to
December 2014 and the specimens provided were used for cytological
diagnosis at the Department of Laboratory Medicine at the Shinshu
University Hospital (Nagano, Japan). BLF specimens were obtained
after transbronchial lung biopsies. PE and CE specimens were
obtained by fine-needle aspiration from patients with advanced lung
cancer. After cytological evaluation of the specimens from the 245
patients, we excluded the specimens that contained no malignant
cells (146 patients) or cytologically diagnosed as malignant
lymphoma (ML) (1 patient), metastasis of renal cell carcinoma (RCC)
(1 patient), small cell carcinoma (SCC) (8 patients) and squamous
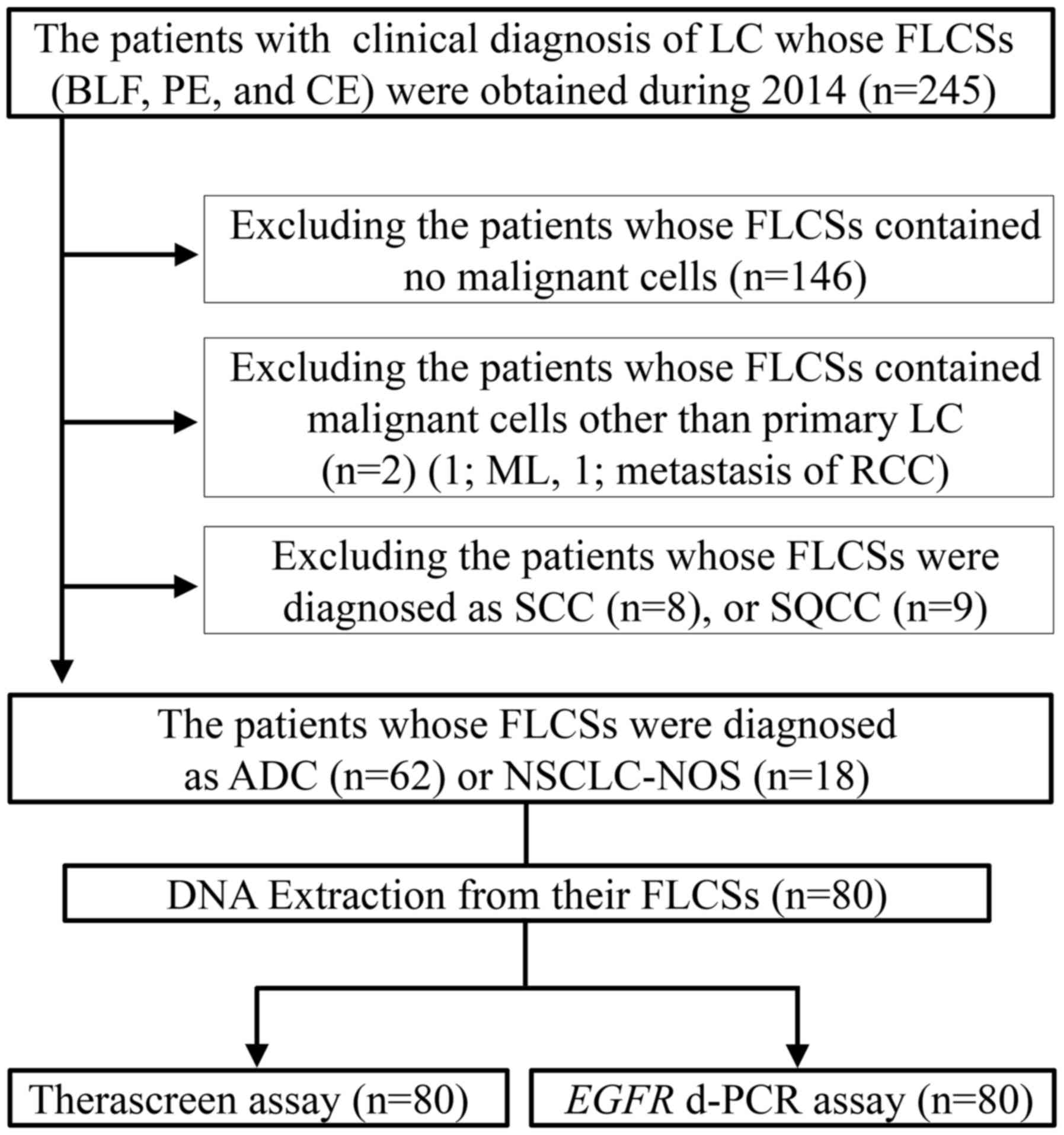
cell carcinoma (SQCC) (9 patients) (Fig. 1). Specimens from the remaining 80
patients, who were cytologically diagnosed with primary lung ADC or
NSCLC-not otherwise specified (NOS), were also evaluated (Fig. 2). The fresh liquid cytology
specimens were centrifuged to create a cell pellet and after
picking up enough of the cell pellet for the routine smear slide
for cytological diagnosis, the remainder of the pellet was used for
the samples in this study. DNA was extracted from the specimens
using the QIAamp DNA Blood Mini kit (Qiagen, Inc., Valencia, CA,
USA) according to the manufacturers instructions.
