Introduction
Lung cancer is the leading cause of death among all
cancer deaths (1). It has the
highest morbidity and mortality among all malignancies worldwide
(1,2). NSCLC accounts for 70–85% of all lung
cancers, and most cases are of advanced stage or metastatic
condition when diagnosed (3,4). As a
targeted therapy, EGFR tyrosine kinase inhibitors (EGFR-TKIs) have
been approved by the FDA for the treatment of advanced NSCLC since
2003 (5). EGFR-TKIs have produced
encouraging results by postponing tumor progression and prolonging
the progression-free survival (PFS) of advanced NSCLC patients for
approximately 5 months compared to platinum-based doublet
chemotherapy (6,7).
However, only 15% of NSCLC patients responded to TKI
(8), and clinical trials on
gefitinib or erlotinib failed in unselected patient populations, as
they were not able to significantly prolong patient overall
survival (OS) compared to traditional chemotherapy (9–11).
Based on known EGFR mutations, researchers eventually discovered
the association between TKI sensitivity and EGFR mutations
(12). Moreover, they also observed
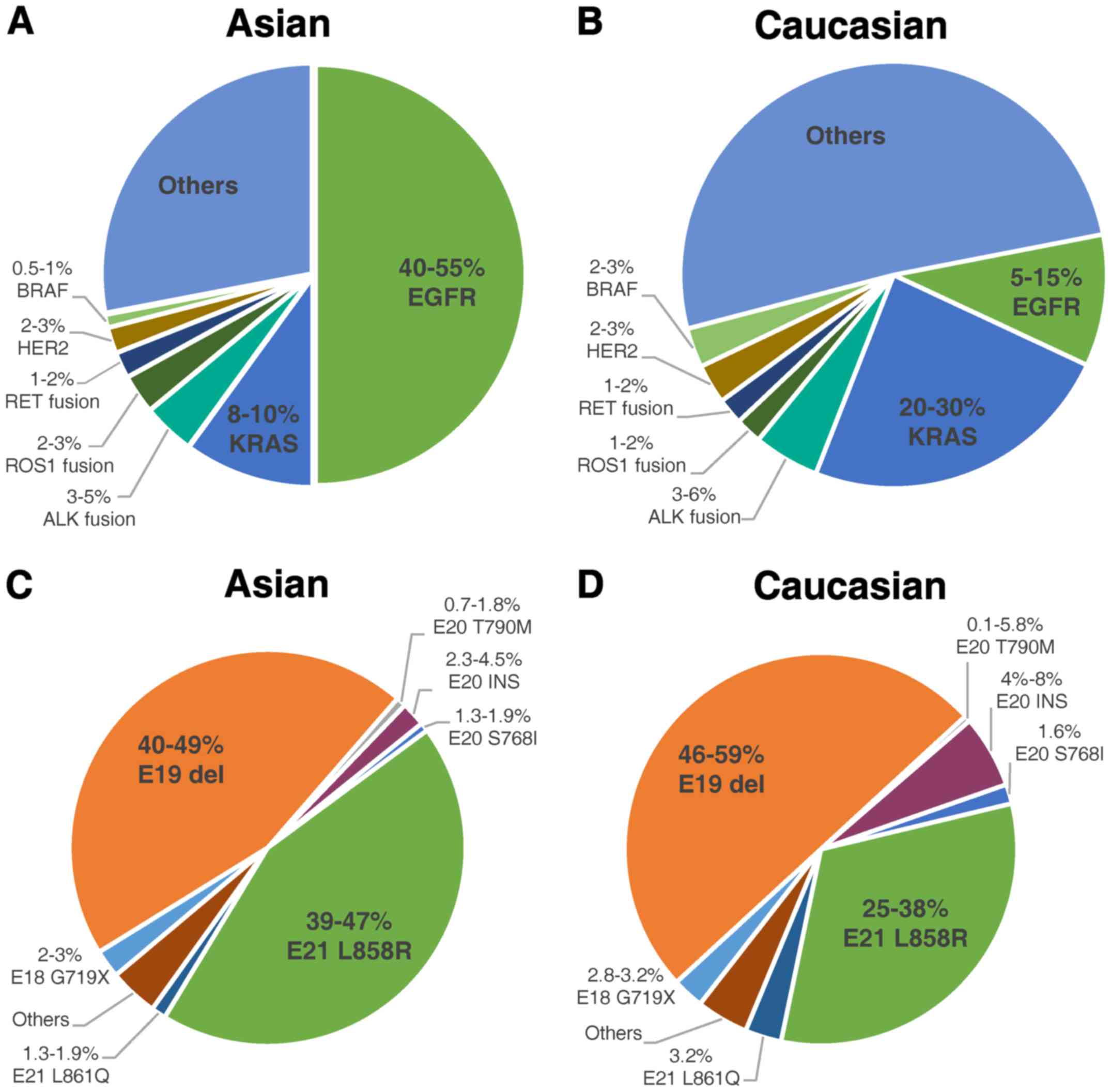
considerable ethnic differences in the frequencies of EGFR
mutations in NSCLC patients. EGFR mutations were detected in
approximately 50% of Asian patients with NSCLC, but only in 10% of
patients in the western world (13–15)
(Fig. 1). Two types of EGFR
mutations, in-frame deletions in exon 19 (Del19) and point
mutations in exon 21 causing a leucine-to-arginine substitution at
codon 858 (L858R), which are established to be definitely sensitive
to TKIs, comprise approximately 90% of all EGFR mutations (15–18).
The remaining 10% of EGFR mutations are defined as uncommon
mutations (Fig. 1). Therefore, for
NSCLC patients with uncommon EGFR mutations of unknown clinical
significance, it is dubious whether they can benefit from TKI
targeted therapy.
To answer this important question, there is an
urgent need to determine the clinical significance of uncommon
mutations in EGFR, particularly their sensitivity to TKIs. Although
they only account for a small proportion of patients with EGFR
mutations, they are still a large population due to the high
incidence of NSCLC.
It seems apparent that we could try to pattern the
methodologies of the Del19 and L858R mutations, which have been
proven to be sensitive, mostly by means of clinical randomized
controlled trials (RCTs) with large sample sizes. However, this is
not a practical approach for uncommon mutations, as cases of
uncommon mutations are too rare to conduct RCTs. Therefore, we have
to take advantage of basic studies or other types of clinical
studies. In other words, the ultimate problem is how we determine
the sensitivity of uncommon EGFR mutations. Then, we can make
clinical decisions regarding whether to apply TKI targeted therapy
to NSCLC patients with certain uncommon mutations.
The G719X mutation in EGFR refers to point mutations
that result in substitutions of the glycine at position 719 to
other residues, primarily alanine (G719A), cysteine (G719C) and
serine (G719S). The G719X mutation accounts for approximately 3%
among all EGFR mutations in both Asian and Caucasian populations
(14–22) (Fig.
1). It is the most commonly seen and most thoroughly studied
EGFR uncommon mutation, and it is considered a sensitive
mutation.
In this study, by reviewing studies of the G719X
mutation, we propose a comprehensive research model to explore the
laboratory and clinical characteristics of uncommon mutations
(Fig. 3). We also systematically
summarized the studies of the G719X mutation and discovered the
missing components to form a complete research system. The
conclusions regarding the G719X mutation would be much more
convincing if the evidence was complete. More importantly, the
research model can be generalized to direct researchers to explore
other uncommon mutations in patients with diseases associated with
gene mutations and to ascertain their pharmacologic properties
efficiently.
