Introduction
Hepatocellular carcinoma (HCC), one of the most
prevalent human cancers, is the third leading cause of
cancer-related deaths worldwide. Although there are diverse
treatments for HCC, such as surgical resection, radiofrequency
ablation, radiotherapy and chemoembolization, the prognosis of
patients with HCC remains poor (1–3). Thus,
it is necessary to develop a deeper understanding of the molecular
mechanisms to identify new molecular markers that can be used for
early diagnosis and therapy of HCC, and to improve the outcome of
these patients. Recent studies have focused on the important role
of the tumor microenvironment in the progression, invasion and
metastasis of HCC (4–6). Tumor-associated macrophages (TAMs) are
the most abundant cellular component of the tumor microenvironment,
and they play essential roles in the treatment of cancers, such as
pancreatic cancer and HCC (7,8).
Previous studies have shown that TAMs are primarily characterized
as alternatively activated M2-like macrophages, with high
expression levels of peroxisome proliferator-activated receptor
(PPAR), CD206 and arginase (Arg)-1, and low expression levels of
tumor necrosis factor-α (TNF-α). In addition, M2-like TAMs play
leading roles in immune suppression by interacting with stromal
cells, which greatly contributes to HCC growth, invasion and
metastasis (9–11). These studies suggest that targeting
the regulation of TAM polarization presents a potential therapeutic
strategy for HCC.
Receptor-interacting protein 140 (RIP140) is a
nuclear receptor co-regulator that affects biological and
pathological processes in the body, including energy metabolism,
inflammatory response and tumorigenesis (12–14).
Previous studies have found that RIP140-mediated macrophage
polarization plays an essential role in regulating the inflammatory
response. Overexpression of RIP140 in macrophages promoted
macrophages to an M1-like polarization and expanded the
inflammatory response. Conversely, lowering the level of RIP140 in
macrophages not only reduced M1-like macrophages, but also expanded
alternative polarization and promoted endotoxin tolerance (ET),
which relieves the inflammatory response (13,15,16).
However, the role of RIP140 in TAMs remains completely unknown.
In the present study, we demonstrated that RIP140
expression in TAMs plays a role in the growth of hepatoma cells,
which is closely related to the prognosis of patients with liver
cancer. Our data, presented in the present study, identified RIP140
as a potential target in HCC immunotherapy that may promote
macrophage-mediated antitumor immunity.
Materials and methods
Cell lines and cell cultures
Human HCC cell lines Huh7 and HepG2, and the mouse
hepatoma cell line H22 were purchased from the American Type
Culture Collection (ATCC, Rockville, MD, USA) and maintained in
Dulbecco's modified Eagle's medium (DMEM; HyClone, Logan, UT, USA)
with 10% fetal bovine serum (FBS) and 1% penicillin G and
streptomycin in 37̊C humidified air containing 5%
CO2.
Human subjects and isolation of
mononuclear cells
Human HCC tissues were obtained from 60 patients.
Peripheral blood specimens were obtained from 5 healthy volunteers
and 5 patients. All the patients were pathologically diagnosed with
HCC at The Second Affiliated Hospital of Chongqing Medical
University. Peripheral blood specimens were used for the isolation
of peripheral mononuclear cells. Mononuclear cell isolation was
performed as previously described (17). None of the individuals were positive
for HCV or HIV. No chemotherapy, radiotherapy or surgical treatment
was performed on these patients prior to obtaining blood specimens.
Informed consent was obtained from all patients before the study
was initiated. The present study was approved by the Research
Ethics Committee of The Second Affiliated Hospital of Chongqing
Medical University.
Macrophage preparation and
lentivirus-mediated overexpression of RIP140
Peritoneal macrophages (PMs) were harvested from
BALB/c mice 72 h after intraperitoneal injection with 2 ml of
sterile 6% starch solution. The lentivirus-mediated PM
overexpression vector containing RIP140 was constructed as
previously described (12).
Lentivirus packaging and cell transduction were carried out as
previously described (18).
HCC-conditioned medium
H22 cells were cultured in serum-free DMEM for 24 h.
The supernatants were collected and used as HCC-conditioned medium
(HCM). PMs were cultured with different amounts of HCM or exposed
to HCM for different time periods in 6-well plates. Briefly,
different amounts of HCM were added into complete medium to keep
the final volume at 2,000 µl. Therefore, the final percentages of
the 0, 100, 200, 300 and 400 µl of HCM were 0, 5, 10, 15 and 20%,
respectively.
In vivo tumorigenicity
Sixteen 4-week-old BALB/c nude mice were housed in a
pathogen-free environment and used for the HCC xenografts.
Xenografts were prepared by subcutaneous injection of H22 cells
along with homologous PMs transfected with Lv-GFP-RIP140 or
Lv-GFP-NC at a ratio of 4:1. Four mice were sacrificed every week,
and the volume and the weight of the tumors were calculated. All
experimental procedures were conducted in accordance with the Guide
for the Care and Use of Laboratory Animals and approved by our
institutional ethical guidelines for animal experiments.
Cell apoptosis assay
H22 cells were harvested and washed with
phosphate-buffered saline (PBS) 3 times after co-culture with PMs
for 24 h. Then, an apoptosis assay was performed using an Annexin
V-FITC/PI Cell Apoptosis kit (KeyGen, Nanjing, Jiangsu, China).
Briefly, a suspension (100 µl) of 5×105 H22 cells was
incubated with 5 µl of Annexin V and 1 µl of propidium iodide (PI)
at room temperature for 15 min. The apoptotic rate was determined
by flow cytometry (BD Pharmingen, San Diego, CA, USA).
Cell invasion assay
Cell invasion assays were performed in numerous
studies. Briefly, 1×105 Huh7 or 1×105 HepG2
cells were added to the upper chamber of an insert coated with
Matrigel (BD Biosciences, San Jose, CA, USA). Cell invasion was
allowed to proceed at 37̊C for 24 h. Then, the upper chambers were
washed with PBS 3 times, and the cells were fixed with methanol at
room temperature for 30 min. After the cells on the inner surface
of the filter membrane were removed, invading cells were stained
with 0.1% crystal violet at room temperature for 15 min, and washed
again with PBS 3 times. Images were captured and cells were counted
using digital microscopy.
RNA extraction and qRT-PCR
analysis
Total RNA was isolated from PMs using TRIzol reagent
(Invitrogen) according to the manufacturer's protocol. qRT-PCR was
performed using SYBR®-Green (Takara, Dalian, China) and
an ABI Prism 7900 Sequence Detection System (Applied Biosystems,
Foster City, CA, USA) according to the manufacturer's protocol. The
primers used were as follows: CD206 forward,
5′-GGGACTCTGGATTGGACTCA-3′ and reverse, 5′-CCAGGCTCTGATGATGGACT-3′;
Arg-1 forward, 5′-CCCCAGTACCAACAGGACTACC-3′ and reverse,
5′-TGAACGTGGCGGAATTTTGT-3′; PPAR forward,
5′-TCCCATACACAACCGCAGTCGC-3′ and reverse,
5′-GGGGTCATTTGGTGACTCTGGGGT-3′; TNF-α forward,
5′-GGATCTCAAAGACAACCAAC-3′ and reverse, 5′-ACAGAGCAATGACTCCAAAG-3′;
NF-κB forward, 5′-AGTGTGGAGGCTGCCTTGCGAATG-3′ and reverse,
5′-TGGGCTTTCAAGACTGGAACGGTC-3′; GAPDH forward,
5′-CACCCACTCCTCCACCTTTG-3′ and reverse, 5′-CCACCACCCTGTTGCTGTAG-3′.
