Introduction
CUB and sushi multiple domains-1 (CSMD1) is a
very large gene which contains 71 exons that span over 2 Mb of
genomic DNA on chromosome 8p23 (1).
Multiple splice variants exist for CSMD1 and these encode
proteins of varying length. The largest transcript is 14.3 kb long
and this encodes a 3,565 amino acid protein (1). The full-length protein is a membrane
protein with an extracellular region containing 14 CUB and 28 sushi
domains, a single transmembrane domain and a short cytoplasmic
domain that contains a putative tyrosine phosphorylation site.
CSMD1 belongs to the CSMD gene family whose members
also include the structurally similar proteins CSMD2 and CSMD3
(1–3). The function of CSMD1 is largely
unknown. Although CSMD1 has been shown to inhibit C3 deposition
onto the surface of cells leading to the inhibition of the
classical complement pathway (4,5). The
structure of CSMD1 predicts that CSMD1 is a receptor for unknown
ligand(s) and is involved in signal transduction (1).
CSMD1 is believed to act as a tumour suppressor
based on a number of observations. CSMD1 is located on
chromosome 8p23, a region that is frequently deleted in different
types of cancer (1,6–10). In
addition, reduced CSMD1 mRNA and protein expression has been
observed in different cancers (11–17).
An array comparative genomic hybridization study revealed a high
rate of loss of 8p23.2 containing the CSMD1 gene in advanced
prostrate cancer samples. A further small real-time PCR study
identified a decrease in CSMD1 mRNA levels in higher stage
prostrate cancer samples (11).
Similarly reduced CSMD1 expression at the mRNA level has been
identified in colorectal cancer associated with reduced patient
survival (12). Furthermore,
somatic mutations were detected in CSMD1 in 11% (6/54) of
colorectal cancer patients along side DNA methylation and allele
loss, predominantly in early onset patients (13). Previously, we demonstrated in a
large series of breast cancers that 79/275 (28.7%) had reduced
CSMD1 protein expression. Low CSMD1 expression was significantly
associated with high tumour grade (p=0.003) and decreased overall
survival (p=0.018). Importantly multivariate analysis showed that
CSMD1 was also an independent predictor of overall survival
(p=0.03) (14).
Recently, in vivo and in vitro studies
using the A375 melanoma cell line have also shown CSMD1 functions
as a tumour suppressor gene (15).
Overexpression of CSMD1 in melanoma cells resulted in reduced
migration and proliferation and induced apoptosis. In addition,
xenografted tumours expressing CSMD1 resulted in reduction
of tumour weight and size (15) Of
note, reduced mRNA CSMD1 expression has been identified in
glioblastoma stem cells compared to neural stem cells (16). Downregulation of CSMD1 was linked to
upregulation of the microRNAs miR-10a and miRNA-10b which formed an
inhibitory complex with the 3′UTR of CSMD1 (16). Similarly high levels of miRNA-10b
associated with low levels of CSMD1 expression were identified in
HepG2 hepatocellular carcinoma cells (17).
In this study, we investigated the consequences of
loss of CSMD1 expression on cell behaviour. We used a
three-dimensional (3D) MCF10A/Matrigel model to study the role of
CSMD1 in mammary cell differentiation. This culture system has been
recently used to address fundamental questions about processes that
disrupt epithelial architecture. It provides culture conditions
that allow mammary epithelial cells to respond to extracellular
matrix (ECM) signals that impact upon proliferation,
differentiation or death (18,19).
We found that loss of CSMD1 expression enhanced cell proliferation,
migration and invasion. Furthermore, it reduced cell-adhesion.
Moreover, in the 3D culture system, reduced CSMD1 expression
disrupted the morphogenesis of epithelial structures and impaired
lumen formation.
Materials and methods
Cell culture
MCF10A cells were obtained from the American Type
Culture Collection and LNCaP cells were obtained from European
Collection of Cell Cultures. MCF10A was cultured in DMEM-F12 medium
(Invitrogen, Paisley, UK) as previously described (18). LNCaP cells were cultured in RPMI
(Invitrogen) containing 10% foetal calf serum (FCS) (Sigma-Aldrich,
Dorset, UK). All cell lines were maintained at 37°C and 5%
CO2 in the tissue culture incubator and were routinely
tested using MycoAlert® Mycoplasma detection assay
(Lonza, Walkersville, MD, USA).
Generation of shCSMD1 stable cell
lines
CSMD1 shRNA pRS vectors (HuSH™, OriGene, Rockville,
MD, USA) were transformed into NEB 5-α competent E. coli
(New England BioLabs, Ipswich, MA, USA) and grown in L broth
containing 50 µg/ml ampilicillin (Sigma-Aldrich). Sequences of
CSMD1 shRNA constructs and their cognate CSMD1 mRNA regions
are shown in Table I. For each cell
line, 1×106 cells/ml were transfected with either
shCSMD1 or shcontrol constructs using Lipofectamine™ 2000
(Invitrogen). Cells were then grown in media that contained the
appropriate dose (0.5 µg/ml, MCF10A; 0.01 µg/ml, LNCaP) of
puromycin (Sigma-Aldrich) for 2 weeks. Single cell colonies were
selected by serial dilutions in 96-well plates.
 | Table I.shCSMD1 construct sequences. |
Table I.
shCSMD1 construct sequences.
| Construct | Sequence | The cognate
CSMD1 mRNA regions |
|---|
| 1 | CCA CAG GCA GAA ATG
CTT ACT GAG ATG A | 5234-5263 bp |
| 2 | GAG GAC ATC CAC AGC
ACC TTC AAC TCA CT | 2505-2534 bp |
| 3 | GGC TTC CTC ATC CAC
TAT GAG AGT GTG AC | 1013-1042 bp |
| 4 | CAT AGC CAT ACC TCT
GAT GGA CAA GCA GT | 8929-8958 bp |
Reverse transcriptase polymerase chain
reaction (RT-PCR)
RNA was extracted from cells using TRIzol
(Invitrogen). First strand cDNA synthesis was then performed using
ThermoScript (Invitrogen) and random hexamers (Promega,
Southampton, UK) following the manufacturer's instructions. To test
the specificity of shRNA constructs, CSMD3 was amplified.
PCR reactions were performed using HotStarTaq (Qiagen, West Sussex,
UK) following the manufacturer's instructions. The final extension
step was for 10 min at 68°C. Annealing temperature of the
CSMD3 primers was 52°C and 50°C for the housekeeping gene
RPLP0. PCR products were analysed in 1.5% agarose gel
containing ethidium bromide. For primer sequences of CSMD3
and the housekeeping gene RPLP0, see Table II.
 | Table II.RT-PCR primer sequences. |
Table II.
RT-PCR primer sequences.
| Gene | Primers 5′-3′ |
|---|
| CSMD3 | Forward:
AGTAGTTCTGTAGCCATTGC |
|
| Reverse:
TGGGATCAAATCGTACCGCC |
| RPLPO | Forward:
ACATGCTCAACATCTCCC |
|
| Reverse:
TTCAACCTTAGCTGGGG |
Quantitative real-time PCR reactions
(qRT-PCR)
CSMD1 and RPLP0 expression levels were
investigated in all shCSMD1 and shcontrol clones using the relative
standard curve method. TaqMan reactions, using sensiMix dT master
mix (Quantace, London, UK), were performed in duplicates and run on
an ABI7500 (Applied Biosystems, Warrington, UK). For sequences of
primers and probes, see Table
III.
 | Table III.qRT-PCR primer and probe
sequences. |
Table III.
qRT-PCR primer and probe
sequences.
| Gene | Primers | Probe |
|---|
| CSMD1 | Forward:
TTCCAGATTTTTATCCAAACTCTCTAA |
CACGTGGACCATTGAAGTGTCTCATGG |
|
| Reverse:
GTGTGAAAGATCATTTGAACTCCTTT |
|
| RPLPO | Forward:
AGATGCAGCAGATCCGCAT |
AGGCTGTGGTGCTGATGGGCAAGAAC |
|
| Reverse:
ATATGAGGAGCAGTTTCTCCAG |
|
Immunohistochemistry (IHC)
IHC was performed to confirm the knockdown of CSMD1
protein expression in shRNA clones. Cells were pelleted, formalin
fixed and paraffin-embedded and CSMD1 was stained using chicken
anti-CSMD1 antibody at a 1:3000 dilution, as previously described
(14). Negative controls, in which
the pre-immune serum was applied, and positive controls of normal
breast tissue were included in each batch of IHC.
Cell viability
Cell viability was evaluated using the MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
assay. Cells (1×104 cells/ml) were incubated with MTT
solution (1 mg/ml) (Sigma-Aldrich) for 3–4 h at 37°C. Crystals were
dissolved in propan-1-ol and their optical densities (OD) were
quantified at 570 nm (Opsys MR Plate reader; Dynex Technologies,
West Sussex, UK).