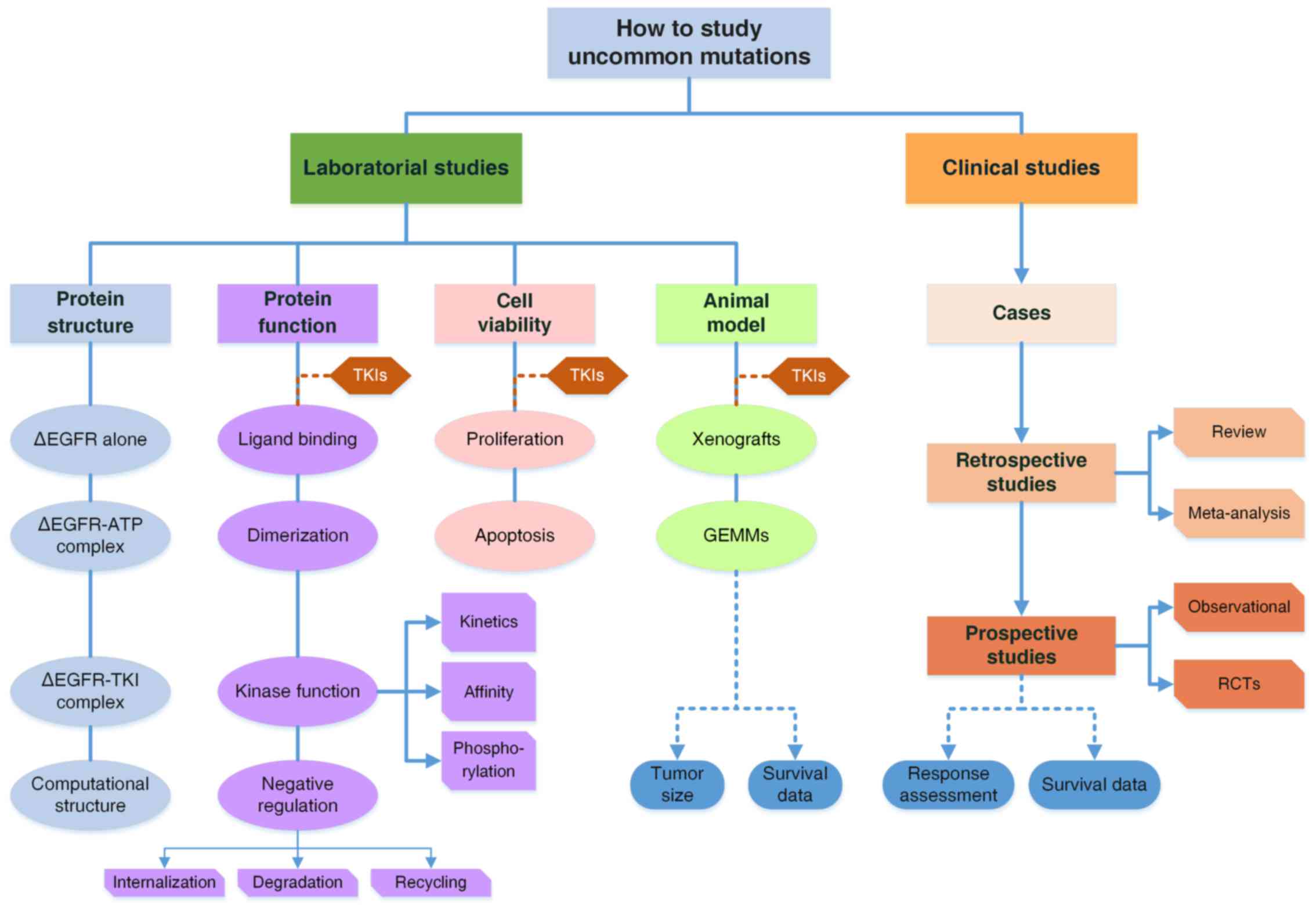 | Figure 3.A comprehensive model to study
uncommon mutations in EGFR. The system is comprised of both basic
and clinical studies. Clinical studies include three parts: Case
reports and series; Retrospective studies are reviews and
meta-analyses to combine the data of RR and survival of patients
with the G719X mutation; Prospective studies of observational ones
or randomized controlled trials. There are two major issues in
clinical studies: response to TKIs and survival data of patients.
Laboratory studies are organized from mutant protein structures to
functional changes, cell viability and animal models. Structural
analyses by crystal diffraction or computational simulation
determine the structures of mutant EGFR, EGFR-ATP complexes and
EGFR-TKI complexes, to elucidate functional changes. Functional
studies are categorized into four parts from the perspective of
EGFR activation, including ligand biding, dimerization, kinase
activity and downregulation, while the latter two are further
subdivided as illustrated. For cell viability, the two aspects of
proliferation and apoptosis are considered. Animal models involve
GEMMs and tumor-cell inoculated xenografts including PDX models,
which are discussed in terms of tumor sizes and animal survival.
TKI treatment is introduced into all experiments on three levels.
With basic and clinical studies integrated together, a complete
evidence system is formed to draw conclusions regarding the
pathogenic and pharmacological properties of uncommon mutations
with high reliability. ∆EGFR, mutant EGFR. |
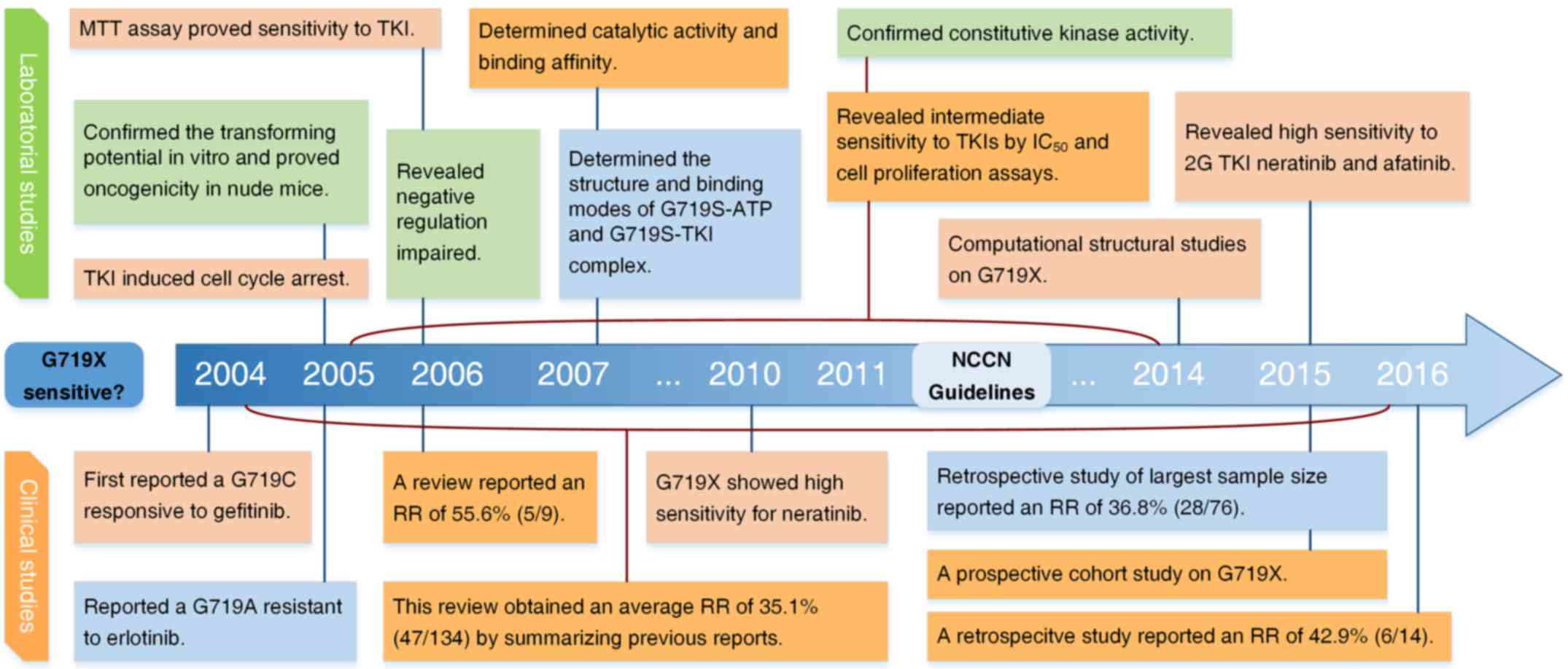
Current studies of the G719X mutation in
EGFR in NSCLC
The first observation of the G719X mutation in EGFR
in NSCLC patients was reported by Lynch et al in 2004
(23). The patient harbored a G719C
mutation and presented with partial response to gefitinib, with an
OS of 17.9 months. Based on studies conducted over the following
two years, the NCCN guidelines for NSCLC (version 2.2011) described
the G719X mutation in EGFR as associated with response to TKIs.
This conclusion was supported by subsequent investigations in
general. Herein, the studies of the G719X mutation are reviewed
comprehensively from both clinical and laboratory perspectives. The
history of studies of the G719X mutation in EGFR is presented in
Fig. 2.
Clinical studies of the G719X mutation in
NSCLC
Case reports and retrospective
studies
Since Lynch reported the first case (23), more and more cases have been
reported either in the form of case reports or retrospective
studies. However, most of them involved no more than ten patients.
