Introduction
Cervical cancer is the second most common malignancy
in women worldwide (1), and it
leads to ~266,000 cancer-related deaths among women worldwide
(2). More than 52,000 new cervical
cancer cases are diagnosed every year worldwide (2). Recently, with the development and
effectiveness of early screening tests for cervical cancer, the
incidence and mortality rates of cervical cancer in developed
countries have decreased (3,4).
However, more than 80% of all new cervical cancer cases are
diagnosed in developing countries (5), and cervical cancer metastasis
negatively impacts the survival of affected cervical cancer
patients directly (3,4). Numerous studies, in recent years
(6–8), have focused on the metastasis of
cervical cancer, but the underlying molecular and cellular
mechanisms remain confusing.
Long non-coding RNAs (lncRNAs) are a group of
endogenous non-coding transcripts with more than 200 nucleotides in
length (9,10). For a long period of time, scientists
believed that lncRNAs had no cellular and molecular functions, and
were considered to be transcriptional noise. However, recently
numerous studies indicate that lncRNAs contribute to different
cellular processes (11–13). During the last decade, scientists
have demonstrated that abnormal expression of lncRNAs is associated
with human diseases (14–16), including different types of human
cancers (17–19). The expression of lncRNA LINC01133
was decreased in human colorectal cancer (CRC) tissues and a low
expression level of LIINC01133 was associated with poor survival in
CRC patients. Overexpression of LINC01133 inhibited
epithelial-mesenchymal transition (EMT) and metastasis by
regulating SRSF6 (20). lncRNA
colon cancer-associated transcript 2 (CCAT2) was found to be
increased in human small cell lung cancer (SCLC) tissues and
knockdown of CCAT2 expression suppressed the ability of SCLC cell
proliferation and metastasis (21).
lncRNA taurine upregulated gene 1 (TUG1) was found to be
significantly increased in human liver cancer tissues and
metastatic liver cancer cell lines and knockdown of TUG1 suppressed
liver cancer growth, metastasis and angiogenesis through the
regulation of miR-34a-5p and the VEGFA regulatory network in
vitro and in vivo (22).
Wang et al performed lncRNA microarray and analyzed the
different expression levels of lncRNAs in pancreatic cancer and
para-carcinoma tissues, and found that lncRNA XLOC_006390 was
significantly increased in tumor tissues (23). lncRNA XLOC_006390 also called lncRNA
HOXA13 was markedly increased in hepatocellular carcinoma (HCC),
and its expression level was associated with HCC patient clinical
progression and survival outcome (24). However, the role of lncRNA
XLOC_006390 in the progression and metastasis of cervical cancer
has not been evaluated and remains unclear. In the present study,
we hypothesized that lncRNA XLOC_006390 is also increased in
cervical cancer, and upregulation of lncRNA XLOC_006390 contributes
to cervical cancer metastasis.
In the present study, we performed quantitative
real-time polymerase chain reaction (qRT-PCR) and analyzed the
expression level of lncRNA XLOC_006390 in human cervical cancer
tissues. Our results found that lncRNA XLOC_006390 was
significantly overexpressed in cervical cancer tissues compared
with matched para-carcinoma tissues, which is consistent with
findings of lncRNA microarray in pancreatic cancer (23). Furthermore, we confirmed that SET
domain containing 8 (SET8) was increased in cervical cancer tissues
and it was positively associated with the expression level of
lncRNA XLOC_006390. In addition, knockdown of lncRNA XLOC_006390
and SET8 suppressed cervical cancer cell proliferation and
metastasis in vitro. Our results suggest that XLOC_006390
promotes cervical cancer proliferation and migration through the
regulation of SET8 expression, at least partly, which indicate
critical roles of XLOC_006390 and SET8 in cervical cancer
progression and metastasis.
Materials and methods
Cervical cancer tissues
Thirty-seven primary cervical cancer tissues and 37
matched adjacent para-carcinoma tissues were obtained from patients
with cervical cancer through surgery at the Department of
Obstetrics and Gynecology of The Affiliated Hospital of Qingdao
University from 2015–2016. All of the patients who participated in
the present study had never received preoperative radiotherapy
and/or chemotherapy. The volunteers were pathologically diagnosed
with infiltrating carcinoma. The tumor stage was confirmed by two
gynecological oncologists based on histopathological evaluation
according to the International Federation of Gynecology and
Obstetrics (FIGO) staging system for cervical cancer.
