Introduction
Gastric cancer (GC) is one of the most common
malignancies, ranking second, and is the fourth leading cause of
cancer-related mortality in China (1). Although the survival time of patients
with GC has improved during the past decade, prognosis remains poor
for patients with advanced GC. The 5-year survival rate for
patients with localized disease is ~60%, and only 3–5% for those
with distant metastasis (2,3). In consideration of the low efficacy of
traditional chemotherapy and radiotherapy, molecular-targeted
therapy, due to its high efficiency, is an attractive therapeutic
strategy (4).
The epidermal growth factor receptor (EGFR)
gene is located at the chromosomal region 7p12 and encodes a 70-kDa
transmembrane tyrosine kinase receptor that contributes to cancer
progression by mediating cellular proliferation, migration,
adhesion and metastasis (5).
Approximately 30% of GC patients are reported to show EGFR protein
overexpression, and thus EGFR signaling pathways serve as
attractive therapeutic targets (6,7).
However, to date, all the agents targeting EGFR have failed to
demonstrate any antitumor efficacy for treating GC patients in the
clinic, presumably since no precise patient inclusion criteria were
set for the clinical trials (8).
Intriguing retrospective biomarker analyses of the clinical trials
suggest that a subpopulation of tumors with EGFR gene copy number
gain or protein overexpression may benefit from anti-EGFR therapy,
implying that refining the EGFR biomarker, may yet yield positive
results (9). EGFR protein
expression or gene amplification may serve as an effective
biomarker for predicting the clinical benefit of anti-EGFR therapy
(9,10). However, various studies have shown
that the levels of EGFR gene amplification are not always
consistent with those of the protein expression in GC which
indicate that a post-transcriptional regulation may exist (11,12).
Further in detail research of the regulatory mechanism underlying
the EGFR protein expression in GC may be of important clinical
significance to guide targeted therapy.
MicroRNAs (miRNAs) are a class of endogenous,
non-coding, single-stranded RNAs (~22 bp) that regulate gene
expression by directly binding with the 3′-untranslated region
(3′-UTR) of target mRNAs, causing degradation of the specific mRNA
sequence or translational inhibition at the post-transcriptional
level (13). Research indicates
that over 30% of human genes are regulated by miRNAs (14,15).
Acting as oncogenes or tumor suppressors, miRNAs can repress the
expression of cancer-related genes (16). Several studies have reported that
alterations in miRNA levels occur in various human tumors,
including GC, where miRNAs are related to several aspects of cancer
pathogenesis including self-renewal, invasion and metastasis by
targeting EGR2 (17), MAPK
(18) and PI3K/AKT signaling
pathways (19). Therefore, we
speculate that specific miRNAs may participate in the regulation of
EGFR protein expression in GC.
In a previous study, miR-370 was reported to
function as a tumor suppressor in colorectal cancer by suppressing
EGFR signaling (20). It has been
reported that miR-370 is also significantly downregulated in GC
(21). However, whether EGFR
protein expression is regulated by miR-370 in GC remains unknown.
In the present study, we used immunohistochemistry (IHC), western
blotting and quantitative RT-PCR (RT-qPCR) assay to study the
relationship between the EGFR protein and the mRNA level in GC
tissues. We also used bioinformatics tools and dual-luciferase
assay to identify and validate the potential miRNA for targeting
EGFR. Finally, a series of functional experiments in vitro
were carried out to confirm the role of miR-370. We aimed to
investigate whether EGFR protein overexpression in GC is modulated
by miR-370 at the post-transcriptional level and how it may impact
the biological behavior of GC cells.
Materials and methods
Human tissues
Human GC and paired adjacent non-cancerous tissues
were derived from patients undergoing radical surgery at the
Tianjin Medical University Cancer Institute and Hospital (Tianjin,
China). Both tumor and non-cancerous tissues were confirmed
histologically and the pathological type of cancer was
adenocarcinoma. Written informed consent was obtained in all cases,
and all aspects of the present study were approved by the Ethics
Committee of Tianjin Medical University Cancer Institute and
Hospital.
Cell line and culture
The human GC cell line MGC-803 was purchased from
the Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). The human embryonic kidney epithelial cell line
HEK293T was obtained from the American Type Culture Collection
(ATCC; Manassas, VA, USA). MGC-803 and HEK293T cells were cultured
in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS) (both from Gibco, Carlsbad, CA, USA), with
routine addition of ampicillin and streptomycin.
Cell transfection
MGC-803 cells were seeded in 6-, 12- or 24-well
plates, and transfected with Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA) 24 h later according to the manufacturer's
instructions. Equal amounts of miR-370 mimics, inhibitors and
negative control (RiboBio, Guangzhou, China) were used for miRNA
overexpression and downregulation. miR-370 mimics are small,
chemically modified double-stranded RNAs that mimic endogenous
miR-370 and enable miR-370 functional analysis by upregulation of
miR-370 activity. miR-370 inhibitors are small, chemically modified
single-stranded RNA molecules designed to specifically bind to and
inhibit endogenous miR-370 molecules and enable miR-370 functional
analysis by downregulation of miR-370 activity. For the purpose of
EGFR upregulation, an EGFR overexpression plasmid without
miR-370-responsive 3′-UTR was put into use and an empty plasmid was
used as a negative control. Two siRNAs targeting human EGFR were
used for EGFR downregulation (EGFR siRNA-1, AACACAGUGGA GCGAAUUCCU;
EGFR siRNA-2, CGCAAAGUGUGUAAC GGAAUA) and also a scrambled siRNA
(both from RiboBio) was used as a negative control.
miRNA target prediction and
dual-lufierase reporter assay
miRNAs potentially targeting the 3′-UTR of EGFR were
identified using 3 internet-based bioinformatic algorithms from
miRanda (http://www.microrna.org/) (22), TargetScan (http://www.targetscan.org/) (23) and PicTar (http://pictar.mdc-berlin.de/) (24). Target recognition is based on
complementarity of the nucleotide sequence to the 3-UTR region of
EGFR mRNA. The miRNAs that were predicted by at least 2 of the
algorithms to bind were accepted as candidates for further study.
miRNA expression data were obtained from microRNA.org (http://www.microrna.org/). The predicted 3′-UTR
miR-370 targeting region was inserted into the pMIR-REPORT plasmid
(Ambion, Austin, TX, USA) and was then affirmed by DNA sequencing.