Only one retrospective study by Chiu et al (42) in 2015 enrolled a relatively large
sample size of 76 patients with the G719X mutation, of which 28
responded to TKIs, indicating a response rate (RR) of 36.8%. To
overcome the limitation of sample size, we summarized all of these
studies and combined the results to obtain an average RR. We
enrolled 22 relative studies from 2004 to 2016 and excluded all
reviews to avoid possible data overlap (18,23–43).
Then, we had a total of 134 G719X patients, of which 47 patients
responded to 1st generation EGFR-TKIs (Table I). The average RR is 35.1% (47/134),
indicating that G719X is a mutation of intermediate sensitivity,
which is in accordance with previous reviews (16,44–46)
(Table II).
 | Table I.Summary of studies of G719X responses
to 1G-TKIs.a |
Table I.
Summary of studies of G719X responses
to 1G-TKIs.a
| Study | Mutation | TKI | Total | Response | RR/% | Sensitivity | Ref. |
|---|
| Lynch 2004 | G719C | G | 1 | 1 | 100 | Sensitive | 23 |
| Han 2005 | G719A | G | 2 | 1 | 50 | Intermediate | 24 |
| Takano 2005 | G719X | G | 2 | 1 | 50 | Intermediate | 25 |
| Eberhard 2005 | G719A | E | 1 | 0 | 0 | Resistant | 26 |
| Janne 2006 | G719C | G | 1 | 1 | 100 | Sensitive | 27 |
| Ichihara 2007 | G719X | G | 1 | 0 | 0 | Resistant | 28 |
| Pallis 2007 | G719D | G | 1 | 0 | 0 | Resistant | 29 |
| Sequist 2008 | G719A | G | 1 | 0 | 0 | Resistant | 30 |
| Wu 2008 | G719A | E/G | 2 | 1 | 50 | Intermediate | 31 |
| Wu 2011 | G719X | E/G | 8 | 4 | 50 | Intermediate | 32 |
| De Pas 2011 | G719S | E | 1 | 1 | 100 | Sensitive | 33 |
| Takahashi 2012 | G719A | G | 1 | 0 | 0 | Resistant | 34 |
| Lee 2013 | G719A | E | 1 | 0 | 0 | Resistant | 35 |
| Umekawa 2013 | G719A | E | 1 | 0 | 0 | Resistant | 36 |
| Locatelli-Sanchez
2013 | G719A | E/G | 1 | 1 | 0 | Resistant | 37 |
| Keam 2014 | G719A | G | 1 | 0 | 0 | Resistant | 38 |
| Beau-Faller
2014 | G719X | E/G | 10 | 1 | 10 | Resistant | 18 |
| Guan 2014 | G719A | E | 1 | 0 | 0 | Resistant | 39 |
| Watanabe 2014 | G719X | G | 3 | 0 | 0 | Resistant | 40 |
| Fukihara 2014 | G719A | E/G | 4 | 1 | 25 | Intermediate | 41 |
| Chiu 2015 | G719X | E/G | 76 | 28 | 36.8 | Intermediate | 42 |
| Xu 2016 | G719X | E/G/I | 14 | 6 | 42.9 | Intermediate | 43 |
| Total |
|
| 134 | 47 | 35.1 | Intermediate |
|
 | Table II.Summary of clinical studies of the
G719X mutation in EGFR. |
Table II.
Summary of clinical studies of the
G719X mutation in EGFR.
| Research type | Article | Mutation | TKI | Total cases | Response | RR/% | Sensitivity | Ref. |
|---|
| Retrospective
studies |
|
|
|
|
|
|
|
|
| Summary
of cases | This article | G719X | E/G | 134 | 47 | 35.1% | Intermediate | Table I |
|
Reviews | Mistudomi 2006 and
2007 | G719X | E/G | 9 | 5 | 55.6 | Intermediate | 44,45 |
|
| Kobayashi 2015 | G719X | G | 3 | 2 | 66.7 | Intermediate | 16 |
|
| Klughammer
2016 | G719X | E | 2 | 1 | 50 | Intermediate | 46 |
|
Meta-analysis | Not found |
|
|
|
|
|
|
|
|
| Prospective
studies |
|
|
|
|
|
|
|
|
|
| Observational | Arrieta 2015 | G719X | E/G/A | 11 | NAa | NA | Not known | 47 |
|
Clinical trials | Not found |
|
|
|
|
|
|
|
Reviews and meta-analyses
Four reviews concerned responses of G719X to TKIs,
with response rates of 50–66.7% (16,44–46)
(Table II). Still, in these
reviews, the numbers of cases were too small to be convincing. No
meta-analyses were found.
Prospective studies
Due to the rarity of uncommon mutations, we could
not enroll enough patients to conduct a prospective randomized
controlled trial. Observational studies are probably a feasible way
to investigate the sensitivity of uncommon mutations in a
prospective manner. Arrieta et al analyzed 188 NSCLC
patients in their cohorts and found 11 patients with the G719X
mutation who received TKIs, including a single G719X mutation and
complex mutations. Although G719X was not discussed separately,
they found the rare mutation group to be intermediately sensitive
with an RR of 32.4% (47).
Summary of clinical studies
All clinical studies enrolled are summarized in
Table II. As stated above, because
of limitations in sample size, it is not adequately convincing to
determine the sensitivity of the G719X mutation based only on
clinical studies. Given the circumstances, it is necessary to seek
supporting evidence from laboratory studies. With both clinical and
basic studies to form a complete evidence system and logic network,
we could have sufficient cause to consider G719X a sensitive
mutation.
Laboratory studies of the G719X
mutation in EGFR in NSCLC
In general, the laboratory studies mainly focused on
alterations caused by the G719X mutation, regarding the protein
structure, protein function, cell viability and animal experiments.
Thus, the laboratory studies were reviewed in these four
perspectives.
Functional alterations
The activation of EGFR is initiated after binding to
its ligand, epidermal growth factor (EGF) or transforming growth
factor-α (TGF-α). The receptor changed its conformation and then
dimerized with another ligand-bound EGFR or other ErbB family
members to form homodimers or heterodimers, respectively. The dimer
harbored kinase activity and would phosphorylate itself at specific
sites (48,49), which could act as catalytic sites to
activate downstream signaling pathways, such as MAPK or PI3K/Akt,
by phosphorylation of the corresponding molecules. Afterwards, the
activated EGFRs were internalized into the cell plasma by
endocytosis, and then they were either recycled onto the cell
membrane or degraded by fusion with lysosomes (50). This is one way of negative
regulation in EGFR signaling pathway. A series of studies revealed
the influences of the G719X mutation and TKI treatment on all of
the functional processes.
Ligand binding and dimerization
Choi et al explored how the G719S mutation
affected ligand binding using a 125I-labelled EGF
binding assay. Moreover, they also used antibodies against the EGFR
extracellular region to label EGFR, and they observed dimerization
of receptors with immunofluorescence microscopy. No differences
were discovered in ligand binding or dimerization between G719S
mutants and wild-type (wt), while they indeed observed
EGF-independent dimerization of EGFRs in Del19 and L858R mutants
(51).
Kinase activity
Greulich et al systematically investigated
the kinase activity of G719S mutants (52). Using immunoblotting, they discovered
the ligand-independent constitutive phosphorylation activity on
both the receptor itself and on downstream signal molecules, such
as Shc, STAT3 and Akt (52). Chen
and Choi further compared the auto-phosphorylation levels of G719S
with that of Del19 and L858R. They found that the
auto-phosphorylation level of G719S was lower, indicating that the
oncogenicity of G719S was weaker than that of the other two common
mutants (51,53). Subsequent studies further confirmed
the conclusions using western blotting and immunofluorescence
staining (51,53–57).
As the transforming potential of G719X was
determined, the question arose as to what extent EGFR-TKI can
inhibit the uncontrolled kinase activity of G719X mutant EGFR.
Jiang et al investigated gefitinib in their kinase assay of
G719X and found that gefitinib was able to inhibit the
auto-phosphorylation of G719S in a dose-dependent manner. However,
compared with L858R, G719S required a higher concentration of
gefitinib (54). Their conclusions
were further validated by subsequent studies using other techniques
(53,57–59).
In general, based on the studies mentioned above, G719X was found
to have moderate oncogenicity and intermediate sensitivity to TKIs
regarding kinase function.