Clinicopathological data are available for all samples in Table I.
 | Table I.The association between XLOC_006390
expression and clinicopathological factors in cervical cancer
patients. |
Table I.
The association between XLOC_006390
expression and clinicopathological factors in cervical cancer
patients.
|
|
| XLOC_006390 |
|
|---|
|
|
|
|
|
|---|
| Clinical
parameter | Total | High (%) | Low (%) |
P-valuea |
|---|
| Age (years) |
|
|
| 0.5148 |
|
≤40 | 17 | 7 | 10 |
|
|
>40 | 20 | 11 | 9 |
|
| Size (cm) |
|
|
| 0.3155 |
| ≥4 | 13 | 8 | 5 |
|
|
<4 | 24 | 10 | 14 |
|
| FIGO stages |
|
|
| 0.0170 |
|
I–II | 14 | 3 | 11 |
|
|
III–IV | 23 | 15 | 8 |
|
| Lymphatic
metastasis |
|
|
| 0.0078 |
|
Yes | 27 | 17 | 10 |
|
| No | 10 | 1 | 9 |
|
| Distant
metastasis |
|
|
| 0.0025 |
|
Yes | 21 | 15 | 6 |
|
| No | 16 | 3 | 13 |
|
Cell lines and culture conditions
Five cervical cancer cell lines, SiHa, HeLa, Caski,
C4-1 and C-33a, were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China) and the American Type Culture
Collection (ATCC; Manassas, VA, USA), respectively. All cell lines
were cultured in RPMI-1640 medium supplemented with 10% fetal
bovine serum (FBS) (both from Gibco-BRL, Gaithersburg, MD, USA).
All the media contained 1% penicillin-streptomycin (100 U/ml
penicillin and 100 µg/ml streptomycin). All cervical cancer cells
were cultured and maintained in a humidified incubator with 5%
CO2 at 37°C.
Cell transfection
The SiHa cell line was used to perform siRNA
knockdown experiments and 4C-1 cells were used in an overexpression
system. Cells (1×105) were seeded into a 24-well culture
plate, and incubated in RPMI-1640 medium with 10% FBS for 24 h.
Then, the cells were transfected with siRNA (100 nM) targeting
XLOC_006390 (siRNA sense, UUCCUAAAAGCUCAGAAACAGUU and antisense,
GUU UCUGAGCUUUUAGGAAGUUU); or SET8 (siRNA sense,
AGGAAUAGAUCUUUGACUGCCUU and antisense, CAGU CAAAGAUCUAUUCCUACUU);
or lncRNA XLOC_006390 overexpression vector (20 µg) or SET8
overexpression plasmids (20 µg) (both from Vipotion, Guangzhou,
China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in
RPMI-1640 medium without FBS in accordance with the manufacturer's
guidelines.
qRT-PCR
Total RNA was extracted from tissues and cells using
TRIzol reagent (Invitrogen). The RNA concentration and quality were
determined by NanoDrop 2000 (Quawell, San Jose, CA, USA). Total RNA
(2 µg) was used for first strand cDNA synthesis with a reverse
transcription reaction using a reverse transcription kit (Takara,
Dalian, China). The corresponding cDNA was used for quantitative
real-time PCR using SYBR-Green Real-Time Master Mix (Takara). GAPDH
was used as the internal control. The primers used for XLOC_006390
were: 5′-CCTTTGAATCCCTGAGAACTGAAC-3′ (forward) and
5′-ACCTTCCTTCCCACTGGACCTTC-3′ (reverse); for SET8,
5′-ACTTACGGATTTCTACCCTGT-3′ (forward) and 5′-CGATGAGGTCAATCTTC-3′
(reverse); for GAPDH, 5′-CCCACTCCTCCACCTTTGAC-3′ (forward) and
5′-ATACCAGGAAATGAGCTTGACAA-3′ (reverse). The qRT-PCR analysis was
performed on Applied Biosystems 7500 Sequence Detection System
(ABI, Foster City, CA, USA). Data were analyzed using the
2−ΔΔCt method.
