Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common solid cancer and the third most common cause of
cancer-related mortality (1). The
current treatment modalities for HCC, including liver
transplantation, surgical resection and local ablation therapy,
have significantly prolonged patient survival (2,3).
However, when liver cancer spreads within the liver or to other
organs, effective therapeutic options are extremely limited, and
the mortality rate is high (4).
Although the risk factors predisposing patients to HCC have been
well established, the precise molecular mechanisms underlying HCC
metastasis are not fully understood. Therefore, a better
understanding of the molecular processes associated with HCC
invasion and metastasis is required for the development of
effective therapeutic strategies to treat metastatic HCC and
prolong patient survival.
Cancer stem cells (CSCs) represent a small
subpopulation of cancer cells in various types of cancer. CSCs are
distinguished by their capacity to self-renew and differentiate
into malignant cells. Despite composing only a small proportion of
the general population of cancer cells, CSCs make a significant
contribution to cancer metastasis and invasion due to their high
metastatic potential (5–7). To date, various cell surface markers
specifically expressed on CSCs, including CD133, CD44, EpCAM and
CD90, have been identified, and each of these markers has been
biologically and clinically characterized (8). The 5-transmembrane protein CD133, also
referred to as human prominin-1, is a well-established protein
marker of multiple types of solid tumors (9). In addition, CD133 has been shown to
mediate HCC metastasis by regulating the expression of
metastasis-associated genes, and CD133 expression in HCC is
associated with a poor prognosis (10–12).
In our previous study, we demonstrated that Huh7CD133+
liver cancer stem cells (LCSCs) exhibit greater metastatic
potential compared with Huh7CD133− cells after
undergoing radiation treatment in vitro and in vivo
(13). The transmembrane
glycoprotein CD44 localizes to the cell surface and plays a key
role in various intracellular interactions, including cell adhesion
and migration (14,15). CD44 is expressed not only in HCC
cells but in other types of cancers as well, such as colorectal and
breast cancer (14,16,17).
Therefore, we focused the present study on the role of the cell
surface markers CD133 and CD44 in HCC metastasis.
ELK3, also referred to as SAP-2, Net or Erp, forms a
ternary complex factor (TCF) along with ELK1 and ELK4 (18). ELK3 is activated by
mitogen-activated protein kinase (MAPK)-associated pathways, such
as the Ras/extracellular signal-regulated kinase (ERK) and p38
pathways, and it plays an important role in various physiological
processes, including cell migration, invasion, wound healing,
angiogenesis and tumorigenesis, via the regulation of c-fos, early
growth response protein 1 (egr-1), and plasminogen activator
inhibitor-1 (PAI-1) (19–23). Mutations in ELK3 disrupt
vasculogenesis, angiogenesis and wound healing in mice during
development and in adulthood (24,25).
In addition, our previous study demonstrated that ELK3 contributes
to epithelial-mesenchymal transition (EMT) of mouse hepatocytes by
regulating egr-1 (26). EMT, a
process whereby epithelial cells reduce their intercellular
adhesions and gain mesenchymal properties, is a critical event in
the process of cancer invasion and metastasis (27,28).
During liver cancer metastasis, epithelial cell-like HCC cells gain
a mesenchymal phenotype characterized by an increase in cell
migration and the ability to degrade the extracellular matrix (ECM)
(27,29). These observations suggest that EMT
may play an important role in HCC metastasis. Moreover, recent
studies revealed that ELK3 is associated with the regulation of
hypoxia-induced factor 1α (HIF-1α) (30,31).
HIF-1α is a transcription factor that plays a well-established role
in cancer development, progression and metastasis. HIF-1α is
associated with the regulation of genes associated with cancer
metastasis, invasion, angiogenesis, cellular proliferation,
apoptosis and glucose metabolism (32–36).
Vascular endothelial growth factor (VEGF) and metalloproteinase-2
(MMP-2), which are target molecules of HIF-1α, are also linked to
the development, invasion and metastasis of HCC (35–38).
Although ELK3 is known to be involved in cell migration and
invasion, the influence of ELK3 on the metastatic potential of
LCSCs remains unclear. Therefore, we investigated the role of ELK3
in LCSC metastasis and found that ELK3 can attenuate the metastatic
potential of LCSCs.
Materials and methods
Cell culture
Huh7 and Hep3B HCC cells were grown in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS), 100 µg/ml penicillin and 0.25 µg/ml streptomycin (all
from Invitrogen, Carlsbad, CA, USA). SKHep1, HepG2 and PLC/PRF/5
cells were grown in minimum essential medium (MEM) supplemented
with 10% FBS, 100 µg/ml penicillin and 0.25 µg/ml streptomycin (all
from Invitrogen). All of the cell lines were maintained at 37°C in
an atmosphere of 5% CO2.
RNA interference
The small interfering RNA (siRNA) method was used to
knock down the expression of ELK3. Three different siRNAs targeting
ELK3 (5′-CCAAGAUCUCCUCUUUAAUtt-3′; 5′-GGACUCAUUAACUGAUGAAtt-3′; and
5′-GGUCUCUAGUAGAAUUUCAtt-3′) (Santa Cruz Biotechnology, Santa Cruz,
CA, USA) were pooled together and used at a final concentration of
30 nM. Control cells were subjected to mock transfection with
scrambled siRNA. The cells were transfected using G-fectin
(Genolution Pharmaceuticals, Seoul, Korea) according to the
manufacturer's protocol.
Cell isolation using
fluorescence-activated cell sorting (FACS) analysis
The cells were harvested using 0.5 mM trypsin/EDTA
(Invitrogen) and subsequently incubated with phycoerythrin
(PE)-conjugated anti-CD133/1 and allophycocyanin (APC)-conjugated
anti-CD44 (Miltenyi Biotec, Auburn, CA, USA) at 4°C for 10 min. The
LCSCs were sorted from the Huh7 cells using flow cytometry (MoFlo
XDP; Beckman Coulter, Miami, FL, USA) with antibodies against
CD133/1 and CD44. An isotype-matched mouse IgG was used as the
negative control.
Sphere formation assay
The cells were seeded in multiple ultra-low
attachment 24-well plates (Corning Costar Corp., Cambridge, MA,
USA) at a density of 2×102 cells/well in serum-free
DMEM/F12K with B27 supplement (both from Invitrogen), basis
fibroblast growth factor (bFGF) (20 ng/ml) and epidermal growth
factor (EGF) (20 ng/ml) (both from PeproTech, Rocky Hill, NJ, USA).
The cells were incubated at 37°C in an atmosphere of 5%
CO2 for 5 days. Then, the spheres (diameter >50 µm)
in each well were counted using an inverted microscope (Olympus,
Tokyo, Japan). The average number of spheres was calculated from
three independent experiments.
Migration assays
Isolated Huh7 cells were grown for 3 days in 10% FBS
complete medium. Then, the cells were harvested using 0.5 mM
trypsin/EDTA (Invitrogen) and resuspended in serum-free medium.
Cell migration was evaluated using 6.5-mm Transwell plates with
8-µm pore size filters (Corning Costar Corp.). The resuspended
cells were seeded into the upper chamber (3×104 cells)
of the Transwell, and 600 µl of 10% FBS complete medium was added
to the bottom chamber and used as a chemoattractant. The cells were
incubated for 48 h at 37°C in an atmosphere of 5% CO2.
The cells that had not migrated were removed using a cotton swab.
The cells that had migrated were stained using a Diff-Quick™
Three-Step Stain kit (Sysmex, Kobe, Japan) and mounted on glass
slides. The cells in 4 randomly selected fields (magnification,
×200) from each slide were quantified using a slide scanner
(Pannoramic MIDI; 3DHISTECH Ltd., Budapest, Hungary).