Reporter assays were performed 36 h post-transfection using the
Dual-Luciferase Assay System (Promega, Madison, WI, USA). The
intensity of firefly luciferase was normalized to Renilla
luciferase.
Protein extraction and western
blotting
Proteins were extracted from the human tissues and
cultured cells in lysis buffer added with a protease inhibitor.
Total cell lysates were separated on SDS-PAGE gels and transferred
to polyvinylidene fluoride (PVDF) membranes (Roche, Mannheim,
Germany). After blocking with 2% bovine serum albumin (BSA), the
membrane was incubated with anti-EGFR (mouse, monocolonal) and
anti-GAPDH (mouse, monocolonal) (both from Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) antibodies overnight at
4°C. Then the membranes were incubated with the secondary
antibodies and visualized with an enhanced chemiluminescencent
horseradish peroxidase (HRP) substrate kit (Millipore, Billerica,
MA, USA) according to the manufacturer's instructions.
RNA isolation and RT-qPCR
Total RNA was isolated from the cultured cells and
tissues using TRIzol reagent (Invitrogen) according to the
manufacturer's protocol. The RNA concentration was quantified using
a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific,
Waltham, MA, USA). The cDNA was synthesized from 1 µg of total RNA
using primers specific to EGFR and GAPDH in conditions as follows:
16°C for 15 min, 42°C for 60 min and 85°C for 5 min. The quantity
of miR-370 was analyzed using TaqMan miRNA probes (Applied
Biosystems, Foster City, CA, USA). The PCR was initiated by a 5-min
hold at 95°C, followed by 40 cycles of denaturation at 95°C for 15
sec and annealing/extension at 60°C for 1 min. All the reactions
were run in triplicate. U6 snRNA was used as an internal control
for miRNA. The mRNA level of EGFR was normalized to GAPDH. The ΔCt
(cycle threshold) method was applied to calculate the relative
level of the target gene that was normalized to the control through
the equation: 2−ΔCt, in which ΔCt = Ctgene -
Ctcontrol. Primer sequences for EGFR and GAPDH were as
follows: EGFR sense, 5′-TTGCCGCAAAGTGTGTAACG-3′ and EGFR antisense,
5′-GTCACCCCTAAATGCCACCG-3′; GAPDH sense, 5′-AGAAGGCTGGGGCTCATTTG-3′
and GAPDH antisense, 5′-AGGGGCCATCCACAGTCTTC-3′.
Cell proliferation assay
MGC-803 cells were seeded in 24-well plates
(4×104 cells/well) and transfected with miR-370 mimics,
inhibitors, EGFR overexpression plasmid, EGFR siRNAs or the
relevant negative controls. Twenty-four hours after transfection, a
Cell-Light 5-ethynyl-2′-deoxyuridine (EdU) Apollo DNA cell kit
(RiboBio) was used to measure the cell proliferation ability
according to the manufacturer's instructions as previously
described (25). Briefly, EdU at a
concentration of 50 µM/ml was added into the medium and cultured
for 5 h. Then, the cells were fixed with 4% paraformaldehyde for 30
min and treated with 0.5% Triton X-100 for 15 min. After being
washed in phosphate-buffered saline (PBS), the cells were incubated
in darkness with a fluorescent dye of Apollo which was incorporated
with EdU for 30 min, and then repeatedly washed. The cell nuclei
were then stained by Hoechst 33342 for another 30 min. Eventually,
EdU-labeled cells were counted manually in 5 fields of view
randomly selected from each well. The EdU-labeling index was
calculated as the ratio of EdU add-in cells to Hoechst
33342-stained cells. All of the stainings were performed in
triplicate.
Transwell cell migration assay
The capability of cell migration was assessed using
a 24-well Boyden chamber with an 8.0-µm pore polycarbonate membrane
insert (Corning, Corning, NY, USA) according to the manufacturer's
instructions. At 24 h post-transfection, ~105 cells were
seeded in the upper chamber with 200 µl serum-free medium.
Simultaneously, 600 µl medium with 10% FBS was added into the lower
chamber for purpose of chemotaxis. Another 24 h later, the
membranes were fixed with methanol and stained with a 3-Step Stain
Set (Thermo Scientific, UK). All assays were performed in
triplicate. To minimize the bias, at least 3 randomly selected
fields were counted at a magnification of ×200, under a
microscope.
Wound healing assay
MGC-803 cells were seeded into 12-well plates, and
transfected using the method as described in ‘Cell transfection’.
The day after transfection, when the density of the cells was over
90%, each well was scratched to create 2 empty linear regions with
a 10-µl pipette tip. Subsequently, the cells were cultured in DMEM
without FBS in a humidified incubator. The wound healing was
observed and imaged at 0, 12 and 24 h after scraping. The distance
of the wound zone was measured for at least 3 randomly selected
fields.
IHC assay
Formalin-fixed paraffin-embedded specimens of both
GC and paired adjacent non-cancerous tissues were sectioned (8-µm
thick) and stained with an anti-EGFR antibody (mouse, monocolonal;
Santa Cruz Biotechnology) at a 1:100 dilution. The
3,3-diaminobenzidine (DAB) system (Zhongshanjinqiao, Beijing,
China) was applied to confirm the positive staining. Six random
fields were selected for each specimen.
Statistical analysis
SPSS Statistics 20.0 (SPSS, Inc., Chicago, IL, USA)
was used to analyze the data. Differences between groups were
tested by analysis of variance and an unpaired Student's t-test.
p<0.05 was considered as statistical significance.