Data were normalized to the expression of GAPDH.
Western blot analysis
PM cells or peripheral blood monocytes were
collected and lysed with radio-immunoprecipitation assay buffer [50
mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) NP-40, 0.1% (w/v) SDS,
0.5% (w/v) sodium deoxycholate] containing protease inhibitors and
phosphatase inhibitors, and cell lysates were centrifuged at 12,000
× g (4̊C for 10 min). Total cellular protein (80 µg) was mixed with
a quarter of loading buffer [62.5 mM Tris-HCl (pH 6.8), 10%
glycerol, 2% SDS, 2% β-mercaptoethanol and bromophenol blue],
boiled for 5 min, and subjected to 8 or 10% SDS-PAGE. Proteins were
transferred to polyvinylidene difluoride membranes (Millipore,
Bedford, MA, USA). After the membranes were blocked with
Tris-buffered saline containing 0.05% Tween-20 (TBST) and 5%
fat-free milk, the membrane was incubated overnight at 4̊C with
primary antibodies in TBST with 5% bovine serum albumin (BSA). The
next day, the membranes were further incubated with the
corresponding horseradish peroxidase-conjugated secondary antibody
at 37̊C for 1 h and then washed 3 times with TBST. Band signals
were analyzed and scanned using Quantity One Software (Bio-Rad,
Hercules, CA, USA) after incubation with an enhanced
chemiluminescence reagent (Millipore). Anti-RIP140 (ab42126),
anti-p-c-jun (ab32385) and anti-TGF-β (ab31013) antibodies were
purchased from Abcam (Cambridge, MA, USA); anti-p-p65 (#3037),
anti-p65 (#8242), anti-TRAF3 (#4729) and anti-β-actin (#3700)
antibodies were all from Cell Signaling Technology (CST; Danvers,
MA, USA).
Immunohistochemical staining
F4/80 antibody is a standard macrophage marker.
Immunohistochemical staining was performed as previously described
(19). Briefly, human liver cancer
(n=60) and subcutaneous tumor tissues (n=4) were fixed in formalin
and embedded in paraffin, and 4-µm-thick consecutive sections were
cut and mounted on glass slides. Then, the slides were first
dewaxed in xylol and rehydrated in a 100, 95 and 85% graded alcohol
series, with antigen retrieval in 0.01 M sodium citrate solution
98̊C for 15 min. Endogenous peroxidase activity was blocked with 3%
H2O2-methanol and normal goat serum for 30
min. The human liver cancer tissues were incubated with RIP140
antibody (1:80; Abcam) or F4/80 antibody (1:50; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) at 4̊C overnight, and the
subcutaneous tumor tissues were incubated with proliferating cell
nuclear antigen (PCNA) antibody (1:100; Cell Signaling, Boston, MA,
USA) at 4̊C overnight. The next day, the slides were washed 3 times
with PBS and incubated with the appropriate biotin-labeled
secondary antibody for 10–15 min. Under high-power magnification
(x400), micrographs of 5 independent microscopic fields of the
stained cells were screened and captured using a Leica DMLA light
microscope (Leica Microsystems, Wetzlar, Germany).
Tissue immunofluorescence
The expression levels of RIP140 and F4/80 in human
liver cancer tissues (n=60) were measured by immunofluorescence.
The experimental procedure was performed as previously described
(5). Cell nuclei were stained with
4,6-diamidino-2-phenylindole (DAPI).
Statistical analysis
Statistical analyses were performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Differences were assessed
for statistical significance by GraphPad Prism. All of the data are
expressed as the means ± SD. Statistical differences between two
groups were determined by Student's t-test. The Kaplan-Meier method
and log-rank test were used to perform the survival analysis.
P-values of <0.05 were considered to indicate a statistically
significant result.
Results
HCC microenvironment inhibits RIP140
expression in TAMs
H22 cells were cultured in serum-free DMEM for 24 h.
The supernatants were collected and used as HCM.
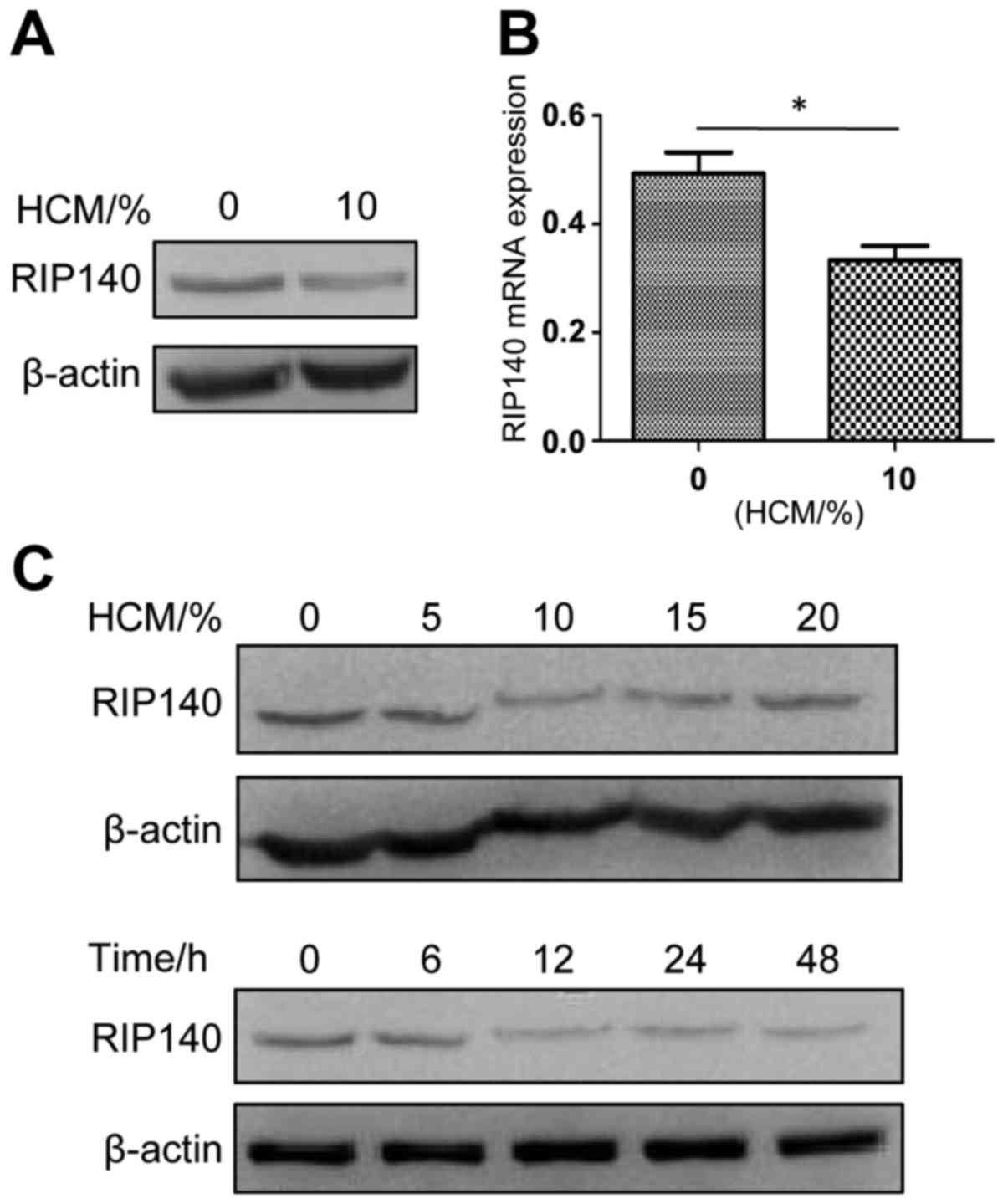
Preliminary experiments revealed that the protein
(Fig. 1A) and mRNA (Fig. 1B) levels of RIP140 in PMs treated
with 10% HCM for 24 h were significantly lower than those of normal
PMs. To evaluate the best length of HCM treatment and best
concentration of HCM to induce the lowest expression of RIP140 in
PMs, the protein level of RIP140 in PMs was examined by western
blotting in concentration gradients (HCM 0, 5, 10, 15 and 20%) and
time gradients (time 0, 6, 12, 24 and 48 h) (Fig. 1C). As shown in Fig. 1C, the lowest level of RIP140 protein
in TAMs was found in PMs treated with 10% HCM for 12 h.