Invasion assays
The Transwell invasion assays were conducted using a
Transwell plate (Corning Costar Corp.) coated with Matrigel (BD
Biosciences, San Jose, CA, USA) according to the manufacturer's
instructions. Briefly, the Matrigel (BD Biosciences) was diluted
with serum-free medium at a dilution of 1:5, added to the upper
chamber, and incubated at 37°C until it had completely solidified.
The unbound Matrigel was removed by washing. Cells in serum-free
medium were seeded into the upper chamber (5×104 cells),
and 600 µl of complete medium was added to the bottom chamber and
used as a chemoattractant. The cells were incubated for 48 h at
37°C in an atmosphere of 5% CO2, and the non-invasive
cells were removed using a cotton swab. The cells that had invaded
the membrane were stained using a Diff-Quick™ Three-Step Stain kit
and mounted on glass slides. The stained cells were counted using a
slide scanner (Pannoramic MIDI) (magnification, ×200).
Western blotting
The protein extracts were separated using 8%
SDS-PAGE and transferred to nitrocellulose membranes (Schleicher
& Schuell, Dassel, Germany). The membranes were blocked in 5%
skim milk and incubated with the indicated primary antibodies
according to the manufacturer's protocol. The following primary
antibodies were used in the western blot assays: anti-ELK3,
anti-VEGF (both from Abcam, Cambridge, MA, USA), anti-HIF-1α (Santa
Cruz Biotechnology) and anti-β-actin (Sigma, St. Louis, MO, USA).
The membranes were subsequently incubated with horseradish
peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary
antibodies (Santa Cruz Biotechnology). The target proteins were
visualized using an enhanced chemiluminescence reagent (Amersham
Biosciences, Cardiff, UK). The density of each band was measured
using Multi Gauge imaging software (Raytest, Straubenhardt,
Germany).
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was purified from
CD133+/CD44+ and
CD133−/CD44− cells using TRIzol reagent
(Invitrogen). The cDNA was synthesized from 2 µg of total RNA, and
ELK3 was amplified using PCR with ELK3-specific primers (forward,
5-ACCCAAAGGCTTGGAAATCT-3 and reverse, 5-TGTATGCTGGAGAGCAGTGG-3′).
The expression level of the target gene was normalized to the
expression of the β-actin internal control. β-actin was amplified
using PCR with β-actin-specific primers (forward,
5′-GGCACCCACCCTTCTACATAGA-3′ and reverse,
5′-CCCTCGTAGATGGGCACACT-3′). The PCR products were separated using
electrophoresis on a 1% agarose gel with 0.5 µg/ml ethidium bromide
(Research Genetics, Huntington, AL, USA) and visualized using a
Gel-Doc system (Bio-Rad, Vienna, Austria).
VEGF enzyme-linked immunosorbent assay
(ELISA)
Human VEGF Quantikine ELISA kits (R&D Systems,
Minneapolis, MN, USA) were used to measure VEGF levels in cell
supernatants according to the manufacturer's instructions. Briefly,
200 µl of the harvested cell supernatants was dispensed into wells
coated with coating buffer and incubated for 2 h at room
temperature (RT). After the wells were completely washed, the VEGF
conjugate was dispensed into each well, and the wells were
incubated at RT. Then, the plates were developed using substrate
solution for 20 min. The absorbance at 450 nm was measured using a
microplate reader.
Gelatin zymography