Results
Biological role of EGFR in GC
cells
We first investigated the effect of EGFR on cell
proliferation and migration in GC cells. Plasmid and 2 siRNAs were
used to overexpress or knock down EGFR in the MGC-803 cells. As
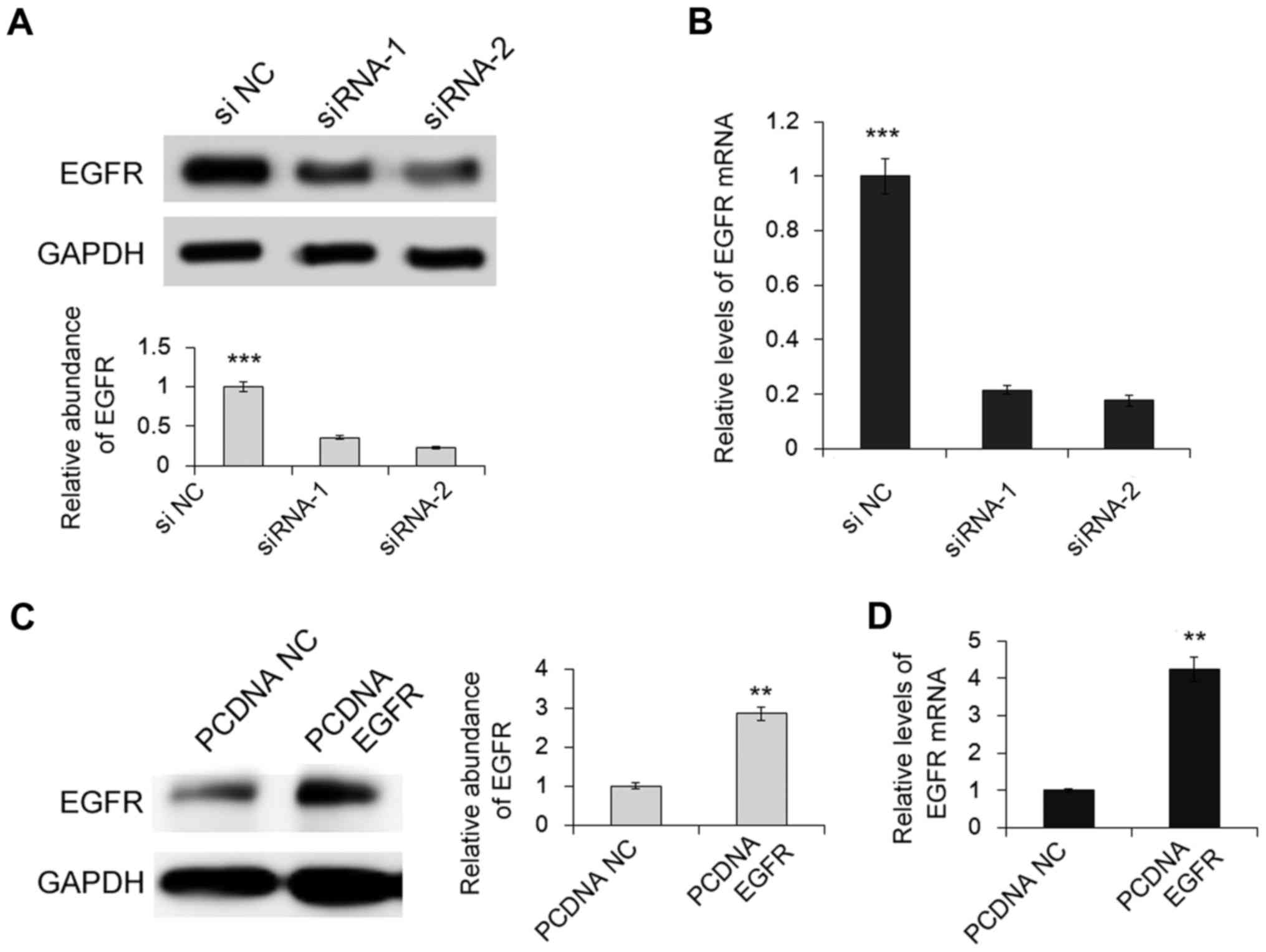
shown in Fig. 1A-D, the expression
levels of both EGFR protein and mRNA were markedly upregulated by
the plasmid and were downregulated by the siRNAs as compared with
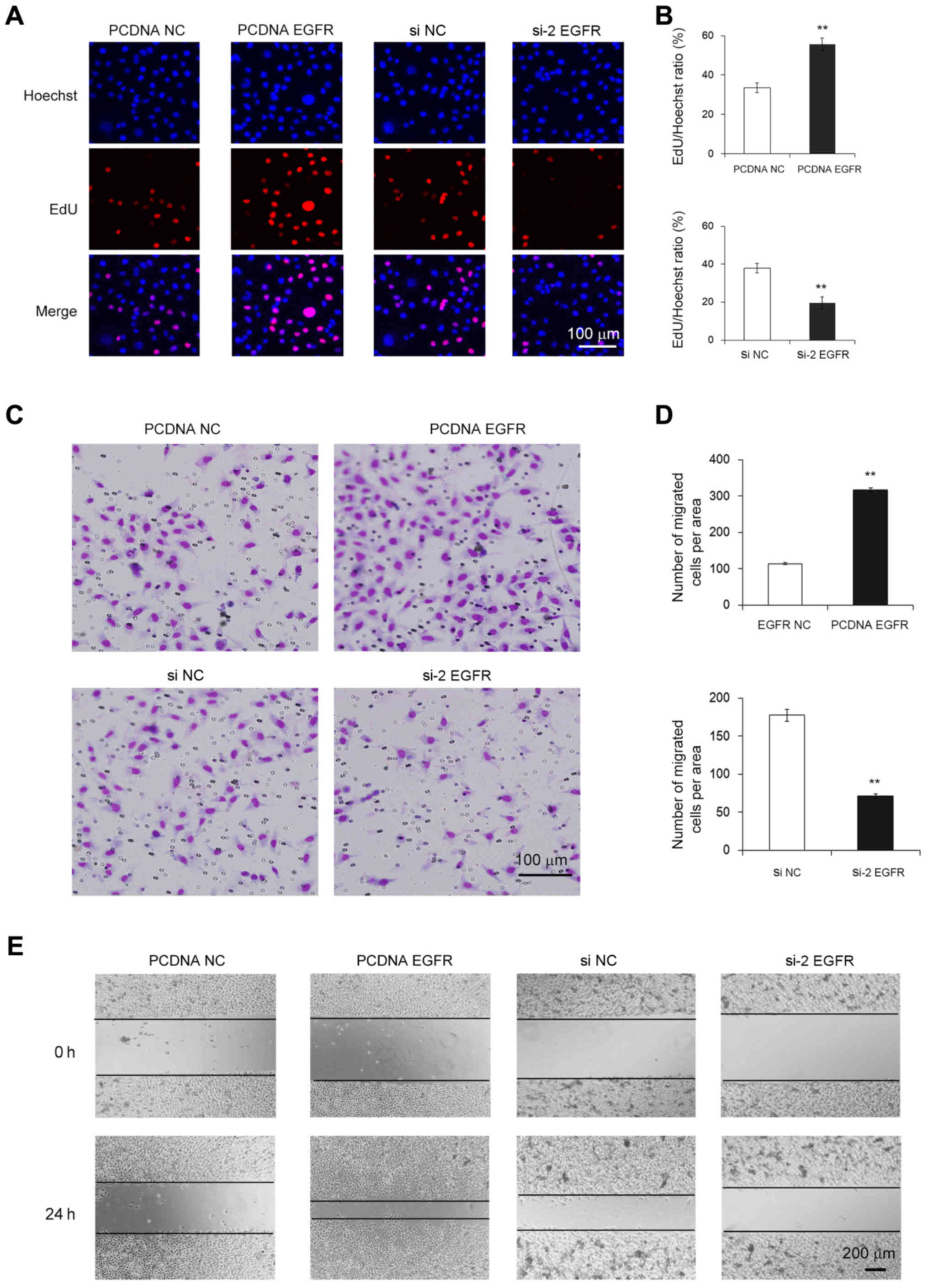
the normal control. MGC-803 cells in which EGFR protein was
overexpression exhibited a higher rate of proliferation and
increased migration, whereas the cells with knockdown of EGFR
exhibited a notably lower rate of proliferation and decreased
migration (Fig. 2A-E). Thus, EGFR
acts as a cancer promoter in GC and its downregulation decreased
the proliferation and migration of cancer cells.
