Introduction
In less developed countries, cervical cancer is the
second most common cancer and the third leading cause of
cancer-related deaths in females. There were an estimated 444,500
new cervical cancer cases and 230,200 deaths in these countries in
2012 (1). Almost 90% of the deaths
occurred in developing countries and for these countries it is a
huge economic health care burden. The highest cervical cancer death
rates are in Asia, with an estimated 144,400 deaths occurring in
2012 (1). Squamous cell carcinoma
(SCC) is the most common histological type of cervical cancer (~80%
of cases) and is closely correlated with human papillomavirus (HPV)
infection (2). Cervical screening
can prevent SCC, as it allows for detection and treatment of
precancerous lesions, and application of the HPV vaccine also
reduces the risk of SCC (3).
However, the high cost is a major barrier to making screening and
vaccination widely available to all women in developing countries,
including China.
Cervical squamous cell carcinoma (CSCC) cases that
are classified as International Federation of Gynecology and
Obstetrics (FIGO) stages IB and IIA have a greater possibility of
being cured and patients have better prognosis (4). Since early detection results in a
better prognosis and cervical screening and vaccines are extremely
expensive, there is an urgent need to identify more efficient early
diagnostic biomarkers and therapeutic targets for CSCC. Therefore,
it is necessary to augment our understanding of the molecular
mechanisms underlying the pathogenesis and development of CSCC.
Although previous studies have identified
chromosomal variations, abnormal expression of oncogenes or
tumor-suppressor genes and aberrant promoter methylation in CSCC,
few are relevant to microRNAs (miRNAs). miRNAs are a class of small
non-coding RNAs (ncRNAs) ~19–25 nucleotides (nts) in length that
can regulate gene expression by known classic mechanisms such as
binding to messenger RNA (mRNA) at the 3′-untranslated region
(3-UTR) to downregulate protein-coding genes by increasing mRNA
degradation (5). They also act
through other newly found mechanisms such as binding to promoters,
proteins, and by directly interacting with other ncRNAs (5,6).
miRNAs play important regulatory roles in various biological
processes e.g. differentiation, development, proliferation,
apoptosis, cell cycle control and metabolism (7,8).
Changes in miRNA expression have been identified in many diseases,
such as cardiac and autoimmune disorders, schizophrenia and cancer
(9). miRNAs have been found to be
differentially expressed between almost all types of analyzed
benign and malignant tumors and non-tumor tissues, including CSCC
(10). In addition, miRNAs
participate in regulating a variety of neoplastic biological
capabilities acquired during the multistep process of tumor
development. Using miRNA microarray, Wilting et al observed
that 46 miRNAs showed significantly differential expression between
normal cervical squamous epithelium and CSCC (11). Both miR-19a and miR-19b were
reported to promote cervical cancer cell proliferation (12), whereas miR-125b inhibited cervical
cancer cell apoptosis (13); both
processes are involved in cervical carcinogenesis. Other studies
reported that miR-9 (14) and
miR-135a (15) contributed to
HPV-induced cervical cancer formation, and that miR-133b (16) and miR-10a (17) promoted the progression and
development of cervical cancer by regulating tumor cell migration,
invasion and metastasis. However, miR-424 (18), miR-124 (19) and miR-218 (20) have opposite roles. All of these
cited studies analyzed the differences in expression of cervical
cancer-associated miRNAs by reverse transcription-polymerase chain
reaction (RT-PCR) and hybridization-based microarray that only
measure known and relatively abundant miRNAs, but are unable to
identify novel miRNAs. To date, no studies have focused on the
miRNA profile in early-stage CSCC. Thus, investigating the miRNA
profile in early-stage CSCC by high-throughput sequencing may
facilitate the analysis of expression of all annotated miRNAs,
detection of novel miRNAs and identification of candidate markers
for early diagnosis, and treatment of CSCC.
In the present study, we applied next-generation
sequencing (NGS) to globally and systematically characterize miRNA
alterations in early-stage CSCC by a comparison with paired normal
samples. Furthermore, we validated these miRNAs by qRT-PCR and
analyzed miRNA target genes and performed function annotation and
pathway analysis.
Materials and methods
Clinical sample collection
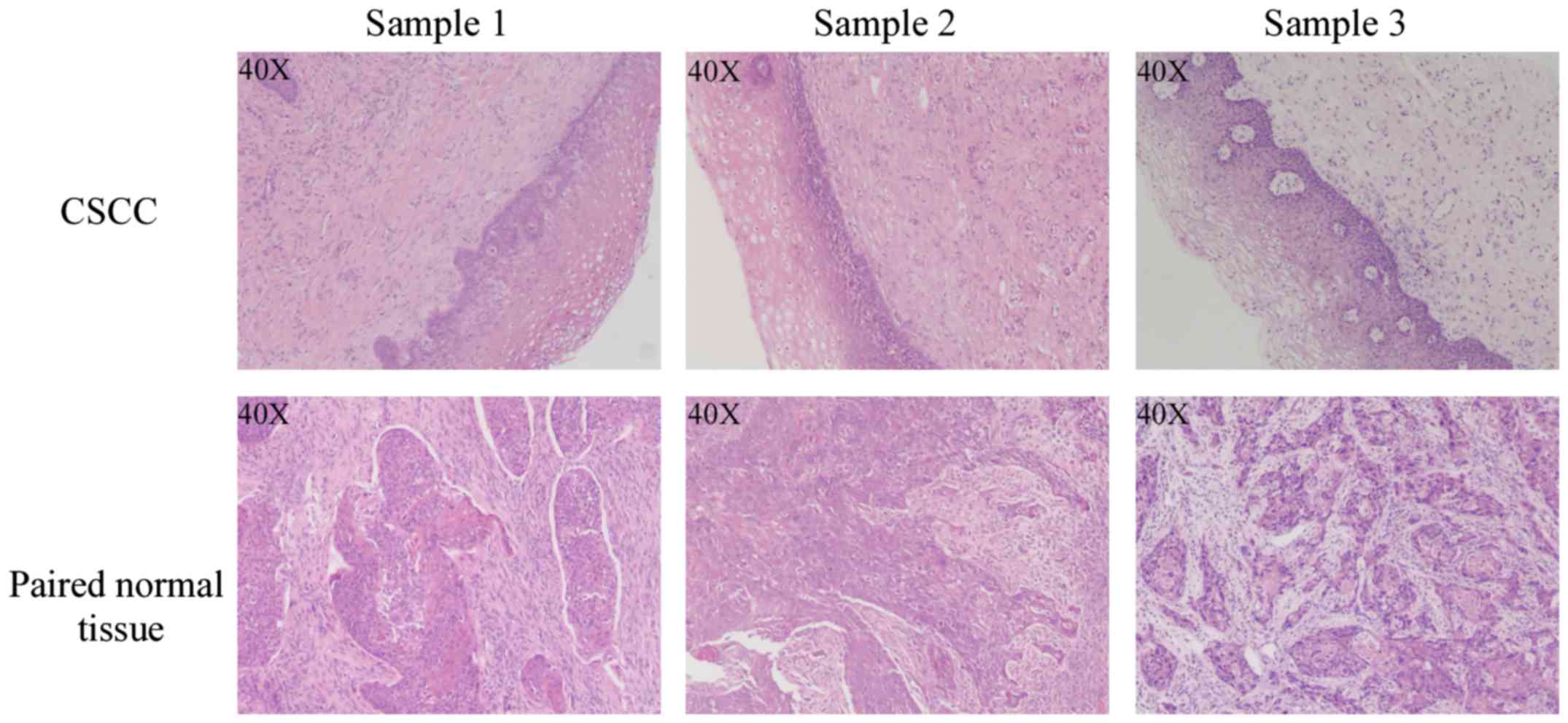
For sequencing, 3 pairs of human CSCC tumor (T) and
paired normal tissue (N) samples in FIGO stages I–II were obtained
from patients undergoing surgery of the cervix. Additional, 20
pairs of CSCC and normal tissues were also collected for
quantitative real-time polymerase chain reaction (qRT-PCR)
validation. All tumor and paired normal tissues were examined by
hematoxylin and eosin (H&E) staining, and all pathologic
diagnosis were confirmed by 2 independent pathologists; tumor
tissues containing >90% tumor cells were selected for further
study (Fig. 1). The study protocol
was approved by the Ethics Committees on Human Research of the
Fujian Cancer Hospital. All patients agreed to join the study and
signed a written informed consent. None of the patients had
received preoperative adjuvant radiotherapy or chemotherapy. All
samples were immediately snap-frozen in liquid nitrogen and then
stored at −80°C until RNA isolation.