The cell supernatants were collected and mixed with
non-denatured sample buffer [1 M Tris-Cl (pH 6.8), 1% bromophenol
blue and 20% SDS]. The resulting cell supernatant solution was
separated on a polyacrylamide gel containing 1 mg/ml gelatin
(Sigma). The gels were renatured with 2.5% Triton X-100 and
incubated in activation buffer [50 mM Tris buffer (pH 7.4), 5 mM
CaCl2 and 1 µM ZnCl2) at 37°C for 12 h. Then,
the gels were stained with Coomassie Brilliant Blue R-250 for 20
min and destained with destaining buffer (45% methanol and 10%
acetic acid). MMP-2 appeared as a clear area and the band density
was measured using Multi Gauge imaging software.
Statistical analysis
The data are presented as the mean ± SD. of at least
3 separate experiments. The statistical analyses were conducted
using a two-tailed Student t-test. p<0.05 and p<0.01 indicate
statistical significance.
Results
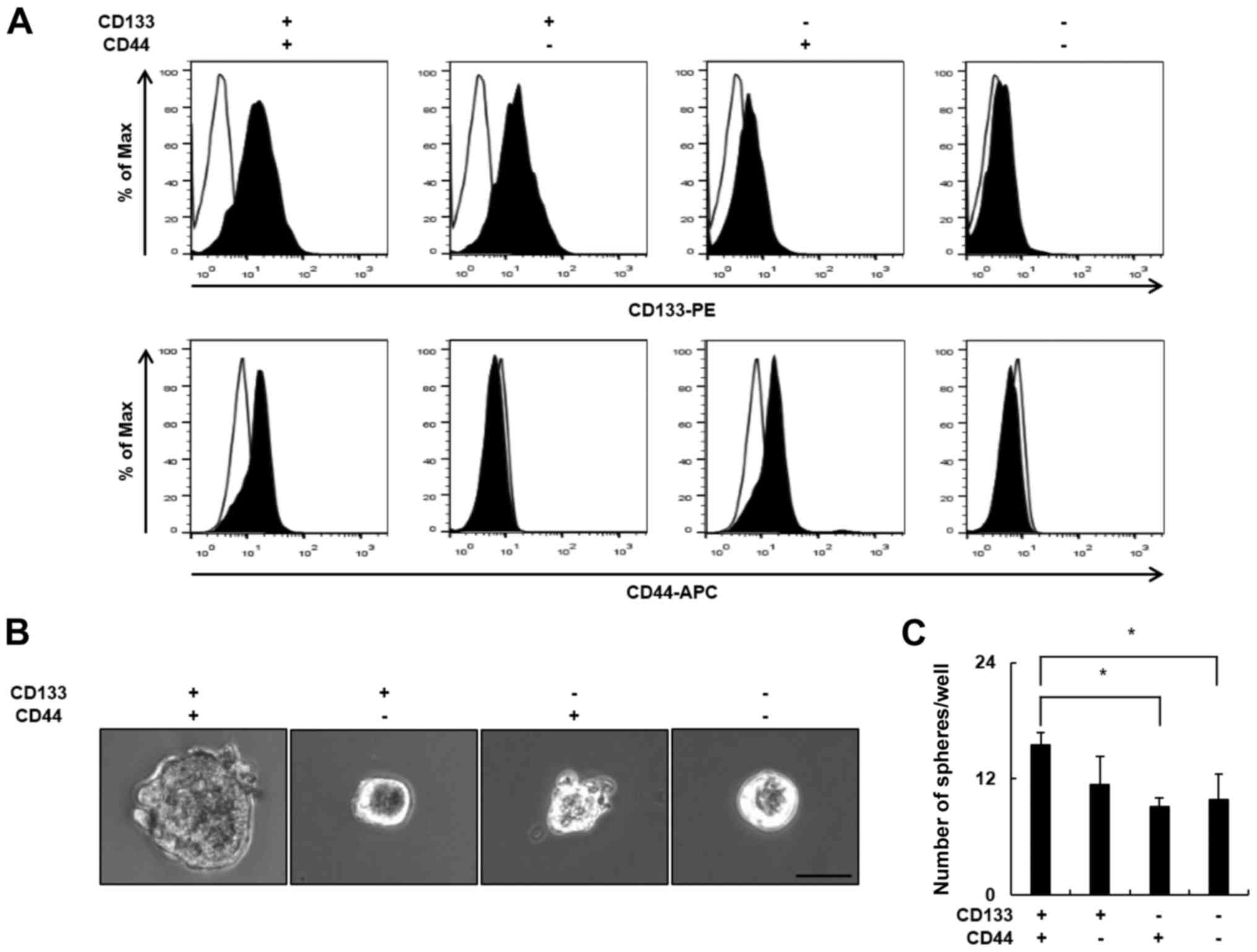
CD133+/CD44+
LCSCs are more clonogenic compared with
non-CD133+/CD44+ cells
To identify a putative marker of LCSCs, we evaluated
the expression of the cell surface markers CD133 and CD44 in 5 HCC
cell lines using flow cytometry. The expression of CD133 and CD44
was significantly different in each cell line. As shown in Table I, the proportion of
CD133+ cells was highest in the PLC/PRF/5 cells (74.6%)
and lowest in the Hep3B cells (0.6%), whereas the proportion of
CD44+ cells was highest in the Hep3B cells (99.6%) and
lowest in the PLC/PRF/5 cells (2.1%). The proportion of
CD133+/CD44+ cells was highest in the Huh7
cells (11.1%) and lowest in the Hep3B cells (0.6%). As the
co-expression of 2 or more CSCs markers is more specific compared
with the presence of a single LCSCs marker, we selected the Huh7
cell line for further experiments. To explore the biological
properties of the CD133+/CD44+ LCSC
subpopulation of Huh7 cells, we compared the clonogenic potential
of CD133+/CD44+ Huh7 cells and
non-CD133+/CD44+ Huh7 cells. Huh7 cells were
sorted into 4 subgroups according to the expression of the CD133
and CD44 surface markers using flow cytometry (Fig. 1A), and the clonogenic potential of
the subgroups was examined using the sphere formation assay. The
CD133+/CD44+ subgroup formed the largest
spheres (Fig. 1B). In addition, the
number of spheres was significantly higher in the
CD133+/CD44+ subgroup (15 spheres/well)
compared with the CD133−/CD44− subgroup (9
spheres/well) (Fig. 1C). These
results demonstrated that CD133+/CD44+ LCSCs
have greater clonogenic potential compared with
non-CD133+/CD44+ cells.
 | Table I.Proportion of CD133+,
CD44+ and CD133+/CD44+ cells in
the HCC cell lines. |
Table I.
Proportion of CD133+,
CD44+ and CD133+/CD44+ cells in
the HCC cell lines.
|
| Cell lines (%) |
|---|
|
|
|
|---|
| Markers | Huh7 | PLC/PRF/5 | HepG2 | SKHep1 | Hep3B |
|---|
|
CD133+ | 42.3±3.9 | 74.6±15.3 | 15.1±4.7 | 3.0±0.6 | 0.6±0.4 |
|
CD44+ | 21.3±2.9 | 2.1±0.7 | 7.1±2.5 | 99.1±0.6 | 99.6±0.3 |
|
CD133+/CD44+ | 11.1±1.3 | 1.8±0.5 | 1.6±0.9 | 2.8±0.6 | 0.6±0.2 |
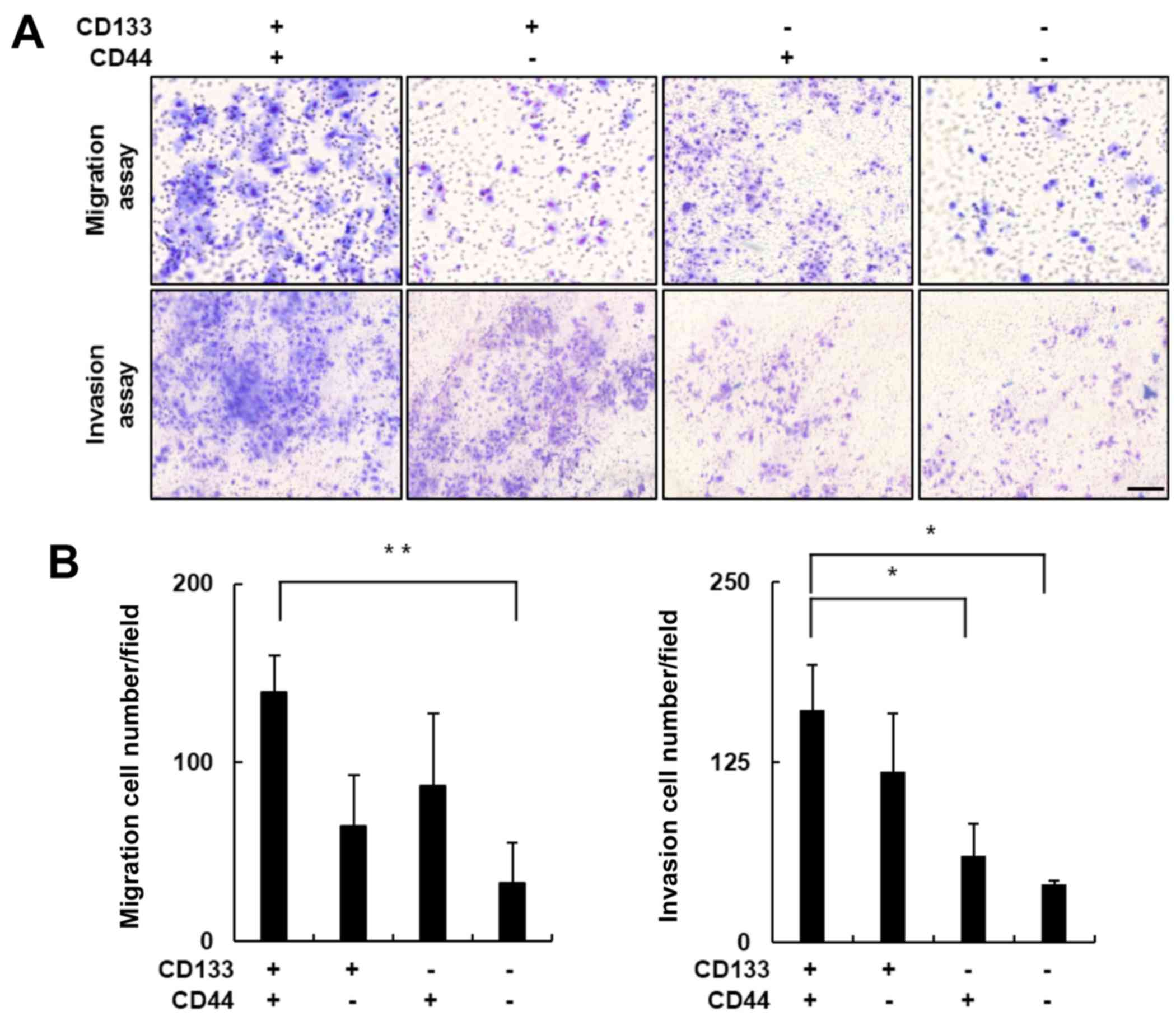
CD133+/CD44+
LCSCs exhibit greater metastatic potential compared with
non-CD133+/CD44+ cells
Recent studies have revealed that high expression
levels of CD133 or CD44 contribute to CSC-associated cancer
metastasis and invasion (10,14,39).