Western blotting
Total protein from tissues and cells were extracted
using SDS lysis buffer (Beyotime, Shanghai, China). Tissues and
cells were homogenized and incubated on ice for 10 min and
centrifuged at 12,000 × g for 15 min. The supernatant was then
collected and its concentration was determined using a BCA protein
assay kit (Pierce, Rockford, IL, USA). Total protein (20 µg) was
separated on 12% SDS polyacrylamide gels and transferred to
polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica,
MA, USA) in 25 mM Tris-base and 190 mM glycine at 60 V for 3 h at
4°C. The membranes were blocked at 4°C in Tris-buffered saline
(TBS) containing 5% non-fat dried milk overnight and washed with
phosphate-buffered saline (PBS) three times. Then, the membranes
were incubated with primary antibodies (SET8; 1:10,000; ab177488)
(GAPDH; 1:2,500; ab9485) (both from Abcam, Cambridge, MA, USA) for
1 h at room temperature and washed with PBS in triplicate. Finally,
the membranes were cultured with goat anti-rabbit IgG-HRP (sc2004;
Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a 1:5,000
dilution for 1 h at 37°C. Proteins were analyzed by enhanced
chemiluminescence (ECL) as described by the manufacturers
instructions (Beyotime).
Immunofluorescence (IF)
For IF, SiHa or C4-1 cells were transfected with
SET8-siRNA or SET8 overexpression vectors and cultured on a
coverslip for 48 h. Then, the coverslips were washed with PBS for 5
min and fixed in 4% paraformaldehyde (Beyotime). Subsequently, the
cells were washed with PBS in triplicate and incubated with a
primary antibody against SET8 at 4°C for 12 h. The cells were then
incubated with Alexa Fluor 594-conjugated goat anti-rabbit
secondary antibody (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). 4′,6-Diamidino-2-phenylindole (DAPI) was used to stain the
nucleus. Images were captured by an FV10i confocal microscope
(Olympus, Tokyo, Japan).
Cell Counting Kit-8 (CCK-8) assay of
cell proliferation
Following transfection with siRNA or overexpression
vectors, 0.5×103 SiHa or C4-1 cells were seeded in a
96-well cell culture plate. At 24, 48 and 72 h, cells were treated
with 10 µl CCK-8 at 37°C for 4 h. The absorbance at 450 nm was
assessed using a microplate reader Thermo Plate (Rayto Life and
Analytical Sciences Co., Ltd., Shenzhen, China). All experiments
were performed at least three times.
Migration and invasion assays
Transwell migration and invasion assays were
performed as previously described (25). SiHa or C4-1 cells/well
(1.5×105) were used for a migration assay, and
1×105 SiHa or C4-1 cells/well were used for an invasion
assay. Cells that migrated or invaded were fixed with 100% methanol
for 30 min and stained using 0.5% crystal violet (Sigma-Aldrich,
St. Louis, MO, USA) for 20 min and counted in five random fields of
each filter under a microscope (IX71; Olympus) at a magnification
of ×200.
Statistical analysis
All data are presented as the mean ± SD. SPSS 20.0
software (SPSS, Inc., Chicago, IL, USA) was used for statistical
analysis. All experiments were performed at least three times. The
statistical significance of differences between groups was assessed
using one-way analysis of variance (ANOVA) or a paired/unpaired
Student's t-test or a Chi-square test, and P<0.05 was considered
to indicate a statistically significant result.