Wound healing assay
Confluent monolayers were incubated in mitomycin C
(10 µg/ml) (ICN Biomedicals, Costa Mesa, CA, USA) for 2 h at 37°C
to inhibit cell division. Wounds were introduced into the monolayer
by scratching with a P200 micropipette tip. Wound closure was
followed for 96 h and phase contrast images were captured using an
Olympus digital still camera attached to an Olympus inverted
microscope with a 4X objective lens. The wound area was imaged
using ImageJ and expressed as a percentage of the wound area
covered at each time point relative to the surface area of the
wound at time zero.
Transwell invasion assay
Invasion assays were carried out as previously
described (20). The upper sides of
12-well format transwell inserts (BD Biosciences, Franklin Lakes,
NJ, USA) were coated with Matrigel at a 1:10 dilution in serum-free
medium (SFM). The undersides of the inserts were coated with
fibronectin (10 µg/ml in PBS). Medium containing FCS (1 ml) was
added to the base of each well. Cells at 5×104 cells/ml
in SFM were placed on the top of inserts, for 16 h. Invasive cells
that penetrated through to the underside of the inserts were
identified by fixation in 4% paraformaldehyde and staining with
crystal violet. Cells were then destained using 2% SDS and the OD
was measured using the Opsys MR Plate reader at 570 nm. The
percentage of invasion was calculated independently using the
following equation: Cell invasion (%) = OD of cells in the
bottom/OD of cells in the bottom + OD of cells in the top.
Cell adhesion assays
Plates (96-well) were coated with fibronectin (10
µg/ml in PBS) or Matrigel (20 µg/ml in SFM). Non-specific adhesion
sites were blocked by pre-incubation in 0.5% bovine serum albumin
in DMEM or RPMI. Cells at 4×105 cells/ml were pipetted
onto the coated plates at 37°C and allowed to attach for 30 min or
2 h. Following this, unattached cells were aspirated and the
remaining attached cells were fixed in 4% paraformaldehyde and
stained with crystal violet. Cells were then destained using 2% SDS
and OD was measured using the Opsys MR Plate reader at 570 nm.
MCF10A Matrigel assay
3D Matrigel cultures of MCF10A cells were performed
as previously described (18).
Briefly, 8-well glass slides (Nunc™; Nalge Nunc International
Corp., Rochester, NY, USA) were coated with growth factor reduced
Matrigel (BD Biosciences). shcontrol or shCSMD1 MCF10A cells (5000
cells/ml in 2% Matrigel assay medium) were seeded onto the slides
and incubated at 37°C for up to 26 days. Cells were fed every 3–4
days with 2% Matrigel assay medium.
Fluorescence analysis and image
acquisition
Fluorescent staining of acinar structures was
performed as previously described (18). Phalloidin (Invitrogen), was used as
a cytoskeletal marker at a 1:50 dilution. Rabbit polyclonal active
caspase-3 antibody (Abcam, Cambridge, UK) was used as an apoptotic
marker at a 1:100 dilution. Fluorescence staining was imaged using
a Nikon confocal microscope (Nikon Eclipse TE2000-E; Nikon UK,
Kingston Upon Thames, UK). Images of acinar structures were
captured by serial confocal sectioning and viewed using confocal
software. Phase contrast images were captured using an Olympus
digital still camera attached to an Olympus inverted microscope
(Olympus CKX41). All images were converted to TIFF format and
figures were assembled and annotated using Adobe Photoshop
7.0®.
Statistical analysis
Initially the distribution of the data from all the
functional assays except wound healing assay was tested using
Shapiro-Wilk or Kolmogorov-Smirnov tests. When the data were
normally distributed, a t-test was used to compare between means. A
Mann-Whitney U test (non-parametric) was used when the data were
not normally distributed. All tests were 2-sided and performed
using SPSS software version 15. A p-value ≤0.05 was considered to
be statistically significant.
Results
Generation of shCSMD1 stable cell
lines
To help determine the function of CSMD1, shRNA gene
silencing methodology was used to create stable cell lines with
suppressed CSMD1 expression. Two different cell lines were
analysed: MCF10A (normal breast) and LNCaP (prostate). Single cell
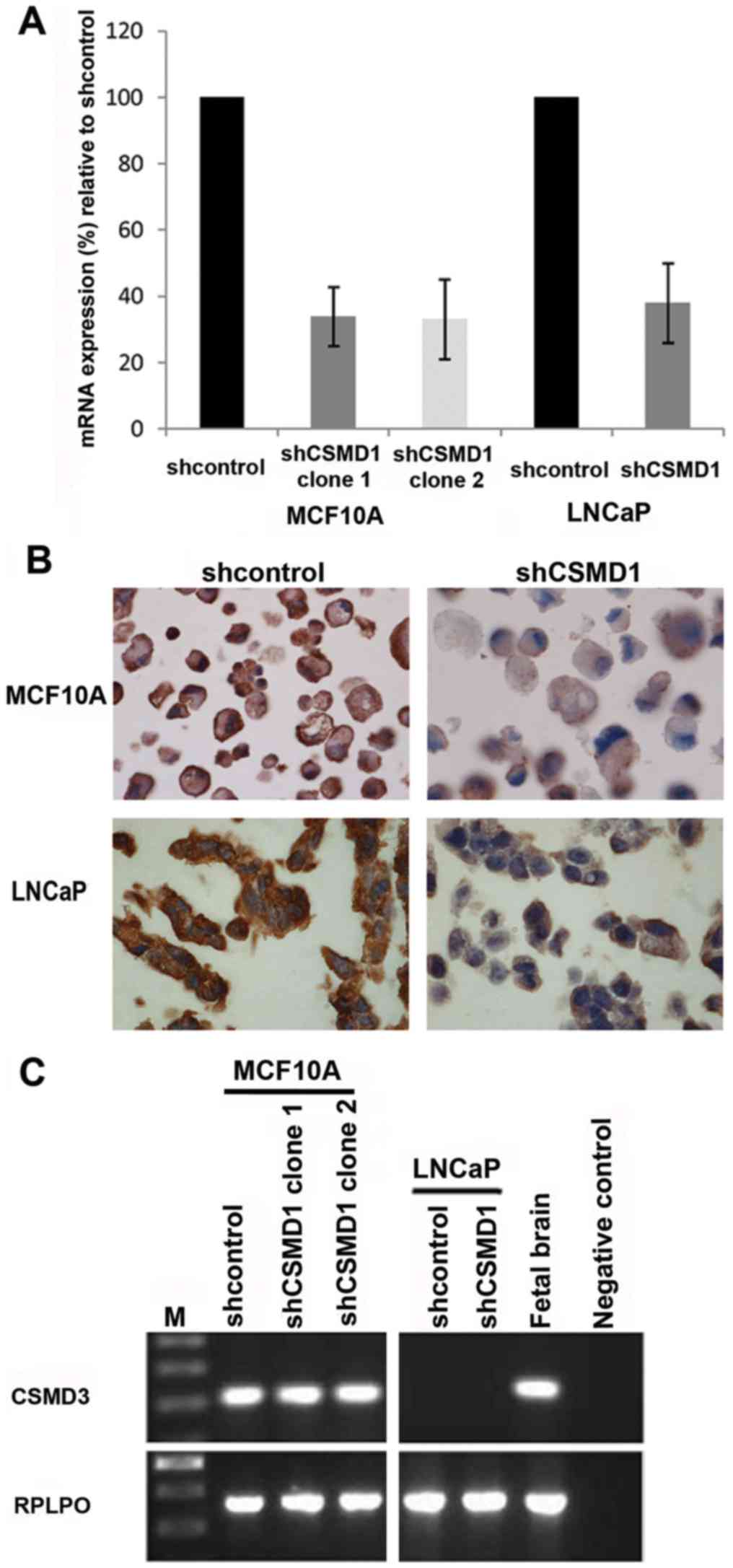
colonies were selected from all cell lines and the mRNA expression
level of CSMD1 was investigated using qRT-PCR. Colonies that showed
the best level of mRNA knockdown were chosen for subsequent
experiments. Two separate shCSMD1 cell lines were generated from
the MCF10A cells (termed clone 1 and 2) and a single cell line was
generated from the LNCaP cells. MCF10A shCSMD1 clones 1 and 2
showed a 66% (SD±8.9%) and 66.9% (SD±12%) reduction in CSMD1
mRNA expression, respectively, when compared to shcontrol cells.
The LNCaP cell line showed a 62% (SD±12%) CSMD1 mRNA
knockdown, relative to shcontrols (Fig.
1A).
To confirm these levels of knockdown at the protein
level, CSMD1 protein expression was investigated using IHC since at
the time a CSMD1 antibody suitable for westerns was not available.