Therefore, we performed in vitro Transwell migration and
invasion assays to evaluate the metastatic potential of the Huh7
cell subpopulations. The migratory cells in each well were stained
using a Diff-Quick Three-Step stain (Fig. 2A, upper panel), and the number of
migratory cells in each group were compared. The number of
migratory cells in the CD133+/CD44+ subgroup
increased ~4-fold compared with the number in the
CD133−/CD44− subgroup (Fig. 2B; p<0.01). The results obtained
from the Matrigel-coated Transwell invasion assay were consistent
with these findings. The number of
CD133+/CD44+ cells that invaded the Matrigel
was significantly greater compared with this number in the
non-CD133+/CD44+ subgroups (Fig. 2A, lower panel). The number of
invasive cells in the CD133+/CD44+ subgroup
was ~4-fold greater when compared with the
CD133−/CD44− subgroup (Fig. 2B; p<0.05). Together, these
results indicated that the cell migration and invasion activities
were significantly elevated in the
CD133+/CD44+ LCSCs.
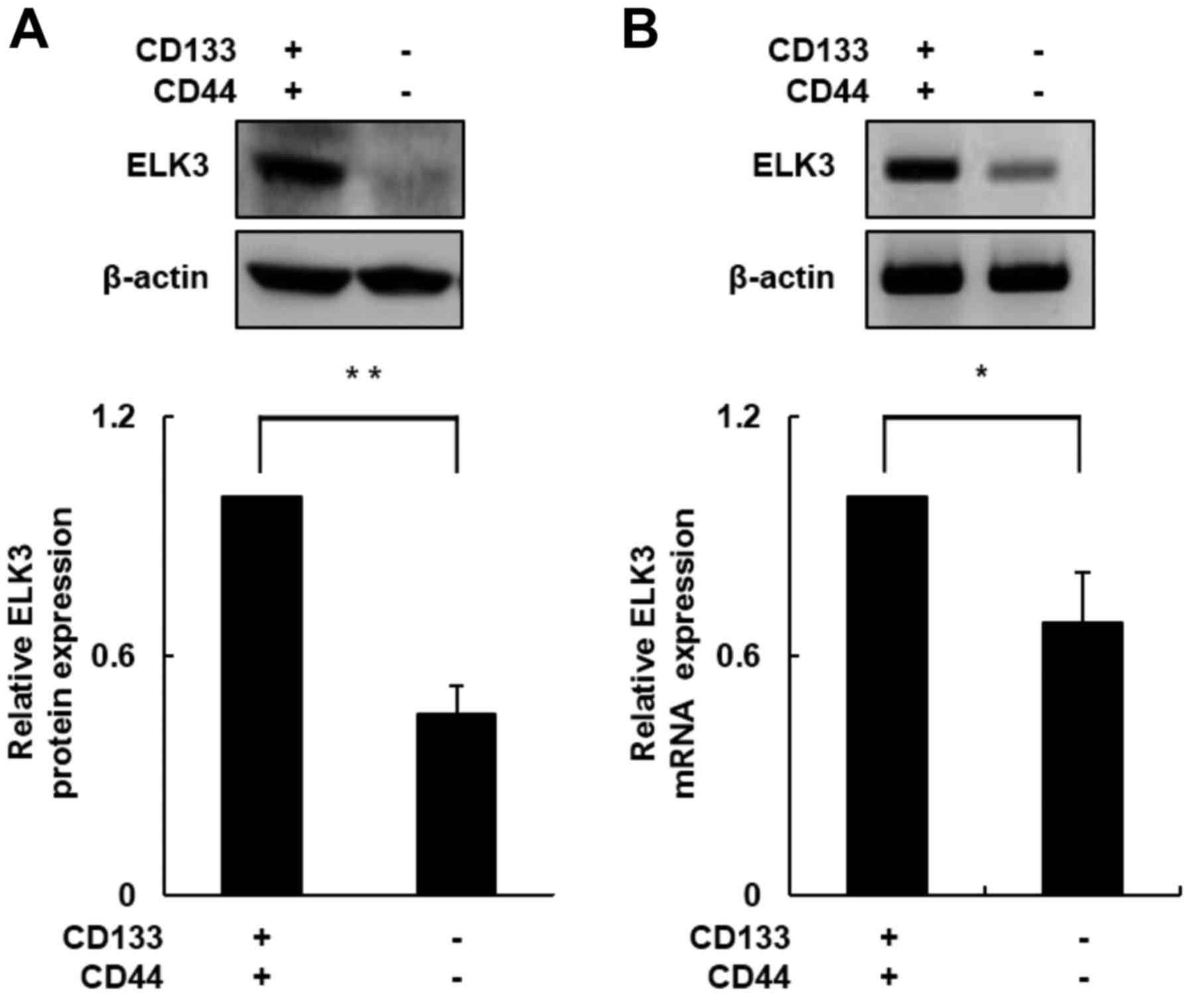
ELK3 expression is elevated in
CD133+/CD44+ cells
In a previous study, we demonstrated that ELK3
expression plays an important role in the process of EMT during
liver fibrogenesis (26). Acquiring
cell motility is necessary for the transition from an epithelial to
a mesenchymal phenotype (27,28).
We used western blot assays to determine whether ELK3 protein
levels differ between CD133+/CD44+ and
CD133−/CD44− cells. As shown in Fig. 3A, the ELK3 protein level was
increased ~2.5-fold in the CD133+/CD44+ cells
compared with the level in the CD133−/CD44−
cells (p<0.01). Next, we conducted RT-PCR to measure mRNA
expression levels of ELK3 in both subpopulations of cells. Similar
to the ELK3 protein level, the ELK3 mRNA expression level was
increased nearly 1.5-fold in the CD133+/CD44+
cells compared with the level in the
CD133−/CD44− cells (Fig. 3B; p<0.05). These results
indicated that ELK3 expression was upregulated in the
CD133+/CD44+ cells and that the upregulation
of ELK3 enhanced HCC cell migration and invasion.