Identification of miR-370 as a
potential regulator of EGFR
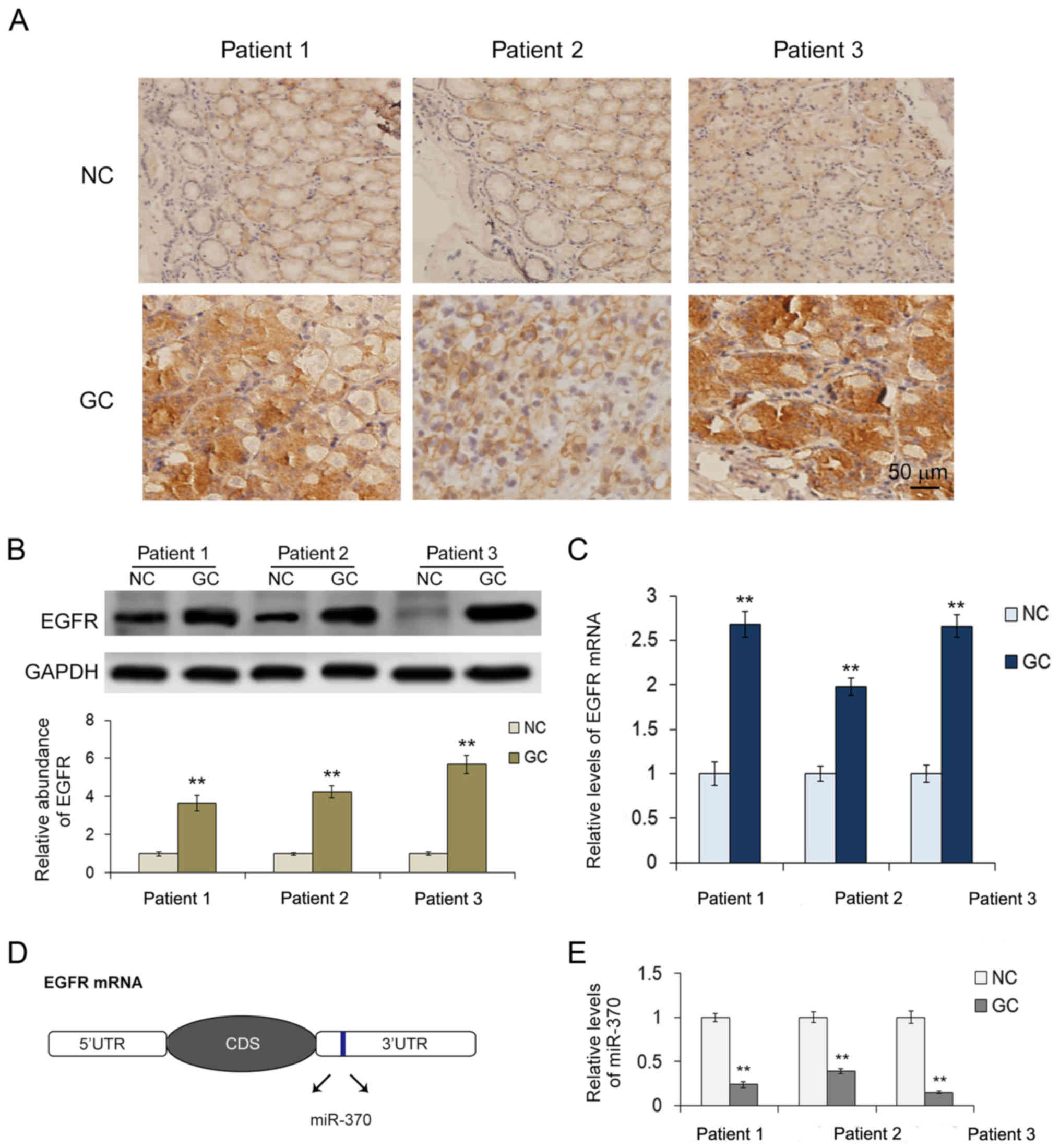
To further understand the expression regulation of
EGFR, IHC was applied to analyze the expression pattern of EGFR
protein in GC. Ten pairs of GC and adjacent non-cancerous tissues
derived from patients undergoing radical gastrectomy were collected
in the present study. Results showed that 3 patients had high EGFR
protein expression in the GC tissues compared with the paired
non-cancerous tissues (Fig. 3A).
These 3 patient tissues were further analyzed using western
blotting and RT-qPCR. As shown in Fig.