Kinetics and binding affinity
Why would the G719X mutation cause weaker
oncogenicity and lower sensitivity to TKIs than L858R? Is it
because the mutation affected the catalytic properties of the
kinase and the interactions between EGFR and TKIs? As TKIs are
competitive inhibitors of ATP, studies comparing the binding
affinities of TKI-EGFR complexes with ATP-EGFR complexes may answer
these questions.
Yun et al investigated the kinetics and
affinities of G719X mutants using a continuous colorimetric assay
and a fluorescence-quenching assay. The mutation in the tyrosine
kinase domain was found to dramatically elevate catalytic activity
by approximately 50-fold in L858R and 10-fold in G719S compared to
wild-type (60). In terms of
affinities, they determined the dissociation constants (Kd) of
EGFR-gefitinib complexes and EGFR-AMPPNP (an analogue of ATP)
complexes. Although the G719S mutation decreased the affinity to
gefitinib, it lowered the affinity to ATP much more, indicating
that the inhibiting potential (KdTKI/KmATP)
of gefitinib was 5-fold stronger than wt, while L858R was
approximately 100 times stronger (60). This might explain why G719S mutants
were less sensitive to gefitinib than L858R mutants.
Negative regulation
There are mainly two ways to negatively regulate the
activated wild-type EGFR after the activated receptor has done its
job to trigger the downstream signaling. Once internalized by
endocytosis, the receptors would either be recycled back to the
cell membrane or be degraded by fusion with lysosomes mediated by
ubiquitination (50,53). Along with persistent positive
activation, impaired downregulation might take part in the
oncogenicity of G719X mutants as well.
Few studies focused on negative
regulation
Chen et al discovered that the G719S mutants
were refractory to ubiquitination and had more sustained tyrosine
phosphorylation than wild-type (53). A similar phenomenon was observed in
Del19 and L858R mutants (61).
Furthermore, internalization was also found to be impaired in Del19
mutants (61).
Structure determination
The determination of kinetic and binding parameters
provided clues to help us understand the mechanisms of the
pathogenic and pharmacological effects of the mutation.
Nonetheless, to further elucidate why the mutation affects
oncogenicity and sensitivity requires determination of the
structures of mutated EGFRs and EGFR-TKI complexes.
Yun et al determined the 3D structures of the
G719S-AMPPNP complex by crystal diffraction (60). The comparison of the structures
between G719S and wild-type EGFR indicated that the G719S mutation
destabilized the inactive conformation and thus promoted the active
conformation of the kinase (60).
This explained the oncogenic potential of G719S mutants on the
basis of structures.
The inactive conformation of EGFR requires the
C-helix to be rotated outward to be displaced from the active site.
The glycine residue at position 719 interacts with several
hydrophobic residues flanking L858, including F723, L747 and L862.
Packing together with the N-terminal portion of the activation
loop, the hydrophobic cluster changed into a helical turn, causing
the C-helix to rotate outward by steric hindrance. Therefore, any
substitution of a glycine residue at position 719 would sabotage
the stable hydrophobic interactions, which are essential for the
receptor to adopt the inactive conformation (60).
Furthermore, they also elucidated the binding mode
of the G719S-gefitinib complex. Unfortunately, they were unable to
resolve the differences in the binding modes among G719S-gefitinib,
L858R-gefitinib and wt-gefitinib complexes, thus failing to explain
the different binding affinities of various EGFRs and the distinct
sensitivities to TKIs (61).
Another structural study of G719X was performed by
Doss et al in 2014, with computational structure simulation,
a new approach to study the structures of proteins. They simulated
the real-time conformational alterations in G719S mutants to
discover that the G719S mutation increased the distance between
residues L718 and G796, forming a wider opening for TKIs to get
into the ATP-binding pocket than wild-type, indicating that this
mutation should respond to TKIs (62). However, they did not include Del19
or L858R mutants in their study.
Cell viability
Constitutive kinase activity of G719X mutants will
persistently activate downstream signaling, resulting in
EGFR-signaling-pathway-mediated cell proliferation in a
ligand-independent manner. Will we observe uncontrolled cell
proliferation when the G719X mutation is introduced into certain
cell lines? And to what extent can TKI inhibit cell
proliferation?
Elevated cell viability in G719S transformed NIH-3T3
and Ba/F3 cell lines were observed in several studies, but it was
lower than that of Del19 and L858R mutants (51,52,56).
Moreover, it can be abrogated by gefitinib, yet is somehow more
resistant than L858R (52). To be
more quantitative, the 50% inhibiting concentration
(IC50) of gefitinib in various mutants was further
measured in series of studies. The IC50 of gefitinib in G719S
mutants was between that of gefitinib in wild-type cells and
Del19/L858R mutants, implying an intermediate sensitivity of G719S
(16,52,54,55,57,63).
The inhibition of cell viability by TKIs was
observed, but the mechanisms for the abrogation remained to be
elucidated. Jiang et al focused on the impact on the cell
cycle of the mutant cells resulting from TKI treatment. Using FACS
and immunoblotting, they found that gefitinib induced cell cycle
arrest in the G1 phase by downregulating the level of
D-type cyclins and CDK4 in G719S mutant Ba/F3 cell lines (54). However, they detected no apoptotic
cells in G719S mutants (54), while
apoptosis induced by gefitinib was observed in L858R mutants
(48,64).
In conclusion, TKIs inhibit the proliferation of
transformed cells to various extents in different mutants. The
transformed cells with the G719S mutation exhibited intermediate
sensitivity to TKI inhibition compared with Del19 and L858R
mutants.
Animal models
The cell culture experiments are still too far from
the authentic situation, thus it is necessary to investigate the
role of the G719X mutation in animal models and to determine
whether TKI treatment would be effective in animals with tumors
driven by the G719X mutation.
Greulich et al injected the G719S transformed
cells into immuno-compromised mice and observed tumorigenesis
(52). Moreover, tumor size varied
among the different mutation groups. The average diameter of tumors
in the G719S group was half of that in the L858R group (52). They confirmed the oncogenic
potential of the G719S mutation in an animal model; however, their
experiment might be more complete if they took the survival
condition of the inoculated animals and TKI treatment into
account.
Summary of basic studies
The laboratory studies of G719X are summarized in
Table III, with a brief overview
of the results and the corresponding methods of the experiments.
The high level of consistency in the results of the laboratory
studies including protein function experiments, cell viability
experiments and animal experiments further authenticate the
oncogenicity and sensitivity of the G719X mutation. However, we
still need a better understanding of the structure leading to
intermediate sensitivity to further complete this research
system.
 | Table III.Summary of laboratory studies of the
G719X mutation in EGFR. |
Table III.
Summary of laboratory studies of the
G719X mutation in EGFR.
| Research type | Conclusion | Main method | Ref. |
|---|
| Protein
structure | Elucidated the
mechanism of constitutive kinase activity of G719S. | Crystal
diffraction | 60 |
|
| Determined the
binding mode of TKI-G719S complex: same as wt and L858R. | Crystal
diffraction | 60 |
|
| Computational
structural studies revealed that G719 caused TKI to move closer to
the binding site and TKI easier to get into the ATP-binding
pocket. | Molecular dynamic
simulation | 62 |
| Protein
function |
|
|
|
| Ligand
binding and dimerization | Basically the same
between G719S and wt, while EGF-independent dimerization found in
Del19 and L858R. |
125I-labelled EGF binding
assay | 51 |
|
Kinetics | Compare the
catalytic activity among EGFRs: L858R>G719S>wt. | Continuous
colorimetric assay and fluorescence-quenching assay | 60 |
|
Affinity | Compare the binding
affinity for TKI versus ATP to EGFRs: L858R>G719S>wt. | Continuous
colorimetric assay | 60 |
| Kinase
activity | Confirmed
constitutive kinase activity in autophosphorylation and downstream
signaling: Wt<G719X<L858R/Del19. | Immunoblotting | 52,54,55 |
|
|
| Western
blotting | 51,53,56 |
|
|
| Immunofluorescence
staining | 57 |
| TKI
inhibition | Determined
IC50 of TKI able to block constitutive kinase activity
of mutants: wt>G719X>L858R/Del19 | Immunoblotting | 54 |
|
|
| Western
blotting | 53 |
|
|
| BRET assay | 58 |
|
|
| Continuous
colorimetric assay | 59 |
|
|
| YFP-EGFR-ICD
relocation assay | 57 |
|
Negative regulation | Negative regulation
of kinase activity is impaired in G719S. | Western
blotting | 53 |
| Cell
proliferation | Confirmed the
transforming potential of G719S mutants. | Colony formation
assay | 52 |
|
|
| 3H
thymidine incorporation assay | 51 |
|
|
| eGFP+ cell
FACS | 56 |
|
| G719X transformed
cells showed intermediate sensitivity to TKI. | Colony formation
assay | 52,56 |
|
|
| Cell viability
assay by trypan/MTT/MTS staining | 54,55,56,63 |
|
|
| Colorimetric
assay | 16 |
|
| G719X transformed
cells show strong sensitivity to TKI. | Cell viability
assay by MTT staining | 53 |
|
| TKI induced cell
cycle arrest in G719S transformed cells. | FACS and
immunoblotting | 54 |
| Animal model | Injection of G719S
transformed cells cause tumorigenesis. | Nude mice
injection | 52 |
A comprehensive model for studying uncommon
mutations
Based on the reviewed studies of the G719X mutation
in EGFR in NSCLC, we propose a systematic model to investigate the
pathogenic and pharmacological characteristics of an uncommon
mutation at both the laboratory and clinical levels (Fig. 3). Due to the small number of
patients with uncommon mutations, it is hardly possible to enroll
enough patients to carry out RCTs, which would provide the most
convincing evidence for the clinical features of a particular
uncommon mutation. For this reason, we need a comprehensive
experimental system.