Conventional real-time PCR assay
For the reference, the conventional method to the
EGFR d-PCR assay in this study, we used the Rotor-Gene Q
5plex HRM instrument with the therascreen® EGFR
RGQ PCR kit (Qiagen, Inc.) (Therascreen assay). This assay is
approved in the United States, Europe, Japan and China and the kit
is based on the amplification-refractory mutation system (ARMS) and
Scorpions PCR technologies, which enable sensitive and selective
site-specific detection of 29 types of somatic mutations in the
EGFR gene (18). The
reaction conditions of the Therascreen assay were as follows: 95°C
for 15 min for 1 cycle, 95°C for 30 sec and then 60°C for 60 sec
for 40 cycles. Analysis was performed using the Rotor-Gene Q series
software, version 2.0.2 (Qiagen, Inc.) and the
manufacturer-supplied ΔCt thresholds were used to analyze the
result of each EGFR mutation reaction. The reaction time was
about 1 h and 45 min.
Real-time droplet-PCR machine used for
the EGFR d-PCR assay
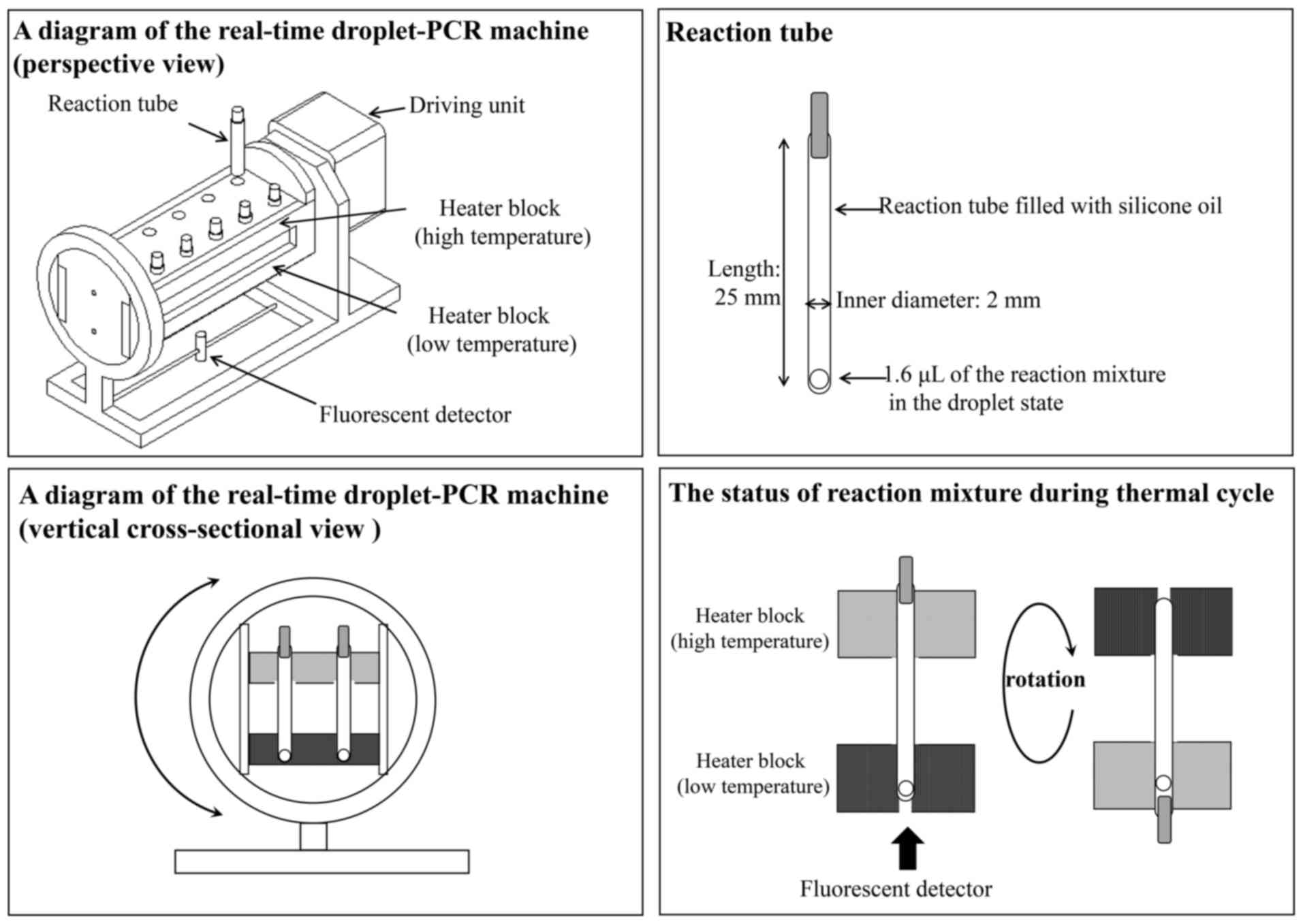
The real-time droplet-PCR machine has two connected
heater blocks, which regulate the temperature of each end of the
reaction tube (Fig. 3). The
reaction temperature of the two heater blocks is consistently
controlled during the denaturation and annealing/extension steps.
To perform a rapid temperature transition, the reaction tube is
filled with silicone oil, allowing the droplet of the PCR mixture
to move easily in the tube during the mechanical rotation of the
machine with the two connected heater blocks. Once the droplet
transfers from the one end to the other owing to gravitation, the
machine with the two heater blocks inverts to return the droplet.