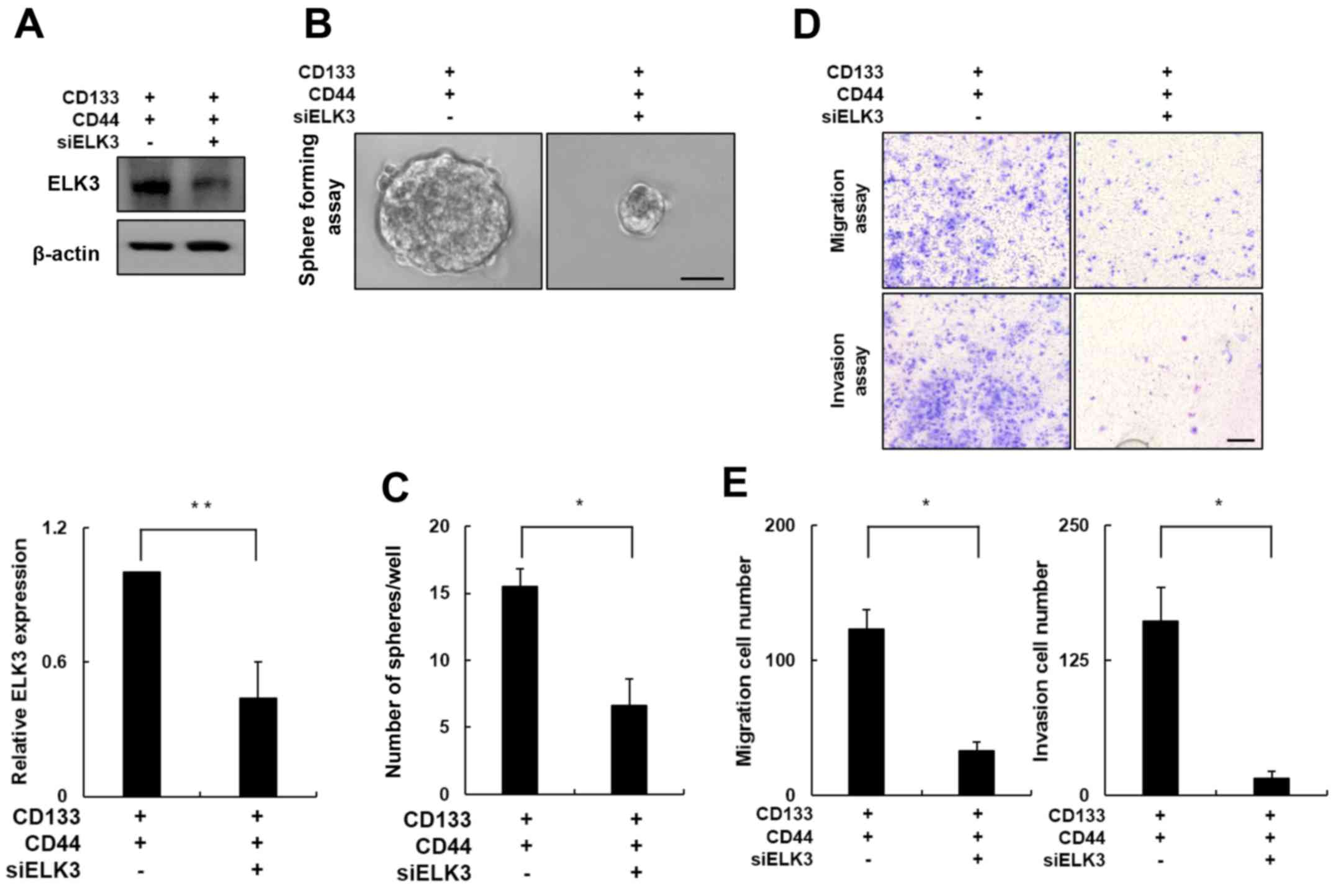
ELK3 knockdown attenuates the
metastatic potential of CD133+/CD44+
cells
To further evaluate the role of ELK3 in
CD133+/CD44+ cell migration and invasion, we
transfected CD133+/CD44+ cells with an siRNA
targeting ELK3. Western blot analysis of the siRNA silencing
efficiency demonstrated that ELK3 expression decreased by ~60% in
the siELK3-transfected cells when compared with the negative
control cells (Fig. 4A; p<0.01).
The clonogenic potential of ELK3-knockdown
CD133+/CD44+ cells was measured using a
sphere formation assay. As shown in Fig. 4B, the size of the spheres formed by
the ELK3-knockdown CD133+/CD44+ cells was
significantly decreased compared with the size of the spheres
formed by the negative control cells. The control
CD133+/CD44+ cells formed an average of 15
spheres, whereas the ELK3-knockdown
CD133+/CD44+ cells formed less than half of
that number (Fig. 4C; p<0.05).
Next, we conducted a Transwell cell migration assay in the
ELK3-knockdown CD133+/CD44+ cells (Fig. 4D, upper panel). Notably, the number
of migratory cells was decreased by ~70% in the ELK3-knockdown
CD133+/CD44+ cells compared with this number
noted in the negative control cells (Fig. 4E; p<0.05). In addition, similar
results were obtained using the Matrigel-coated Transwell invasion
assay (Fig. 4D, lower panel). The
number of cells that invaded the Matrigel was decreased by ~90% in
the ELK3-knockdown CD133+/CD44+ cells
compared with this number in the negative control group (Fig. 4E; p<0.05). Collectively, these
results indicate that ELK3 expression is associated with the
migration and invasion of CD133+/CD44+
cells.
ELK3 attenuates the metastatic
potential of CD133+/CD44+ cells by regulating
the HIF-1α signaling pathway
To investigate the molecular mechanism underlying
the ELK3-mediated properties of CD133+/CD44+
cells, we focused on the relationship between ELK3 and expression
of its target gene HIF-1α. Previous studies revealed that several
genes including Egr-1, PAI-1 and HIF-1α are ELK3 target genes by
forming ternary complexes. Among these target genes, HIF-1α is
known to stimulate angiogenic and metastatic responses by
activating transcription of the genes encoding several growth
factors, including VEGF and MMP-2 (19–23,35,36).
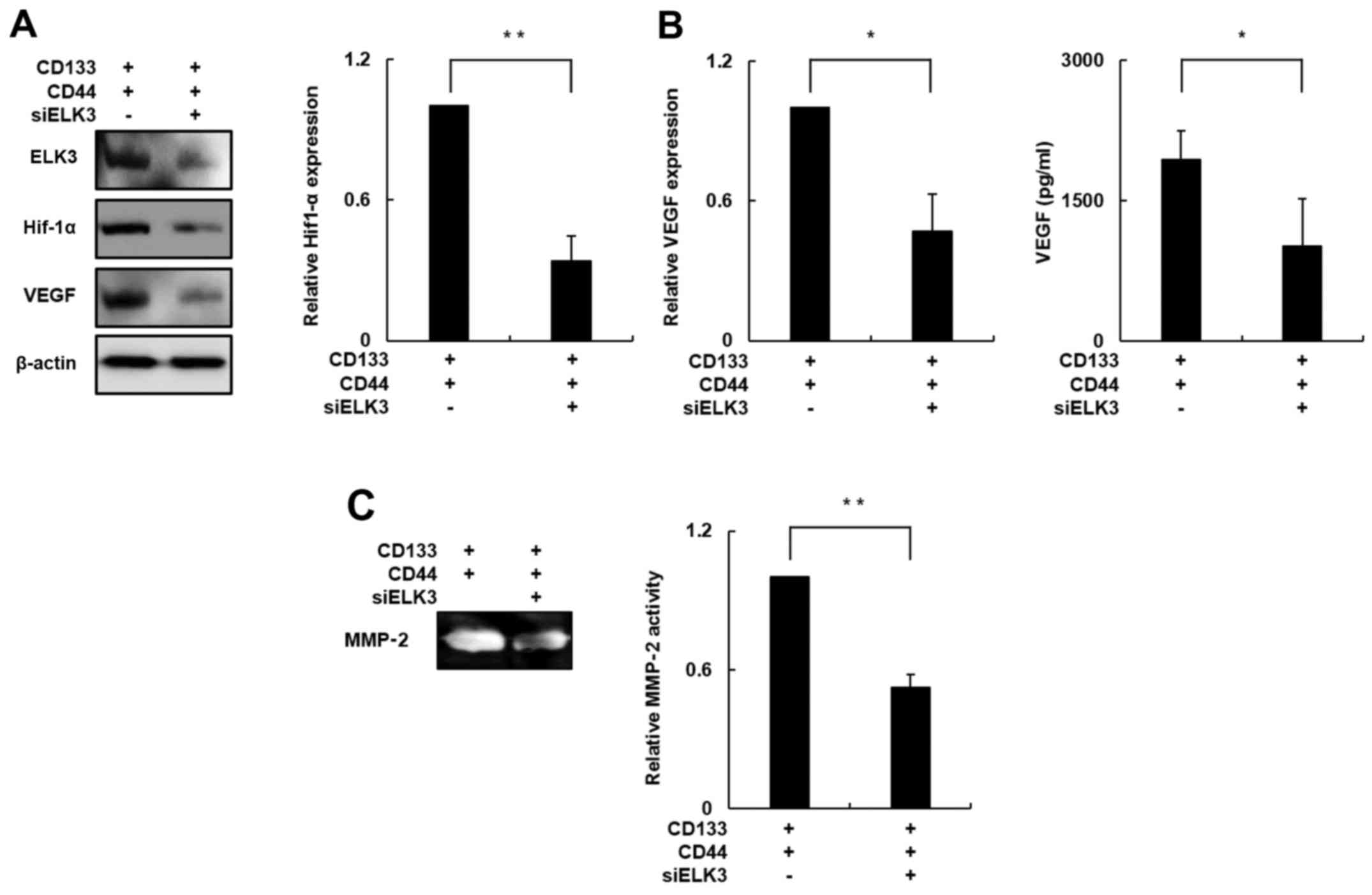
To this end, we evaluated the expression of HIF-1α
in the ELK3-knockdown CD133+/CD44+ cells. The
HIF-1α protein level was decreased by ~70% in the ELK3-knockdown
CD133+/CD44+ cells compared with this level
noted in the negative control cells (Fig. 5A; p<0.01). The VEGF protein level
was significantly decreased by ~60% in the ELK3-knockdown
CD133+/CD44+ cells compared with this level
in the negative control cells (Fig.