3B, the levels of EGFR protein in the GC tissues were ~4 times
higher than that in the corresponding paired non-cancerous tissues.
However, the mRNA levels of EGFR were only ~2-fold as high as that
of the normal control (Fig. 3C).
The disparity between the protein and the mRNA suggests that
expression of EGFR in GC may be regulated at a post-transcription
level. As miRNA-mediated regression of mRNA transcripts is thought
to be one of the most important modes of post-transcriptional
regulation, 3 publicly available computational algorithms were then
used to predict the potential miRNAs targeting EGFR. Both miRanda
and TargetScan predictions showed that the 3′-UTR of EGFR mRNA
contains a potential miR-370-binding site (Fig. 3D). To identify the relationship
between miR-370 and EGFR in the GC, we further explored the levels
of miR-370 in the 3 pairs of GC and the corresponding paired
non-cancerous tissues. As expected, miR-370 showed an obvious
decrease in all of the tumor tissues (Fig. 3E). Thus, we proposed that miR-370 is
a regulator of EGFR in GC and we selected miR-370 for further
experiments.
Validation of miR-370 as a regulator
of EGFR
The levels of miR-370 and EGFR protein showed an
inverse correlation in the GC tissues, and the prediction by
bioinformatics suggested that miR-370 is a potential regulator of
EGFR. We then further validated miR-370 as a regulator of EGFR. As
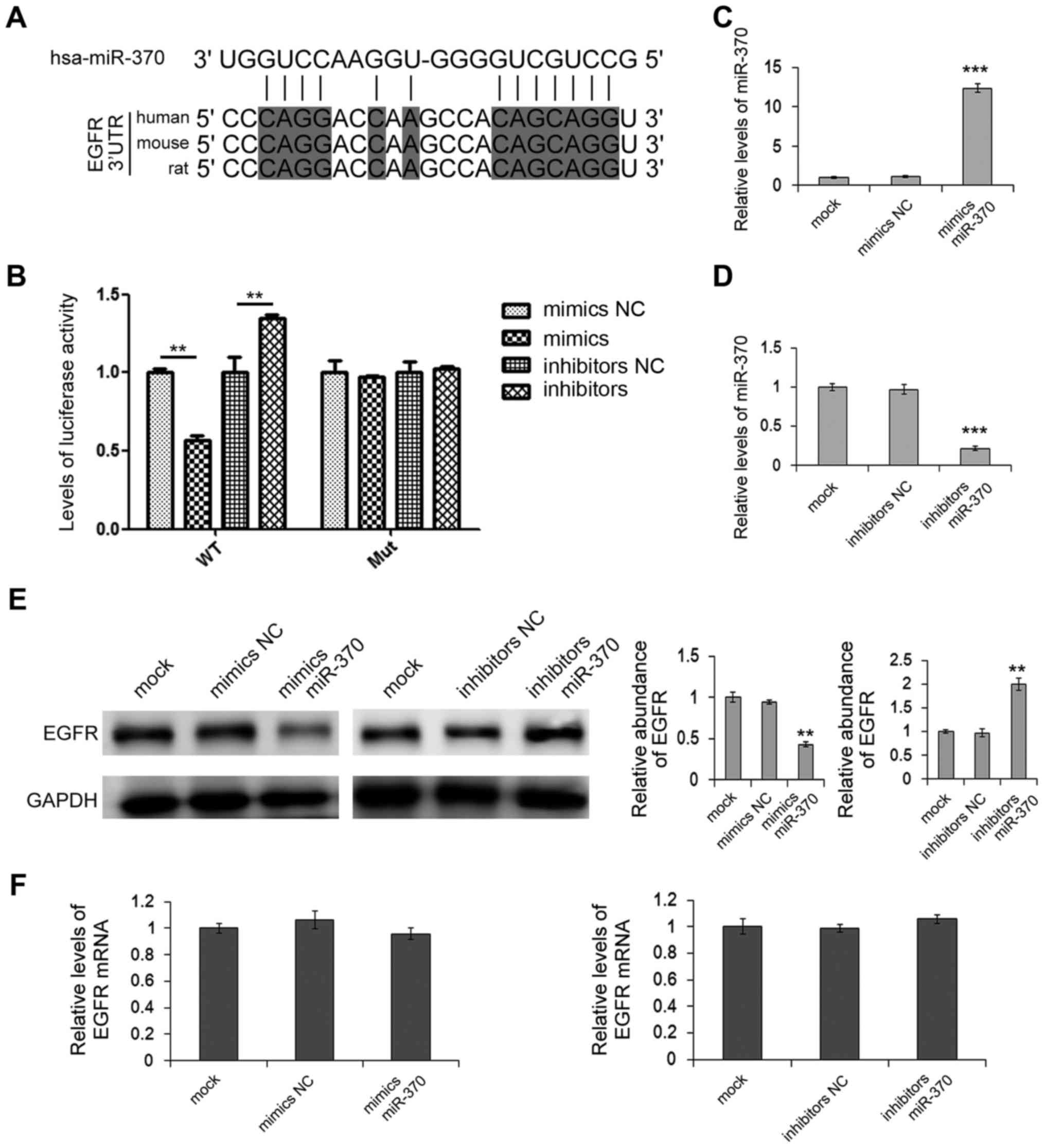
shown in Fig. 4A, bioinformatics
results showed that miR-370 can bind with 3′-UTR of EGFR mRNA with
7-mer seeds and this putative miR-370-binding site and its adjacent
sequences are highly conserved among vertebrates. Dual-luciferase
assay showed that the relative levels of luciferase activity of the
reporter gene were significantly inhibited when HEK293T cells were
cotransfected with miR-370 mimics and the luciferase reporters
containing the predicted target regions of EGFR mRNA, while the
inhibition was lost when the binding sites in 3′-URT were mutated
(Fig. 4B). Next, relative levels of
miR-370 and the expression of EGFR protein and mRNA in the MGC-803
cells were also respectively detected, using RT-qPCR or western
blot analysis following transfection of mimics or inhibitors. As
shown in Fig. 4C and D, miR-370
mimics significantly increased the miR-370 levels in the MGC-803
cells, while miR-370 inhibitors markedly downregulated the miR-370
levels. Overexpression of miR-370 led to a reduction in EGFR
protein, whereas inhibition of miR-370 led to enhanced expression
of the EGFR protein (Fig. 4E).