Therefore, the PCR mixture in the droplet is able to perform
shuttle PCR in the reaction tube. The real-time droplet-PCR machine
integrates a fluorescence detector and it allows monitoring of the
amount of PCR products after each extension step.
Target mutations of EGFR
As for the target mutations of EGFR in this
study, we selected the two most frequent mutations. One was the
point mutation L858R (c.2573T>G) in exon 21 and the other was
the 15-bp/5-amino acid deletion of E746_A750del (c.2235_2249del and
c.2236_2250del) in exon 19. These two major mutations cover about
90% of oncogenic EGFR mutations (19). In addition, we also selected the
point mutation of T790M (c.2369>T) in exon 20, which is the most
common resistance mutation for EGFR-TKIs (20).
Reaction conditions of the EGFR d-PCR
assay
We designed specific primers based on ARMS that
induced a mutation- matched nucleotide in the 3-end and a
template-mismatched nucleotide at the −2 position from the 3-end.
The primers and probes used in this study are as follows (5′-3′):
L858R forward, GCTTGGTGCACCGCGACCTG; reverse, CGCACCCAGCAGTTTGGCAC;
probe, 6FAM-AGCCAGGAACGTACTGGTGAAAACACCGCA-BHQ-1; E746_A750del
forward, GGCAGCATGTGGCACCATC; reverse, GTTGGCTTTCGGAGATGTAT; probe,
6FAM-TCTCACCTTCTGGGATCCAGAGTCCCT-BHQ-1; T790M forward,
CCCCACGTGTGCCGCCTG; reverse, GCCGAAGGGCATGAGCTGTA; probe,
6FAM-TGGGCATCTGCCTCACCTCCACCGTG CA-BHQ-1.
The reaction mixture contained genomic DNA (10
ng/µl), Platinum® Taq DNA polymerase (Life
Technologies, Grand Island, NY, USA), 800 nmol/l of primer designed
as aforementioned, 300 nmol/l of TaqMan probe and reaction buffer
composed of Tris-HCl pH 9.0, KCl and MgCl2, in a total
volume of 10 µl. From the reaction mixtures, 1.6 µl was used for
one reaction tube.
Each reaction condition was as follows: for L858R,
98°C for 10 sec, 40 cycles at 98°C for 5 sec and 60°C for 6 sec;
for E746_A750del and T790M, 98°C for 10 sec, 40 cycles at 98°C for
5 sec and 55°C for 6 sec. The reaction time was 8 min and 10
sec.
Evaluation of the EGFR d-PCR assay
results
To evaluate the PCR results as EGFR
mutation-positive or -negative, we determined the arbitrary cut-off
values of fluorescence level in 40 cycles, which were based on the
receiver operating characteristic (ROC) curve analysis and using
the Therascreen assay as ‘gold standard’. Each cut-off value was as
follows: 4.7 for L858R, 4.7 for E746_A750del and 6.8 for T790M.
Compared to the EGFR d-PCR assay, which was
designed to detect only E746_A750del in exon 19, the Therascreen
assay can detect 19 types of deletions in exon 19 including
E746_A750del, however, it cannot tell which mutation was detected
among the 19 types. To examine the exact locus of the deletion in
all the specimens determined as deletion positive by Therascreen
assay, we performed direct sequencing for those specimens.
Sensitivity analysis of EGFR d-PCR
assay
To evaluate the sensitivity of the EGFR d-PCR
assay, we performed sensitivity assays using DNA mixtures extracted
from the following cell lines: PC9 [E746_A750del (c.2235_2249del)],
H1975 [L858R (c.2573T>G) and T790M (c.2369>T)] and A549
[EGFR wild-type]. The PC9 cell line was obtained directly
from the RIKEN Cell Bank (Tsukuba, Japan). The H1975 cell line was
obtained directly from the American Type Culture Collection (ATCC,
Rockville, MD, USA). The A549 cell line was obtained directly from
the Japanese Cancer Research Bank (Tokyo, Japan). PC9 and A549 and
H1975 and A549 were mixed respectively in ratios of 1:0, 0.5:0.5,
0.1:0.9, 0.05:0.95, 0.01:0.99, 0.005:0.995, 0.001:0.999,
0.0005:0.9995, 0.0001:0.9999 and 0:1. As a result, each percentage
of EGFR mutant cells in these mixtures was 100, 50, 10, 5,
1, 0.5, 0.1, 0.05, 0.01 and 0%. EGFR mutations of these
mixtures were analyzed by the EGFR d-PCR assay.
Results
Characteristics of the patients and
specimens
Fresh liquid cytology specimens from 80 lung cancer
patients, including 77 BLF specimens, 2 PE specimens and 1 CE
specimen were used in this study and the patient clinical
characteristics are shown in Table
I. The median age of the 80 patients was 68 years (range, 41–85
years); all of the patients were Japanese and 57 patients (71.3%)
were men while 23 patients (28.7%) were women. Cytologically, 62
patients (77.5%) were diagnosed with ADC and 18 patients (22.5%)
were diagnosed with NSCLC-NOS. Radiologically, the median diameter
of the tumors was 34.2 mm (range, 7–132 mm). The distribution of
the clinical stages at the time of diagnosis was as follows: 37
patients (46.3%) had stage I cancer, 5 patients (6.3%) had stage
II, 9 patients (11.3%) had stage III and 27 patients (33.8%) had
stage IV.
 | Table I.Clinical characteristics of the
patients. |
Table I.