5A; p<0.05), and a VEGF-specific ELISA demonstrated that
VEGF secretion was decreased by ~50% in the ELK3-knockdown
CD133+/CD44+ cells compared with the
secretion observed in the negative control group (Fig. 5B; p<0.05). These result suggested
that ELK3 knockdown has an inhibitory effect on VEGF-induced
angiogenesis.
Therefore, we performed a gelatin zymography assay
to determine the enzymatic activity of MMP-2 in the ELK3-knockdown
CD133+/CD44+ cells since recent studies have
demonstrated that MMP-2 participates in the cancer metastasis
process and is also known to correlate with HIF-1α expression
(36,38). As shown in Fig. 5C, the enzymatic activity of MMP-2
was decreased by ~50% in the ELK3-knockdown
CD133+/CD44+ cells compared with that in the
negative control group (p<0.05). Together, these results
indicate that silencing of ELK3 expression in the
CD133+/CD44+ cells can decrease the activity
of metastasis-associated molecules by inhibiting HIF-1α
expression.
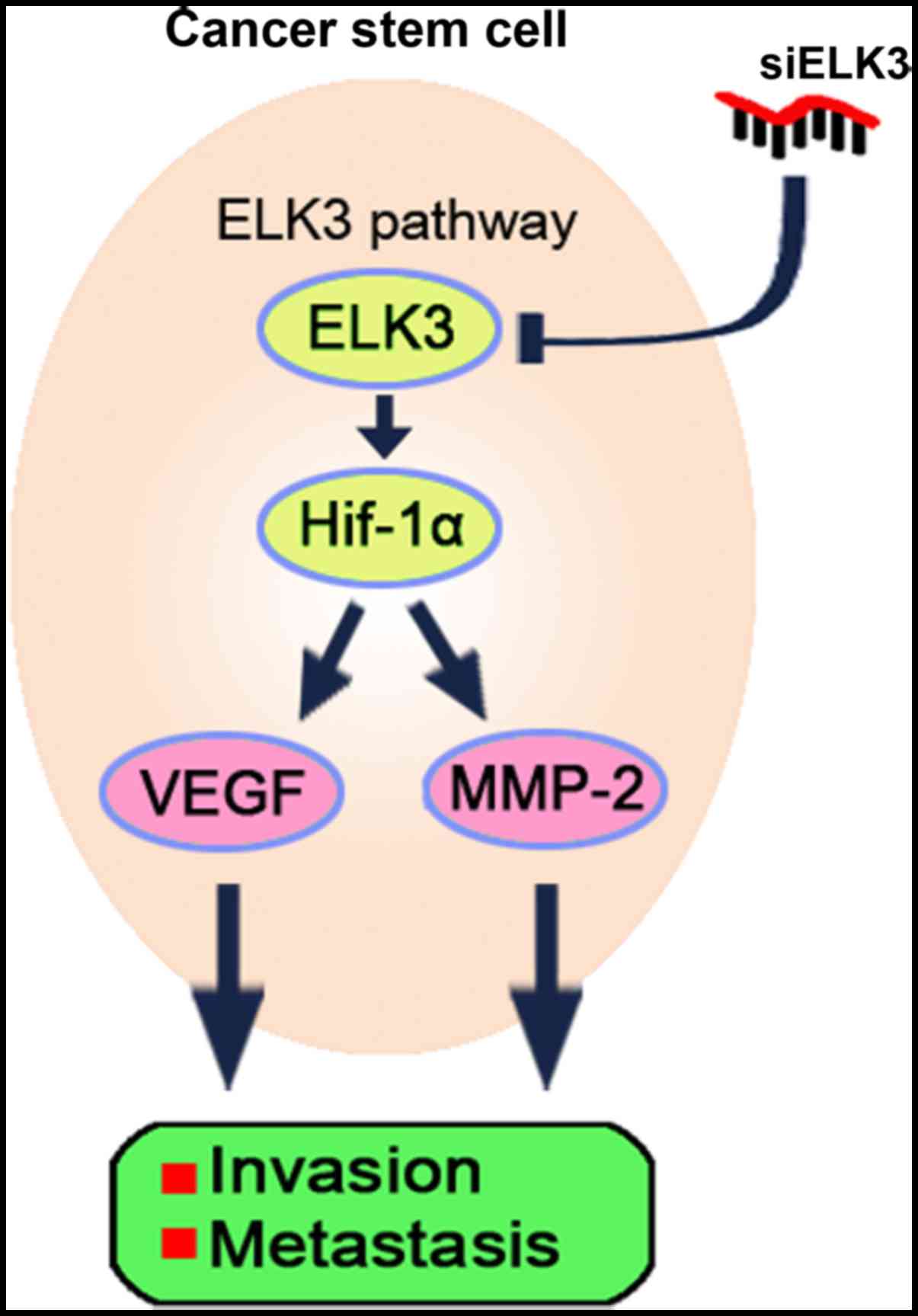
Collectively, these data demonstrated that ELK3
overexpression promotes metastasis in
CD133+/CD44+ cells and that inhibiting ELK3
expression attenuates the metastatic potential of
CD133+/CD44+ LCSCs (Fig. 6).
Discussion
HCC metastasis occurs through a series of several
complex steps. Briefly, metastatic cancer cells escape from the
primary cancer, enter the blood circulation and ultimately colonize
a new organ and form metastasis (40). As multiple complex steps are
required for this process, the majority of cancer cells rarely
metastasize to other organs. Recent studies have shown that CSCs
comprise an extremely small subpopulation of cancer cells and have
high metastatic potential (5–7).
Therefore, several cancers including HCC, are very difficult to
cure using standard cancer chemotherapy treatments due to the
persistence of CSCs (41). Cancer
cells that survive anticancer therapy can initiate metastasis,
thereby contributing to a poor prognosis. Therefore, identifying
and specifically targeting CSCs may represent a promising
therapeutic strategy for treating cancer. Consistent with this
hypothesis, numerous studies have investigated methods for
identifying and targeting CSCs among the general population of
cancer cells.
Many specific cell surface markers have been
identified and characterized at the biological and clinical level
(8). Among these markers, CD133 and
CD44 play a well-established role in the metastatic potential of
LCSCs (8,10,14,42).
Notably, several recent studies reported that the expression of
CD133 or CD44 alone is not sufficient to account for all of the
biological properties of CSCs (43,44).