Furthermore, miR-370 did not affect the mRNA levels of EGFR in the
MGC-803 cells (Fig. 4F). These data
demonstrated that miR-370 is a vital post-transcriptional regulator
of EGFR in the GC cells, and miR-370 regulates EGFR protein
expression by directly targeting the 3′-UTR of EGFR mRNA.
miR-370 suppresses cell proliferation
and migration
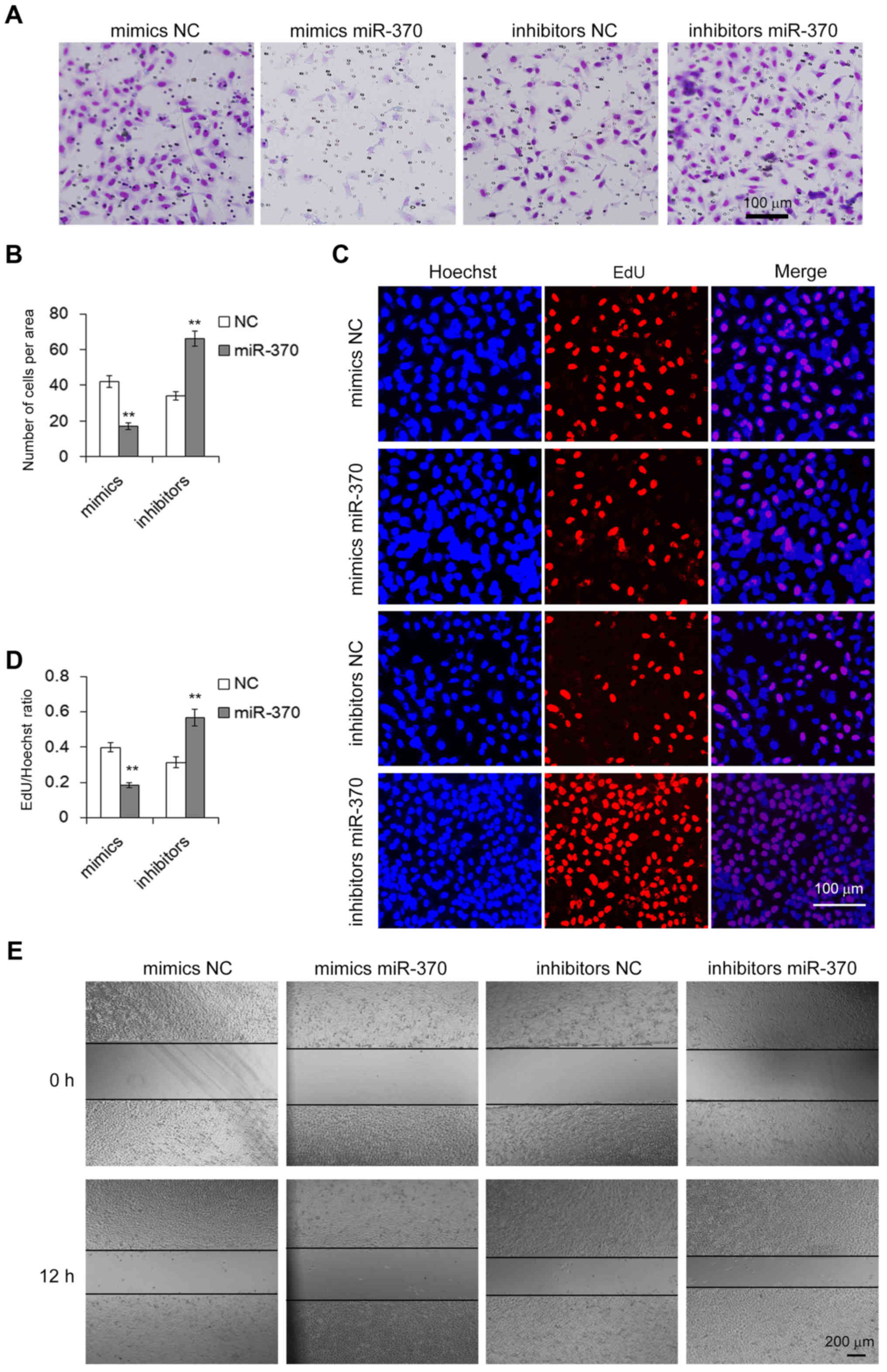
We next assessed the biological effects of miR-370
in the GC cells in vitro. Transwell assay was performed to
evaluate the migration capacity of MGC-803 cells transfected with
the miR-370 mimics or inhibitors. We found that upregulation of
miR-370 inhibited cell migration, whereas downregulation of miR-370
promoted migration (Fig. 5A and B).
The cell proliferation activity was determined by the Cell-Light
EdU Apollo DNA cell kit. As expected, a high level of miR-370
resulted in a significant suppression of cell growth capacity,
while inhibition of miR-370 led to an increase in cell
proliferation (Fig. 5C and D). We
also conducted a wound healing assay to evaluate the migration
ability of the transfected cells. In accordance with the results of
the Transwell assay, overexpression of miR-370 showed a lower rate
of migration and knockdown of miR-370 caused an increase in cell
migration (Fig. 5E). These results
demonstrated that miR-370 is an oncosuppressor-miRNA and plays an
important role in the biological process in GC cells.
Discussion
In the present study, we obtained a number of
results regarding EGFR and miR-370 in gastric cancer (GC). EGFR
protein expression was controlled by post-transcriptional
regulation. An miRNA regulatory mechanism was involved in this
process. The biological behavior of GC cells was manipulated by
altering the levels of miR-370. Our results supported these
suggestions. The expression level of EGFR protein in GC tissues was
not consistent with that of the mRNA level in comparison to paired
adjacent non-cancerous tissues. Bioinformatics tools and
dual-luciferase assay validated that miR-370 is a regulator of
EGFR. Overexpression of miR-370 suppressed the proliferation and
migration of the GC cells. In addition, downregulation of miR-370
promoted the proliferation and migration of the GC cells.