Overexpression of RIP140 in TAMs
inhibits HCC microenvironment-mediated TAM M2-like
polarization
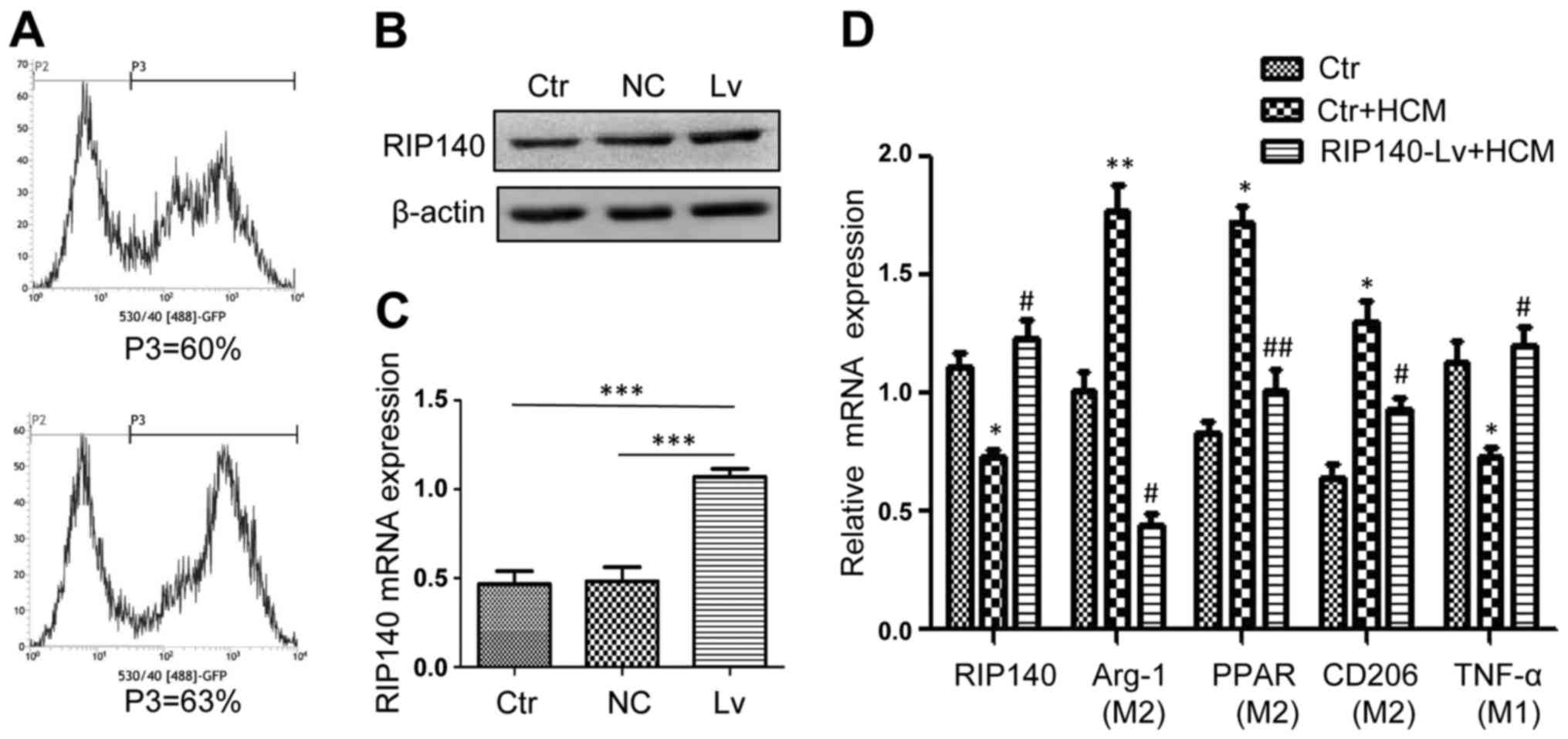
Lentivirus transfection mediated RIP140
overexpression in PMs. To detect the efficiency of the lentivirus
transfection, flow cytometry was used to analyze the rate of
lentivirus transfection 96 h after PMs were transfected. A
transfection rate of ~60% was observed in the negative control (NC)
lentiviral transfection, and a transfection rate of 63% was
observed in the targeted lentiviral transfection (Fig. 2A). Western blotting was used to
evaluate the protein expression level of RIP140 in PMs. As
expected, RIP140 protein expression in PMs with targeted lentivirus
transfection was significantly higher than that noted in the
control groups (Fig. 2B). Using
qRT-PCR, RIP140 mRNA expression was determined in the PMs. As
expected, RIP140 mRNA expression in the PMs was also markedly
increased in the targeted lentivirus transfection group (Fig. 2C).
Previous studies indicate that TAMs are primarily
polarized to an M2-like phenotype in response to the tumor
microenvironment (20). In
addition, TAMs were reported to express high levels of Arg-1, PPAR
and CD206 and low levels of the pro-inflammatory molecule TNF-α
(8). To analyze the effects of
RIP140 on TAMs, we overexpressed RIP140 in TAMs. qRT-PCR was used
to detect the mRNA expression of RIP140, Arg-1, PPAR, CD205 and
TNF-α. As expected, overexpression of RIP140 in TAMs inhibited the
expression of Arg-1, PPAR and CD206, which are closely related to
the M2-like polarization phenotype of macrophages, and promoted the
expression of TNF-α (associated with the M1 phenotype) (Fig. 2D).