Therefore, several studies have investigated whether CSCs that
co-express CD133 and CD44 represent a more potent CSC subpopulation
(45,46). These studies revealed that the
co-expression of CD133 and CD44 is a more specific LSCS biomarker
and a more valuable prognostic indicator in HCC (44–46).
In the present study, we demonstrated that
CD133+/CD44+ cells have increased metastatic
potential compared with non-CD133+/CD44+
cells using cell migration and invasion assays.
After evaluating the metastatic potential of
CD133+/CD44+ LCSCs, we investigated the
molecular mechanisms underlying these properties. In a previous
study, we revealed that CD133+ LCSCs mediate radiation
resistance in human HCC by regulating MAPK signaling pathways
(47). In addition, a recent study
revealed that CD133 expression was modulated by the ERK1/2 and Src
signaling pathways in pancreatic cancer and that CD133 promoted
cancer invasion and metastasis by activating an EMT regulatory
feedback loop (48). CD44 maintains
the high basal level of motility in breast cancer cells by forming
a complex with ERK1 and 2, and CD168 (15). Both of these studies concluded that
CD133 and CD44 are associated with the MAPK signaling pathways such
as the ERK pathway. Furthermore, the results of our previous study
revealed that ELK3 contributes to the process of EMT via
MAPK/ELK3/egr-1 signaling (26).
Moreover, as previous studies have demonstrated that EMT promotes a
mesenchymal phenotype and consequently enhances cell motility, EMT
is regarded as a contributing factor to metastasis and invasion in
various types of cancer. As ELK3 expression is associated with cell
migration, vasculogenesis and wound healing (24,25),
we examined the potential role of ELK3 in the mechanisms underlying
metastasis and invasion in LCSCs.
In the present study, ELK3 expression levels in
CD133+/CD44+ cells were significantly
elevated compared with CD133−/CD44− cells.
These results suggest that ELK3 plays a specific role in cancer
metastasis and invasion in LCSCs. Furthermore, we investigated the
molecular mechanisms underlying the function of ELK3 in
CD133+/CD44+ LCSC metastasis by transfecting
CD133+/CD44+ LCSCs with ELK3-specific siRNA.
Notably, we found that silencing ELK3 expression inhibited LCSC
metastasis and invasion. Previous studies demonstrated that ELK3
participates in the regulation of various genes, including c-fos,
egr-1, and PAI-1. In addition, ELK3 is known as a regulator of
HIF-1α expression by modulating HIF-1α stability (21–23,30,31).
HIF-1α plays a key role in the induction of cancer invasion,
metastasis and angiogenesis, as well as in cancer growth, glucose
metabolism, and other metastasis-associated signaling pathways
(32–36). Notably, HIF-1α is a key regulator of
cancer metastasis and invasion in HCC (35,36).
In the present study, the HIF-1α protein level was decreased by
~60% in the ELK3-knockdown CD133+/CD44+
cells. Consistent with this observation, the activity of VEGF and
MMP-2 was significantly decreased in the ELK3-knockdown
CD133+/CD44+ LCSCs. These results indicate
that ELK3 expression, and the activity of VEGF and MMP-2 are
directly associated with the co-expression of the cell surface
markers CD133 and CD44 and that the co-expression of these markers
is a more specific and reliable marker of HCC compared to the
expression of either CD133 or CD44 alone.
According to these results, we conclude that Huh7
CD133+/CD44+ LCSCs have high metastatic and
invasive potential due to the overexpression of ELK3. Moreover, we
found that silencing of ELK3 expression in the
CD133+/CD44+ LCSCs attenuated the metastatic
and invasive potential via modulation of the HIF-1α signaling
pathway (Fig. 6).
A better understanding of the cellular and
molecular mechanisms underlying LCSC metastasis and invasion is
essential to improving the efficacy of therapies designed to
suppress HCC metastasis in the clinical setting. In this regard,
the novel findings reported in the present study indicate that ELK3
expression contributes to the metastatic potential of LCSCs and
that silencing of ELK3 expression can reduce the metastatic and
invasive potential of LCSCs by regulating HIF-1α. In conclusion,
modulation of ELK3 expression may represent a novel therapeutic
approach for preventing the process of cancer metastasis and
invasion in HCC.
Acknowledgements
The present study was supported by grants from the
National Research Foundation of Korea funded by the Korea
government (NRF-2015R1A2A1A15052783) and of the Ministry of
Science, ICT and Future Planning through the National Research
Foundation.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
CSCs
|
cancer stem cells
|
|
HIF-1α
|
heat shock induced factor-1α
|
|
TCF
|
ternary complex factor
|
|
MAPK
|
mitogen-activated protein kinase
|
|
ERK
|
extracellular signal-regulated
kinase
|
|
LCSCs
|
liver cancer stem cells
|
|
egr-1
|
early growth response protein-1
|
|
PAI-1
|
plasminogen activator inhibitor-1
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ECM
|
extracellular matrix
|
|
VEGF
|
vascular endothelial growth
factor
|
|
MMP-2
|
metalloproteinase-2
|
|
siRNA
|
small interfering RNA
|
|
bFGF
|
basis fibroblast growth factor
|
|
EGF
|
epidermal growth factor
|
|
RT-PCR
|
reverse transcription-polymerase
chain reaction
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo Z, Zhong JH, Jiang JH, Zhang J, Xiang
BD and Li LQ: Comparison of survival of patients with BCLC stage A
hepatocellular carcinoma after hepatic resection or transarterial
chemoembolization: A propensity score-based analysis. Ann Surg
Oncol. 21:3069–3076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ueno M, Uchiyama K, Ozawa S, Hayami S,
Shigekawa Y, Tani M and Yamaue H: Adjuvant chemolipiodolization
reduces early recurrence derived from intrahepatic metastasis of
hepatocellular carcinoma after hepatectomy. Ann Surg Oncol.
18:3624–3631. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB, Marrero JA, Rudolph L and
Reddy KR: Diagnosis and treatment of hepatocellular carcinoma.
Gastroenterology. 134:1752–1763. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on Cancer Stem Cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamashita T and Wang XW: Cancer stem cells
in the development of liver cancer. J Clin Invest. 123:1911–1918.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee TK, Cheung VC and Ng IO: Liver
tumor-initiating cells as a therapeutic target for hepatocellular
carcinoma. Cancer Lett. 338:101–109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grosse-Gehling P, Fargeas CA, Dittfeld C,
Garbe Y, Alison MR, Corbeil D and Kunz-Schughart LA: CD133 as a
biomarker for putative cancer stem cells in solid tumours:
Limitations, problems and challenges. J Pathol. 229:355–378. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kohga K, Tatsumi T, Takehara T, Tsunematsu
H, Shimizu S, Yamamoto M, Sasakawa A, Miyagi T and Hayashi N:
Expression of CD133 confers malignant potential by regulating
metalloproteinases in human hepatocellular carcinoma. J Hepatol.
52:872–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO,
Zheng BJ and Guan XY: Identification and characterization of
tumorigenic liver cancer stem/progenitor cells. Gastroenterology.
132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan M, Li H, Zhu M, Zhao F, Zhang L, Chen
T, Jiang G, Xie H, Cui Y, Yao M, et al: G protein-coupled receptor
87 (GPR87) promotes the growth and metastasis of CD133+
cancer stem-like cells in hepatocellular carcinoma. PLoS One.
8:e610562013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hong SW, Hur W, Choi JE, Kim JH, Hwang D
and Yoon SK: Role of ADAM17 in invasion and migration of
CD133-expressing liver cancer stem cells after irradiation.