The role of EGFR in the pathogenesis of human
malignancy has been established in a variety of cancers (26). Previous studies have shown that EGFR
signaling pathways are frequently dysregulated in GC and anti-EGFR
is a potential therapeutic target (6,7). The
present study also showed that the suppression of EGFR can inhibit
the proliferation and migration of GC cells. Although, to date, all
the randomized controlled phase III trials have failed to
demonstrate any efficacy of anti-EGFR therapy, subpopulation
analysis of the recruited patients showed that patients with EGFR
gene copy number gain or protein overexpression may benefit from
anti-EGFR therapy in GC (8–10). Therefore, EGFR is still a strong
candidate for molecular-targeted therapy. A better understanding of
the molecular mechanism that regulates EGFR protein expresion, may
provide new biomarkers for predicting the clinical benefits of
anti-EGFR therapy.
The expression of EGFR protein in GC is regulated at
a post-transcription level. Previous studies have shown that the
level of EGFR protein expression in GC is inconsistent with that of
the gene. EGFR protein overexpression was detected in ~30% of GC
samples and one of the main causes was EGFR gene amplification
(12). Kim et al found that
27.4% of GC tissues from 511 patients showed EGFR protein
overexpression by IHC. However, only 2.3% of cases had gene
amplification by FISH (12).
Similar result from Higaki et al showed that EGFR gene
amplification was not completely identical to that of IHC (2+-3+)
and EGFR gene copy number gain was a more accurate prognostic
bimomarker than EGFR protein overexpression in patients with GC
(11). This phenomenon of
inconsistency suggests that post-transcriptional regulation may
exist in the expression of EGFR, although promoter methylation has
been proved to be one of the mechanisms for the high expression of
EGFR protein in GC (27).
As miRNA-mediated regression of mRNA transcripts is
thought to be one of the most important modes of
post-transcriptional regulation, we speculated that specific miRNAs
may be involved in the process of EGFR protein expression in GC and
we found that miR-370 acted as a negative regulator of EGFR in the
present study. Actually, various previous studies have confirmed
that the expression of EGFR protein is regulated by several miRNAs
in lung (28), breast (29) and also in colorectal cancer
(30). In addition, miR-370 was
reported to function as a tumor suppressor in colorectal cancer by
suppressing EGFR signaling (20).
However, to the best of our knowledge, this is the first attempt to
investigate the miR-370-EGFR pathway in GC tissues and cells.
Expression of miR-370 has been found to be
dysregulated in several types of cancers. Low expression of miR-370
was found in non-small cell lung cancer and upregulation of miR-370
inhibited cell proliferation and induced cell apoptosis (31). Wu et al reported that miR-370
was aberrantly expressed in laryngeal squamous cell carcinoma
(LSCC) and miR-370 may function as a tumor suppressor in LSCC
through downregulation of FoxM1 (32). In addition, downregulation of
miR-370 has also been noted in other malignancies such as
cholangiocellular carcinoma (33),
ovarian cancer (34) and oral
carcinoma (35). Our research found
that miR-370 was also downregulated in GC and could function as a
tumor suppressor via targeting EGFR. This is consistent with the
result from one previous study in which upregulation of miR-370
promoted cell apoptosis and inhibited proliferation via targeting
PTEN in human GC (21). However,
studies also found that upregulation of miR-370 contributed to the
progression of gastric carcinoma (36,37).
The explanation for this may be due to the different functions of
the targeted gene. Furthermore, one gene could be modulated by
several miRNAs. For instance, miR-7 (38) and miR-34a (39) were both verified to be suppressors
of EGFR in GC. Therefore, regulatory mechanisms of the targeted
genes and the relevant miRNAs in GC need further investigation.
In conclusion, we demonstrated that EGFR protein
expression is regulated by miR-370 and the miR-370-EGFR pathway is
involved in the process of cell growth and migration in GC. The
present study may provide new information concerning the molecular
mechanism that regulates EGFR protein expression in GC and may be
of important clinical significance to guide targeted therapy.
Acknowledgements
The present study was funded by grants from the
National Natural Science Foundation of China (nos. 81372394,
81602158, 81572321 and 81602156), and Tianjin Health and Family
Planning Commission Foundation of Science and Technology (15KG142).
This work was also funded by Tianjin Science Foundation (no.
15JCYBJC28200) and Doctoral Foundation of Tianjin Medical
University Cancer Institute and Hospital (B1502).
References
|
1
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al CONCORD Working Group: Global surveillance of cancer survival
1995–2009: Analysis of individual data for 25,676,887 patients from
279 population-based registries in 67 countries (CONCORD-2).
Lancet. 385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dixon M, Mahar AL, Helyer LK,
Vasilevska-Ristovska J, Law C and Coburn NG: Prognostic factors in
metastatic gastric cancer: Results of a population-based,
retrospective cohort study in Ontario. Gastric Cancer. 19:150–159.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Strong VE, Wu AW, Selby LV, Gonen M, Hsu
M, Song KY, Park CH, Coit DG, Ji JF and Brennan MF: Differences in
gastric cancer survival between the U.S. and China. J Surg Oncol.
112:31–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu W, Yang Z and Lu N: Molecular targeted
therapy for the treatment of gastric cancer. J Exp Clin Cancer Res.
35:12016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yarden Y and Pines G: The ERBB network: At
last, cancer therapy meets systems biology. Nat Rev Cancer.
12:553–563. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Terashima M, Kitada K, Ochiai A, Ichikawa
W, Kurahashi I, Sakuramoto S, Katai H, Sano T, Imamura H and Sasako
M: ACTS-GC Group: Impact of expression of human epidermal growth
factor receptors EGFR and ERBB2 on survival in stage II/III gastric
cancer. Clin Cancer Res. 18:5992–6000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang W, Raufi A and Klempner SJ: Targeted
therapy for gastric cancer: Molecular pathways and ongoing
investigations. Biochim Biophys Acta. 1846:232–237. 2014.PubMed/NCBI
|
|
8
|
Fontana E and Smyth EC: Novel targets in
the treatment of advanced gastric cancer: A perspective review.