Clinical characteristics of the
patients.
|
Characteristics | Data |
|---|
| Age (years) |
|
|
Average | 68 |
|
Range | 41–85 |
| Gender, n (%) |
|
|
Male | 57 (71.3) |
|
Female | 23 (28.7) |
| Diameter of the
tumor (mm) |
|
|
Average | 34.2 |
|
Range | 7–132 |
| Stage, n (%) |
|
| I | 37 (46.3) |
| II | 5 (6.3) |
|
III | 9 (11.3) |
| IV | 27 (33.8) |
|
Unknown | 2 (2.5) |
Comparison between the Therascreen
assay and the EGFR d-PCR assay
Table II shows the
comparison between the Therascreen assay and the EGFR d-PCR
assay. Among the specimens from 80 patients, the Therascreen assay
detected exon 21 L858R point mutations in 16 patients (20.0%), exon
19 deletions in 11 patients (13.8%), exon 20 insertion in one
patient (1.3%) and exon 20 T790M point mutation in one patient
(1.3%). In contrast, the EGFR d-PCR assay detected L858R in
16 patients (20.0%), E746_A750del in 8 patients (10.0%) and T790M
in one patient (1.3%). The Therascreen assay but not the
EGFR d-PCR assay detected exon 19 deletions in three
specimens. Direct sequencing confirmed L747_S752del P753S mutation
in one specimen and L747_S752del mutation in two specimens. As a
result, in regards to the L858R, E746_A750del and T790M mutations,
the results of the EGFR d-PCR assay were in complete
concordance with those of the Therascreen assay.
 | Table II.Comparison between Therascreen assay
and EGFR d-PCR assay |
Table II.
Comparison between Therascreen assay
and EGFR d-PCR assay
| EGFR
mutation type | Therascreen assay,
n (%) | EGFR d-PCR
assay, n (%) |
|---|
| Positive | 28 (35.0) | 25 (31.3) |
| Exon 21 |
|
|
|
L858R | 16 (20.0) | 16 (20.0) |
| Exon 19 | 11 (13.8) | 8 (10.0) |
|
E746_A750del | [8] (10.0) | 8 (10.0) |
|
L747_S752del | [2] (2.5) | N/D |
|
L747_S752del
P753Sa | [1] (1.3) | N/D |
| Exon 20 |
|
|
|
|
T790Ma | 1 (1.3) | 1 (1.3) |
|
Insertions | 1 (1.3) | N/D |
| Negative | 52 (65.0) | 55 (68.8) |
| Total | 80 | 80 |
Sensitivity of the novel EGFR d-PCR
assay
With the EGFR d-PCR assay, the L858R mutation
was detected in L858R-positive cell line mixtures (H1975 and A549)
at 100, 50, 10, 5, 1 and 0.5% H1975 cells (Fig. 4A). Similarly, E746_A750del was
detected in E746_A750del-positive cell line mixtures (PC9 and A549)
at 100, 50, 10, 5, 1, 0.5, 0.1 and 0.05% PC9 cells (Fig. 4B). T790M was detected in
T790M-positive cell line mixtures (H1975 and A549) at 100, 50, 10,
5, 1 and 0.5% H1975 cells (Fig.
4C). These results suggest that the detection limits of the
EGFR d-PCR assay were 0.5% for L858R, 0.05% for E746_A750del
and 0.5% for T790M.
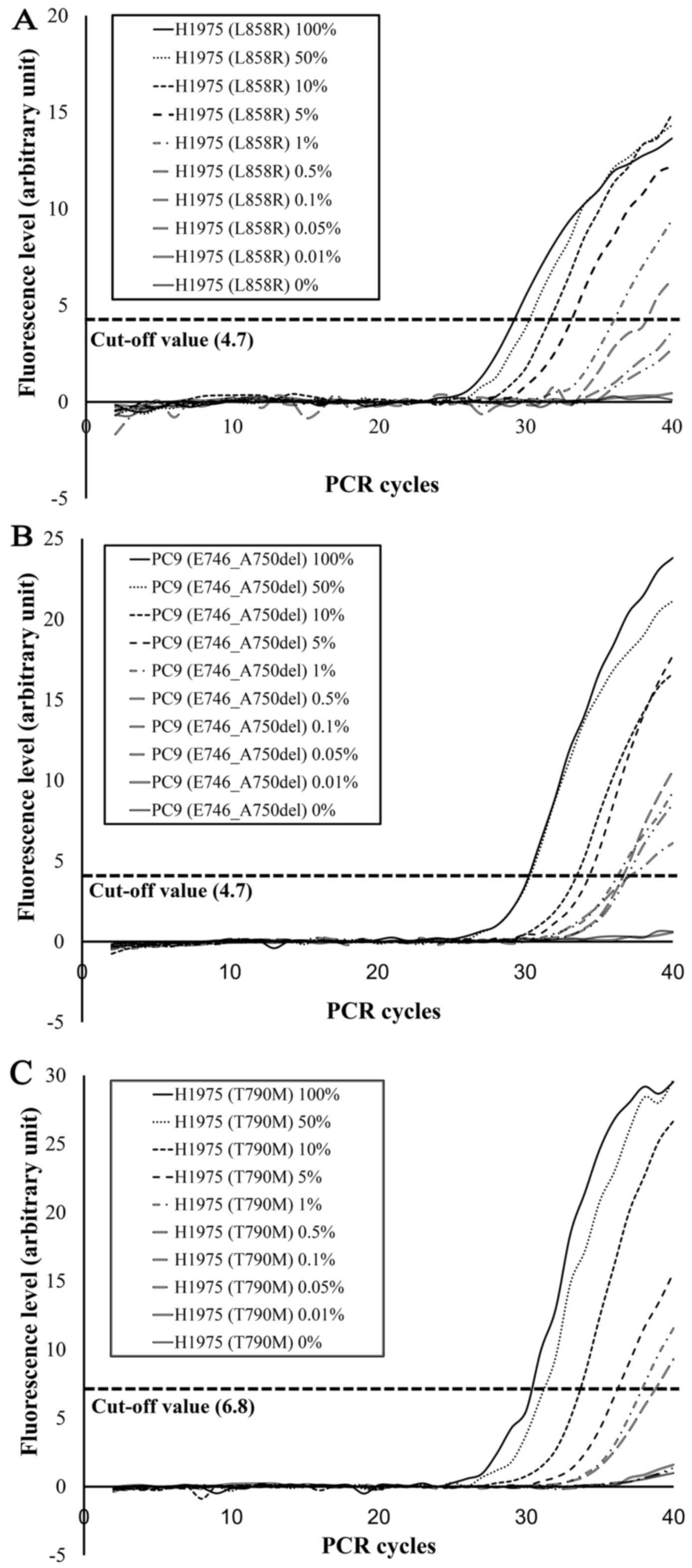 | Figure 4.Sensitivity assessment of the EGFR
d-PCR assay. (A) L858R, (B) E746_A750del and (C) T790M mutations
were detected in mutation-positive cell line mixtures at respective
percentages of 100, 50, 10, 5, 1 and 0.5% of H1975 cells for L858R
(A), 100, 50, 10, 5, 1, 0.5, 0.1 and 0.05% of PC9 cells for
E746_A750del (B), and 100, 50, 10, 5, 1 and 0.5% of H1975 cells for
T790M (C). |
Discussion
Numerous methods are available for EGFR
mutation testing: direct sequencing, PCR-based assays such as
single-strand conformation polymorphism (PCR-SSCP) (21), peptide nucleic acid-locked nucleic
acid (PNA-LNA) PCR clamp assays (22), PCR-Invader (23), cycleave PCR (24), the Scorpions
amplification-refractory mutation system (ARMS) (25), immunohistochemistry with EGFR
mutation-specific antibodies (26),