MCF10A shCSMD1 colonies showed lower CSMD1 protein expression
compared to the shcontrol. However, unexpectedly, the LNCaP shCSMD1
cells did not show a reduced level of protein expression compared
to shcontrols cells in contrast to the high reduction in
CSMD1 mRNA expression observed by qRT-PCR (Fig. 1B).
To test the specificity of the shCSMD1 constructs
the expression of CSMD3 mRNA was investigated using RT-PCR.
Since normally none of the tested cell lines express CSMD2
its expression was not tested. The shCSMD1 constructs did not
affect the expression pattern of CSMD3 in either of the cell
lines examined. Moreover, the shcontrol plasmids had no effect on
the expression pattern of CSMD3 (Fig. 1C).
Downregulation of CSMD1 disrupts cell
morphology
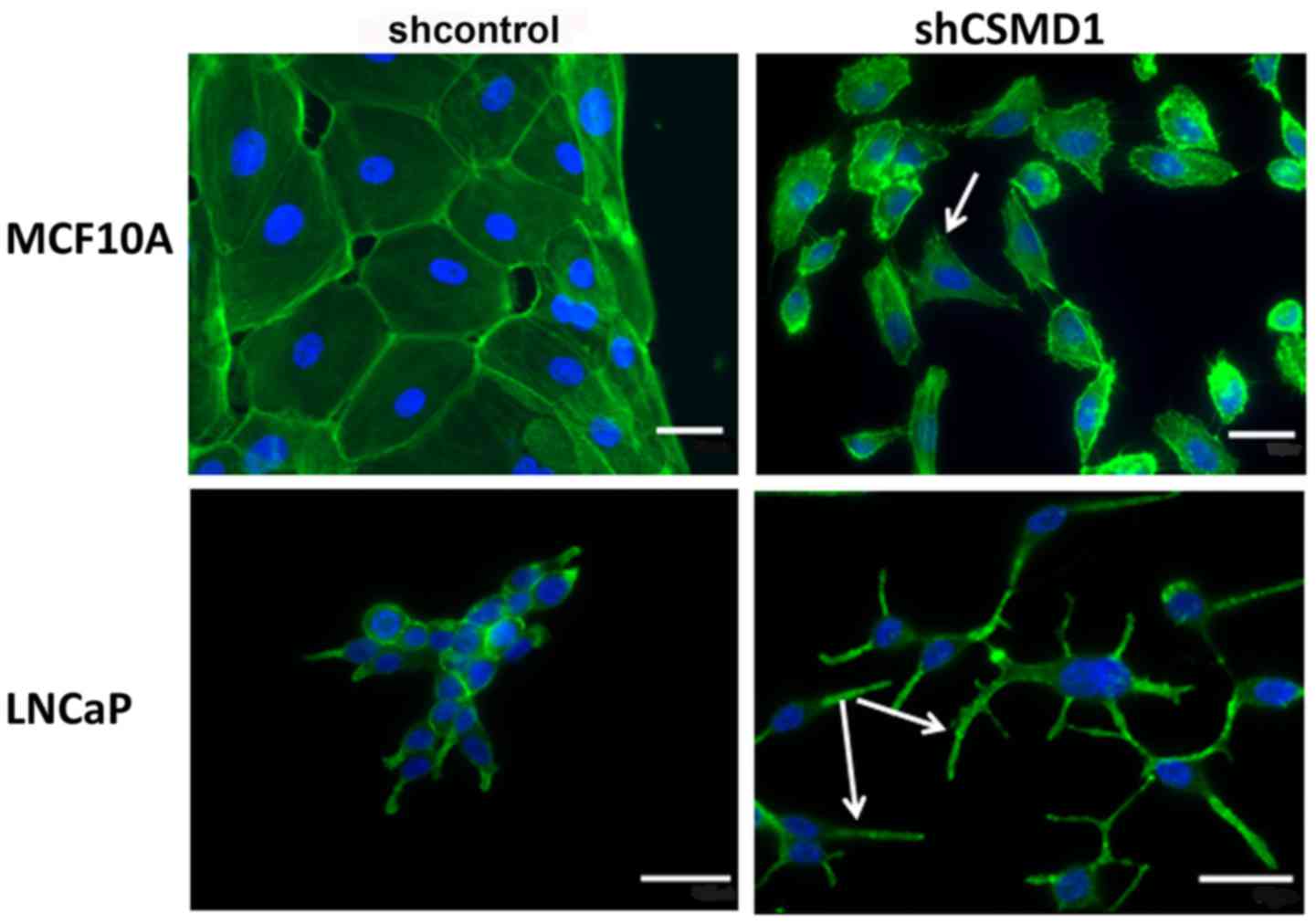
To investigate the morphology of the shCSMD1 cells,
the F-actin stain Phalloidin was used to examine the cells.
Staining revealed that in all lines examined, loss of CSMD1
expression resulted in misshapen cells that tended to grow
individually. In contrast, shcontrol cells mainly grew as colonies.
In MCF10A, many shCSMD1 cells possessed lamellipodia-like
protrusions suggestive of a motile phenotype (Fig. 2, arrow). LNCaP shCSMD1 cells
displayed very long filopodia-like protrusions (Fig. 2, arrows). In all cases these
features were not observed in the appropriate shcontrol cell
lines.
Effect of CSMD1 suppression on cell
proliferation, migration and invasion
Having shown that CSMD1 silencing affected cell
morphology, using MCF10A cells as a model we focused on
understanding its functional effects by investigating a number of
important cellular processes. Since minimal knockdown at the
protein level was observed in the LNCap cell line further
functional work was not performed in these cells. Cell
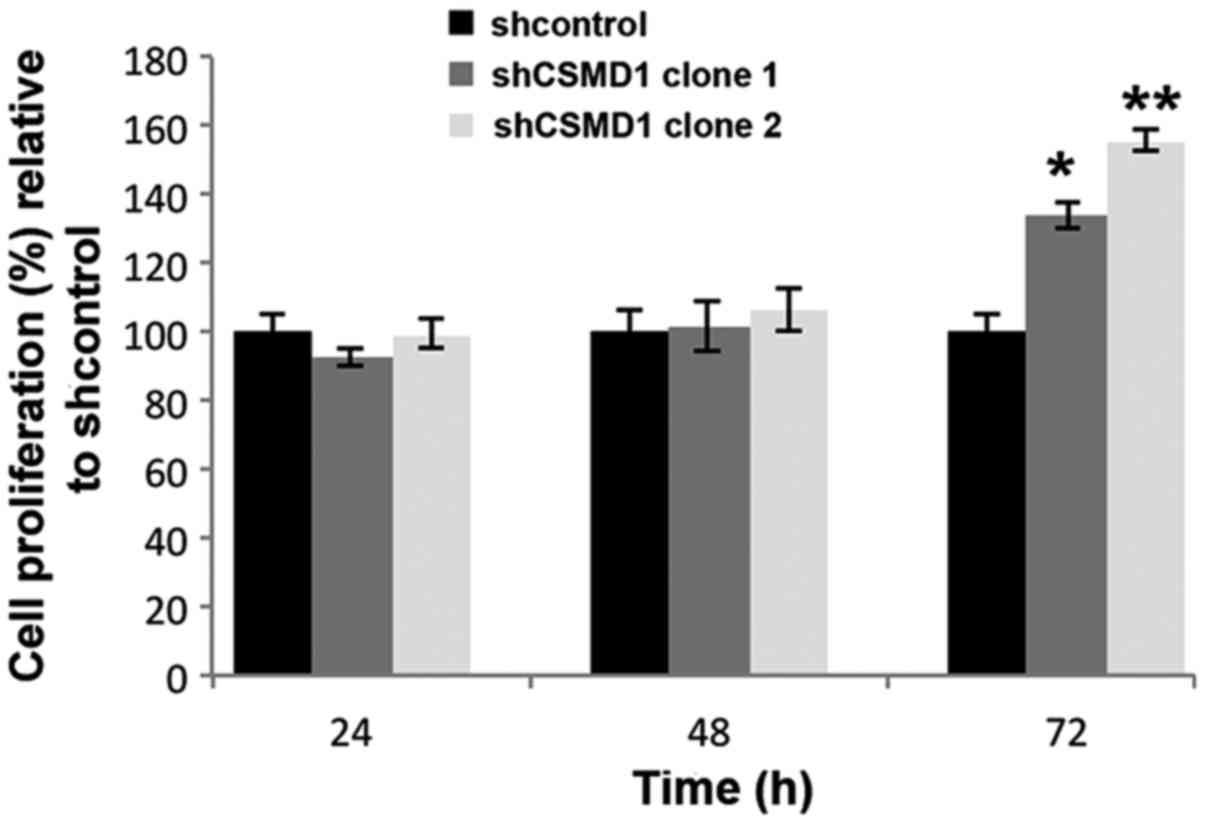
proliferation was assayed using the MTT protocol. Silencing CSMD1
expression did not significantly affect cell proliferation until 72
h post-plating when increases of 34% (SD±4%, p=0.003) and 56%
(SD±3.3%, p=0.001) were observed in MCF10A shCSMD1 clones 1 and 2
(Fig. 3).