Ther Adv Med Oncol. 8:113–125. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang Z, Li C, Li F and Wang X: EGFR gene
copy number as a prognostic marker in colorectal cancer patients
treated with cetuximab or panitumumab: A systematic review and meta
analysis. PLoS One. 8:e562052013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, De Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Higaki E, Kuwata T, Nagatsuma AK, Nishida
Y, Kinoshita T, Aizawa M, Nitta H, Nagino M and Ochiai A: Gene copy
number gain of EGFR is a poor prognostic biomarker in gastric
cancer: Evaluation of 855 patients with bright-field dual in situ
hybridization (DISH) method. Gastric Cancer. 19:63–73. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim MA, Lee HS, Lee HE, Jeon YK, Yang HK
and Kim WH: EGFR in gastric carcinomas: Prognostic significance of
protein overexpression and high gene copy number. Histopathology.
52:738–746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedman JM and Jones PA: MicroRNAs:
Critical mediators of differentiation, development and disease.
Swiss Med Wkly. 139:466–472. 2009.PubMed/NCBI
|
|
16
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M,
et al: Human microRNA genes are frequently located at fragile sites
and genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Zhang Z, Yu M, Li L, Du G, Xiao W
and Yang H: Involvement of miR-20a in promoting gastric cancer
progression by targeting early growth response 2 (EGR2). Int J Mol
Sci. 14:16226–16239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Xie S, Liu X, Wu H, Lin X, Gu J,
Wang H and Duan Y: Matrine alters microRNA expression profiles in
SGC-7901 human gastric cancer cells. Oncol Rep. 32:2118–2126.
2014.PubMed/NCBI
|
|
19
|
Riquelme I, Tapia O, Leal P, Sandoval A,
Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM,
et al: miR-101-2, miR-125b-2 and miR-451a act as potential tumor
suppressors in gastric cancer through regulation of the
PI3K/AKT/mTOR pathway. Cell Oncol. 39:23–33. 2016. View Article : Google Scholar
|
|
20
|
El-Daly SM, Abba ML, Patil N and Allgayer
H: miRs-134 and −370 function as tumor suppressors in colorectal
cancer by independently suppressing EGFR and PI3K signaling. Sci
Rep. 6:247202016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng Y, Fu M, Wu GW, Zhang AZ, Chen JP,
Lin HY, Fu YA, Jia J, Cai ZD, Wu XJ, et al: Upregulation of
microRNA-370 promotes cell apoptosis and inhibits proliferation by
targeting PTEN in human gastric cancer. Int J Oncol. 49:1589–1599.
2016.PubMed/NCBI
|
|
22
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang S, Luo A, Hao X, Lai Z, Ding T, Ma X,
Mayinuer M, Shen W, Wang X, Lu Y, et al: Peroxiredoxin 2 inhibits
granulosa cell apoptosis during follicle atresia through the NFKB
pathway in mice. Biol Reprod. 84:1182–1189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ciardiello F and Tortora G: EGFR
antagonists in cancer treatment. N Engl J Med. 358:1160–1174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weng X, Zhang H, Ye J, Kan M, Liu F, Wang
T, Deng J, Tan Y, He L and Liu Y: Hypermethylated Epidermal growth
factor receptor (EGFR) promoter is associated with gastric cancer.
Sci Rep. 5:101542015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weiss GJ, Bemis LT, Nakajima E, Sugita M,
Birks DK, Robinson WA, Varella-Garcia M, Bunn PA Jr, Haney J,
Helfrich BA, et al: EGFR regulation by microRNA in lung cancer:
Correlation with clinical response and survival to gefitinib and
EGFR expression in cell lines. Ann Oncol. 19:1053–1059. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Uhlmann S, Mannsperger H, Zhang JD, Horvat
EA, Schmidt C, Küblbeck M, Henjes F, Ward A, Tschulena U, Zweig K,
et al: Global microRNA level regulation of EGFR-driven cell-cycle
protein network in breast cancer. Mol Syst Biol. 8:5702012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mlcochova J, Faltejskova P, Nemecek R,
Svoboda M and Slaby O: MicroRNAs targeting EGFR signalling pathway
in colorectal cancer. J Cancer Res Clin Oncol. 139:1615–1624. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen T, Gao F, Feng S, Yang T and Chen M:
MicroRNA-370 inhibits the progression of non-small cell lung cancer
by downregulating oncogene TRAF4. Oncol Rep. 34:461–468.
2015.PubMed/NCBI
|
|
32
|
Yungang W, Xiaoyu L, Pang T, Wenming L and
Pan X: miR-370 targeted FoxM1 functions as a tumor suppressor in
laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother.
68:149–154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng F, Wehbe-Janek H, Henson R, Smith H
and Patel T: Epigenetic regulation of microRNA-370 by interleukin-6
in malignant human cholangiocytes. Oncogene. 27:378–386. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen XP, Chen YG, Lan JY and Shen ZJ:
MicroRNA-370 suppresses proliferation and promotes endometrioid
ovarian cancer chemosensitivity to cDDP by negatively regulating
ENG. Cancer Lett. 353:201–210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang KW, Chu TH, Gong NR, Chiang WF, Yang
CC, Liu CJ, Wu CH and Lin SC: miR-370 modulates insulin receptor
substrate-1 expression and inhibits the tumor phenotypes of oral
carcinoma. Oral Dis. 19:611–619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lo SS, Hung PS, Chen JH, Tu HF, Fang WL,
Chen CY, Chen WT, Gong NR and Wu CW: Overexpression of miR-370 and
downregulation of its novel target TGFβ-RII contribute to the
progression of gastric carcinoma. Oncogene. 31:226–237. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan C, Liu S, Zhao Y, Han Y, Yang L, Tao
G, Li Q and Zhang L: Upregulation of miR-370 contributes to the
progression of gastric carcinoma via suppression of FOXO1. Biomed
Pharmacother. 67:521–526. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie J, Chen M, Zhou J, Mo MS, Zhu LH, Liu
YP, Gui QJ, Zhang L and Li GQ: miR-7 inhibits the invasion and
metastasis of gastric cancer cells by suppressing epidermal growth
factor receptor expression. Oncol Rep. 31:1715–1722.
2014.PubMed/NCBI
|
|
39
|
Liu G, Jiang C, Li D, Wang R and Wang W:
MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in
gastric cancer. Tumour Biol. 35:9801–9806. 2014. View Article : Google Scholar : PubMed/NCBI
|