RNA isolation
Total RNA was extracted using TRIzol (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's recommendations,
and quantified using a ND-1000 spectrophotometer (NanoDrop,
Rockland, DE, USA). The RNA quality was determined using an Agilent
2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and
the 28S/18S ratio was determined on 1% agarose gel. Only RNA
extracts with an RNA integrity number (RIN) value ≥7 and 28S/18S
ratio >1.8 were used for further analysis.
Deep sequencing and bioinformatic
analysis
RNA sequencing and bioinformatic analysis were
performed at the Beijing Genomics Institute (BGI; Shenzhen, China).
In brief, total RNA was ligated to the 5′- and 3′-end adaptors, and
sequentially amplified by an RT-PCR reaction. The fragments of
~62–75 bp (small RNA plus adaptors) were isolated from the agarose
gel and then purified. These products were directly sequenced with
Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) according to the
manufacturer's instructions. After trimming adaptor sequences,
removing contaminated reads and filtering low quality reads, the
remaining clean reads were mapped to the human genome using short
oligonucleotide alignment program (SOAP). The other small RNAs
(rRNA, tRNA, snRNA, snoRNA and piRNA) were detected by screening
against Rfam 10.1 and the GenBank database. The known miRNAs were
identified by aligning them to the miRBase. The expression
differences of the known miRNAs between the CSCCs and paired normal
samples were evaluated by comparing the log2 ratio of
the 2 groups. The miRNAs with at least a 2-fold-change of
expression and a P-value <0.05 between the 2 groups in all three
patients were selected for further study. The novel miRNAs were
predicted using MIREAP software.
Validation of mature miRNA expression
by qRT-PCR
For validation of the selected miRNAs that showed
differential expression and predicted novel miRNAs, qRT-PCR was
applied using the Applied Biosystems 7500 Sequence Detection System
(Applied Biosystems, Foster City, CA, USA) according to the
manufacturer's instructions. In brief, the miRNAs were reverse
transcribed into cDNA using the miScript II RT kit (Qiagen,
Valencia, CA, USA) with 5X miScript HiSpec buffer. The mixture was
incubated at 37°C for 60 min, and then at 95°C for 5 min and then
placed on ice. Quantitative PCR was performed using the miScript
SYBR-Green PCR kit and 10X miScript Primer Assay (both from
Qiagen). The reaction consisted of 1 cycle at 95°C for 15 min,
followed by 40 cycles at 94°C for 15 sec and 55°C for 30 sec and
70°C for 30 sec. All reactions were repeated 3 times. The relative
expression levels of miRNAs were calculated using the
2−ΔΔCt method with snRNA U6 as the internal reference.
Quantitative PCR data were analyzed by SPSS statistics software
(version 19.0; IBM, Armonk, NY, USA). A Student's t-test was used
to compare the difference in expression between CSCC and normal
tissues. P<0.05 was considered to indicate a statistically
significant result.
miRNA target prediction, functional
annotation and pathway analysis
The target genes of the selected miRNAs with
differential expression and novel miRNAs were predicted using
RNAhybrid. For determination of the main biological functions of
the candidate target genes, Gene Ontology (GO) enrichment analysis
was applied. We mapped all candidates to GO terms in the database
(http://www.geneontology.org/), using a
hypergeometric test to find significantly enriched GO terms. The GO
terms with corrected P-values with an enrichment ratio ≤0.05 were
defined as significantly enriched target gene candidates. For
identification of significantly enriched metabolic pathways or
signal transduction pathways in target gene candidates, we mapped
these candidates to the Kyoto Encyclopedia of Genes and Genomes
(KEGG) databases (21). The
calculation method is the same as that in the GO analysis. Pathways
with corrected P-values with an enrichment ratio ≤0.05 were
considered as significantly enriched target gene candidates.
Statistical analysis
All quantitative data are expressed as the mean ±
standard deviation (mean ± SD) from at least 3 samples or
experiments/data point. Significant differences were analyzed by
the Student's t-test to compare 2 groups of independent
samples.
Results
Overview of small RNA sequencing
results
To determine the miRNA profile of early-stage CSCC,
we performed high-throughput NGS in 3 CSCC and their paired normal
samples using Illumina HiSeq 2500. After removing adaptors, low
quality tags and contaminants, an average of 11,478,411 clean reads
(ranging from 10,313,021 to 11,820.021) remained (Table I). The majority of these clean reads
were 18–28 nt in length, with 22 nt RNAs the most abundant. These
clean reads were mapped to the Human Genome and rRNA, tRNA and
snRNA were identified by the alignment to Rfam 10.1 and GenBank
databases. Subsequently, an average of 1,726 mature miRNAs (ranging
from 1,571 to 1,881) were successfully annotated in the miRBase,
and thus identified as known miRNAs (Table I). The sequences which did not match
with any database were considered as unannotated sRNAs (Table I).
 | Table I.Summary of the small RNA
sequencing. |
Table I.
Summary of the small RNA
sequencing.