Since the morphology of the MCF10A shCSMD1 cells
suggested that they were more motile than their shcontrol
counterparts (Fig. 2), we next
investigated the role of CSMD1 in cell migration. Wound healing
assays were performed after inhibiting MCF10A proliferation by
treating cells with mitomycin C prior to the experiment. In the
shcontrol cells 14.15% (SD±3.6%) and 31.55% (SD±4.3%) of the wound
were covered during the first 16 h and 24 h after wounding
respectively. In the shCSMD1 clones 1 and 2, 37.02% (SD±8.5%,
p=0.07) and 56.15% (SD±7.1%, p=0.018) of the wound were covered
during the first 16 h. Moreover, after 24 h, cells in shCSMD1
clones 1 and 2 covered 69.9% (SD±6.6%, p=0.02) and 88.2% (SD±0.9%,
p=0.003) of the wound (Fig. 4A and
B), confirming that suppression of CSMD1 expression promotes a
more motile phenotype in MCF10A cells.
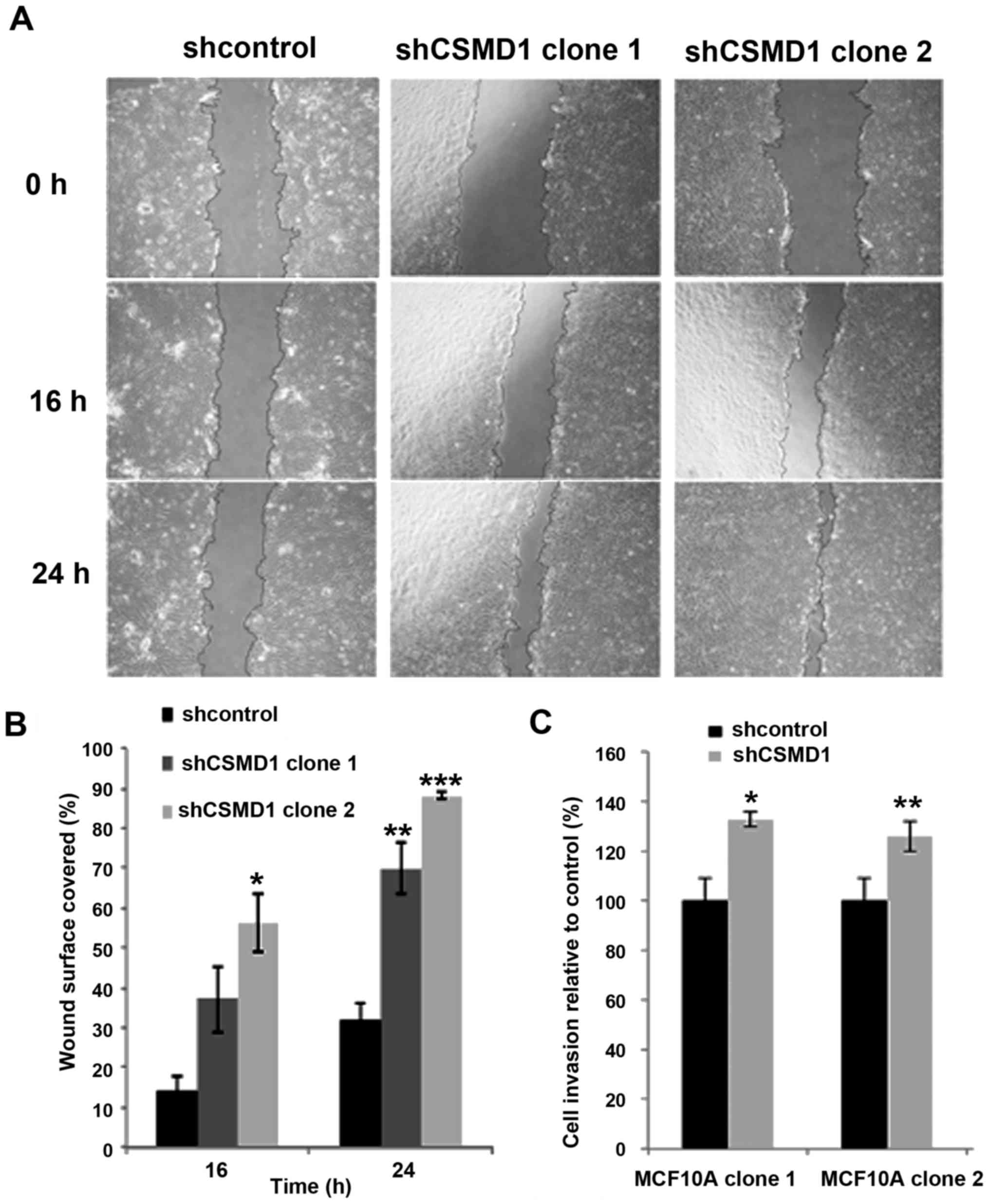 | Figure 4.Loss of CSMD1 expression increases
cell migration and invasion. (A) Wound healing assays, after
inhibiting cell proliferation by mitomycin C, revealed that loss of
CSMD1 expression enhanced migration of MCF10A cells. Images were
taken, using phase contrast microscopy every 8 h. Magnification,
×4; scale bar, 500 µm. (B) In the MCF10A shcontrol cells 14.15 and
31.55% of the wound were covered after 16 and 24 h, respectively.
In the MCF10A shCSMD1 clones 1 and 2, 37.02% (p=0.07) and 56.15%
(*p=0.018) of the wound were covered after 16 h and 69.9%
(**p=0.02) and 88.2% (***p=0.003) after 24 h, respectively. The
percentages of the surface area of the wound covered relative to
time point zero, are presented as the mean ± SD of two independent
experiments. (C) MCF10A shCSMD1 clones 1 and 2 showed 33%
(*p<0.001) and 26% (**p<0.001) increase in invasion,
respectively. The percentages of cell invasion, relative to
shcontrols, are presented as the mean ± SD of at least 3
independent experiments. |
Since loss of CSMD1 promoted cell migration, we next
studied its role in cell invasion. At 16 h after plating,
suppression of CSMD1 expression resulted in 33% (SD±3%, p<0.001)
and 26% (SD±6%, p<0.001) increases in cell invasion of MCF10A
shCSMD1 clones 1 and 2, respectively, compared to shcontrol
(Fig. 4C).