The laboratory experiments are comprised of studies
of protein structure, studies of functional alterations, cell
viability assays and animal experiments, providing an understanding
of the features of the mutations from the level of molecules to
cells and then to animal models, microcosmically to
macroscopically. In terms of clinical studies, although it is not
feasible to perform RCTs, prospective observational studies might
be a possible way to enroll more patients. On the other hand,
summarizing cases retrospectively enables us to obtain the average
response rates and survival data from a relatively large sample.
The results would be more reliable if we enrolled adequate studies
of low heterogeneity to conduct meta-analyses.
This model was organized logically. Structure
determination elucidates the mechanisms of the functional
alterations and pharmacologic effects of TKIs, which are further
verified in cells and animals, and even in patients in clinical
settings, thus providing sufficient evidence to determine the
oncogenicity and sensitivity to TKI of uncommon mutations in EGFR
in NSCLC.
Discussion
Basic studies of G719X are far from
enough
Laboratory studies of G719X have included almost all
processes in the EGFR signaling pathway; however, they are still
far from being complete or satisfactory.
Dimerization of G719S differs from that of Del19 and
L858R mutants in terms of the dependence on its ligand EGF,
indicating a lower activating level of G719S. This may explain its
lower oncogenicity. However, it remains to be investigated whether
mutations alter the mode of dimerization and how TKI will affect
ligand binding and dimerization.
Regarding negative regulation, the effects of TKI on
impaired negative regulation were not explored. If TKI treatment
could reverse the impairment of negative regulation by the G719S
mutation, this could be another explanation of the sensitivity to
TKIs. However, we found no studies regarding the internalization or
recycling of G719S mutated EGFR. In addition, the negative
regulation of a pathogenic mutation might be a new target for drug
development.
For structural studies, apart from real 3D structure
determination by crystal diffraction, computational structure
studies are another more economical and efficient way to study the
structures of proteins and the effects of mutations on protein
structure. Computational structure studies can be used to determine
the structures of mutants using real-time computation and can
simulate the conformational alterations caused by specific
mutations without having to purify the mutated proteins and obtain
crystals. However, neither type of study explained the lower
sensitivity of G719X compared to Del19 and L858R mutants.
The structural analyses by either crystal
diffraction or computational structural studies provide a
structural basis for oncogenicity and sensitivity to TKIs.
Moreover, fully understanding the structures of mutants and the
binding features of drug-mutant complexes will help us to predict
the sensitivity of a mutation to a particular potential targeted
drug, and our ultimate purpose is to design a tailored targeted
drug for people with specific gene mutation-related diseases using
computational biology and bioinformatics. These are important goals
of precision medicine.
Cell viabilities are determined by both cell
proliferation and apoptosis. Tumorigenesis and response to TKIs
could also be studied according to these two measures.
Proliferation mainly refers to experiments concerning the cell
cycle, and apoptosis is basically about the caspase-mediated
apoptotic pathway. The studies of proliferation are relatively
clear; however, few studies focused on apoptosis in G719X mutants.
The L858R mutation was found to enable cells to escape from
apoptosis (48), yet no similar
studies on G719X were found. Furthermore, G719X and L858R mutants
showed distinct responses to TKIs in terms of apoptosis (48,54,64),
providing a possible mechanism for the difference in their
sensitivities. Unlike common mutations, there are no cell lines
bearing a single G719X mutation that are established for research
on cell viability, while plenty of cell lines harboring Del19 or
L858R mutations are available to be used directly, such as H3255
and PC-9 (65–68). This might be the reason for the
scarcity of relative studies of the G719X mutation concerning cell
viability. Establishing cell lines harboring uncommon mutations
would provide a foundation for studying these mutations.
For xenograft-type models, apart from these
transformed cell-inoculated animals, the patient-derived xenograft
(PDX) model, also known as ‘Mouse Avatars’, is a new type of animal
model used to study neoplasms (69). The PDX model has demonstrated
substantial clinical potential in predicting drug sensitivity,
including sensitivity to TKI targeted therapies. As the tumor
sample is obtained from the actual patient, it is the closest
representation of the individual's authentic situation.
Another type of animal model is genetically
engineered mouse model (GEMM). Distinct from xenografts, they are
generated using genetic engineering techniques to harbor specific
mutations for driver mutations. To the best of our knowledge, there
are no GEMMs for the G719X mutation in EGFR available for basic or
clinical studies. Gazdar et al discussed the comparison
between these two types (70).
Is G719X a TKI-sensitive mutation
The NCCN guidelines for NSCLC first mentioned G719X
as significantly associated with response to TKIs in 2011 (version
2.2011) and then referred to G719X as a drug-sensitive mutation in
2012 (version 2.2012), mainly based on laboratory studies of
Greulich et al (52) and
clinical studies of Lynch et al and Han et al
(23,24), which each reported a single case of
G719X with partial response to gefitinib. The newest NCCN
guidelines for NSCLC (version 4.2016) (71) remained almost the same. In a word,
the G719X mutation in EGFR is basically considered to be sensitive
according to the NCCN guidelines.
Nevertheless, based on subsequent clinical studies
involving more cases and a series of laboratory experiments, we
have drawn the conclusion that it would be more accurate to define
the G719X mutation as intermediately sensitive to first-generation
TKIs. The high heterogeneity of NSCLC might explain the ambiguity
of the G719X mutation in terms of sensitivity to TKIs.
Moreover, we still need to integrate some deeper and
broader studies of G719X into the model to form a complete evidence
system in order to be more confident about its sensitivity. Based
on the research model, the following issues remain to be
investigated. In the laboratory, the conformational differences
between complexes of TKIs with various mutants requires more
analysis to explain the differences in sensitivity between mutants
on the basis of 3D structure. The influences of the G719X mutation
and TKI treatment on negative regulation are far from clear. In
terms of cell viability, more studies are required to determine
whether G719X mutants could escape from apoptosis such as L858R
mutants. In terms of animal experiments, we still need more data
regarding tumor sizes and survival data from TKI treatment on
animals with tumors driven by the G719X mutation or in PDX models
from patients with the G719X mutation. Still, there is an urgent
need to establish cell lines harboring the G719X mutation. At the
clinical level, the major problem and difficulty lies in the sample
sizes of the corresponding studies. It would be feasible to
systematically review the clinical data in large-scale trial
projects to retrieve information about responses and survival of
patients with the G719X mutation (72). Based on these data, we could further
conduct a meta-analysis on the sensitivity of the G719X
mutation.
Complex mutations
Another interesting phenomenon to be noted is that
G719X often occurs along with other mutations in EGFR, especially
with S768I and L861Q with frequencies of 24.5 and 8.2% in all G719X
mutations (15). They were also
found to co-exist with mutations in other genes, for instance KRAS,
BRAF and PIK3CA (15). It is
referred to as complex mutation. Other uncommon mutations also tend
to occur in the form of complex mutations. A possible explanation
is that these uncommon mutations harbor inadequate tumor-driving
ability and, therefore, must co-occur with another mutation to
initiate tumorigenesis. Compared with the G719X single mutation,
complex mutations are scarcer and their sensitivities to TKI are
more obscure. Our research model provides practical approaches for
studying these complex mutations.