|
| Number |
|---|
|
|
|
|---|
| Category | T-1 | T-2 | T-3 | N-1 | N-2 | N-3 |
|---|
| Total reads | 12,000,000 | 10,523,729 | 12,000,000 | 12,000,000 | 12,000,000 | 12,000,000 |
| Clean reads | 11,713,704 | 10,313,021 | 11,519,252 | 11,799,688 | 11,820,021 | 11,704,777 |
| % | 98.40 | 98.73 | 96.70 | 99.02 | 99.18 | 98.25 |
| Unique sRNAs | 884,315 | 673,581 | 867,499 | 615,904 | 466,839 | 658,536 |
| miRNAs | 4,196 | 4,545 | 4,346 | 3,634 | 4,035 | 4,173 |
| Known miRNAs in
miRBase | 1,708 | 1,881 | 1,810 | 1,571 | 1,659 | 1,724 |
| miRNAs |
200 |
225 |
229 |
171 |
182 |
200 |
| miRNA-5p |
414 |
431 |
418 |
385 |
394 |
400 |
| miRNA-3p |
306 |
351 |
325 |
298 |
330 |
335 |
| miRNA
precursors |
788 |
874 |
838 |
717 |
753 |
789 |
| Unannotated
sRNAs | 232,043 | 205,460 | 255,885 | 291,651 | 156,254 | 240,371 |
| Novel miRNA
candidates | 28 | 24 | 31 | 20 | 22 | 26 |
Identification and validation of
differentially expressed known miRNAs between CSCC and normal
tissues
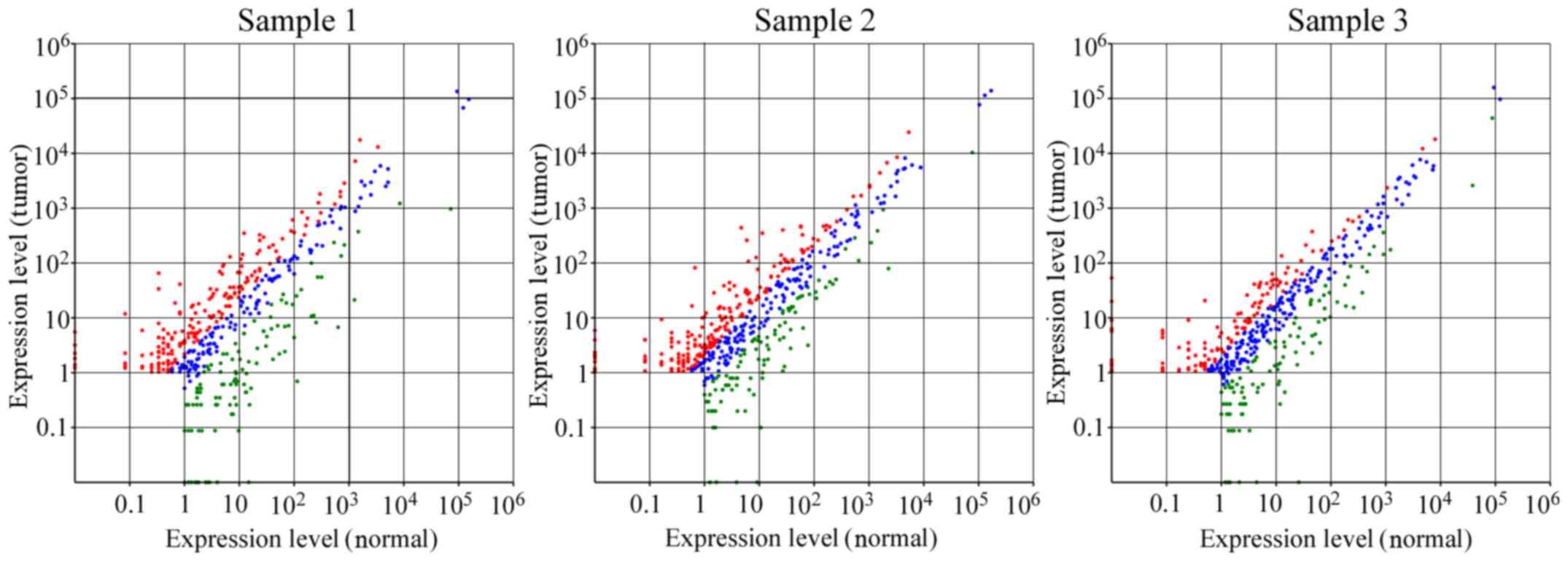
To detect the known miRNAs which were differentially
expressed between the CSCC and normal tissues, the expression level
of each miRNA was first normalized to provide the expression of
transcripts per million (TPM), and then the fold-change and
P-values were calculated from the normalized expression. There were
424 differentially expressed miRNAs between T1 and N1, 480 between
T2 and N2 and 440 between T3 and N3 (Fig. 2). Subsequently, on the basis of the
cut-off criteria (fold-change >2 and p-value <0.05), 186, 149
and 126 miRNAs were selected from the data from each of the 3 pairs
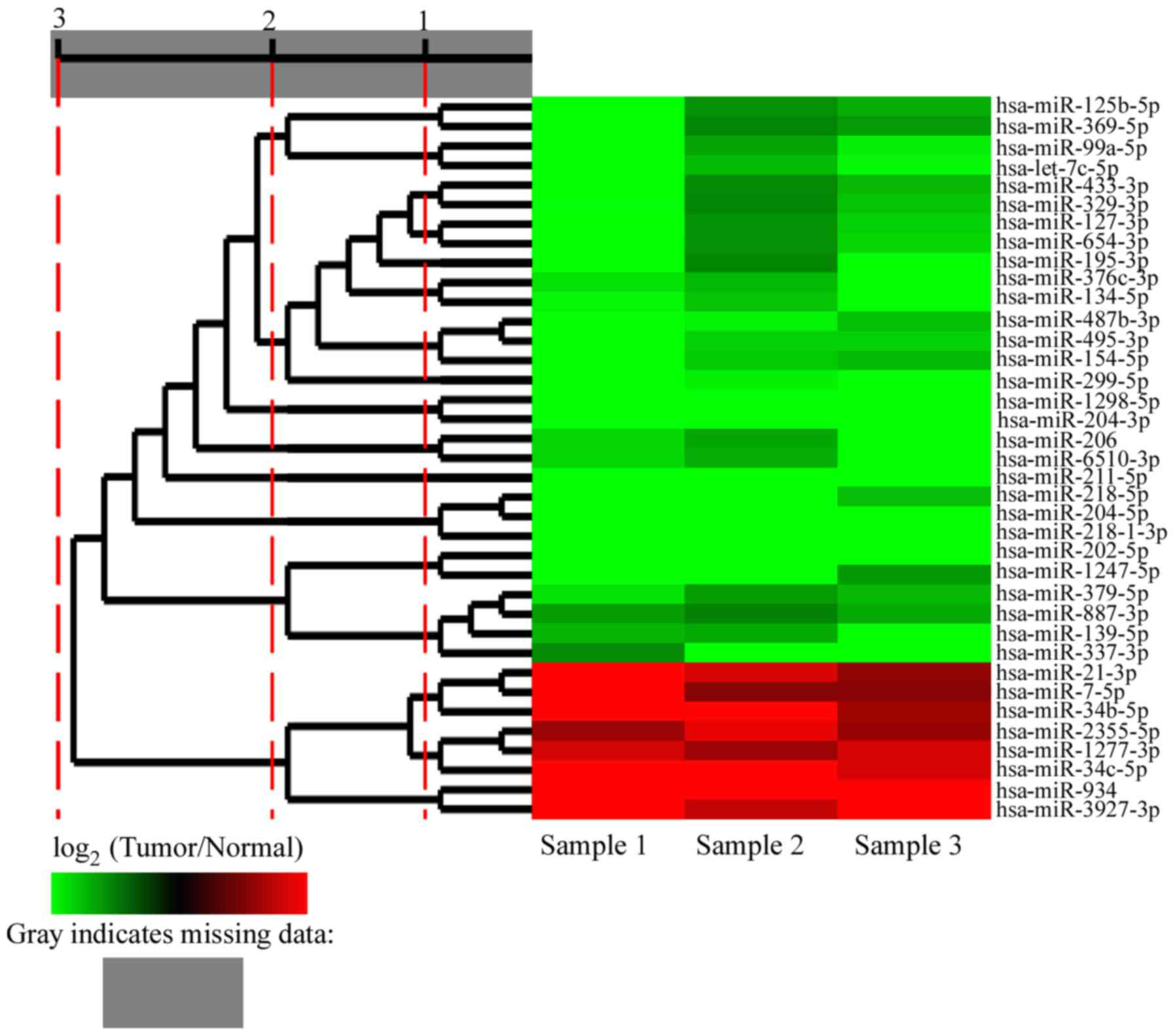
of patient samples. Finally, only 37 miRNAs exhibited consistent
changes in all 3 pairs of samples; among these 37 differentially
expressed miRNAs, 8 were significantly upregulated and 29 were
significantly downregulated (Fig.