Effect of CSMD1 suppression on cell
matrix adhesion
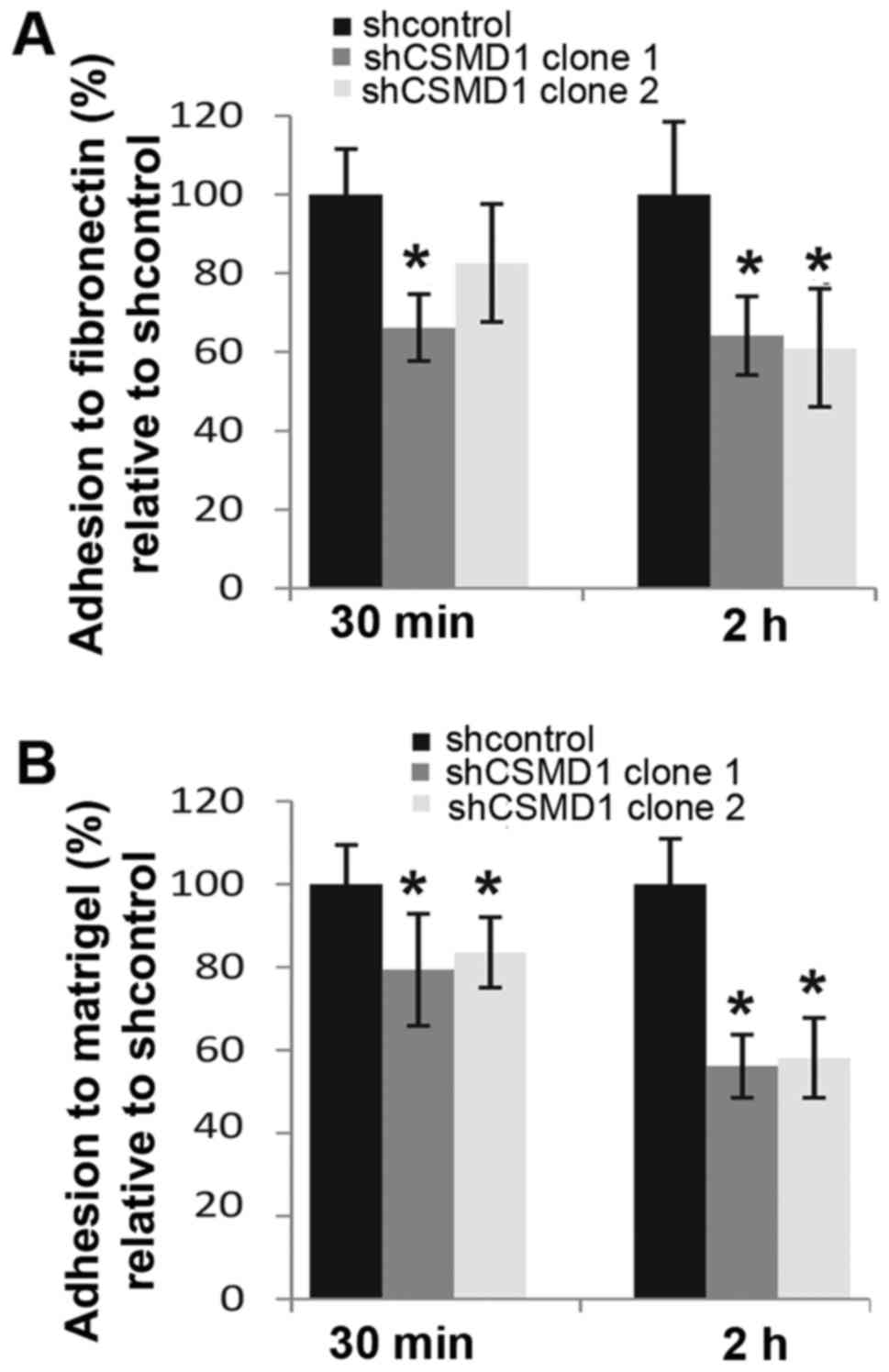
Shared homology with Cub domain containing protein 1
(CDCP1), which is involved in cell-matrix interactions (21,22),
suggested that CSMD1 could play a role in cell adhesion. To examine
this possibility adhesion assays using Matrigel and fibronectin
were performed 30 min and 2 h after cell plating. Loss of CSMD1
expression caused ~34% (SD±8.5%, p=0.001) and ~17% (SD±15%, p=0.06)
decreases in the adhesion of MCF10A shCSMD1 clones 1 and 2 to
fibronectin after 30 min, compared to shcontrol. After 2 h, this
difference had increased to ~36% (SD±10%, p=0.01) and ~40% (SD±15%,
p=0.004) respectively (Fig. 5A).
Following CSMD1 silencing, adhesion of MCF10A shCSMD1 clones 1 and
2 to Matrigel decreased by 20% (SD±13.5%, p=0.02) and 16% (SD±8.5%,
p=0.05) after 30 min compared to shcontrol. Strikingly, 2 h after
plating these differences had increased to 44% (SD±7.6%, p=0.0006)
and ~42% (SD±9.6%, p=0.001) (Fig.
5B).
Loss of CSMD1 expression disrupts
acini formation
We have previously observed that CSMD1 is highly
expressed in well-differentiated areas of breast cancer samples,
suggesting that CSMD1 might play a role in mammary duct formation
(14). To examine this further, an
MCF10A 3D Matrigel model was established and the effects of CSMD1
knockdown on the morphogenesis of mammary acini were investigated.
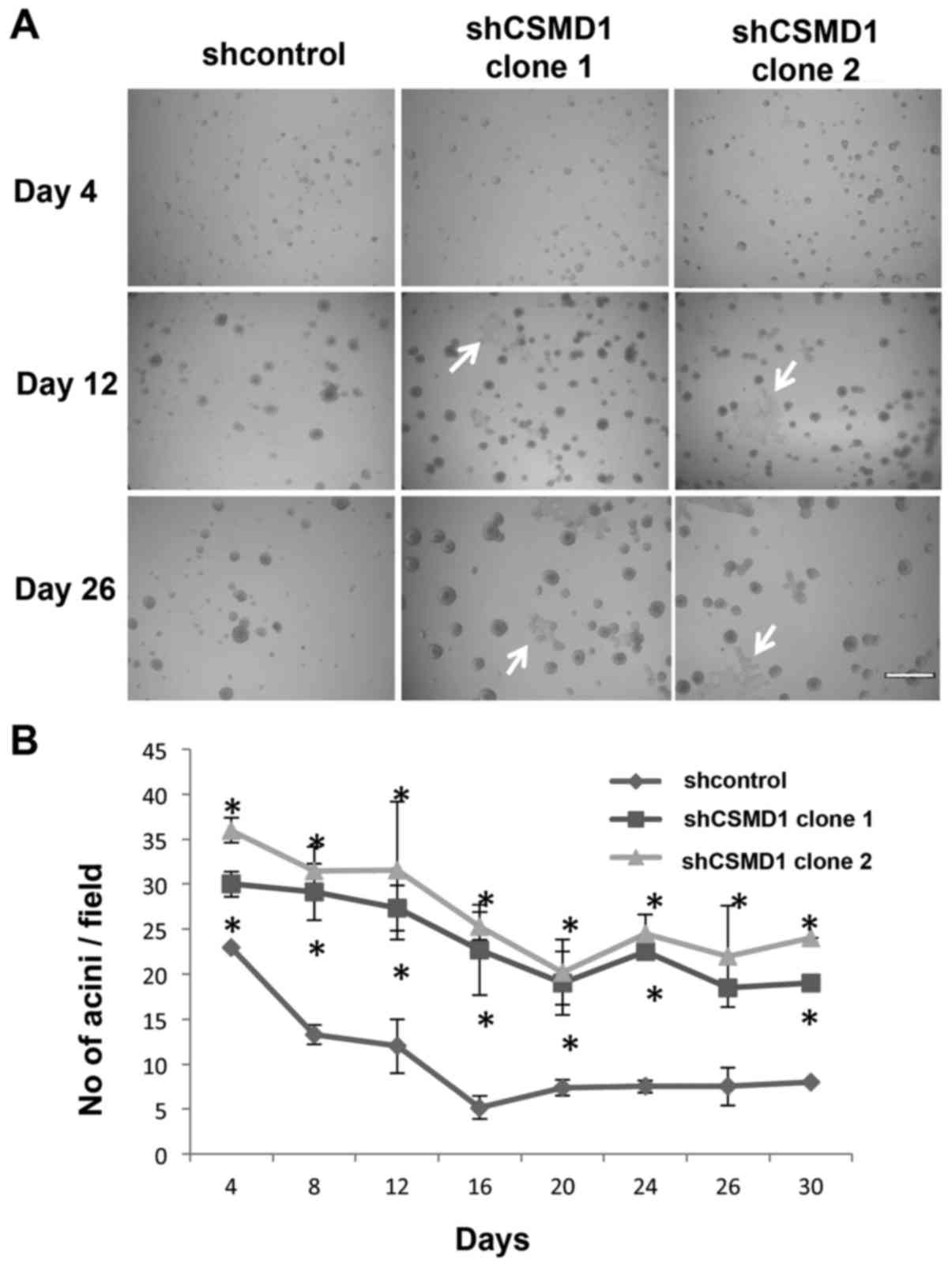
Loss of CSMD1 expression resulted in a larger number of acini and
larger areas of flat cells (Fig.
6A, arrows). Acini in 3 fields from 3–5 independent experiments
were counted and averaged every 2–4 days. There was a statistically
significant increase in the average number of shCSMD1 acini
compared to shcontrol at all time points (p<0.05) (Fig. 6B).
Phalloidin staining and confocal microscopy revealed
that shCSMD1 acini were irregular in shape and heterogeneous in
size (Fig. 7A). The diameter of
each acinus was measured and the mean diameter of acini from
shcontrol cells determined (85 µm). This was then used as a cut-off
to distinguish between large and small acini. Noteworthy, the
proportion of large acini in MCF10A shCSMD1 clones 1 and 2 were 33%
(SD±21%, p=0.1) and 43% (SD±15%, p=0.03) higher than in the
shcontrol cell line (Fig. 7B).
Furthermore, confocal imaging revealed that in shCSMD1 clones 1 and
2, respectively, 55% (SD±9.8%, p=0.045) and 90% (SD±8%, p=0.008) of
acini failed to form a lumen compared to shcontrol cells (Fig. 7C).