Oncotarget. 7:23482–23497. 2016.PubMed/NCBI
|
|
14
|
Xie Z, Choong PF, Poon LF, Zhou J, Khng J,
Jasinghe VJ, Palaniyandi S and Chen CS: Inhibition of CD44
expression in hepatocellular carcinoma cells enhances apoptosis,
chemosensitivity, and reduces tumorigenesis and invasion. Cancer
Chemother Pharmacol. 62:949–957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hamilton SR, Fard SF, Paiwand FF, Tolg C,
Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J and Turley
EA: The hyaluronan receptors CD44 and Rhamm (CD168) form complexes
with ERK1,2 that sustain high basal motility in breast cancer
cells. J Biol Chem. 282:16667–16680. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fillmore C and Kuperwasser C: Human breast
cancer stem cell markers CD44 and CD24: Enriching for cells with
functional properties in mice or in man? Breast Cancer Res.
9:3032007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y, et al: CD44 is of functional
importance for colorectal cancer stem cells. Clin Cancer Res.
14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Buchwalter G, Gross C and Wasylyk B: Ets
ternary complex transcription factors. Gene. 324:1–14. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ducret C, Maira SM, Lutz Y and Wasylyk B:
The ternary complex factor Net contains two distinct elements that
mediate different responses to MAP kinase signalling cascades.
Oncogene. 19:5063–5072. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giovane A, Pintzas A, Maira SM,
Sobieszczuk P and Wasylyk B: Net, a new ets transcription factor
that is activated by Ras. Genes Dev. 8:1502–1513. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Buchwalter G, Gross C and Wasylyk B: The
ternary complex factor Net regulates cell migration through
inhibition of PAI-1 expression. Mol Cell Biol. 25:10853–10862.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ayadi A, Zheng H, Sobieszczuk P,
Buchwalter G, Moerman P, Alitalo K and Wasylyk B: Net-targeted
mutant mice develop a vascular phenotype and up-regulate egr-1.
EMBO J. 20:5139–5152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wasylyk C, Zheng H, Castell C, Debussche
L, Multon MC and Wasylyk B: Inhibition of the Ras-Net (Elk-3)
pathway by a novel pyrazole that affects microtubules. Cancer Res.
68:1275–1283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng H, Wasylyk C, Ayadi A, Abecassis J,
Schalken JA, Rogatsch H, Wernert N, Maira SM, Multon MC and Wasylyk
B: The transcription factor Net regulates the angiogenic switch.
Genes Dev. 17:2283–2297. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ayadi A, Suelves M, Dollé P and Wasylyk B:
Net, an Ets ternary complex transcription factor, is expressed in
sites of vasculogenesis, angiogenesis, and chondrogenesis during
mouse development. Mech Dev. 102:205–208. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li TZ, Kim SM, Hur W, Choi JE, Kim JH,
Hong SW, Lee EB, Lee JH and Yoon SK: Elk-3 contributes to the
progression of liver fibrosis by regulating the
epithelial-mesenchymal transition. Gut Liver. Aug 19–2016.(Epub
ahead of print). doi: 10.5009/gnl15566.
|
|
27
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Książkiewicz M, Markiewicz A and Zaczek
AJ: Epithelial-mesenchymal transition: A hallmark in metastasis
formation linking circulating tumor cells and cancer stem cells.
Pathobiology. 79:195–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gross C, Dubois-Pot H and Wasylyk B: The
ternary complex factor Net/Elk-3 participates in the
transcriptional response to hypoxia and regulates HIF-1 alpha.
Oncogene. 27:1333–1341. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gross C, Buchwalter G, Dubois-Pot H, Cler
E, Zheng H and Wasylyk B: The ternary complex factor net is
downregulated by hypoxia and regulates hypoxia-responsive genes.
Mol Cell Biol. 27:4133–4141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xia L, Mo P, Huang W, Zhang L, Wang Y, Zhu
H, Tian D, Liu J, Chen Z, Zhang Y, et al: The
TNF-α/ROS/HIF-1-induced upregulation of FoxMI expression promotes
HCC proliferation and resistance to apoptosis. Carcinogenesis.
33:2250–2259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu Z, Liu E, Peng C, Li Y, He Z, Zhao C
and Niu J: Role of hypoxia-inducible-1α in hepatocellular carcinoma
cells using a Tet-on inducible system to regulate its expression in
vitro. Oncol Rep. 27:573–578. 2012.PubMed/NCBI
|
|
34
|
Denko NC: Hypoxia, HIF1 and glucose
metabolism in the solid tumour. Nat Rev Cancer. 8:705–713. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo D, Wang Z and Wu J, Jiang C and Wu J:
The role of hypoxia inducible factor-1 in hepatocellular carcinoma.
Biomed Res Int. 2014:4092722014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zeng HY
and Gao DM: Gene expression profiling of fixed tissues identified
hypoxia-inducible factor-1α, VEGF, and matrix metalloproteinase-2
as biomarkers of lymph node metastasis in hepatocellular carcinoma.
Clin Cancer Res. 17:5463–5472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin D and Wu J: Hypoxia inducible factor
in hepatocellular carcinoma: A therapeutic target. World J
Gastroenterol. 21:12171–12178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Choi SH, Shin HW, Park JY, Yoo JY, Kim DY,
Ro WS, Yun CO and Han KH: Effects of the knockdown of hypoxia
inducible factor-1α expression by adenovirus-mediated shRNA on
angiogenesis and tumor growth in hepatocellular carcinoma cell
lines. Korean J Hepatol. 16:280–287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J and
Li J: Cancer stem/progenitor cells are highly enriched in
CD133+CD44+ population in hepatocellular
carcinoma. Int J Cancer. 126:2067–2078. 2010.PubMed/NCBI
|
|
40
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pardal R, Clarke MF and Morrison SJ:
Applying the principles of stem-cell biology to cancer. Nat Rev
Cancer. 3:895–902. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of
CD90+ cancer stem cells in human liver cancer. Cancer
Cell. 13:153–166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Salnikov AV, Kusumawidjaja G, Rausch V,
Bruns H, Gross W, Khamidjanov A, Ryschich E, Gebhard MM,
Moldenhauer G, Büchler MW, et al: Cancer stem cell marker
expression in hepatocellular carcinoma and liver metastases is not
sufficient as single prognostic parameter. Cancer Lett.
275:185–193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu LL, Fu D, Ma Y and Shen XZ: The power
and the promise of liver cancer stem cell markers. Stem Cells Dev.
20:2023–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hou Y, Zou Q, Ge R, Shen F and Wang Y: The
critical role of CD133+CD44+/high tumor cells
in hematogenous metastasis of liver cancers. Cell Res. 22:259–272.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen KL, Pan F, Jiang H, Chen JF, Pei L,
Xie FW and Liang HJ: Highly enriched
CD133+CD44+ stem-like cells with
CD133+CD44high metastatic subset in HCT116
colon cancer cells. Clin Exp Metastasis. 28:751–763. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Piao LS, Hur W, Kim TK, Hong SW, Kim SW,
Choi JE, Sung PS, Song MJ, Lee BC, Hwang D, et al:
CD133+ liver cancer stem cells modulate radioresistance
in human hepatocellular carcinoma. Cancer Lett. 315:129–137. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ding Q, Miyazaki Y, Tsukasa K, Matsubara
S, Yoshimitsu M and Takao S: CD133 facilitates
epithelial-mesenchymal transition through interaction with the ERK
pathway in pancreatic cancer metastasis. Mol Cancer. 13:152014.
View Article : Google Scholar : PubMed/NCBI
|