Overexpression of RIP140 in TAMs
suppresses invasion and induces apoptosis of HCC cells
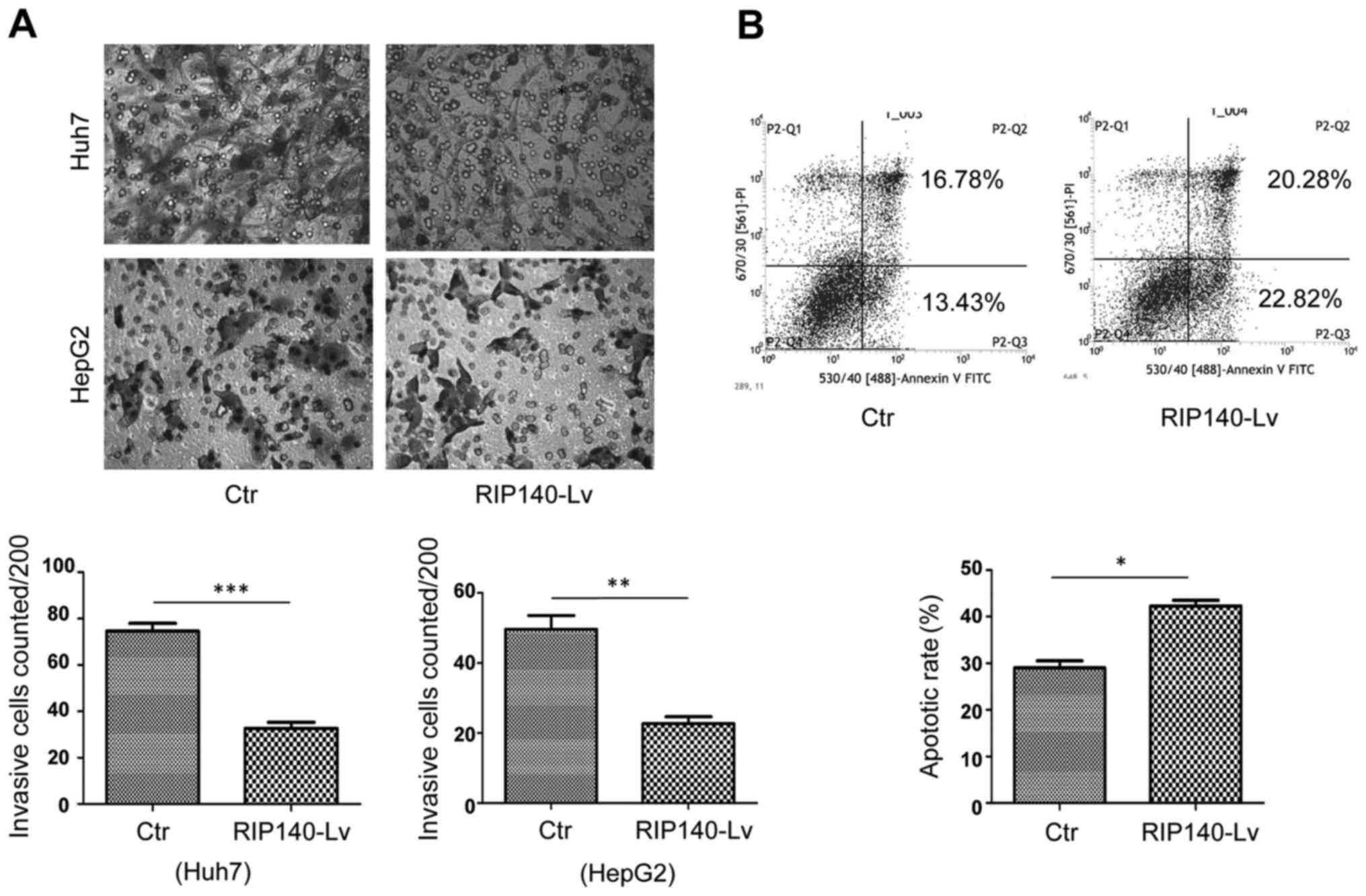
It has been well documented that M2-like TAMs
promote HCC proliferation and metastasis (8,9). In
the present study, we found that overexpression of RIP140 in TAMs
suppressed HCC microenvironment-mediated M2-like polarization of
TAMs. To investigate whether RIP140 overexpressed in TAMs could
inhibit migration and promote apoptosis of HCC cells, we performed
Transwell assays and flow cytometric analysis. The Transwell assays
further demonstrated that overexpression of RIP140 in TAMs greatly
inhibited the invasion of HepG2 and Huh7 cells (Fig. 3A). Flow cytometry showed that
overexpression of RIP140 in TAMs induced apoptosis in the H22 cells
(Fig. 3B).
Overexpression of RIP140 in TAMs
suppresses the growth of H22 subcutaneous tumors in BALB/c nude
mice
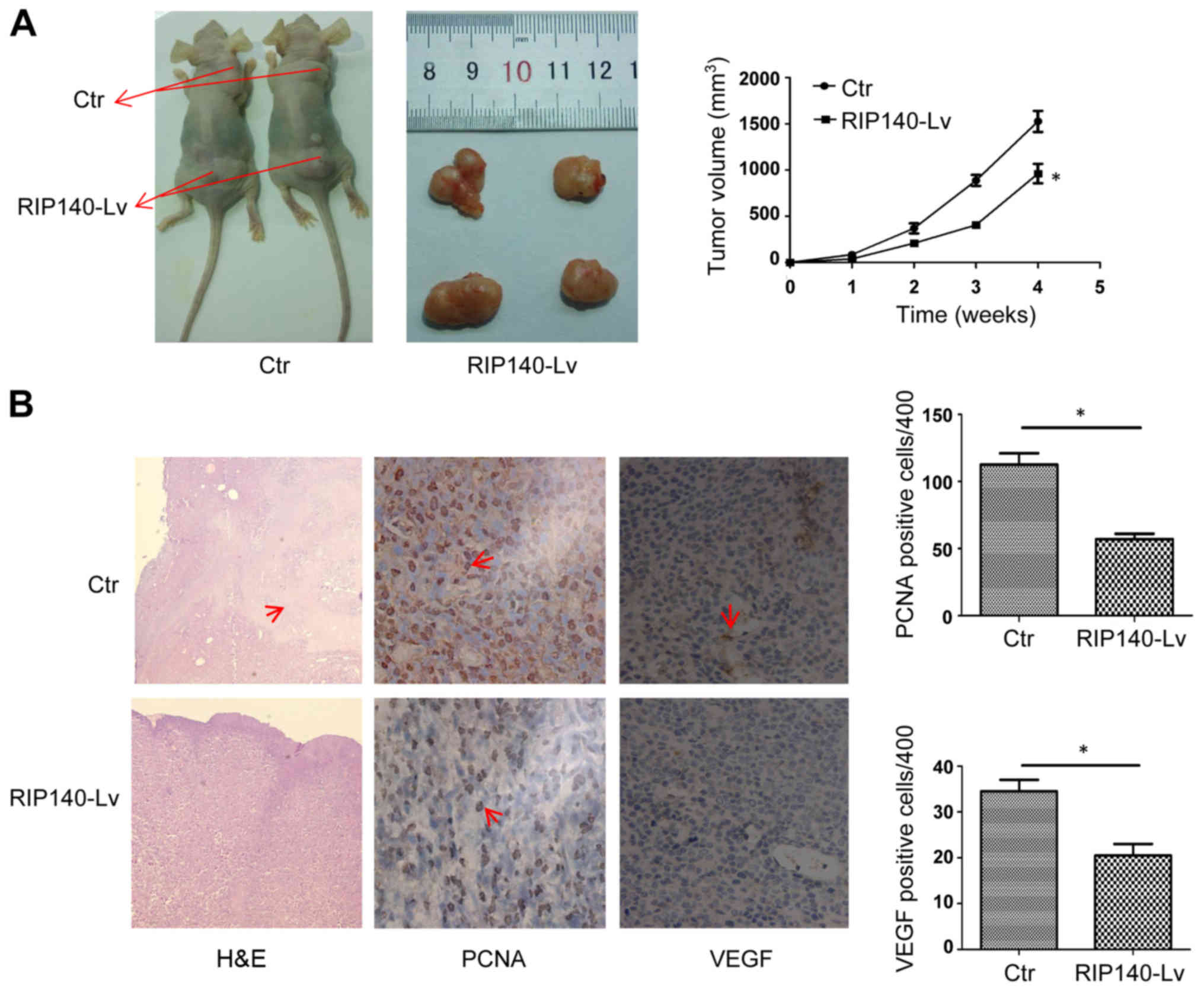
To determine the effects of RIP140 in macrophages on
tumor growth, we established a subcutaneous tumor model in mice
(n=16). Subcutaneous tumor models were prepared by subcutaneous
injection of H22 cells combined with homologous PMs at a ratio of
4:1. As shown in Fig. 4A, compared
with control macrophages, macrophages overexpressing RIP140
significantly inhibited the growth of H22 subcutaneous tumors.