Results
Expression level of lncRNA XLOC_006390
in cervical cancer tissues and cell lines
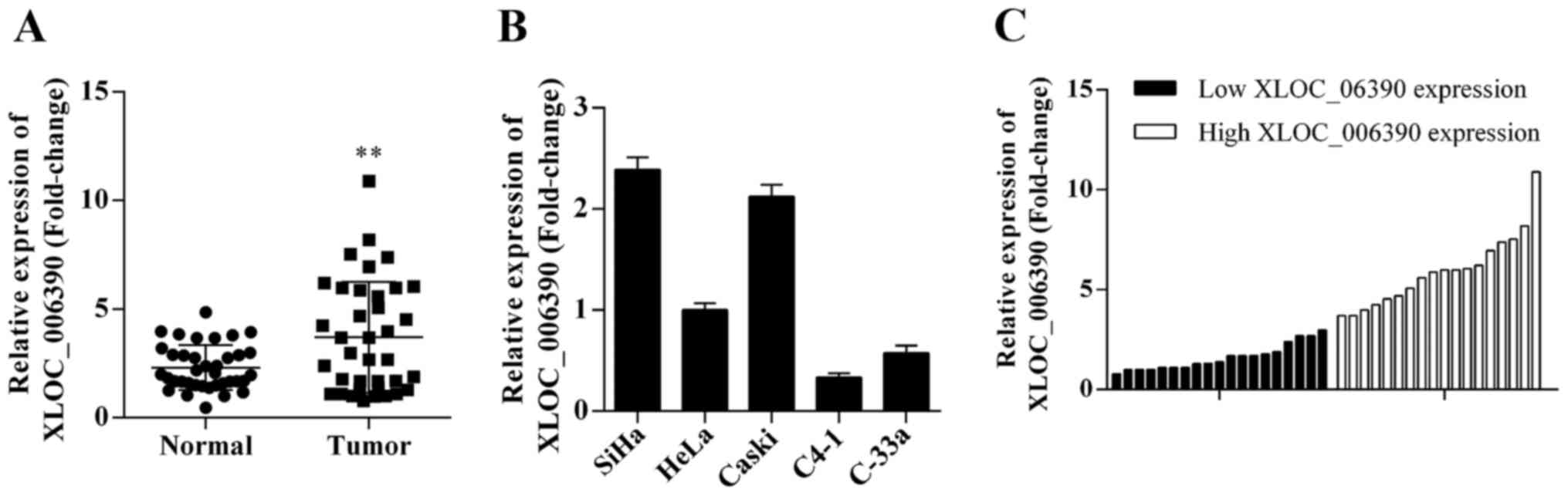
To investigate the expression level of lncRNA
XLOC_006390 in human cervical cancer tissues, qRT-PCR was performed
to detect the expression of XLOC_006390 in 37 cervical cancer
samples. lncRNA XLOC_006390 was significantly upregulated in tumor
tissues compared with matched adjacent para-carcinoma tissues
(Fig. 1A). Furthermore, we also
analyzed the expression of XLOC_006390 in five cervical cancer cell
lines and the results revealed that XLOC_006390 was increased in
SiHa cells, and decreased in C4-1 cells compared with the HeLa cell
line (Fig. 1B). Based on the
results, the SiHa and C4-1 cell lines were selected for further
study.
Correlation of lncRNA XLOC_006390
expression with clinicopathological factors in cervical cancer
patients
To investigate the relationship between lncRNA
XLOC_006390 expression and clinicopathological factors in cervical
cancer, 37 patients were divided into two groups according to the
median expression level of XLOC_006390 (2.98): a low XLOC_006390
expression group (n=19, XLOC_006390 expression ratio ≤ median
ratio) and a high XLOC_006390 expression group (n=18, XLOC_006390
expression ratio ≥ median ratio) (Fig.
1C). The relationship between the expression level of
XLOC_006390 and clinicopathological factors are shown in Table I. High XLOC_006390 expression was
observed to be associated with FIGO stage (P=0.0170), lymphatic
(P=0.0078) and distant metastasis (P=0.0025). In contrast, there
was no association between XLOC_006390 expression with age
(P=0.5148) and tumor size (P=0.3155).