Second generation EGFR-TKIs and
G719X
New generations of EGFR-TKIs are developed to
improve selectivity and efficacy and, thus, to reduce toxicity.
First generation-TKIs (1G-TKIs) refer to reversible TKIs such as
gefitinib, erlotinib and icotinib, while afatinib, dacomtinib and
neratinib are categorized as irreversible 2G-TKIs (73,74).
3G-TKIs, including AZD-9291 (Osimertinib) and CO-1686 (75), are highly specific irreversible
inhibitors for mutated receptors only, which are primarily used to
target the secondary resistant mutation T790M (75,76).
The G719X mutation showed intermediate sensitivity to 1G-TKIs;
however, it was notable that 2G-TKIs demonstrated markedly high
efficacy in patients with the G719X mutation. Although neratinib
showed barely satisfactory efficacy in treating NSCLC patients in
its phase 2 clinical trial (77),
it exhibited great potential in patients with the G719X mutation.
Three out of four patients with the G719X mutation exhibited a
partial response with the tumor shrinking more than 50% in
diameter, and one exhibited stable disease, indicating an RR of 75%
and a DCR (disease control rate) of 100% (77). In addition, Yang et al
reviewed patients with the G719X mutation who were treated with
afatinib in the LUX-LUNG trial series and found that the overall
response rate was 77.8% (14/18) (78). In laboratory studies, the G719A
mutation exhibited higher sensitivity than Del19 in terms of both
kinase activity and cell viability (16).
Although 2G-TKIs are not widely used clinically due
to their high toxicity, some studies did explore their potential in
treating patients with mutations in exon 18, especially the G719X
mutation. These findings also shed light on TKI selection in
patients with uncommon mutations. Although some types of TKIs
failed to treat NSCLC patients with common sensitive mutations,
they might have great potential against uncommon mutations.
Innovations and limitations
We systematically reviewed studies of G719X from
both laboratory and clinical settings, and we sorted out the
history of these studies. The basic studies are summarized
regarding conclusions and corresponding experimental techniques. In
terms of the clinical studies, we summarized 22 studies (18,23–43)
investigating the response of the G719X mutation and determined an
average response rate based on a relatively larger sample size. To
the best of our knowledge, this is one of the most comprehensive
systematic reviews of the G719X mutation in EGFR in NSCLC.
Furthermore, the comprehensive research model we proposed provides
researchers with a practical approach to determine the clinical
significance of uncommon mutations of NSCLC or other diseases
associated with gene mutation.
Nevertheless, there are some limitations of this
review. Although we have included considerable studies of G719X in
our review, it is inevitable that we may still have omitted some
important studies. Moreover, due to the heterogeneity of the
studies that we enrolled to calculate the average response rate,
there might be a bias in the result. The heterogeneities of the
studies mainly consist of demographic features, stage and
histological classification of the tumor, different treatment lines
and inconsistency in the criteria for response assessment.
Additionally, the exclusion of 3 studies of which the original data
were inaccessible in our calculation might also cause bias
(47,79,80).
Although we combined several studies to increase the sample size,
it was still far from enough to draw a convincing conclusion on the
sensitivity. By properly combining the response and survival data
extracted from several large-scale trial projects, we can obtain
more accurate and persuasive results.
Acknowledgements
We would like to thank Jie Zhou, Dongdong Wang, Jun
Chen, Yuan Xu and Chao Guo from Peking Union Medical College
Hospital for their generous help. We would also like to express our
gratitude to American Journal Experts for their language
assistance.
Glossary
Abbreviations
Abbreviations:
|
Del19
|
in-frame deletions in exon 19 of
EGFR
|
|
EGF
|
epidermal growth factor
|
|
EGFR
|
epidermal growth factor receptor
|
|
G719X
|
point mutations that result in
substitutions of the glycine at position 719 to other residues
|
|
L858R
|
point mutations that result in
substitutions of the leucine at position 858 to arginine
|
|
NSCLC
|
non-small cell lung cancer
|
|
RCT
|
randomized controlled trial
|
|
RR
|
response rate
|
|
TKI
|
tyrosine kinase inhibitor
|
|
wt
|
wild-type
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parums DV: Current status of targeted
therapy in non-small cell lung cancer. Drugs Today (Barc).
50:503–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herbst RS, Fukuoka M and Baselga J:
Gefitinib - a novel targeted approach to treating cancer. Nat Rev
Cancer. 4:956–965. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: North-East Japan Study Group: Gefitinib or chemotherapy for
non-small-cell lung cancer with mutated EGFR. N Engl J Med.
362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee CK, Wu YL, Ding PN, Lord SJ, Inoue A,
Zhou C, Mitsudomi T, Rosell R, Pavlakis N, Links M, et al: Impact
of specific epidermal growth factor receptor (EGFR) mutations and
clinical characteristics on outcomes after treatment with EGFR
tyrosine kinase inhibitors versus chemotherapy in EGFR-mutant lung
cancer: A meta-analysis. J Clin Oncol. 33:1958–1965. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Miller VA, Kris MG, Shah N, Patel J,
Azzoli C, Gomez J, Krug LM, Pao W, Rizvi N, Pizzo B, et al:
Bronchioloalveolar pathologic subtype and smoking history predict
sensitivity to gefitinib in advanced non-small-cell lung cancer. J
Clin Oncol. 22:1103–1109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maruyama R, Nishiwaki Y, Tamura T,
Yamamoto N, Tsuboi M, Nakagawa K, Shinkai T, Negoro S, Imamura F,
Eguchi K, et al: Phase III study, V-15-32, of gefitinib versus
docetaxel in previously treated Japanese patients with
non-small-cell lung cancer. J Clin Oncol. 26:4244–4252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee CK, Brown C, Gralla RJ, Hirsh V,
Thongprasert S, Tsai CM, Tan EH, Ho JC, Chu T, Zaatar A, et al:
Impact of EGFR inhibitor in non-small cell lung cancer on
progression-free and overall survival: A meta-analysis. J Natl
Cancer Inst. 105:595–605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kris MG: How today's developments in the
treatment of non-small cell lung cancer will change tomorrow's
standards of care. Oncologist. 10 Suppl 2:23–29. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hirsch FR and Bunn PA Jr.: EGFR testing in
lung cancer is ready for prime time. Lancet Oncol. 10:432–433.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kohno T, Nakaoku T, Tsuta K, Tsuchihara K,
Matsumoto S, Yoh K and Goto K: Beyond ALK-RET ROS1 and other
oncogene fusions in lung cancer. Transl Lung Cancer Res. 4:156–164.
2015.PubMed/NCBI
|
|
15
|
Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H,
Ren-Heidenreich L, Shi B, Ren H, Chu X, et al: Coexistence of EGFR
with KRAS or BRAF or PIK 3CA somatic mutations in lung cancer: A
comprehensive mutation profiling from 5125 Chinese cohorts. Br J
Cancer. 110:2812–2820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi Y, Togashi Y, Yatabe Y, Mizuuchi
H, Jangchul P, Kondo C, Shimoji M, Sato K, Suda K, Tomizawa K, et
al: EGFR exon 18 mutations in lung cancer: Molecular predictors of
augmented sensitivity to afatinib or neratinib as compared with
first- or third-generation TKIs. Clin Cancer Res. 21:5305–5313.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han B, Tjulandin S, Hagiwara K, Normanno
N, Wulandari L, Konstantinovich L Konstantin, Hudoyo A, Ratcliffe
M, McCormack R and Reck M: Determining the prevalence of EGFR
mutations in Asian and Russian patients (PTS) with advanced
non-small-cell lung cancer (aNSCLC) of adenocarcinoma (ADC) and
non-ADC histology: Ignite study. Ann Oncol. 26 Suppl 1:i29–i30.
2015. View Article : Google Scholar
|
|
18
|
Beau-Faller M, Prim N, Ruppert AM,
Nanni-Metéllus I, Lacave R, Lacroix L, Escande F, Lizard S, Pretet
JL, Rouquette I, et al: Rare EGFR exon 18 and exon 20 mutations in
non-small-cell lung cancer on 10 117 patients: A multicentre
observational study by the French ERMETIC-IFCT network. Ann Oncol.