Consistently, the weight and volume of the tumors with
RIP140-overexpressing macrophages were less than the weight and
volume of tumors with control macrophages (Fig. 4A). Additionally, hematoxylin and
eosin (H&E) staining further demonstrated that overexpression
of RIP140 in TAMs inhibited tumor growth compared with tumors in
the control mice. We observed a large necrotic area (red arrows) in
control tumor sections, which indicates that H22 cells rapidly
proliferated (Fig. 4B). In
addition, PCNA and vascular endothelial growth factor (VEGF)
immunohistochemical staining in the RIP140-overexpressing tumor
sections was significantly less than that in tumors of the control
mice. The red arrows point to the PCNA- or VEGF-positive cells
(Fig. 4B). Taken together, our data
suggest that overexpression of RIP140 in TAMs inhibits HCC growth
in vivo.
RIP140 overexpression in TAMs inhibits
the growth of HCC cell lines by suppressing the NF-κB/IL-6 axis in
TAMs
Numerous studies have reported that the tumor
microenvironment activates the NF-κB pathway in TAMs, which is the
well-known transcriptional controller of IL-6. The activation of
the NF-κB/IL-6 pathway in TAMs is closely related to tumor growth
(21–23). Therefore, we investigated the
expression of the NF-κB/IL-6 axis in RIP140-overexpressing TAMs. As
expected, the tumor microenvironment activated the NF-κB/IL-6 axis
in TAMs and increased the protein expression of phosphorylated p65
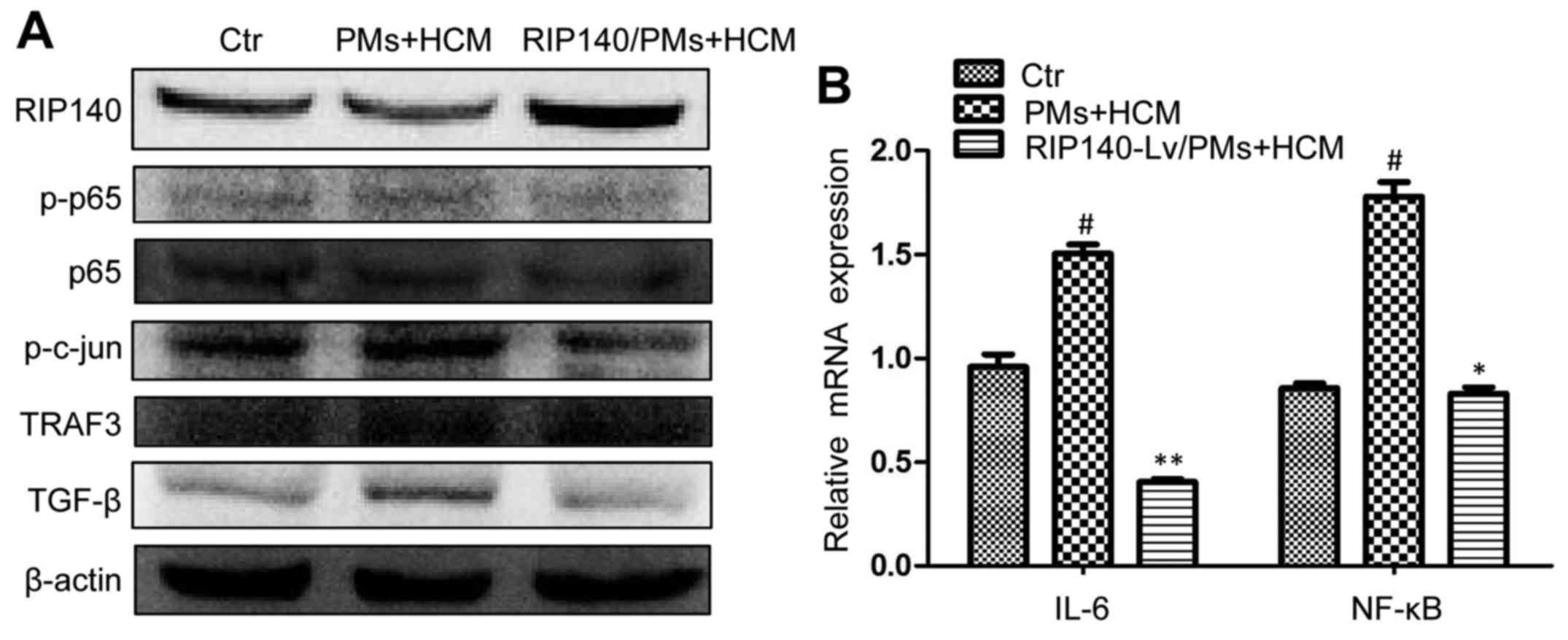
(p-p65), phosphorylated c-Jun (p-c-Jun) and TRAF3 (Fig. 5A). Previous studies suggest that
p-p65, p-c-Jun and TRAF3 are closely related to the NF-κB/IL-6 axis
and macrophage polarization (24,25).
Conversely, RIP140 overexpression in TAMs inhibited the NF-κB-/IL-6
axis and decreased the protein level of p-p65, p-c-Jun and TRAF3
(Fig. 5A). The IL-6 results were
essentially in agreement with the above figures (Fig. 5B). This finding indicates that
RIP140 overexpression in TAMs inhibited the growth of HCC cell
lines probably by suppressing the NF-κB/IL-6 signaling pathway in
TAMs. Transforming growth factor (TGF)-β is closely related to
tumor progression and metastasis (26). However, whether TGF-β is a tumor
promoter or suppressor is still unknown (27,28).