real-time PCR methods (27) and
next generation sequencing (28).
Each method has different sensitivity and specificity and they need
at least a few hours to effectively detect EGFR mutations.
In this study, we demonstrated that the EGFR d-PCR assay
markedly reduced the detection time of major EGFR mutations
with high sensitivity compared with conventional methods. To the
best of our knowledge, the novel EGFR d-PCR assay is the
most rapid detection method and a reliable test to assess the
dynamics of key EGFR mutations.
In 2013, to establish an evidence-based
recommendation of the molecular testing of EGFR and
ALK for patients with lung cancer, three professional
societies (the College of American Pathologists, the International
Associations for the Study of Lung Cancer and the Association for
Molecular Pathology) developed a guideline (29). The guideline recommends performing
EGFR and ALK molecular testing for all patients with
advanced-stage lung ADC or tumor with an ADC component. Regarding
testing samples, the guideline suggested the use of cytology
specimens for EGFR and ALK molecular testing,
although FFPE, fresh frozen, or alcohol-fixed tissue specimens
should be prioritized. In addition, it recommends that FFPE cell
blocks were preferable than smear preparations when cytology
specimens were used. However, it requires several days to prepare
FFPE specimens for pathological diagnosis and several hours to
extract DNA from those specimens because they need
deparaffinization and reversal of formaldehyde-induced modification
of nucleic acids. Some studies have reported the utility and the
reliability of fresh cytology specimens for EGFR mutation
genotyping in NSCLC patients and their detection sensitivity is
similar to or even better than that of FFPE tissue or cell block
samples (30–32). Thus, we attempted to use fresh
liquid cytology specimens, such as BLF, PE and CE, for the
detection of EGFR mutations in this study, because it can
shorten the time to prepare smear slides for pathological diagnosis
to 30 min and the time to extract DNA from the specimens to 30
min.
This study demonstrated that the EGFR d-PCR
assay could markedly reduce the reaction time to 8 min and 10 sec,
while the conventional method, the Therascreen assay, takes 1 h and
45 min. In the EGFR d-PCR assay, the volume of the droplet
including the sample and reagents is only 1.6 µl and is surrounded
by silicone oil, which has good heat conduction characteristics.
Therefore, the temperature of the droplet can quickly reach the
temperature regulated by the two heater blocks. In addition, the
rapid transition of the droplet between the two set temperatures
decreases the time for each cycle in PCR. Few studies have reported
rapid assays for detecting EGFR gene mutations in lung
cancer patients within 10 min. Sakamoto et al (33,34)
developed a high-speed EGFR real-time PCR assay known as the
ultra-rapid PCR assay, which can detect L858R and E746_A750del in
<10 min. Our EGFR d-PCR assay is, however, more rapid and
sensitive than the ultra-rapid PCR assay. Moreover, it can also
detect the T790M mutation in addition to the L858R and E746_A750del
mutations in the same reaction. As a result, the entire procedure
of the EGFR d-PCR assay is accomplished within 1 h and 30
min; thus, EGFR gene mutational status can be reported on
the day the specimens are obtained from the patients.