25:126–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
An SJ, Chen ZH, Su J, Zhang XC, Zhong WZ,
Yang JJ, Zhou Q, Yang XN, Huang L, Guan JL, et al: Identification
of enriched driver gene alterations in subgroups of non-small cell
lung cancer patients based on histology and smoking status. PLoS
One. 7:e401092012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gahr S, Stoehr R, Geissinger E, Ficker JH,
Brueckl WM, Gschwendtner A, Gattenloehner S, Fuchs FS, Schulz C,
Rieker RJ, et al: EGFR mutational status in a large series of
Caucasian European NSCLC patients: Data from daily practice. Br J
Cancer. 109:1821–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Au JS, Thongprasert S, Srinivasan
S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G and Yang PC: A
prospective, molecular epidemiology study of EGFR mutations in
Asian patients with advanced non-small-cell lung cancer of
adenocarcinoma histology (PIONEER). J Thorac Oncol. 9:154–162.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Skov BG, Høgdall E, Clementsen P, Krasnik
M, Larsen KR, Sørensen JB, Skov T and Mellemgaard A: The prevalence
of EGFR mutations in non-small cell lung cancer in an unselected
Caucasian population. APMIS. 123:108–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han SW, Kim TY, Hwang PG, Jeong S, Kim J,
Choi IS, Oh DY, Kim JH, Kim DW, Chung DH, et al: Predictive and
prognostic impact of epidermal growth factor receptor mutation in
non-small-cell lung cancer patients treated with gefitinib. J Clin
Oncol. 23:2493–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takano T, Ohe Y, Sakamoto H, Tsuta K,
Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine
I, et al: Epidermal growth factor receptor gene mutations and
increased copy numbers predict gefitinib sensitivity in patients
with recurrent non-small-cell lung cancer. J Clin Oncol.
23:6829–6837. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eberhard DA, Johnson BE, Amler LC, Goddard
AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson
DH, et al: Mutations in the epidermal growth factor receptor and in
KRAS are predictive and prognostic indicators in patients with
non-small-cell lung cancer treated with chemotherapy alone and in
combination with erlotinib. J Clin Oncol. 23:5900–5909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jänne PA, Borras AM, Kuang Y, Rogers AM,
Joshi VA, Liyanage H, Lindeman N, Lee JC, Halmos B, Maher EA, et
al: A rapid and sensitive enzymatic method for epidermal growth
factor receptor mutation screening. Clin Cancer Res. 12:751–758.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ichihara S, Toyooka S, Fujiwara Y, Hotta
K, Shigematsu H, Tokumo M, Soh J, Asano H, Ichimura K, Aoe K, et
al: The impact of epidermal growth factor receptor gene status on
gefitinib-treated Japanese patients with non-small-cell lung
cancer. Int J Cancer. 120:1239–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pallis AG, Voutsina A, Kalikaki A,
Souglakos J, Briasoulis E, Murray S, Koutsopoulos A, Tripaki M,
Stathopoulos E, Mavroudis D, et al: ‘Classical’ but not ‘other’
mutations of EGFR kinase domain are associated with clinical
outcome in gefitinib-treated patients with non-small cell lung
cancer. Br J Cancer. 97:1560–1566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sequist LV, Martins RG, Spigel D, Grunberg
SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky
A, et al: First-line gefitinib in patients with advanced
non-small-cell lung cancer harboring somatic EGFR mutations. J Clin
Oncol. 26:2442–2449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu SG, Gow CH, Yu CJ, Chang YL, Yang CH,
Hsu YC, Shih JY, Lee YC and Yang PC: Frequent epidermal growth
factor receptor gene mutations in malignant pleural effusion of
lung adenocarcinoma. Eur Respir J. 32:924–930. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY
and Yang PC: Effectiveness of tyrosine kinase inhibitors on
‘uncommon’ epidermal growth factor receptor mutations of unknown
clinical significance in non-small cell lung cancer. Clin Cancer
Res. 17:3812–3821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Pas T, Toffalorio F, Manzotti M,
Fumagalli C, Spitaleri G, Catania C, Delmonte A, Giovannini M,
Spaggiari L, De Braud F, et al: Activity of epidermal growth factor
receptor-tyrosine kinase inhibitors in patients with non-small cell
lung cancer harboring rare epidermal growth factor receptor
mutations. J Thorac Oncol. 6:1895–1901. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takahashi R, Hirata H, Tachibana I,
Shimosegawa E, Inoue A, Nagatomo I, Takeda Y, Kida H, Goya S,
Kijima T, et al: Early [18F]fluorodeoxyglucose positron emission
tomography at two days of gefitinib treatment predicts clinical
outcome in patients with adenocarcinoma of the lung. Clin Cancer
Res. 18:220–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee JK, Shin JY, Kim S, Lee S, Park C, Kim
JY, Koh Y, Keam B, Min HS, Kim TM, et al: Primary resistance to
epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors
(TKIs) in patients with non-small-cell lung cancer harboring
TKI-sensitive EGFR mutations: An exploratory study. Ann Oncol.
24:2080–2087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Umekawa K, Kimura T, Kudoh S, Suzumura T,
Oka T, Nagata M, Mitsuoka S, Matsuura K, Nakai T, Yoshimura N, et
al: Plasma RANTES IL-10, and IL-8 levels in non-small-cell lung
cancer patients treated with EGFR-TKIs. BMC Res Notes. 6:1392013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Locatelli-Sanchez M, Couraud S, Arpin D,
Riou R, Bringuier PP and Souquet PJ: Routine EGFR molecular
analysis in non-small-cell lung cancer patients is feasible: Exons
18–21 sequencing results of 753 patients and subsequent clinical
outcomes. Lung. 191:491–499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Keam B, Kim DW, Park JH, Lee JO, Kim TM,
Lee SH, Chung DH and Heo DS: Rare and complex mutations of
epidermal growth factor receptor, and efficacy of tyrosine kinase
inhibitor in patients with non-small cell lung cancer. Int J Clin
Oncol. 19:594–600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guan Y, Zhao H, Meng J, Yan X and Jiao S:
Dramatic response to high-dose icotinib in a lung adenocarcinoma
patient after erlotinib failure. Lung Cancer. 83:305–307. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Watanabe S, Minegishi Y, Yoshizawa H,
Maemondo M, Inoue A, Sugawara S, Isobe H, Harada M, Ishii Y, Gemma
A, et al: Effectiveness of gefitinib against non-small-cell lung
cancer with the uncommon EGFR mutations G719X and L861Q. J Thorac
Oncol. 9:189–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fukihara J, Watanabe N, Taniguchi H,
Kondoh Y, Kimura T, Kataoka K, Matsuda T, Yokoyama T and Hasegawa
Y: Clinical predictors of response to EGFR tyrosine kinase
inhibitors in patients with EGFR-mutant non-small cell lung cancer.
Oncology. 86:86–93. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chiu CH, Yang CT, Shih JY, Huang MS, Su
WC, Lai RS, Wang CC, Hsiao SH, Lin YC, Ho CL, et al: Epidermal
growth factor receptor tyrosine kinase inhibitor treatment response
in advanced lung adenocarcinomas with G719X/L861Q/S768I mutations.
J Thorac Oncol. 10:793–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu J, Jin B, Chu T, Dong X, Yang H, Zhang
Y, Wu D, Lou Y, Zhang X, Wang H, et al: EGFR tyrosine kinase
inhibitor (TKI) in patients with advanced non-small cell lung
cancer (NSCLC) harboring uncommon EGFR mutations: A real-world
study in China. Lung Cancer. 96:87–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mitsudomi T, Kosaka T and Yatabe Y:
Biological and clinical implications of EGFR mutations in lung
cancer. Int J Clin Oncol. 11:190–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Klughammer B, Brugger W, Cappuzzo F,
Ciuleanu T, Mok T, Reck M, Tan EH, Delmar P, Klingelschmitt G, Yin
AY, et al: Examining treatment outcomes with erlotinib in patients
with advanced non-small cell lung cancer whose tumors harbor
uncommon EGFR mutations. J Thorac Oncol. 11:545–555. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Arrieta O, Cardona AF, Corrales L,
Campos-Parra AD, Sánchez-Reyes R, Amieva-Rivera E, Rodríguez J,
Vargas C, Carranza H, Otero J, et al: CLICaP: The impact of common
and rare EGFR mutations in response to EGFR tyrosine kinase
inhibitors and platinum-based chemotherapy in patients with
non-small cell lung cancer. Lung Cancer. 87:169–175. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sordella R, Bell DW, Haber DA and
Settleman J: Gefitinib-sensitizing EGFR mutations in lung cancer
activate anti-apoptotic pathways. Science. 305:1163–1167. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hynes NE and Lane HA: ERBB receptors and
cancer: The complexity of targeted inhibitors. Nat Rev Cancer.