It has been reported that TGF-β in the tumor microenvironment
facilitates the alternative activation of macrophages and promotes
tumor growth (8,29). In the present study, we detected
TGF-β protein expression in TAMs. As shown, RIP140 overexpression
decreased the tumor microenvironment-stimulated TGF-β expression in
TAMs (Fig. 5A). Therefore, we
hypothesize that the TGF-β-promoted tumor growth was not only
related to the TGF-β secreted by tumor cells but was also related
to the high expression of TGF-β in TAMs. Of course, this hypothesis
needs further confirmation.
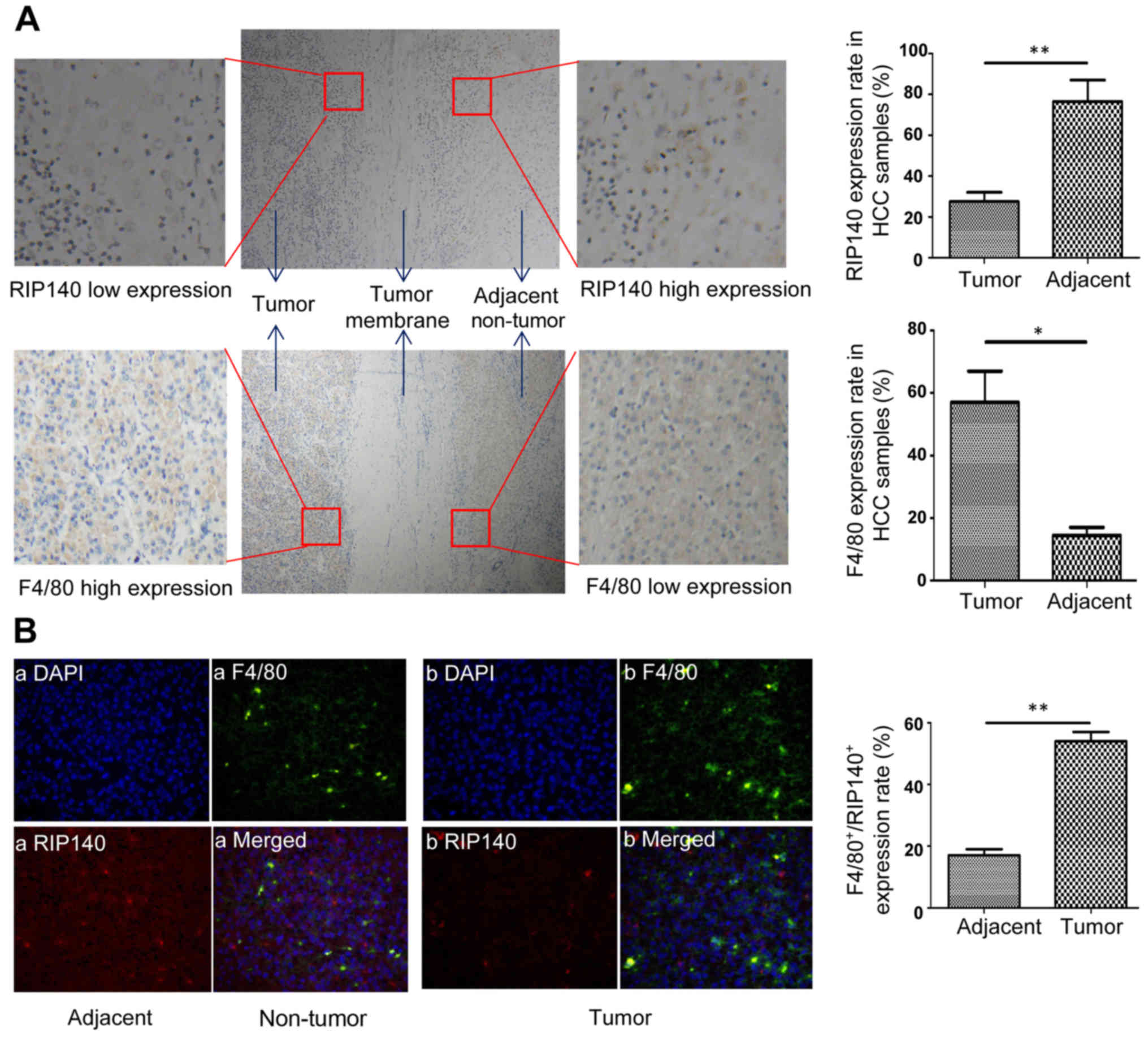
Low expression of RIP140 in TAMs of
human HCC tissues
To detect the expression of RIP140 and F4/80 in
human HCC samples, the paraffin sections of human HCC tissues
(n=60) were assessed by immunohistochemistry and immunofluorescence
(Fig. 6A and B). As shown in
Fig. 6A, the protein expression of
RIP140 in the cancer tissues was lower than that noted in adjacent
non-tumor tissue. In contrast, the protein expression of F4/80 in
cancer tissues was higher than that noted in adjacent non-tumor
tissue. The expression level of RIP140 in macrophages was scored by
evaluating the number of RIP140+-F4/80+ cells
(both positive for RIP140 and F4/80) in relation to the total
number of cells. As shown in Fig.
6B, the expression of RIP140 in macrophages (F4/80-positive
cells) was significantly lower in the HCC specimens than that noted
in adjacent non-tumor specimens, indicating low expression of
RIP140 in TAMs of human HCC samples.