3; Table II).
 | Table II.Summary of the significantly
upregulated and downregulated miRNAs in CSCC. |
Table II.
Summary of the significantly
upregulated and downregulated miRNAs in CSCC.
|
| Fold-change
(log2 tumor/normal) |
|---|
|
|
|
|---|
| miRNA | T1 vs. N1 | T2 vs. N2 | T3 vs. N3 |
|---|
| hsa-miR-934 | 8.30318711 | 4.2842649 | 7.20535341 |
|
hsa-miR-3927-3p | 4.51831602 | 3.10372182 | 9.11787682 |
| hsa-miR-34b-5p | 9.09373446 | 4.03811041 | 2.54681543 |
| hsa-miR-34c-5p | 4.78205816 | 4.92246647 | 3.3850871 |
| hsa-miR-21-3p | 6.62522824 | 3.4080715 | 2.26011983 |
| hsa-miR-7-5p | 5.56817204 | 2.07123043 | 2.11570518 |
|
hsa-miR-1277-3p | 3.30345745 | 2.463592 | 3.34499026 |
|
hsa-miR-2355-5p | 2.42581593 | 3.65621776 | 2.39229062 |
| hsa-miR-211-5p | −7.08321336 | −6.80271289 | −11.34275801 |
| hsa-miR-204-5p | −6.85930564 | −9.83557679 | −5.26254955 |
|
hsa-miR-218-1-3p | −7.33109609 | −7.40258575 | −6.9021942 |
| hsa-miR-202-5p | −10.57502857 | −4.68366909 | −5.0642676 |
| hsa-miR-218-5p | −7.941165 | −8.60425671 | −2.97717439 |
|
hsa-miR-1298-5p | −5.92810297 | −4.54163652 | −7.7387003 |
| hsa-miR-204-3p | −6.66817587 | −4.05060043 | −7.09486942 |
|
hsa-miR-6510-3p | −3.36793893 | −2.73610137 | −8.822762 |
| hsa-miR-299-5p | −5.52895357 | −3.78516379 | −5.511115 |
|
hsa-miR-1247-5p | −7.39024513 | −4.67666656 | −2.41593802 |
| hsa-miR-206 | −3.37648276 | −2.61057144 | −7.18229503 |
| hsa-let-7c-5p | −6.26600709 | −2.93147207 | −3.93093655 |
| hsa-miR-99a-5p | −6.62465962 | −2.58200581 | −3.74953697 |
|
hsa-miR-487b-3p | −5.00445576 | −3.79678741 | −3.01875682 |
| hsa-miR-154-5p | −5.43273636 | −3.17579365 | −2.93133666 |
| hsa-miR-337-3p | −2.21193294 | −4.38841479 | −4.62100525 |
| hsa-miR-495-3p | −4.59233357 | −3.30958264 | −3.28815361 |
| hsa-miR-134-5p | −3.89631011 | −3.08449386 | −4.1468506 |
|
hsa-miR-125b-5p | −5.94077162 | −2.27910623 | −2.73045243 |
|
hsa-miR-376c-3p | −3.55711379 | −2.91544225 | −4.19191414 |
| hsa-miR-195-5p | −4.33482863 | −2.13708218 | −4.13398935 |
| hsa-miR-654-3p | −4.63279944 | −2.26265207 | −3.39215912 |
| hsa-miR-127-3p | −4.56838942 | −2.30793519 | −3.28638674 |
| hsa-miR-369-5p | −5.41522226 | −2.08867671 | −2.46223868 |
| hsa-miR-139-5p | −2.84273417 | −2.67661687 | −4.37453647 |
| hsa-miR-433-3p | −4.37189471 | −2.16954783 | −2.89020567 |
| hsa-miR-329-3p | −3.95914876 | −2.12515371 | −3.0926029 |
| hsa-miR-379-5p | −3.57432673 | −2.49126887 | −2.8998286 |
| hsa-miR-887-3p | −2.49199079 | −2.07624607 | −2.66140231 |
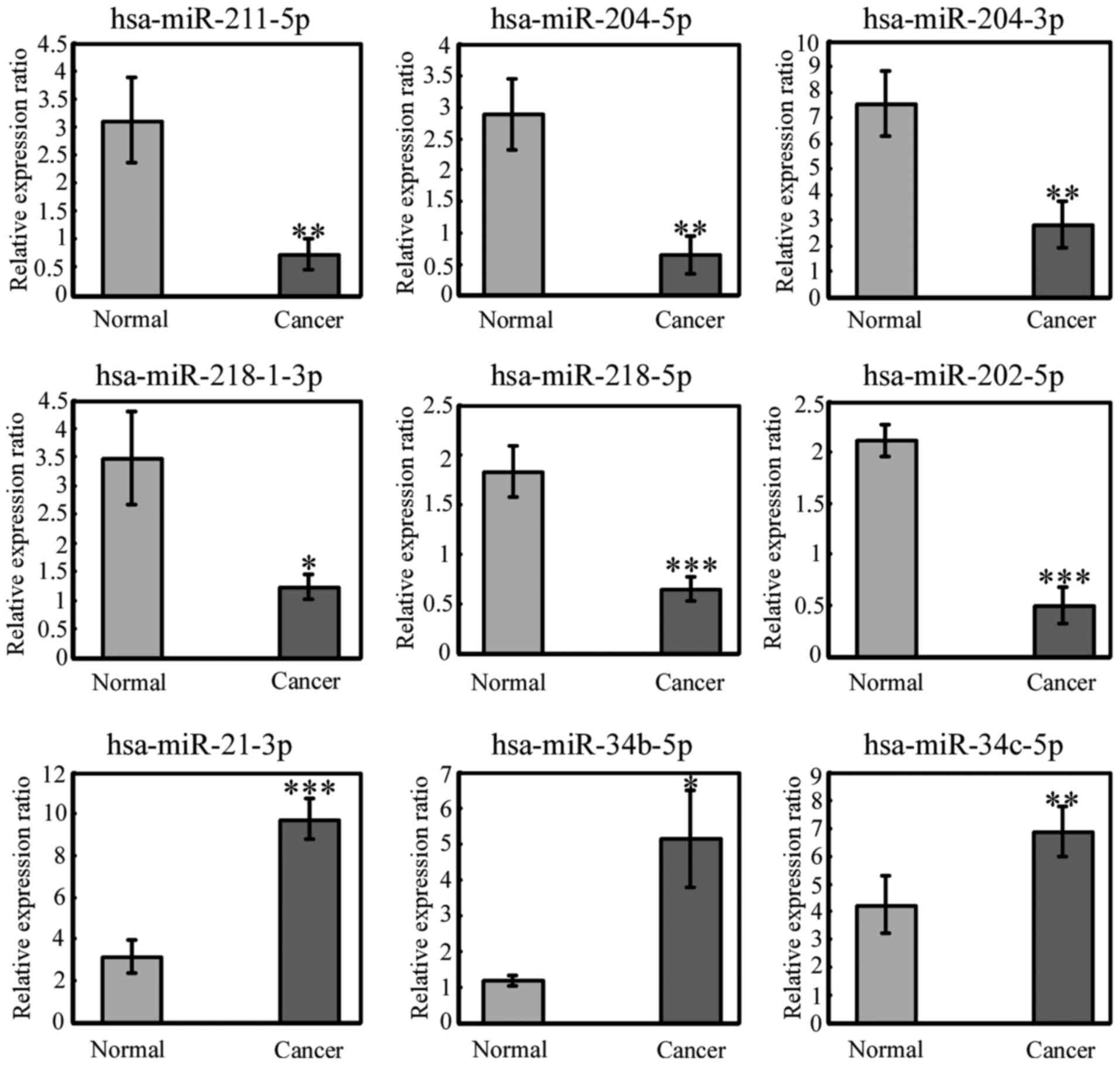
To further validate the deep sequencing results,
qRT-PCR was applied to assess the significantly differentially
expressed miRNAs in 3 pairs of samples used for sequencing and the
additional 20 pairs of samples. Nine miRNAs were selected from the
miRNAs with significantly modified expression and measured by
qRT-PCR. All of the 9 miRNAs displayed the same expression pattern
consistent with the sequencing data (Fig. 4). This indicated that our high
through-put sequencing results were reliable.