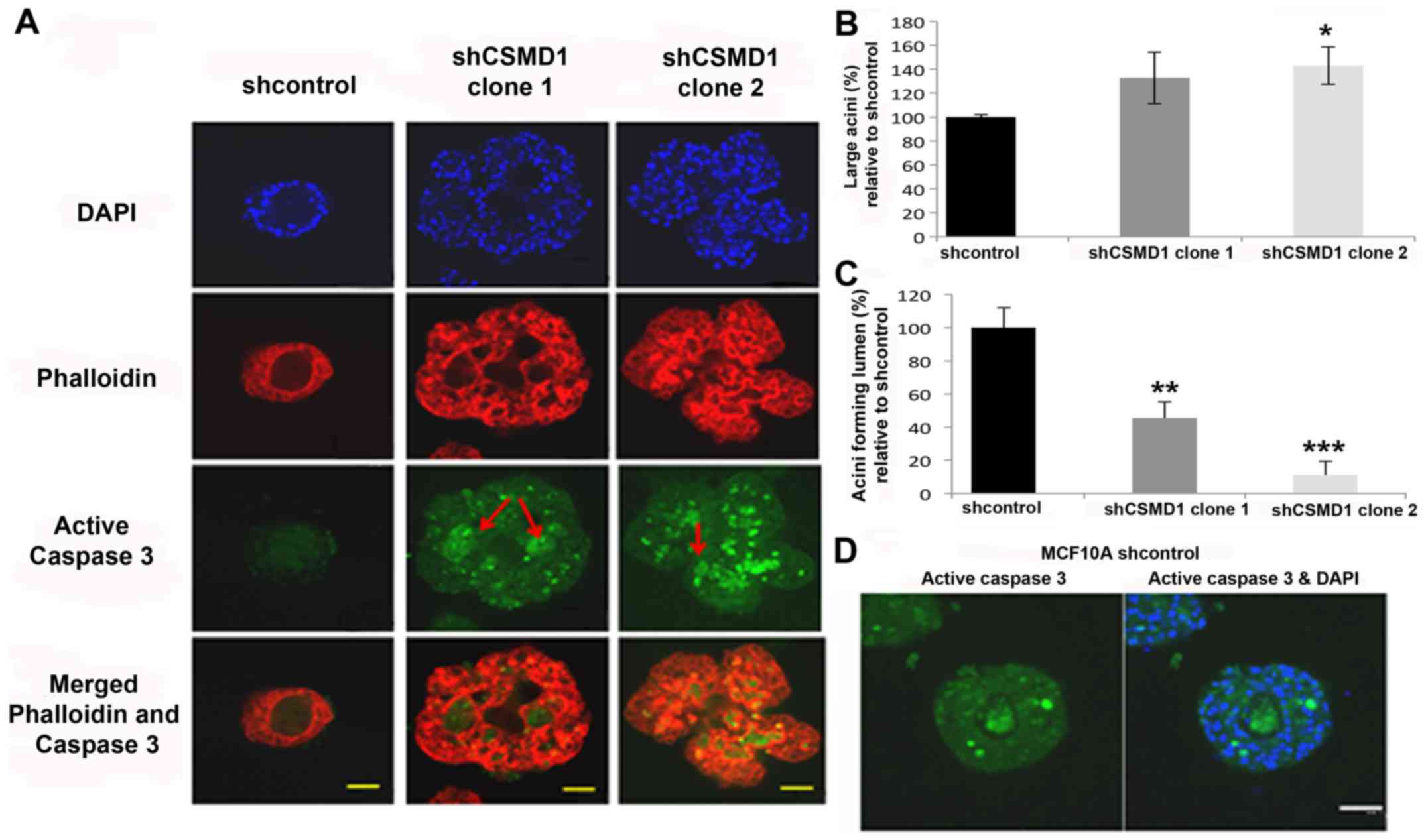 | Figure 7.Silencing CSMD1 expression disrupts
mammary acini morphology and inhibits lumen formation in the MCF10A
3D model. (A) Acini, at day 26, stained with DAPI (blue),
phalloidin (red), and active caspase-3 antibody (green).
Magnification, ×40; scale bar, 50 µm. shCSMD1 acini are irregular
in shape and heterogeneous in size with no lumen. shcontrol acini
showed weak staining for active caspase-3, while, lumens of shCSMD1
acini exhibit strong staining (arrows). (B) The percentages of
large acini, relative to shcontrol, are presented as the mean ± SD
of at least 3 independent experiments. Reduced CSMD1 expression
resulted in 33% (p=0.1) and 43% (*p=0.03) increase in the
percentage of large acini in MCF10A shCSMD1 clones 1 and 2,
respectively. (C) The percentages of lumen forming acini, relative
to shcontrol, are presented as the mean ± SD of at least 3
independent experiments. Reduced CSMD1 expression resulted in 55%
(**p=0.045) and 90% (***p=0.008) decrease in the percentage of
lumen forming acini in MCF10A shCSMD1 clones 1 and clone 2,
respectively. (D) shcontrol acini, at day 12, stained with DAPI
(blue) and active caspase-3 antibody (green). Magnification, ×40;
scale bar, 50 µm. |
In this model system lumen formation depends on
apoptosis, raising the possibility that a failure to generate acini
with a lumen might be due to differences in developmentally
regulated programmed cell death in cells where CSMD1 expression is
inhibited. To test this, acini at day 12 (before the completion of
lumen formation in shcontrol cells) and day 26 were stained with
antibodies specific for active caspase-3. At day 26, lumens of
shcontrol acini showed very weak staining compared to shCSMD1
lumens (Fig. 7A, arrows). However,
at day 12, shcontrol acini showed strong staining of active
caspase-3 (Fig. 7D). These
observations confirm that during normal acinar development in this
model system a lumen is generated by temporally coordinated
apoptotic processes. Apoptosis is then severely suppressed or
absent after the completion of lumen in shcontrol acini. In
contrast, because shCSMD1 acini failed to form a lumen, apoptotic
processes remain active until day 26.
Discussion
CSMD1 encodes a transmembrane protein and is
thought to function as a tumour suppressor. The function of CSMD1
is largely unknown. However, it is suggested to be a receptor or
co-receptor involved in signal transduction (1). Herein, we investigated the biological
consequences of loss of CSMD1 expression in cell line models. CSMD1
expression was suppressed in the cell lines MCF10A and LNCaP, using
CSMD1 shRNA pRS vectors. The specificity of these vectors was
validated, demonstrating that CSMD3 expression was not affected.
This is an important consideration since it is possible that
functional overlap between the CSMD proteins may modify the
phenotype resulting from the loss of any one of them in tumours
(1).
In both cell lines examined, downregulation of CSMD1
expression caused cell dissociation reflecting a more motile
phenotype with enhanced formation of lamellipodia and
filopodia-like protrusions. Formation of such structures is an
indicator of extensive actin polymerisation (23). Previous studies have revealed that
the rat ortholog of CSMD1 colocalizes with F-actin (4,24).
Thus CSMD1 may have an inhibitory effect on actin assembly or the
signalling processes that impact upon the regulation of this
process (25).
Further functional assays were focused on the MCF10A
cell line which demonstrated high CSMD1 protein knockdown by IHC
compared to the lower levels of protein knockdown observed in the
LNCaP cells. Loss of CSMD1 expression increased cell proliferation.
This agrees with studies that showed expression of the rat ortholog
of CSMD1 is low in brain regions exhibiting high levels of cell
proliferation (4). Similarly, a
recent study has demonstrated that increasing miRNA-10b expression
in HepG2 hepatocellular carcinoma cells resulted in decreased CSMD1
expression and increased proliferation (17). Our results also complement a study
where increased expression of CSMD1 in A375 melanoma cells reduced
proliferation (15). The inhibitory
role of CSMD1 on cell proliferation is also in line with a
potential role in cell differentiation, where CSMD1 enhances
mammary duct formation. In normal development, growth arrest
precedes differentiation and defects in the processes that control
differentiation and proliferation are associated with
carcinogenesis (26).
Reduced CSMD1 expression also enhanced cell
migration in MCF10A CSMD1 knockdown cells. A similar
increase in migration was identified in HepG2 hepatocellular
carcinoma cells with increased miRNA-10b expression resulting in
reduced CSMD1 expression (17).
These findings also complement work in A375 melanoma cells where
overexpression of CSMD1 reduced cell migration compared to control
cells (15). Migration is a process
of 4 steps; cell polarization, formation of lamellipodia, focal
adhesions, and detachment of the cell rear as the cell front
advances. Our data suggest that CSMD1 normally suppresses the
formation of cellular morphologies associated with motility (such
as lamellipodia). At focal adhesions integrins interact with actin
providing a link between the cytoskeleton and the extracellular
environment (27). The
co-localisation of CSMD with both α3-integrin and F-actin suggests
that CSMD1 may also be involved in the formation of these
structures (24). Previous work has
shown that the strength of focal adhesions determines the
translocation velocity of the cell, with maximal velocities reached
at intermediate adhesion strengths (23,28–31).
This might suggest that the presence of CSMD1 strengthens focal
adhesions, which in turn impairs migration.