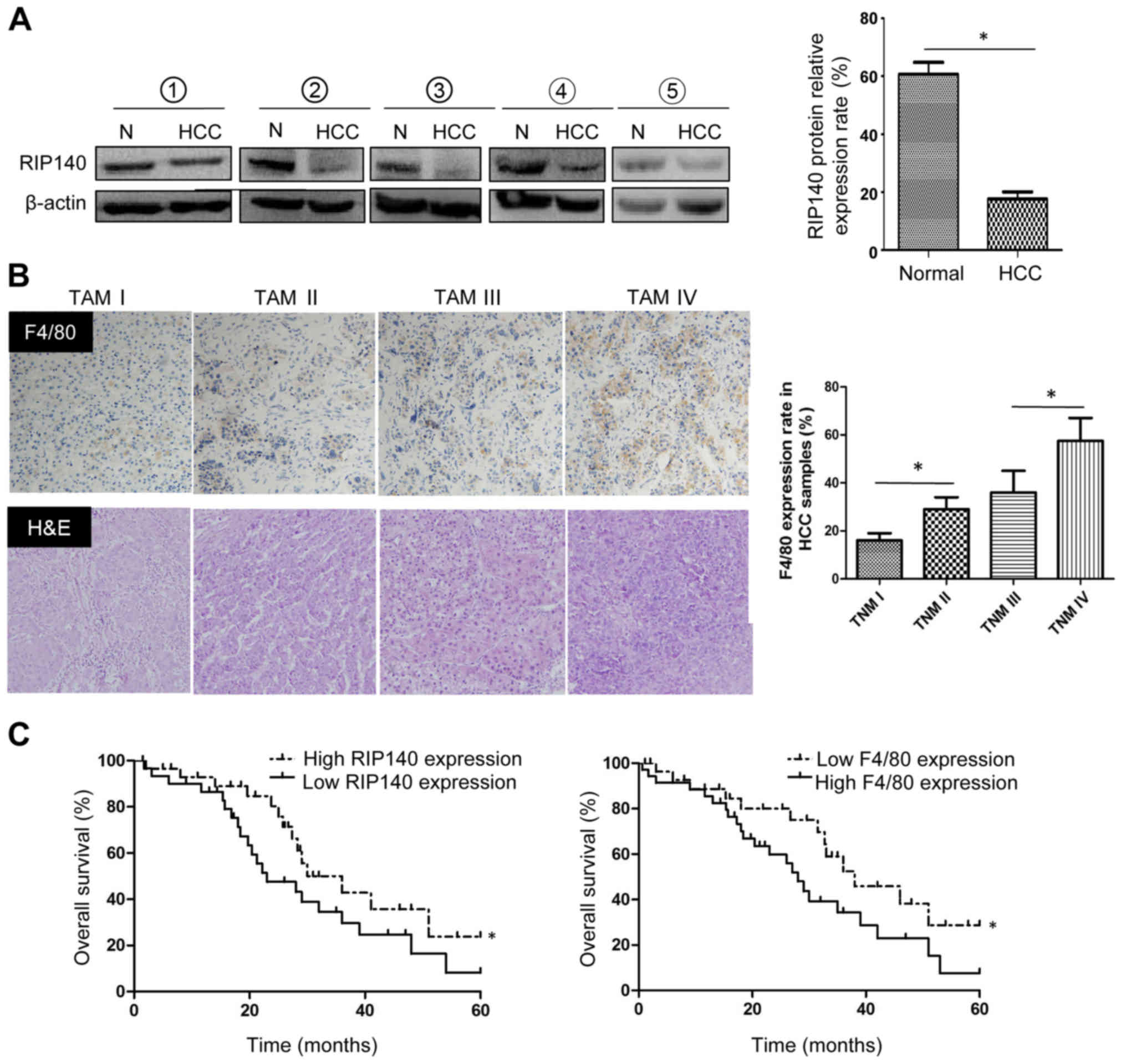
Low expression levels of RIP140 in
peripheral mononuclear cells of patients with HCC predict poor
patient survival
To detect the RIP140 protein expression in TAMs from
HCC samples, we isolated the peripheral mononuclear cells from 5
patients with pathologically confirmed HCC at the Second Affiliated
Hospital of Chongqing Medical University and 5 healthy volunteers.
Western blot analyses were used to evaluate the RIP140 protein
level in peripheral mononuclear cells. As expected, RIP140 protein
expression in peripheral mononuclear cells from patients with HCC
was significantly lower than that from healthy subjects (Fig. 7A). In fact, the positive expression
rate of F4/80 in human HCC samples increased with the TNM stage
(Fig. 7B). This indicates that a
large number of macrophages with low RIP140 expression accumulate
in HCC tissues. In addition, low RIP140 expression and high F4/80
expression were found to be closely correlated with shorter
survival time (Fig. 7C).
Discussion
Tumor-associated macrophages (TAMs) and their
alternative activation contribute greatly to the development of HCC
(8–10). RIP140 is a nuclear receptor
co-regulator that is widely expressed in macrophages and regulates
macrophage-mediated energy metabolism, inflammatory response and
tumorigenesis (12–16). However, whether RIP140 is involved
in the activation and function of TAMs has not yet been reported.
In the present study, for the first time, we found decreased
expression of RIP140 in TAMs after treatment with an HCC
microenvironment, which facilitated alternative activation of the
macrophages and accelerated tumor growth. Our data provide a
previously unrecognized link between RIP140 and TAM-related
inflammation in HCC, and mark RIP140 as a potential target in HCC
immunotherapy that may promote macrophage-mediated antitumor
immunity. The present study presents the following evidence to
support these conclusions.
Firstly, we found that HCC-conditioned medium (HCM)
inhibited RIP140 expression and fostered the alternative activation
of macrophages. Previous studies have found that RIP140 mediated
macrophage polarization, which plays essential roles in regulating
the inflammatory response (13,14).
Overexpression of RIP140 in macrophages promoted macrophages to
M1-like polarization and expand the inflammatory response.
Conversely, lowering the level of RIP140 in macrophages not only
reduces M1-like macrophages but also expands alternative
polarization, which promotes endotoxin tolerance (ET) and relieves
inflammation (13,15,16).
We found that HCM decreased the expression of RIP140 in TAMs.
Therefore, we suspected that the HCC microenvironment-mediated
macrophage M2-like polarization may be closely related to the low
expression of RIP140 in macrophages. To verify the above
hypothesis, lentiviral-mediated transfection was performed to
induce RIP140 overexpression in TAMs.
Secondly, RIP140 overexpression in TAMs
significantly inhibited the alternative activation of macrophages
and suppressed HCC cell growth both in vitro and in
vivo. Visibly, the effect of the tumor microenvironment
promoted TAMs to M2-like polarization and was closely related to
decrease RIP140 expression in TAMs. Therefore, one important
question is how RIP140 regulates TAM polarization and further
affects tumor growth. Previous studies have shown that the TAM
effect on the growth of tumors is associated with a TAM-mediated
tumor microenvironment immune response, particularly related to the
activation of the NF-κB/IL-6 axis. The high level of IL-6 that
induces ‘parainflammation’ in the tumor microenvironment is the
most important reason for the promotion of tumor growth by TAMs
(30–32). Our data revealed that overexpression
of RIP140 in TAMs suppressed the NF-κB/IL-6 axis and reduced the
release of IL-6 into the tumor microenvironment, which is one of
the important reasons why RIP140 inhibited the growth of HCC in
vivo and in vitro.
Finally, in order to evaluate RIP140 clinical
values, we performed various related experiments using tissue and
blood samples of patients with HCC or healthy volunteers. Our data
indicated that RIP140 was poorly expressed in TAMs of human HCC
tissues, which predicted poor patient survival. In the present
study, we reproduced the findings regarding human peripheral
mononuclear cells in TAMs (9).
However, we think that our methods may not be precise. The best
method is to directly extract the TAMs from fresh HCC tissues or
adjacent non-tumor tissues to perform further studies (11). Due to technical reasons and limited
experimental conditions, this research was not able to be
satisfactorily completed.
In conclusion, our results demonstrated that the
tumor microenvironment inhibits the transcription of RIP140 in
TAMs, which promotes M2-like TAM polarization and HCC growth.
Overexpression of RIP140 in TAMs suppresses HCC growth in
vivo and in vitro. In addition, we found that there is a
low expression level of RIP140 in TAMs of human HCC tissues, which
predicts poor patient survival. These findings suggest that RIP140
plays a role in TAMs and may provide a new strategy for HCC
treatment.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 31370753, 81470899 and
81401622), the Key Item of Chongqing Health and Family Planning
Commission of China (2015zdxm026), and the Basic Science and
Frontier Technology Research Foundation of Chongqing Science and
Technology Commission (cstc2015jcy jBX0070).