Target genes of the significantly
altered miRNAs
To demonstrate the differences in gene expression
associated with the miRNA profile in CSCC, the putative target
genes of the miRNAs that were expected to be significantly
differently expressed between CSCC and normal tissues were
predicted using RNAhybrid. GO and KEGG pathway analyses were
performed to enrich the main biological processes and pathways in
which the target gene candidates were involved. However, the
P-value of the enrichment ratio failed to achieve filter conditions
and the analysis was not continued.
Prediction of novel miRNAs, their
target genes and pathways
To detect novel miRNAs, unannotated sRNAs were
further processed using MIREAP. The number of novel miRNA
candidates obtained from the 6 small RNA libraries ranged from 20
to 31 (Table I). There were 5
common novel miRNA candidates in the 3 pairs of CSCC and normal
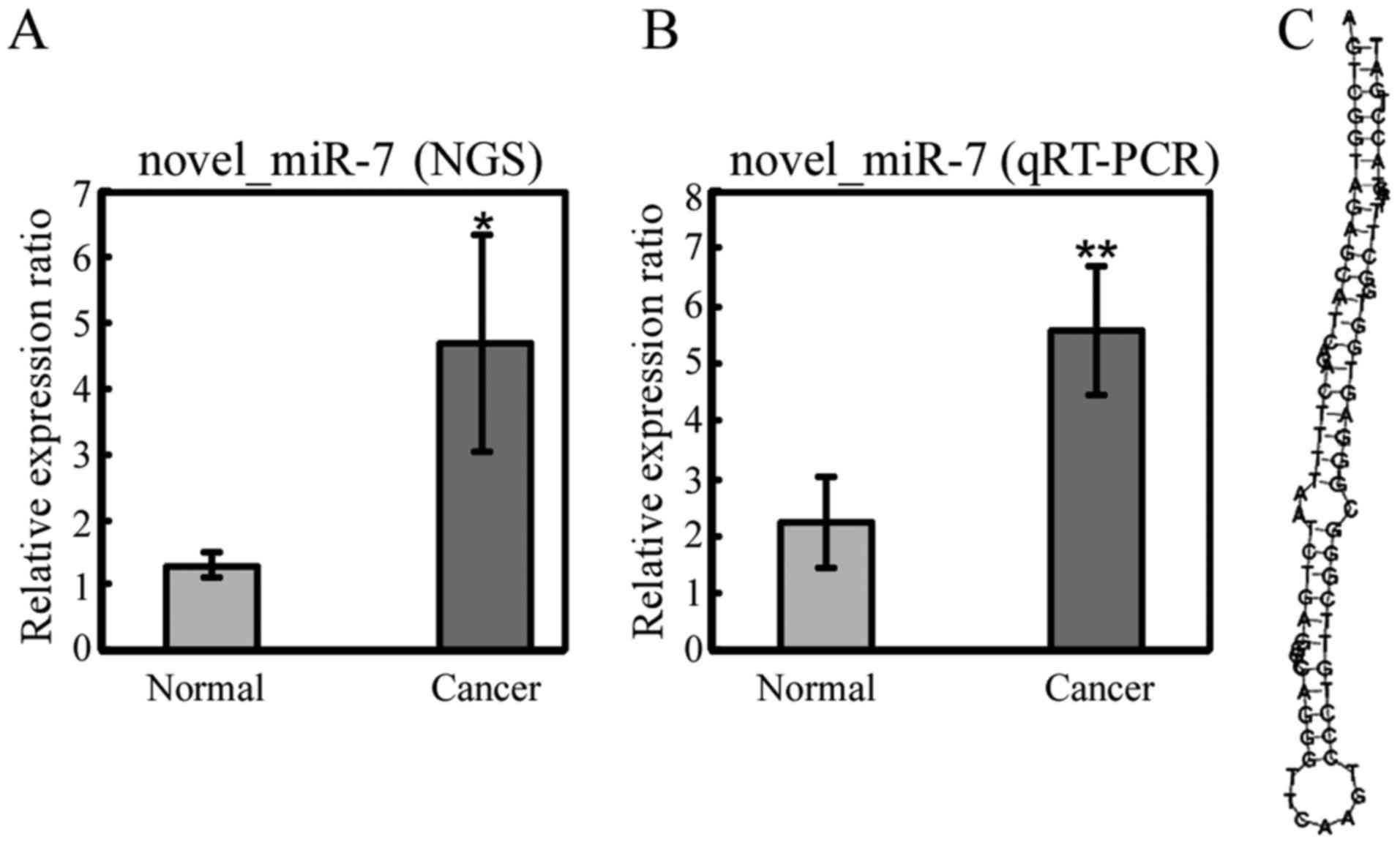
samples. Among the 5 common miRNAs, only 1 miRNA, novel_miR_7,
displayed a significant difference in expression (fold-change >1
and P-value <0.05) in all 3 pairs of samples: novel_miR_7 was
selected as a putative novel miRNA and verified by qRT-PCR
(Fig. 5). Its sequence and
stem-loop secondary structure are shown in Fig. 5. To further understand the function
of the novel miRNA, its target genes were predicted using RNAhybrid
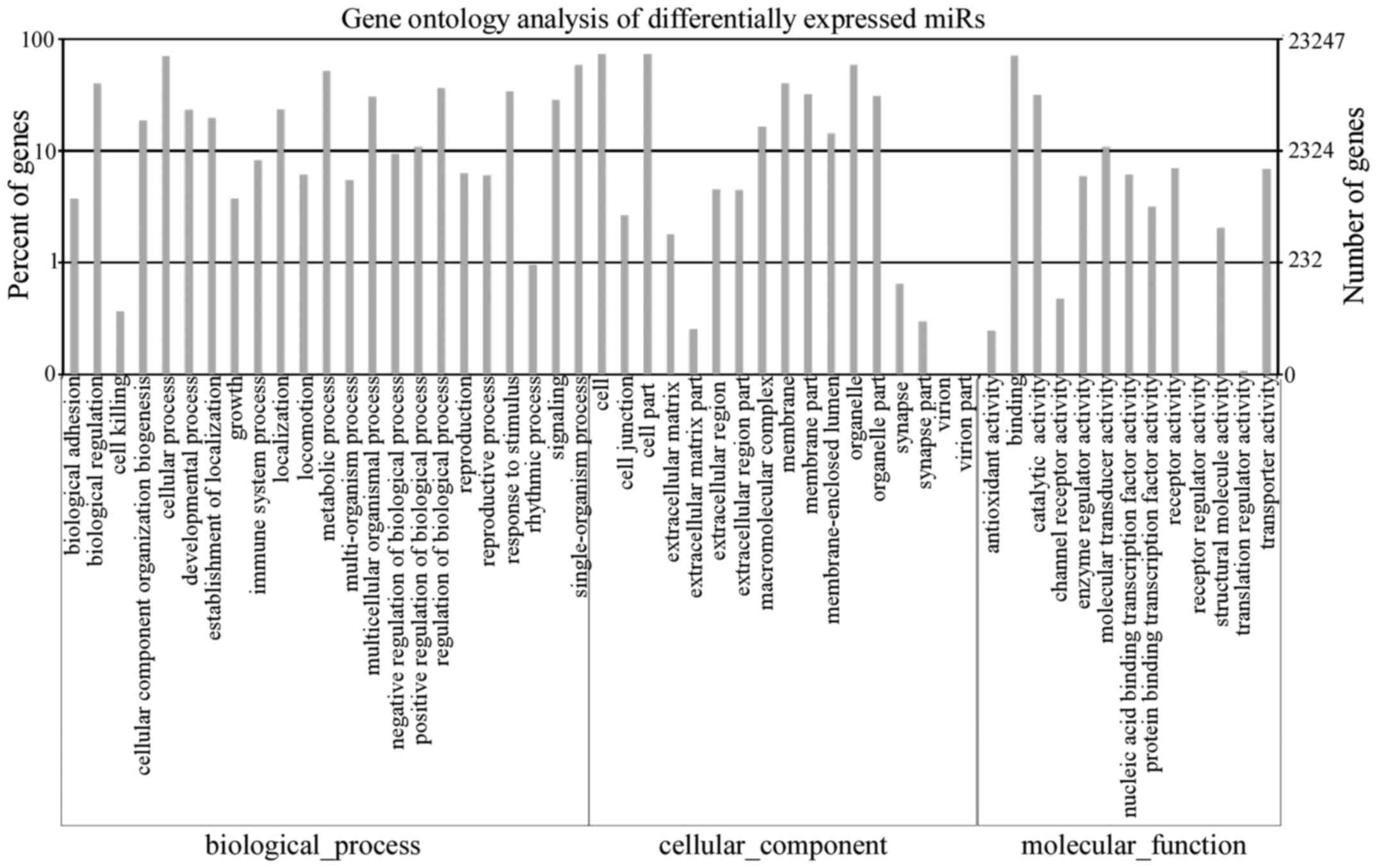
and also assessed by GO and KEGG pathway analyses. The GO analysis
revealed the main biological processes in which the target genes
are involved and is shown in Fig.
6. The KEGG pathway analysis indicated that 5 pathways were
enriched; including the MAPK, calcium and adherent junction
signaling pathways (Table
III).
 | Table III.KEGG pathway analysis for predicted
target genes of the novel miRNA in CSCC. |
Table III.
KEGG pathway analysis for predicted
target genes of the novel miRNA in CSCC.
| KEGG no. | KEGG pathway | Significance |
| KO:04010 | MAPK signaling
pathway | a |
| KO:04020 | Calcium signaling
pathway | b |
| KO:04520 | Adherent
junction | b |
| KO:05217 | Basal cell
carcinoma | b |
| KO:04730 | Long-term
depression | b |
Discussion
Although CSCC caused by HPV infection can be
prevented, it is still a common cancer in women. There is
accumulating evidence to indicate that HPV infection alone is
insufficient to cause the disease and miRNAs may play critical
roles in carcinogenesis (22–24).
Therefore, global analysis of miRNA expression comparing CSCC with
normal tissue may aid in the better understanding of the function
of miRNAs in the course of the disease as well as finding novel
biomarkers for diagnosis, treatment and prognostic evaluation. To
the best of our knowledge, this is the first study to profile miRNA
expression in early-stage CSCC on the NGS platform. We identified
37 significantly differentially regulated known miRNAs in CSCC
compared with normal tissue and confirmed a sample of 9 by qRT-PCR.
Furthermore, a novel miRNA candidate, novel_miR_7, was identified
for the first time and its target genes are involved in MAPK,
calcium and adherent junction signaling pathways.
miRNAs are rather stable, even in body fluids such
as serum, plasma, urine and saliva (25). The expression of miRNAs is unique to