Effects of XLOC_006390 on cervical
cancer cell proliferation
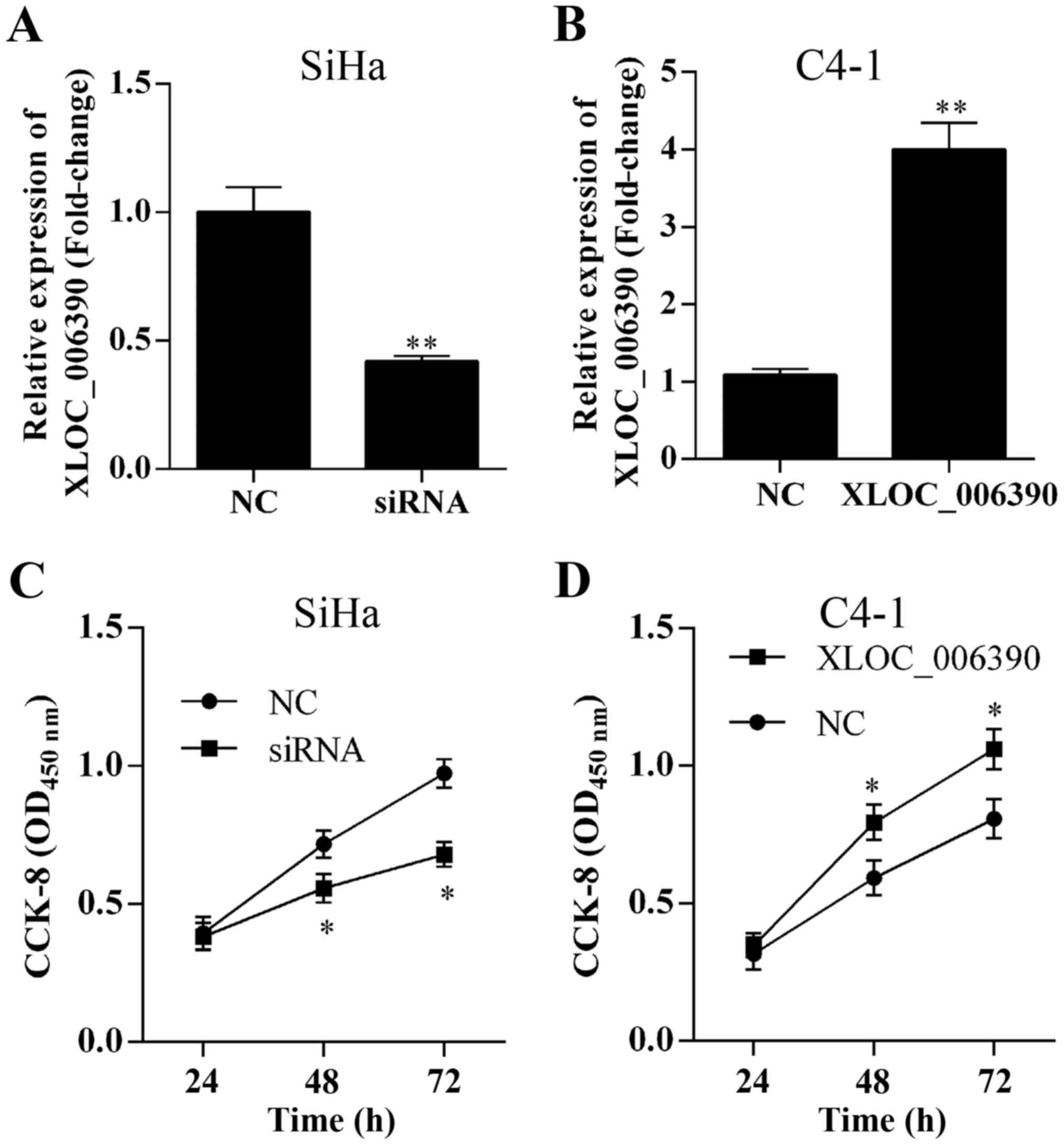
To evaluate the role of lncRNA XLOC_006390 in
cervical cancer cell proliferation, the expression of XLOC_006390
was knocked down or overexpressed by siRNA or lncRNA LXOC_006390
overexpression vector transfection, respectively. The expression of
XLOC_006390 was decreased in the SiHa cells after transfection with
XLOC_006390-siRNA (Fig. 2A) and
increased in C4-1 cells after transfection with XLOC_006390
overexpression vectors (Fig. 2B).
CCK-8 assay results revealed that suppression or overexpression of
XLOC_006390 expression inhibited or promoted the proliferation of
the SiHa and C4-1 cell lines, respectively (Fig. 2C and D).