References
|
1
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schlachterman A, Craft WW Jr, Hilgenfeldt
E, Mitra A and Cabrera R: Current and future treatments for
hepatocellular carcinoma. World J Gastroenterol. 21:8478–8491.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng AL, Thongprasert S, Lim HY,
Sukeepaisarnjaroen W, Yang TS, Wu CC, Chao Y, Chan SL, Kudo M,
Ikeda M, et al: Randomized, open-label phase 2 study comparing
frontline dovitinib versus sorafenib in patients with advanced
hepatocellular carcinoma. Hepatology. 64:774–784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z,
Chen EB, Fan J, Cao Y, Dai Z and Zhou J: Tumor-associated
neutrophils recruit macrophages and T-regulatory cells to promote
progression of hepatocellular carcinoma and resistance to
sorafenib. Gastroenterology. 150:1646–1658.e17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ye LY, Chen W, Bai XL, Xu XY, Zhang Q, Xia
XF, Sun X, Li GG, Hu QD, Fu QH, et al: Hypoxia-induced
epithelial-to-mesenchymal transition in hepatocellular carcinoma
induces an immunosuppressive tumor microenvironment to promote
metastasis. Cancer Res. 76:818–830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hernandez-Gea V, Toffanin S, Friedman SL
and Llovet JM: Role of the microenvironment in the pathogenesis and
treatment of hepatocellular carcinoma. Gastroenterology.
144:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nywening TM, Wang-Gillam A, Sanford DE,
Belt BA, Panni RZ, Cusworth BM, Toriola AT, Nieman RK, Worley LA,
Yano M, et al: Targeting tumour-associated macrophages with CCR2
inhibition in combination with FOLFIRINOX in patients with
borderline resectable and locally advanced pancreatic cancer: A
single-centre, open-label, dose-finding, non-randomised, phase 1b
trial. Lancet Oncol. 17:651–662. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flecken T and Sarobe P: Tim-3 expression
in tumour-associated macrophages: A new player in HCC progression.
Gut. 64:1502–1503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan W, Liu X, Ma H, Zhang H, Song X, Gao
L, Liang X and Ma C: Tim-3 fosters HCC development by enhancing
TGF-β-mediated alternative activation of macrophages. Gut.
64:1593–1604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou W, Ke SQ, Huang Z, Flavahan W, Fang
X, Paul J, Wu L, Sloan AE, McLendon RE, Li X, et al: Periostin
secreted by glioblastoma stem cells recruits M2 tumour-associated
macrophages and promotes malignant growth. Nat Cell Biol.
17:170–182. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heusinkveld M and van der Burg SH:
Identification and manipulation of tumor associated macrophages in
human cancers. J Transl Med. 9:2162011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lapierre M, Bonnet S, Bascoul-Mollevi C,
Ait-Arsa I, Jalaguier S, Del Rio M, Plateroti M, Roepman P, Ychou
M, Pannequin J, et al: RIP140 increases APC expression and
controls intestinal homeostasis and tumorigenesis. J Clin Invest.
124:1899–1913. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ho PC, Tsui YC, Feng X, Greaves DR and Wei
LN: NF-κB-mediated degradation of the coactivator RIP140 regulates
inflammatory responses and contributes to endotoxin tolerance. Nat
Immunol. 13:379–386. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nautiyal J, Christian M and Parker MG:
Distinct functions for RIP140 in development, inflammation, and
metabolism. Trends Endocrinol Metab. 24:451–459. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu PS, Lin YW, Burton FH and Wei LN:
M1-M2 balancing act in white adipose tissue browning - a new role
for RIP140. Adipocyte. 4:146–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin YW, Lee B, Liu PS and Wei LN:
Receptor-interacting protein 140 orchestrates the dynamics of
macrophage m1/m2 polarization. J Innate Immun. 8:97–107. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moreli JB, Santos JH, Lorenzon-Ojea AR,
Corrêa-Silva S, Fortunato RS, Rocha CR, Rudge MV, Damasceno DC,
Bevilacqua E and Calderon IM: Hyperglycemia differentially affects
maternal and fetal DNA integrity and DNA damage response. Int J
Biol Sci. 12:466–477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng X, Krogh KA, Wu CY, Lin YW, Tsai HC,
Thayer SA and Wei LN: Receptor-interacting protein 140 attenuates
endoplasmic reticulum stress in neurons and protects against cell
death. Nat Commun. 5:4487–4495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao
L and Zhang S: Critical role for the long non-coding RNA AFAP1-AS1
in the proliferation and metastasis of hepatocellular carcinoma.
Tumour Biol. 37:9699–9707. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mancino A and Lawrence T: Nuclear
factor-kappaB and tumor-associated macrophages. Clin Cancer Res.
16:784–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang CP, Su YC, Lee PH and Lei HY:
Targeting NFKB by autophagy to polarize hepatoma-associated
macrophage differentiation. Autophagy. 9:619–621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wan S, Zhao E, Kryczek I, Vatan L,
Sadovskaya A, Ludema G, Simeone DM, Zou W and Welling TH:
Tumor-associated macrophages produce interleukin 6 and signal via
STAT3 to promote expansion of human hepatocellular carcinoma stem
cells. Gastroenterology. 147:1393–1404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hefetz-Sela S, Stein I, Klieger Y, Porat
R, Sade-Feldman M, Zreik F, Nagler A, Pappo O, Quagliata L, Dazert
E, et al: Acquisition of an immunosuppressive protumorigenic
macrophage phenotype depending on c-Jun phosphorylation. Proc Natl
Acad Sci USA. 111:17582–17587. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lalani AI, Moore CR, Luo C, Kreider BZ,
Liu Y, Morse HC III and Xie P: Myeloid cell TRAF3 regulates immune
responses and inhibits inflammation and tumor development in mice.
J Immunol. 194:334–348. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu H, Luo H, Shen Z, Hu X, Sun L and Zhu
X: Transforming growth factor-β1 in carcinogenesis, progression,
and therapy in cervical cancer. Tumour Biol. 37:7075–7083. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
David CJ, Huang YH, Chen M, Su J, Zou Y,
Bardeesy N, Iacobuzio-Donahue CA and Massagué J: TGF-β tumor
suppression through a lethal EMT. Cell. 164:1015–1030. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu J, Acharya S, Sahin O, Zhang Q, Saito
Y, Yao J, Wang H, Li P, Zhang L, Lowery FJ, et al: 14-3-3ζ turns
TGF-β's function from tumor suppressor to metastasis promoter in
breast cancer by contextual changes of Smad partners from p53 to
Gli2. Cancer Cell. 27:177–192. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gratchev A: TGF-β signalling in tumour
associated macrophages. Immunobiology. 222:75–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu L, Zhang X, Zhang B, Shi H, Yuan X, Sun
Y, Pan Z, Qian H and Xu W: Exosomes derived from gastric cancer
cells activate NF-κB pathway in macrophages to promote cancer
progression. Tumour Biol. 37:12169–12180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Karin M: NF-kappaB as a critical link
between inflammation and cancer. Cold Spring Harb Perspect Biol.
1:a0001412009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ara T and Declerck YA: Interleukin-6 in
bone metastasis and cancer progression. Eur J Cancer. 46:1223–1231.
2010. View Article : Google Scholar : PubMed/NCBI
|