tissues or organs and they can be easily detected (26). These characteristics make them ideal
cancer biomarkers. Juan et al (27) performed deep sequencing on the serum
pools of cervical cancer patients and healthy controls to identify
putative novel miRNAs. They found that only 1 of 17 common novel
miRNA candidates may distinguish cervical cancer patients from
healthy controls. Wang et al (28) also constructed expression profiles
of miRNAs in the serum of CSCC patients by microarray. They
discovered that the levels of 291 of 338 detectable circulating
miRNAs were changed >2-fold in serum samples from CSCC patients
vs. controls. However, they did not validate these results through
any other methodologies. Although detection of miRNAs in serum has
been proposed to be a simpler and less invasive diagnostic method
than sampling tissue, the consequential biological and technical
variability, which is mainly assessed by a series of different
normalization processes based on the expression of reference genes,
is difficult to avoid. Currently, there is no known suitable
extracellular reference RNA for a proper normalization,
particularly in cell-free miRNA analysis (26). Commonly used references, including
U6 small nuclear RNA (RNU6B) and 5S ribosomal RNA, may be degraded
or expressed less stably than others in serum (29). Therefore, we decided to analyze
miRNA expression in CSCC tissues. In previous studies, a microarray
technique, which is biased towards abundantly expressed known
miRNAs, was the major platform for miRNA expression profiling
(11,28). We chose a more sensitive and
specific method, NGS, which enables high-throughput analysis and
novel miRNA discovery, to measure miRNA expression.
In the present study, we identified 9 miRNAs that
exhibited differential expression in CSCC using deep sequencing and
confirmed by qRT-PCR evaluation. Six were downregulated, including
miR-211-5p, miR-204-5p/3p, miR-218-1-3p/5p and miR-202-5p, and 3
were upregulated, including miR-34b-5p, miR-34c-5p and miR-21-3p.
Among these miRNAs, miR-218, miR-21 and miR-34b-5p have previously
been discovered and their functions had been verified to be
associated with cervical cancer. In comparison to normal cervical
tissues the expression of miR-218 was reported to be lower in
cervical cancer tissues (20), and
that finding was concordant with our results. Its downregulation
was significantly associated with worse overall survival, worse
disease-free survival and pelvic/aortic lymph node recurrence.
miR-218 was found to target survivin to inhibit cervical cancer
progression by regulating clonogenicity, migration and invasion.
Increased miR-21 expression was found to be correlated with poorer
histological diagnosis in cervical cancer (30). Meanwhile, miR-21 was found to be
mainly expressed in the tumor stromal microenvironment where it may
be involved in cervical cancer progression. In addition, expression
of miR-21 in HPV-positive samples and SCC was significantly higher
than that noted in HPV-negative samples (31). Constitutive activation of aberrant
STAT3 signaling via miR-21, which is regulated by the HPV
oncoprotein E6, plays a pivotal role in the initiation and
progression of HPV-induced cervical carcinogenesis (32,33).