Since downregulation of CSMD1 expression affected
cell migration, further investigations into whether changes in
migration were concomitant with changes in cell invasion were
undertaken by performing invasion assays using the shCSMD1
stable cell lines. Knock-down of CSMD1 expression significantly
enhanced cell invasion of MCF10A cells. The inhibitory effect of
CSMD1 on cell invasion agrees with work in HepG2 hepatocellular
carcinomas cells where reduced expression of CSMD1 due to
upregulation of miRNA-10b caused increased invasion (17). This study provides evidence of a
role for CSMD1 in cell migration and invasion. However, the
mechanisms underlying these roles have not been investigated.
Together with its effects on cell morphology, it is interesting to
speculate that the role of CSMD1 in migration and invasion is
through its ability to modulate the cell cytoskeleton. This would
be in agreement with a study by Tang et al in melanoma cells
which demonstrated CSMD1 interacts with Smad3, activates Smad1,
Smad2 and Smad3 and increase the expression of Smad4 (15). Smad3 has been shown to activate Rho
signalling and cytoskeletal reorganisation (32).
Downregulation of CSMD1 expression in MCF10A cells
decreased adhesion to Matrigel and fibronectin. The stimulatory
role of CSMD1 in cell-matrix adhesion is consistent with the
effects of other structurally similar proteins such as neuropilin-1
(33), bone morphogenetic protein 1
and TNF-stimulating gene 6 (34,35) in
this process.
In the 3D MCF10A model of duct formation, loss of
CSMD1 expression resulted in a larger number of acini and an
increase in the proportion of large acini. This may be due to the
inhibitory effect of CSMD1 on cell proliferation. Moreover, shCSMD1
acini are irregular in shape, which may be attributed to the effect
of CSMD1 on the morphology of individual cells. This finding is
similar to a study that demonstrated distorted MCF10A 3D structures
as a result of overexpressed Akt or reduced expression of PDLIM2
(36,37).
Notably, a higher proportion of shCSMD1 acini failed
to form a lumen. A filled in lumen is a hallmark feature of breast
cancer/ductal carcinoma in situ. In breast development
apoptosis plays a well-established role in lumen formation
(38,39). The MCF10A 3D model has been used
extensively to investigate lumen formation in vitro and
numerous studies have clearly demonstrated that MCF10A 3D acini
failure to form a lumen is due to increased cell proliferation
combined with inhibition of apoptosis (18,36,40–42).
In our model we show that this appears to be the case as caspase-3
activity is observed in both shCSMD1 and shcontrol acini lumen at
day 12. However, whereas in the shcontrol cells the expression of
caspase-3 is reduced after the formation of the lumen, in the
shCSMD1 acini caspase-3 expression is still strong and lumen
formation was incomplete. It maybe that this is due to the increase
in cell proliferation observed in cells with reduced CSMD1
expression. This idea agrees with studies suggesting that the
maintenance of acinar architecture relies on the ability of
increased cell death to counterbalance aberrant proliferation
(18,36,40–42).
Contrasting results were shown in a recent study in melanoma cell
where CSMD1 overexpression resulted in reduced proliferation and
increased apoptosis (15).
In future studies, it would be interesting to
investigate mammary gland morphogenesis using an available CSMD1
knockout mouse model (43). Whole
mounts of mouse fat pads would be prepared at 1, 14, 21, 28, 35
days after parturition and the ductal network and terminal buds
compared between Csmd1 (−/-) and wild type (+/+) litter mates. IHC
staining would also be performed for the proliferation marker Ki67
and apoptosis marker active caspase-3 to identify a potential
regulatory imbalance between the two processes during mammary
morphogenesis. Future studies would also address a limitation of
the current study where CSMD1 protein knockdown was detected by
IHC. Further experiments would involve the validation of the MCF10A
knockdown cell lines by western blot using the newly available
anti-CSMD1 antibody ab166908 from Abcam.
The effects of suppressing CSMD1 expression in our
cell culture models suggest that CSMD1 may achieve its widespread
effects on cell behaviour by playing a role in regulating cell
morphology. Disruption of cell morphology affects processes as
disparate as cell adhesion, proliferation, differentiation and
motility (44–46). Cell morphology is mainly regulated
by the actin cytoskeleton and CSMD1 has a potential role as a
regulator for actin polymerisation via its potential interaction
with F-actin. Furthermore, the broad effects of CSMD1 on cell
functions may be due to its role in cell-ECM adhesion and its
potential interaction with α3-integrin, which have similarly broad
effects on the regulation of actin polymerization (47,48),
cell migration, formation of membrane protrusions (49), invasion and proliferation (50).
Loss of CSMD1 expression resulted in hallmarks of
transformation including increased proliferation, migration and
invasion. Our data support the proposed role of CSMD1 as a tumour
suppressor. Potentially CSMD1 is involved in a signaling cascade
regulating a wide range of cell processes. Future work will be
aimed at dissecting the precise pathways involved.
Acknowledgements
This study was supported by an Egyptian Government
scholarship to M.K. and grants from Yorkshire Cancer Research grant
number L292 (S.M.B.) and Breast Cancer Research Campaign grant
numbers 2007NovPR53 (S.M.B. and V.S.) and 2008NovPR04 (D.H. and
V.S.).
Glossary
Abbreviations
Abbreviations:
|
CDCP1
|
CUB domain containing protein 1
|
|
CSMD1
|
CUB and sushi multiple domains 1
|
|
CSMD2
|
CUB and sushi multiple domains 2
|
|
CSMD3
|
CUB and sushi multiple domains 3
|
|
ECM
|
extracellular matrix
|
|
FCS
|
foetal calf serum
|
|
IHC
|
immunohistochemistry
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
|
|
qRT-PCR
|
quantitative real-time polymerase
chain reaction
|
|
RPLP0
|
60S acidic ribosomal protein P0
|
|
RT-PCR
|
reverse transcriptase polymerase chain
reaction
|
|
SFM
|
serum-free medium
|
|
shRNA
|
short hairpin RNA
|
References
|
1
|
Sun PC, Uppaluri R, Schmidt AP, Pashia ME,
Quant EC, Sunwoo JB, Gollin SM and Scholnick SB: Transcript map of
the 8p23 putative tumor suppressor region. Genomics. 75:17–25.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lau WL and Scholnick SB: Identification of
two new members of the CSMD gene family. Genomics. 82:412–415.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shimizu A, Asakawa S, Sasaki T, Yamazaki
S, Yamagata H, Kudoh J, Minoshima S, Kondo I and Shimizu N: A novel
giant gene CSMD3 encoding a protein with CUB and sushi multiple
domains: A candidate gene for benign adult familial myoclonic
epilepsy on human chromosome 8q23.3-q24.1. Biochem Biophys Res
Commun. 309:143–154. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kraus DM, Elliott GS, Chute H, Horan T,
Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA and
Holers VM: CSMD1 is a novel multiple domain complement-regulatory
protein highly expressed in the central nervous system and
epithelial tissues. J Immunol. 176:4419–4430. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Escudero-Esparza A, Kalchishkova N,
Kurbasic E, Jiang WG and Blom AM: The novel complement inhibitor
human CUB and Sushi multiple domains 1 (CSMD1) protein promotes
factor I-mediated degradation of C4b and C3b and inhibits the
membrane attack complex assembly. FASEB J. 27:5083–5093. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toomes C, Jackson A, Maguire K, Wood J,
Gollin S, Ishwad C, Paterson I, Prime S, Parkinson K, Bell S, et
al: The presence of multiple regions of homozygous deletion at the
CSMD1 locus in oral squamous cell carcinoma question the role of
CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer.
37:132–140. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Macoska JA, Trybus TM, Benson PD, Sakr WA,
Grignon DJ, Wojno KD, Pietruk T and Powell IJ: Evidence for three
tumor suppressor gene loci on chromosome 8p in human prostate
cancer. Cancer Res. 55:5390–5395. 1995.PubMed/NCBI
|
|
8
|
Tørring N, Borre M, Sørensen KD, Andersen
CL, Wiuf C and Ørntoft TF: Genome-wide analysis of allelic
imbalance in prostate cancer using the Affymetrix 50K SNP mapping
array. Br J Cancer. 96:499–506. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blaveri E, Brewer JL, Roydasgupta R,
Fridlyand J, DeVries S, Koppie T, Pejavar S, Mehta K, Carroll P,
Simko JP, et al: Bladder cancer stage and outcome by array-based
comparative genomic hybridization. Clin Cancer Res. 11:7012–7022.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma C, Quesnelle KM, Sparano A, Rao S, Park
MS, Cohen MA, Wang Y, Samanta M, Kumar MS, Aziz MU, et al:
Characterization of CSMD1 in a large set of primary lung, head and
neck, breast and skin cancer tissues. Cancer Biol Ther. 8:907–916.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paris PL, Andaya A, Fridlyand J, Jain AN,
Weinberg V, Kowbel D, Brebner JH, Simko J, Watson JE, Volik S, et
al: Whole genome scanning identifies genotypes associated with
recurrence and metastasis in prostate tumors. Hum Mol Genet.