In addition to the benefit of shortening the testing
time of EGFR mutations, the sample results of the
EGFR d-PCR assay were highly concordant with those of the
Therascreen assay and EGFR d-PCR had excellent detection
limits. The sensitivity of the Therascreen assay is well documented
(18,35) and its detectable mutation
percentages are 1.26% for L858R, 1.64% for deletions and 7.02% for
T790M according to the manufacturers database concerning
performance characteristics. Our EGFR d-PCR assay had
detection limits of 0.5% for L858R, 0.05% for E746_A750 and 1.0%
for T790M of mutation-positive cells, respectively. These
mutation-positive cell lines have two copies of the EGFR
gene: one is mutated and the other is wild-type. Thus, the
EGFR d-PCR assay may have much lower detectable mutation
percentages than the Therascreen assay.
Regarding the cost of the EGFR d-PCR assay,
the assay is a cost-effective method compared to conventional
assays. This is due to the fact that the reaction mixture volume of
EGFR d-PCR assay is only 1.6 µl in each reaction tube and it
needs a smaller volume of primers and probes. Moreover, it also
needs a smaller volume of samples from the patients, so the
remnants from the cytology specimens for the diagnosis were
sufficient and thus used for the assays in this study. This facet
could lead to using the samples from patients for other additional
molecular tests.
EGFR mutations detectable by the novel assay
were limited to the two major EGFR mutations, L858R and
E746_A750del and the most common resistance mutation for
EGFR-TKIs, T790M. However, it has been reported that the
response rates for EGFR-TKIs in patients with the two major
mutations were higher than those with minor mutations such as
insertions in exon 20 (36), L861Q
in exon 21 and exon 19 deletions starting codon L747 (37); the effectiveness of EGFR-TKIs
in patients with minor EGFR mutations is limited (38,39).
Therefore, after examining the major EGFR mutations by
EGFR d-PCR assay, we can examine the minor EGFR
mutations by multiplex PCR assay or direct sequencing after the
precise histological tumor evaluation. Thus, the EGFR d-PCR
assay may help detect frequent and effective mutations for
EGFR-TKI therapy right after obtaining specimens and
contribute to a more rapid start of therapy for advanced-stage lung
cancer patients. Although the EGFR d-PCR assay needs two PCR
reactions for one sample at this point, once for L858R and once for
E746_A750del and T790M, we are planning to adjust the PCR
conditions to detect all three EGFR mutations simultaneously
and currently apply newly designed primers to other EGFR
mutations to increase the variety of the types it can detect in the
future.
In conclusion, our EGFR d-PCR assay can
detect major EGFR mutations rapidly, correctly, sensitively
and cost-effectively with a small amount of specimen. By means of
using fresh liquid cytology specimens, which require fewer and
easier steps to obtain DNA, the TAT can be markedly reduced
compared with conventional assays using FFPE specimens. This assay
is expected to contribute to an expedited advent of EGFR-TKI
therapy for NSCLC patients. Furthermore, this assay can be a
point-of-care test for such patients in the very near future.
Acknowledgements
This study was supported by the Japan Society for
the Promotion of Science (JSPS) Grants-in-Aid for Scientific
Research, commonly called KAKENHI (grant no. 25460434). This study
was also supported in-kind by the Seiko Epson Corporation (Suwa,
Japan) and Akemi Yamaguchi, one of the authors in this study, was
an employee of this company. The authors acknowledge Masayuki
Uehara (Corporate Research and Development Div., Seiko Epson
Corporation), Akane Sueki, Yukihiro Kobayashi and Mitsutoshi Sugano
(Department of Laboratory Medicine, Shinshu University Hospital,
Nagano, Japan) for the technical assistance. We would like to thank
Editage (www.editage.jp) for the English language
editing.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
ADC
|
adenocarcinoma
|
|
EGFR
|
epidermal growth factor receptor
gene
|
|
ALK
|
anaplastic lymphoma kinase gene
|
|
EGFR-TKIs
|
EGFR tyrosine kinase
inhibitors
|
|
TAT
|
turnaround time
|
|
PCR
|
polymerase chain reaction
|
|
PML
|
promyelocytic leukemia
|
|
RARA
|
retinoic acid receptor α
|
|
BCR
|
breakpoint cluster region
|
|
ABL
|
Abelson murine leukemia viral
oncogene
|
|
EGFR d-PCR assay
|
rapid EGFR mutation assay by
using real-time droplet-PCR machine
|
|
BLF
|
bronchial lavage fluid
|
|
PE
|
pleural effusion
|
|
CE
|
cardiac effusion
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
ML
|
malignant lymphoma
|
|
RCC
|
renal cell carcinoma
|
|
SCC
|
small cell carcinoma
|
|
SQCC
|
squamous cell carcinoma
|
|
NOS
|
not otherwise specified
|
|
ARMS
|
amplification-refractory mutation
system
|
|
ROC
|
receiver operating characteristic
|
|
PCR-SSCP
|
PCR-based assays such as single-strand
conformation polymorphism
|
|
PNA-LNA
|
peptide nucleic acid-locked nucleic
acid
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mano H: Non-solid oncogenes in solid
tumors: EML4-ALK fusion genes in lung cancer. Cancer Sci.