Our sequencing results found that miR-21 was upregulated in CSCC
and it may act as an oncogene; which is consistent with previous
studies (30,31). miR-34b-5p was found to be
downregulated in minimal deviation adenocarcinoma (MDA) compared
with normal proliferative endocervical tissues (34), but in the present study, it was
found to be upregulated in CSCC. The discrepancy suggests different
expression patterns of miRNAs in different histological subtypes of
tumors.
The other differentially expressed miRNAs we
described have seldom been reported in previous studies of cervical
cancer, but they have been demonstrated to be correlated with other
malignant tumors. Trends seen in the expression of certain miRNAs
that were determined by sequencing were in accord with published
functional studies. For example, miRNA-211 functions as a
tumor-suppressor in melanoma (35,36)
and glioma (37). It could block
tumor invasion and migration by targeting NUAK1 (38), BRN2 (39), KCNMA1 (40) or MMP-9 (37). Both miR-204-5p/3p and miR-202 have
tumor-suppressive function as well. Downregulation of miR-204-5p in
colorectal cancer tissues was associated with poor prognosis: It
suppresses colorectal cancer cell growth, migration, and invasion
as well as promotes tumor sensitivity to chemotherapy by inhibiting
RAB22A (41). In endometrial
carcinoma, miR-204-5p exhibits a similar function through the
TrkB-STAT3-miR-204-5p regulatory pathways (42) and miR-204-3p inhibits the growth of
hepatocellular carcinoma tumor endothelial cells by blocking the
adhesion function of FN1 (43).
miR-202 can suppress osteosarcoma cell proliferation and induce
cell apoptosis by downregulating Gli2 expression (44). While miR-34c is considered to be a
putative tumor suppressor in a majority of tumors (45–48),
we found it to be upregulated in CSCC, which is in agreement with
previous results that determined expression by miRNA microarray in
cervical cancer (11,28). Thus, we hypothesized that miR-34c
may play different roles in cervical cancer and other types of
malignancy.
NGS technology allows the rapid discovery of novel
miRNAs, for some of which the biological function is unknown. We
identified 5 common novel miRNA candidates with differential
expression between CSCC and normal tissues. Our focus was narrowed
to miRNAs that exhibited a significant change of expression in all
3 pairs of samples. We identified novel_miR_7 as a candidate novel
miRNA and confirmed its presence. Functional annotation revealed
that its predicted target genes are in the MAPK signaling pathway,
which is closely related to cell proliferation, cell death, and the
balance between them. Early in 2006, Engelbrecht et al
indicated the involvement of MAPK in cervical cancer and
demonstrated the correlation between the activity of MAPK and
apoptosis in the disease process (49). The HPV 16 E2 protein may induce
apoptosis by inhibiting MAPK signaling in CSCC (50). However, playing a role in cervical
cancer carcinogenesis, the expression level of MAPK may serve as a
marker for cervical cancer progression (51,52)
and its inhibitors have shown the potential to be useful for
cervical cancer therapy (53).
Therefore, our results provide a solid basis for the
characterization of the role of novel_miR_7 in the pathogenesis and
development of CSCC and in the treatment of this disease.
In summary, for the first time we identified known
and novel miRNAs in early-stage CSCC by high-throughput NGS.
Moreover, we validated these miRNAs in other independent samples by
qRT-PCR. Our analysis of differentially expressed miRNAs not only
support existing findings but also uncovered various differentially
expressed miRNAs which had not previously been reported in CSCC,
thus providing new molecular signatures of the disease. The
prediction of novel miRNAs found that novel_miR_7 may be a
promising biomarker for CSCC. These results have implications for
exploring the regulatory network of miRNAs, provide new targets for
molecular mechanistic studies of CSCC as well as for early
detection, targeted therapy and prognostic evaluation of CSCC
patients in the future.
Acknowledgements
The present study was supported by grants from the
Fujian Province Science and Technology Plan Key Project (no.
2013Y0030) and the National Clinical Key Specialty Construction
Program of China.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bosch FX, Lorincz A, Muñoz N, Meijer CJ
and Shah KV: The causal relation between human papillomavirus and
cervical cancer. J Clin Pathol. 55:244–265. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaccarella S, Lortet-Tieulent J, Plummer
M, Franceschi S and Bray F: Worldwide trends in cervical cancer
incidence: Impact of screening against changes in disease risk
factors. Eur J Cancer. 49:3262–3273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cole L and Stoler MH: Issues and
inconsistencies in the revised gynecologic staging systems. Semin
Diagn Pathol. 29:167–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berindan-Neagoe I, Monroig PC, Pasculli B
and Calin GA: MicroRNAome genome: A treasure for cancer diagnosis
and therapy. CA Cancer J Clin. 64:311–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rossi JJ: New hope for a microRNA therapy
for liver cancer. Cell. 137:990–992. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wilting SM, Snijders PJ, Verlaat W,
Jaspers A, van De Wiel MA, van Wieringen WN, Meijer GA, Kenter GG,
Yi Y, le Sage C, et al: Altered microRNA expression associated with
chromosomal changes contributes to cervical carcinogenesis.
Oncogene. 32:106–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu XM, Wang XB, Chen MM, Liu T, Li YX, Jia
WH, Liu M, Li X and Tang H: MicroRNA-19a and −19b regulate cervical
carcinoma cell proliferation and invasion by targeting CUL5. Cancer
Lett. 322:148–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang YD, Cai N, Wu XL, Cao HZ, Xie LL and
Zheng PS: OCT4 promotes tumorigenesis and inhibits apoptosis of
cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis.
4:e7602013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu W, Gao G, Hu X, Wang Y, Schwarz JK,
Chen JJ, Grigsby PW and Wang X: Activation of miR-9 by human
papillomavirus in cervical cancer. Oncotarget. 5:11620–11630. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leung CO, Deng W, Ye TM, Ngan HY, Tsao SW,
Cheung AN, Pang RT and Yeung WS: miR-135a leads to cervical cancer
cell transformation through regulation of β-catenin via a
SIAH1-dependent ubiquitin proteosomal pathway. Carcinogenesis.
35:1931–1940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qin W, Dong P, Ma C, Mitchelson K, Deng T,
Zhang L, Sun Y, Feng X, Ding Y, Lu X, et al: MicroRNA-133b is a key
promoter of cervical carcinoma development through the activation
of the ERK and AKT1 pathways. Oncogene. 31:4067–4075. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Long MJ, Wu FX, Li P, Liu M, Li X and Tang
H: MicroRNA-10a targets CHL1 and promotes cell growth, migration
and invasion in human cervical cancer cells. Cancer Lett.
324:186–196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F,
Xie X, Zhou C and Lu W: Suppressed miR-424 expression via
upregulation of target gene Chk1 contributes to the progression of
cervical cancer. Oncogene. 32:976–987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wan HY, Li QQ, Zhang Y, Tian W, Li YN, Liu
M, Li X and Tang H: MiR-124 represses vasculogenic mimicry and cell
motility by targeting amotL1 in cervical cancer cells. Cancer Lett.
355:148–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T,
et al: KEGG for linking genomes to life and the environment.