13:1303–1313. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang R and Song C: Loss of CSMD1 or 2 may
contribute to the poor prognosis of colorectal cancer patients.
Tumour Biol. 35:4419–4423. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shull AY, Clendenning ML, Ghoshal-Gupta S,
Farrell CL, Vangapandu HV, Dudas L, Wilkerson BJ and Buckhaults PJ:
Somatic mutations, allele loss, and DNA methylation of the Cub and
Sushi Multiple Domains 1 (CSMD1) gene reveals association with
early age of diagnosis in colorectal cancer patients. PLoS One.
8:e587312013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kamal M, Shaaban AM, Zhang L, Walker C,
Gray S, Thakker N, Toomes C, Speirs V and Bell SM: Loss of CSMD1
expression is associated with high tumour grade and poor survival
in invasive ductal breast carcinoma. Breast Cancer Res Treat.
121:555–563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang M-R, Wang Y-X, Guo S, Han S-Y and
Wang D: CSMD1 exhibits antitumor activity in A375 melanoma cells
through activation of the Smad pathway. Apoptosis. 17:927–937.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lang M-F, Yang S, Zhao C, Sun G, Murai K,
Wu X, Wang J, Gao H, Brown CE, Liu X, et al: Genome-wide profiling
identified a set of miRNAs that are differentially expressed in
glioblastoma stem cells and normal neural stem cells. PLoS One.
7:e362482012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang
JR, Han XJ, Zhu SJ, Wang SM, Li YH, et al: miR-10b exerts oncogenic
activity in human hepatocellular carcinoma cells by targeting
expression of CUB and sushi multiple domains 1 (CSMD1). BMC Cancer.
16:8062016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Debnath J, Muthuswamy SK and Brugge JS:
Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini
grown in three-dimensional basement membrane cultures. Methods.
30:256–268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shaw KR, Wrobel CN and Brugge JS: Use of
three-dimensional basement membrane cultures to model
oncogene-induced changes in mammary epithelial morphogenesis. J
Mammary Gland Biol Neoplasia. 9:297–310. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Holliday DL, Hughes S, Shaw JA, Walker RA
and Jones JL: Intrinsic genetic characteristics determine
tumor-modifying capacity of fibroblasts: Matrix metalloproteinase-3
5A/5A genotype enhances breast cancer cell invasion. Breast Cancer
Res. 9:R672007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scherl-Mostageer M, Sommergruber W,
Abseher R, Hauptmann R, Ambros P and Schweifer N and Schweifer N:
Identification of a novel gene, CDCP1, overexpressed in human
colorectal cancer. Oncogene. 20:4402–4408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benes CH, Poulogiannis G, Cantley LC and
Soltoff SP: The SRC-associated protein CUB Domain-Containing
Protein-1 regulates adhesion and motility. Oncogene. 31:653–663.
2012.PubMed/NCBI
|
|
23
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kraus DM, Pfenninger KH, Sanford SD and
Holers VM: CSMD1 is expressed as a membrane protein on neuronal
growth cones that colocalizes with F-actin and alpha-3 integrin.
Mol Immunol. 44:1982007. View Article : Google Scholar
|
|
25
|
Ulrich F and Heisenberg C-P: Trafficking
and cell migration. Traffic. 10:811–818. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scott RE, Tzen CY, Witte MM, Blatti S and
Wang H: Regulation of differentiation, proliferation and cancer
suppressor activity. Int J Dev Biol. 37:67–74. 1993.PubMed/NCBI
|
|
27
|
Ziober BL, Silverman SS Jr and Kramer RH:
Adhesive mechanisms regulating invasion and metastasis in oral
cancer. Crit Rev Oral Biol Med. 12:499–510. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huttenlocher A, Sandborg RR and Horwitz
AF: Adhesion in cell migration. Curr Opin Cell Biol. 7:697–706.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lambrechts A, Van Troys M and Ampe C: The
actin cytoskeleton in normal and pathological cell motility. Int J
Biochem Cell Biol. 36:1890–1909. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Palecek SP, Huttenlocher A, Horwitz AF and
Lauffenburger DA: Physical and biochemical regulation of integrin
release during rear detachment of migrating cells. J Cell Sci.
111:929–940. 1998.PubMed/NCBI
|
|
31
|
Cox EA, Sastry SK and Huttenlocher A:
Integrin-mediated adhesion regulates cell polarity and membrane
protrusion through the Rho family of GTPases. Mol Biol Cell.
12:265–277. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee J, Moon HJ, Lee JM and Joo CK: Smad3
regulates Rho signaling via NET1 in the transforming growth
factor-beta-induced epithelial-mesenchymal transition of human
retinal pigment epithelial cells. J Biol Chem. 285:26618–26627.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Valdembri D, Caswell PT, Anderson KI,
Schwarz JP, König I, Astanina E, Caccavari F, Norman JC, Humphries
MJ, Bussolino F, et al: Neuropilin-1/GIPC1 signaling regulates
alpha5beta1 integrin traffic and function in endothelial cells.
PLoS Biol. 7:e252009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang G, Zhang Y, Kim B, Ge G, Annis DS,
Mosher DF and Greenspan DS: Fibronectin binds and enhances the
activity of bone morphogenetic protein 1. J Biol Chem.
284:25879–25888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuznetsova SA, Mahoney DJ, Martin-Manso G,
Ali T, Nentwich HA, Sipes JM, Zeng B, Vogel T, Day AJ and Roberts
DD: TSG-6 binds via its CUB_C domain to the cell-binding domain of
fibronectin and increases fibronectin matrix assembly. Matrix Biol.
27:201–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Debnath J, Walker SJ and Brugge JS: Akt
activation disrupts mammary acinar architecture and enhances
proliferation in an mTOR-dependent manner. J Cell Biol.
163:315–326. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deevi RK, Cox OT and O'Connor R: Essential
function for PDLIM2 in cell polarization in three-dimensional
cultures by feedback regulation of the β1-integrin-RhoA signaling
axis. Neoplasia. 16:422–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Humphreys RC, Krajewska M, Krnacik S,
Jaeger R, Weiher H, Krajewski S, Reed JC and Rosen JM: Apoptosis in
the terminal endbud of the murine mammary gland: A mechanism of
ductal morphogenesis. Development. 122:4013–4022. 1996.PubMed/NCBI
|
|
40
|
Debnath J, Mills KR, Collins NL, Reginato
MJ, Muthuswamy SK and Brugge JS: The role of apoptosis in creating
and maintaining luminal space within normal and oncogene-expressing
mammary acini. Cell. 111:29–40. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hebner C, Weaver VM and Debnath J:
Modeling morphogenesis and oncogenesis in three-dimensional breast
epithelial cultures. Annu Rev Pathol. 3:313–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yanochko GM and Eckhart W: Type I
insulin-like growth factor receptor over-expression induces
proliferation and anti-apoptotic signaling in a three-dimensional
culture model of breast epithelial cells. Breast Cancer Res.
8:R182006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Steen VM, Nepal C, Ersland KM, Holdhus R,
Nævdal M, Ratvik SM, Skrede S and Håvik B: Neuropsychological
deficits in mice depleted of the schizophrenia susceptibility gene
CSMD1. PLoS One. 8:e795012013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Boivin D, Bilodeau D and Béliveau R:
Regulation of cytoskeletal functions by Rho small GTP-binding
proteins in normal and cancer cells. Can J Physiol Pharmacol.
74:801–810. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rao KM and Cohen HJ: Actin cytoskeletal
network in aging and cancer. Mutat Res. 256:139–148. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Trump BF, Heatfield BM, Phelps PC,
Sanefuji H and Shamsuddin AK: Cell surface changes in preneoplastic
and neoplastic epithelium. Scan Electron Microsc. 3:43–60.
1980.
|
|
47
|
Vermeulen K, Van Bockstaele DR and
Berneman ZN: The cell cycle: A review of regulation, deregulation
and therapeutic targets in cancer. Cell Prolif. 36:131–149. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kharitonova MA and Vasiliev JM:
Controlling cell length. Semin Cell Dev Biol. 19:480–484. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Frame MC and Brunton VG: Advances in
Rho-dependent actin regulation and oncogenic transformation. Curr
Opin Genet Dev. 12:36–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Alexandrova AY: Evolution of cell
interactions with extracellular matrix during carcinogenesis.
Biochemistry (Mosc). 73:733–741. 2008. View Article : Google Scholar : PubMed/NCBI
|