5:341–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Oda K, Matsuoka Y, Funahashi A and Kitano
H: A comprehensive pathway map of epidermal growth factor receptor
signaling. Mol Syst Biol. 1:2005.00102005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Choi SH, Mendrola JM and Lemmon MA:
EGF-independent activation of cell-surface EGF receptors harboring
mutations found in gefitinib-sensitive lung cancer. Oncogene.
26:1567–1576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Greulich H, Chen TH, Feng W, Jänne PA,
Alvarez JV, Zappaterra M, Bulmer SE, Frank DA, Hahn WC, Sellers WR,
et al: Oncogenic transformation by inhibitor-sensitive and
-resistant EGFR mutants. PLoS Med. 2:e3132005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen Y-R, Fu Y-N, Lin C-H, Yang ST, Hu SF,
Chen YT, Tsai SF and Huang SF: Distinctive activation patterns in
constitutively active and gefitinib-sensitive EGFR mutants.
Oncogene. 25:1205–1215. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jiang J, Greulich H, Jänne PA, Sellers WR,
Meyerson M and Griffin JD: Epidermal growth factor-independent
transformation of Ba/F3 cells with cancer-derived epidermal growth
factor receptor mutants induces gefitinib-sensitive cell cycle
progression. Cancer Res. 65:8968–8974. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kancha RK, von Bubnoff N, Peschel C and
Duyster J: Functional analysis of epidermal growth factor receptor
(EGFR) mutations and potential implications for EGFR targeted
therapy. Clin Cancer Res. 15:460–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kancha RK, Peschel C and Duyster J: The
epidermal growth factor receptor-L861Q mutation increases kinase
activity without leading to enhanced sensitivity toward epidermal
growth factor receptor kinase inhibitors. J Thorac Oncol.
6:387–392. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Furuyama K, Harada T, Iwama E, Shiraishi
Y, Okamura K, Ijichi K, Fujii A, Ota K, Wang S, Li H, et al:
Sensitivity and kinase activity of epidermal growth factor receptor
(EGFR) exon 19 and others to EGFR-tyrosine kinase inhibitors.
Cancer Sci. 104:584–589. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Schiffer HH, Reding EC, Fuhs SR, Lu Q, Piu
F, Wong S, Littler PL, Weiner DM, Keefe W, Tan PK, et al:
Pharmacology and signaling properties of epidermal growth factor
receptor isoforms studied by bioluminescence resonance energy
transfer. Mol Pharmacol. 71:508–518. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yoshikawa S, Kukimoto-Niino M, Parker L,
Handa N, Terada T, Fujimoto T, Terazawa Y, Wakiyama M, Sato M, Sano
S, et al: Structural basis for the altered drug sensitivities of
non-small cell lung cancer-associated mutants of human epidermal
growth factor receptor. Oncogene. 32:27–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yun CH, Boggon TJ, Li Y, Woo MS, Greulich
H, Meyerson M and Eck MJ: Structures of lung cancer-derived EGFR
mutants and inhibitor complexes: Mechanism of activation and
insights into differential inhibitor sensitivity. Cancer Cell.
11:217–227. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shtiegman K, Kochupurakkal BS, Zwang Y,
Pines G, Starr A, Vexler A, Citri A, Katz M, Lavi S, Ben-Basat Y,
et al: Defective ubiquitinylation of EGFR mutants of lung cancer
confers prolonged signaling. Oncogene. 26:6968–6978. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Doss GP, Rajith B, Chakraborty C,
NagaSundaram N, Ali SK and Zhu H: Structural signature of the
G719S-T790M double mutation in the EGFR kinase domain and its
response to inhibitors. Sci Rep. 4:58682014.PubMed/NCBI
|
|
63
|
Yuza Y, Glatt KA, Jiang J, Greulich H,
Minami Y, Woo MS, Shimamura T, Shapiro G, Lee JC, Ji H, et al:
Allele-dependent variation in the relative cellular potency of
distinct EGFR inhibitors. Cancer Biol Ther. 6:661–667. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tracy S, Mukohara T, Hansen M, Meyerson M,
Johnson BE and Jänne PA: Gefitinib induces apoptosis in the
EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res.
64:7241–7244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Paez JG, Janne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Helfrich BA, Raben D, Varella-Garcia M,
Gustafson D, Chan DC, Bemis L, Coldren C, Barón A, Zeng C, Franklin
WA, et al: Antitumor activity of the epidermal growth factor
receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839,
Iressa) in non-small cell lung cancer cell lines correlates with
gene copy number and EGFR mutations but not EGFR protein levels.
Clin Cancer Res. 12:7117–7125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sharma SV, Gajowniczek P, Way IP, Lee DY,
Jiang J, Yuza Y, Classon M, Haber DA and Settleman J: A common
signaling cascade may underlie ‘addiction’ to the Src, BCR-ABL, and
EGF receptor oncogenes. Cancer Cell. 10:425–435. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Malaney P, Nicosia SV and Davé V: One
mouse, one patient paradigm: New avatars of personalized cancer
therapy. Cancer Lett. 344:1–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Gazdar AF, Hirsch FR and Minna JD: From
mice to men and back: An assessment of preclinical model systems
for the study of lung cancers. J Thorac Oncol. 11:287–299. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
National Comprehensive Cancer Network, .
Clinical Practice Guidelines in Oncology (NCCN
Guidelines®): Non-Small Cell Lung Cancer (Version 4.
2016). 2016 https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdfJuly
5–2016.
|
|
72
|
Sebastian M, Schmittel A and Reck M:
First-line treatment of EGFR-mutated nonsmall cell lung cancer:
Critical review on study methodology. Eur Respir Rev. 23:92–105.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Subramaniam D, He AR, Hwang J, Deeken J,
Pishvaian M, Hartley ML and Marshall JL: Irreversible multitargeted
ErbB family inhibitors for therapy of lung and breast cancer. Curr
Cancer Drug Targets. 14:775–793. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Stasi I and Cappuzzo F: Second generation
tyrosine kinase inhibitors for the treatment of metastatic
non-small-cell lung cancer. Transl Respir Med. 2:22014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cross DAE, Ashton SE, Ghiorghiu S,
Eberlein C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA,
Mellor MJ, et al: AZD9291, an irreversible EGFR TKI overcomes T
790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer
Discov. 4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Jänne PA, Yang JC, Kim DW, Planchard D,
Ohe Y, Ramalingam SS, Ahn MJ, Kim SW, Su WC, Horn L, et al: AZD9291
in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J
Med. 372:1689–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Sequist LV, Besse B, Lynch TJ, Miller VA,
Wong KK, Gitlitz B, Eaton K, Zacharchuk C, Freyman A, Powell C, et
al: Neratinib, an irreversible pan-ErbB receptor tyrosine kinase
inhibitor: Results of a phase II trial in patients with advanced
non-small-cell lung cancer. J Clin Oncol. 28:3076–3083. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yang JC, Sequist LV, Geater SL, Tsai CM,
Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C, et al:
Clinical activity of afatinib in patients with advanced
non-small-cell lung cancer harbouring uncommon EGFR mutations: A
combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung
6. Lancet Oncol. 16:830–838. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mitsudomi T, Kosaka T, Endoh H, Horio Y,
Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T and Yatabe Y:
Mutations of the epidermal growth factor receptor gene predict
prolonged survival after gefitinib treatment in patients with
non-small-cell lung cancer with postoperative recurrence. J Clin
Oncol. 23:2513–2520. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Baek JH, Sun JM, Min YJ, Cho EK, Cho BC,
Kim JH, Ahn MJ and Park K: Efficacy of EGFR tyrosine kinase
inhibitors in patients with EGFR-mutated non-small cell lung cancer
except both exon 19 deletion and exon 21 L858R: A retrospective
analysis in Korea. Lung Cancer. 87:148–154. 2015. View Article : Google Scholar : PubMed/NCBI
|