99:2349–2355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravon P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: North-East Japan Study Group: Gefitinib or chemotherapy for
non-small-cell lung cancer with mutated EGFR. N Engl J Med.
362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Spanish Lung Cancer Group in
collaboration with Groupe Français de Pneumo-Cancérologie and
Associazione Italiana Oncologia Toracica: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pao W and Ladanyi M: Epidermal growth
factor receptor mutation testing in lung cancer: Searching for the
ideal method. Clin Cancer Res. 13:4954–4955. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsuda K, Yamaguchi A, Taira C, Sueki A,
Koeda H, Takagi F, Sugano M and Honda T: A novel high-speed
droplet-polymerase chain reaction can detect human influenza virus
in less than 30 min. Clin Chim Acta. 413:1742–1745. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sueki A, Matsuda K, Yamaguchi A, Uehara M,
Sugano M, Uehara T and Honda T: Evaluation of saliva as diagnostic
materials for influenza virus infection by PCR-based assays. Clin
Chim Acta. 453:71–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sueki A, Matsuda K, Taira C, Yamaguchi A,
Koeda H, Takagi F, Kobayashi Y, Sugano M and Honda T: Rapid
detection of PML-RARA fusion gene by novel high-speed
droplet-reverse transcriptase-polymerase chain reaction:
Possibility for molecular diagnosis without lagging behind the
morphological analyses. Clin Chim Acta. 415:276–278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shigeto S, Matsuda K, Yamaguchi A, Sueki
A, Uehara M, Sugano M, Uehara T and Honda T: Rapid diagnosis of
acute promyelocytic leukemia with the PML-RARA fusion gene using a
combination of droplet-reverse transcription-polymerase chain
reaction and instant-quality fluorescence in situ hybridization.
Clin Chim Acta. 453:38–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taira C, Matsuda K, Yamaguchi A, Sueki A,
Koeda H, Takagi F, Kobayashi Y, Sugano M and Honda T: Novel
high-speed droplet-allele specific-polymerase chain reaction:
Application in the rapid genotyping of single nucleotide
polymorphisms. Clin Chim Acta. 424:39–46. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taira C, Matsuda K, Yamaguchi A, Uehara M,
Sugano M, Okumura N and Honda T: Rapid single nucleotide
polymorphism based method for hematopoietic chimerism analysis and
monitoring using high-speed droplet allele-specific PCR and
allele-specific quantitative PCR. Clin Chim Acta. 445:101–106.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uehara M, Matsuda K, Sugano M and Honda T:
A new high-speed droplet-real-time polymerase chain reaction method
can detect bovine respiratory syncytial virus in less than 10 min.
J Vet Med Sci. 76:477–480. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamaguchi A, Matsuda K, Sueki A, Taira C,
Uehara M, Saito Y and Honda T: Development of a rapid and sensitive
one-step reverse transcription-nested polymerase chain reaction in
a single tube using the droplet-polymerase chain reaction machine.
Clin Chim Acta. 448:150–154. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vallée A, Le Loupp AG and Denis MG:
Efficiency of the Therascreen® RGQ PCR kit for the
detection of EGFR mutations in non-small cell lung carcinomas. Clin
Chim Acta. 429:8–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitsudomi T, Kosaka T and Yatabe Y:
Biological and clinical implications of EGFR mutations in lung
cancer. Int J Clin Oncol. 11:190–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu HA, Arcila ME, Rekhtman N, Sima CS,
Zakowski MF, Pao W, Kris MG, Miller VA, Ladanyi M and Riely GJ:
Analysis of tumor specimens at the time of acquired resistance to
EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers.
Clin Cancer Res. 19:2240–2247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marchetti A, Martella C, Felicioni L,
Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli
F, Mezzetti A, et al: EGFR mutations in non-small-cell lung cancer:
Analysis of a large series of cases and development of a rapid and
sensitive method for diagnostic screening with potential
implications on pharmacologic treatment. J Clin Oncol. 23:857–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nagai Y, Miyazawa H, Huqun Tanaka T,
Udagawa K, Kato M, Fukuyama S, Yokote A, Kobayashi K, Kanazawa M,
et al: Genetic heterogeneity of the epidermal growth factor
receptor in non-small cell lung cancer cell lines revealed by a
rapid and sensitive detection system, the peptide nucleic
acid-locked nucleic acid PCR clamp. Cancer Res. 65:7276–7282. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hall JG, Eis PS, Law SM, Reynaldo LP,
Prudent JR, Marshall DJ, Allawi HT, Mast AL, Dahlberg JE,
Kwiatkowski RW, et al: Sensitive detection of DNA polymorphisms by
the serial invasive signal amplification reaction. Proc Natl Acad
Sci USA. 97:8272–8277. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yatabe Y, Hida T, Horio Y, Kosaka T,
Takahashi T and Mitsudomi T: A rapid, sensitive assay to detect
EGFR mutation in small biopsy specimens from lung cancer. J Mol
Diagn. 8:335–341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kimura H, Kasahara K, Kawaishi M, Kunitoh
H, Tamura T, Holloway B and Nishio K: Detection of epidermal growth
factor receptor mutations in serum as a predictor of the response
to gefitinib in patients with non-small-cell lung cancer. Clin
Cancer Res. 12:3915–3921. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kawahara A, Taira T, Azuma K, Tominaga M,
Hattori S, Kawahara M, Abe H, Yamaguchi T, Akiba J, Takamori S, et
al: A diagnostic algorithm using EGFR mutation-specific antibodies
for rapid response EGFR-TKI treatment in patients with non-small
cell lung cancer. Lung Cancer. 78:39–44. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lewandowska MA, Jóźwicki W, Jochymski C
and Kowalewski J: Application of PCR methods to evaluate EGFR, KRAS
and BRAF mutations in a small number of tumor cells in cytological
material from lung cancer patients. Oncol Rep. 30:1045–1052.