Nucleic Acids Res. 36:D480–D484. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.PubMed/NCBI
|
|
23
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gilabert-Estelles J, Braza-Boils A, Ramon
LA, Zorio E, Medina P, Espana F and Estelles A: Role of microRNAs
in gynecological pathology. Curr Med Chem. 19:2406–2413. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body fluids -
the mix of hormones and biomarkers. Nat Rev Clin Oncol. 8:467–477.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Etheridge A, Lee I, Hood L, Galas D and
Wang K: Extracellular microRNA: A new source of biomarkers. Mutat
Res. 717:85–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Juan L, Tong HL, Zhang P, Guo G, Wang Z,
Wen X, Dong Z and Tian YP: Identification and characterization of
novel serum microRNA candidates from deep sequencing in cervical
cancer patients. Sci Rep. 4:62772014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang WT, Zhao YN, Yan JX, Weng MY, Wang Y,
Chen YQ and Hong SJ: Differentially expressed microRNAs in the
serum of cervical squamous cell carcinoma patients before and after
surgery. J Hematol Oncol. 7:62014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deftereos G, Corrie SR, Feng Q, Morihara
J, Stern J, Hawes SE and Kiviat NB: Expression of mir-21 and
mir-143 in cervical specimens ranging from histologically normal
through to invasive cervical cancer. PLoS One. 6:e284232011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bumrungthai S, Ekalaksananan T, Evans MF,
Chopjitt P, Tangsiriwatthana T, Patarapadungkit N, Kleebkaow P,
Luanratanakorn S, Kongyingyoes B, Worawichawong S, et al:
Up-regulation of miR-21 is associated with cervicitis and human
papillomavirus infection in cervical tissues. PLoS One.
10:e01271092015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shishodia G, Verma G, Srivastava Y,
Mehrotra R, Das BC and Bharti AC: Deregulation of microRNAs let-7a
and miR-21 mediate aberrant STAT3 signaling during human
papillomavirus-induced cervical carcinogenesis: Role of E6
oncoprotein. BMC Cancer. 14:9962014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shishodia G, Shukla S, Srivastava Y,
Masaldan S, Mehta S, Bhambhani S, Sharma S, Mehrotra R, Das BC and
Bharti AC: Alterations in microRNAs miR-21 and let-7a correlate
with aberrant STAT3 signaling and downstream effects during
cervical carcinogenesis. Mol Cancer. 14:1162015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee H, Kim KR, Cho NH, Hong SR, Jeong H,
Kwon SY, Park KH, An HJ, Kim TH, Kim I, et al: Gynecological
Pathology Study Group of the Korean Society of Pathologists:
MicroRNA expression profiling and Notch1 and Notch2 expression in
minimal deviation adenocarcinoma of uterine cervix. World J Surg
Oncol. 12:3342014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levy C, Khaled M, Iliopoulos D, Janas MM,
Schubert S, Pinner S, Chen PH, Li S, Fletcher AL, Yokoyama S, et
al: Intronic miR-211 assumes the tumor suppressive function of its
host gene in melanoma. Mol Cell. 40:841–849. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu Y, Brenn T, Brown ER, Doherty V and
Melton DW: Differential expression of microRNAs during melanoma
progression: miR-200c, miR-205 and miR-211 are downregulated in
melanoma and act as tumour suppressors. Br J Cancer. 106:553–561.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Asuthkar S, Velpula KK, Chetty C, Gorantla
B and Rao JS: Epigenetic regulation of miRNA-211 by MMP-9 governs
glioma cell apoptosis, chemosensitivity and radiosensitivity.
Oncotarget. 3:1439–1454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bell RE, Khaled M, Netanely D, Schubert S,
Golan T, Buxbaum A, Janas MM, Postolsky B, Goldberg MS, Shamir R,
et al: Transcription factor/microRNA axis blocks melanoma invasion
program by miR-211 targeting NUAK1. J Invest Dermatol. 134:441–451.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Boyle GM, Woods SL, Bonazzi VF, Stark MS,
Hacker E, Aoude LG, Dutton-Regester K, Cook AL, Sturm RA and
Hayward NK: Melanoma cell invasiveness is regulated by miR-211
suppression of the BRN2 transcription factor. Pigment Cell Melanoma
Res. 24:525–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mazar J, DeYoung K, Khaitan D, Meister E,
Almodovar A, Goydos J, Ray A and Perera RJ: The regulation of
miRNA-211 expression and its role in melanoma cell invasiveness.
PLoS One. 5:e137792010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bao W, Wang HH, Tian FJ, He XY, Qiu MT,
Wang JY, Zhang HJ, Wang LH and Wan XP: A TrkB-STAT3-miR-204-5p
regulatory circuitry controls proliferation and invasion of
endometrial carcinoma cells. Mol Cancer. 12:1552013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cui ZH, Shen SQ, Chen ZB and Hu C: Growth
inhibition of hepatocellular carcinoma tumor endothelial cells by
miR-204-3p and underlying mechanism. World J Gastroenterol.
20:5493–5504. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun Z, Zhang T, Hong H, Liu Q and Zhang H:
miR-202 suppresses proliferation and induces apoptosis of
osteosarcoma cells by downregulating Gli2. Mol Cell Biochem.
397:277–283. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu Z, Kim J, He L, Creighton CJ, Gunaratne
PH, Hawkins SM and Matzuk MM: Functional analysis of miR-34c as a
putative tumor suppressor in high-grade serous ovarian cancer. Biol
Reprod. 91:1132014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang S, Li WS, Dong F, Sun HM, Wu B, Tan
J, Zou WJ and Zhou DS: KITLG is a novel target of miR-34c that is
associated with the inhibition of growth and invasion in colorectal
cancer cells. J Cell Mol Med. 18:2092–2102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Achari C, Winslow S, Ceder Y and Larsson
C: Expression of miR-34c induces G2/M cell cycle arrest in breast
cancer cells. BMC Cancer. 14:5382014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
van der Deen M, Taipaleenmäki H, Zhang Y,
Teplyuk NM, Gupta A, Cinghu S, Shogren K, Maran A, Yaszemski MJ,
Ling L, et al: MicroRNA-34c inversely couples the biological
functions of the runt-related transcription factor RUNX2 and the
tumor suppressor p53 in osteosarcoma. J Biol Chem. 288:21307–21319.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Engelbrecht AM, Gebhardt S and Louw L: Ex
vivo study of MAPK profiles correlated with parameters of apoptosis
during cervical carcinogenesis. Cancer Lett. 235:93–99. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gao LJ, Gu PQ, Zhao W, Ding WY, Zhao XQ,
Guo SY and Zhong TY: The role of globular heads of the C1q receptor
in HPV 16 E2-induced human cervical squamous carcinoma cell
apoptosis is associated with p38 MAPK/JNK activation. J Transl Med.
11:1182013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen TP, Chen CM, Chang HW, Wang JS, Chang
WC, Hsu SI and Cho CL: Increased expression of SKP2 and
phospho-MAPK/ERK1/2 and decreased expression of p27 during tumor
progression of cervical neoplasms. Gynecol Oncol. 104:516–523.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Su PH, Lin YW, Huang RL, Liao YP, Lee HY,
Wang HC, Chao TK, Chen CK, Chan MW, Chu TY, et al: Epigenetic
silencing of PTPRR activates MAPK signaling, promotes metastasis
and serves as a biomarker of invasive cervical cancer. Oncogene.
32:15–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Meira DD, de Almeida VH, Mororó JS,
Nóbrega I, Bardella L, Silva RL, Albano RM and Ferreira CG:
Combination of cetuximab with chemoradiation, trastuzumab or MAPK
inhibitors: Mechanisms of sensitisation of cervical cancer cells.
Br J Cancer. 101:782–791. 2009. View Article : Google Scholar : PubMed/NCBI
|