2013.PubMed/NCBI
|
|
28
|
Tuononen K, Mäki-Nevala S, Sarhadi VK,
Wirtanen A, Rönty M, Salmenkivi K, Andrews JM, Telaranta-Keerie AI,
Hannula S, Lagström S, et al: Comparison of targeted
next-generation sequencing (NGS) and real-time PCR in the detection
of EGFR, KRAS, and BRAF mutations on formalin-fixed,
paraffin-embedded tumor material of non-small cell lung carcinoma -
superiority of NGS. Genes Chromosomes Cancer. 52:503–511. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lindeman NI, Cagle PT, Beasley MB, Chitale
DA, Dacic S, Giaccone G, Jenkins RB, Kwiatkowski DJ, Saldivar JS,
Squire J, et al: College of American Pathologists International
Association for the Study of Lung Cancer and Association for
Molecular Pathology: Molecular testing guideline for selection of
lung cancer patients for EGFR and ALK tyrosine kinase inhibitors:
Guideline from the College of American Pathologists, International
Association for the Study of Lung Cancer, and Association for
Molecular Pathology. J Mol Diagn. 15:415–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smouse JH, Cibas ES, Jänne PA, Joshi VA,
Zou KH and Lindeman NI: EGFR mutations are detected comparably in
cytologic and surgical pathology specimens of nonsmall cell lung
cancer. Cancer. 117:67–72. 2009.PubMed/NCBI
|
|
31
|
Reynolds JP, Tubbs RR, Minca EC, MacNamara
S, Almeida FA, Ma PC, Pennell NA and Cicenia JC: EGFR mutational
genotyping of liquid based cytology samples obtained via fine
needle aspiration (FNA) at endobronchial ultrasound of non-small
cell lung cancer (NSCLC). Lung Cancer. 86:158–163. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lozano MD, Labiano T, Echeveste J, Gurpide
A, Martín- Algarra S, Zhang G, Sharma A and Palma JF: Assessment of
EGFR and KRAS mutation status from FNAs and core-needle biopsies of
non-small cell lung cancer. Cancer Cytopathol. 123:230–236. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sakamoto T, Kodani M, Takata M, Chikumi H,
Nakamoto M, Nishii-Ito S, Ueda Y, Izumi H, Makino H, Touge H, et
al: A novel point-of-care system for high-speed real-time
polymerase chain reaction testing for epidermal growth factor
receptor mutations in bronchial lavage fluids after transbronchial
biopsy in patients with non-small cell lung cancer. Int J Oncol.
46:1473–1480. 2015.PubMed/NCBI
|
|
34
|
Takata M, Chikumi H, Matsunami K, Kodani
M, Sakamoto T, Hashimoto K, Nakamoto M, Okada K, Kitaura T,
Matsumoto S, et al: A new rapid method for detecting epidermal
growth factor receptor mutations in non-small cell lung cancer.
Oncol Rep. 33:1040–1048. 2015.PubMed/NCBI
|
|
35
|
Angulo B, Conde E, Suárez-Gauthier A,
Plaza C, Martínez R, Redondo P, Izquierdo E, Rubio-Viqueira B,
Paz-Ares L, Hidalgo M, et al: A comparison of EGFR mutation testing
methods in lung carcinoma: Direct sequencing, real-time PCR and
immunohistochemistry. PLoS One. 7:e438422012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Naidoo J, Sima CS, Rodriguez K, Busby N,
Nafa K, Ladanyi M, Riely GJ, Kris MG, Arcila ME and Yu HA:
Epidermal growth factor receptor exon 20 insertions in advanced
lung adenocarcinomas: Clinical outcomes and response to erlotinib.
Cancer. 121:3212–3220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee VHF, Tin VPC, Choy TS, Lam KO, Choi
CW, Chung LP, Tsang JW, Ho PP, Leung DK, Ma ES, et al: Association
of exon 19 and 21 EGFR mutation patterns with treatment outcome
after first-line tyrosine kinase inhibitor in metastatic
non-small-cell lung cancer. J Thorac Oncol. 8:1148–1155. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY
and Yang PC: Effectiveness of tyrosine kinase inhibitors on
‘uncommon’ epidermal growth factor receptor mutations of unknown
clinical significance in non-small cell lung cancer. Clin Cancer
Res. 17:3812–3821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Otsuka T, Mori M, Yano Y, Uchida J,
Nishino K, Kaji R, Hata A, Hattori Y, Urata Y, Kaneda T, et al:
Effectiveness of tyrosine kinase inhibitors in Japanese patients
with non-small cell lung cancer harboring minor epidermal growth
factor receptor mutations: Results from a multicenter retrospective
study (HANSHIN Oncology Group 0212). Anticancer Res. 35:3885–3891.
2015.PubMed/NCBI
|