Effects of XLOC_006390 on cervical
cancer cell migration and invasion
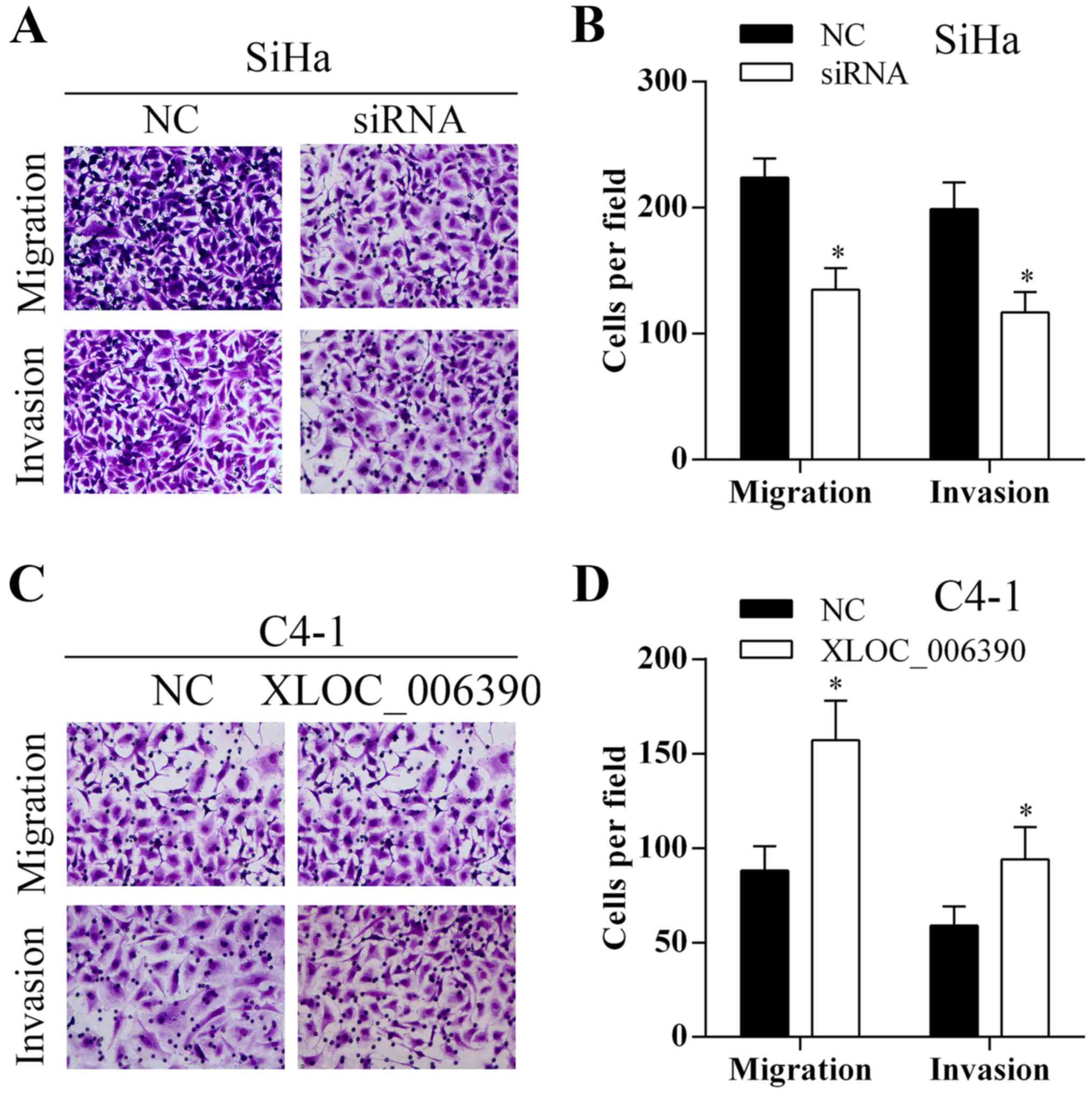
To investigate the effect of lncRNA XLOC_006390 on
cervical cancer cell migration and invasion, Transwell assays were
performed. As shown in Fig. 3A and
B, knockdown of XLOC_006390 inhibited the migration and
invasion abilities of the SiHa cells. Overexpression of XLOC_006390
promoted the migration and invasion abilities of the C4-1 cells
(Fig. 3C and D).
SET8 is upregulated in cervical cancer
tissues and correlated with XLOC_006390 expression
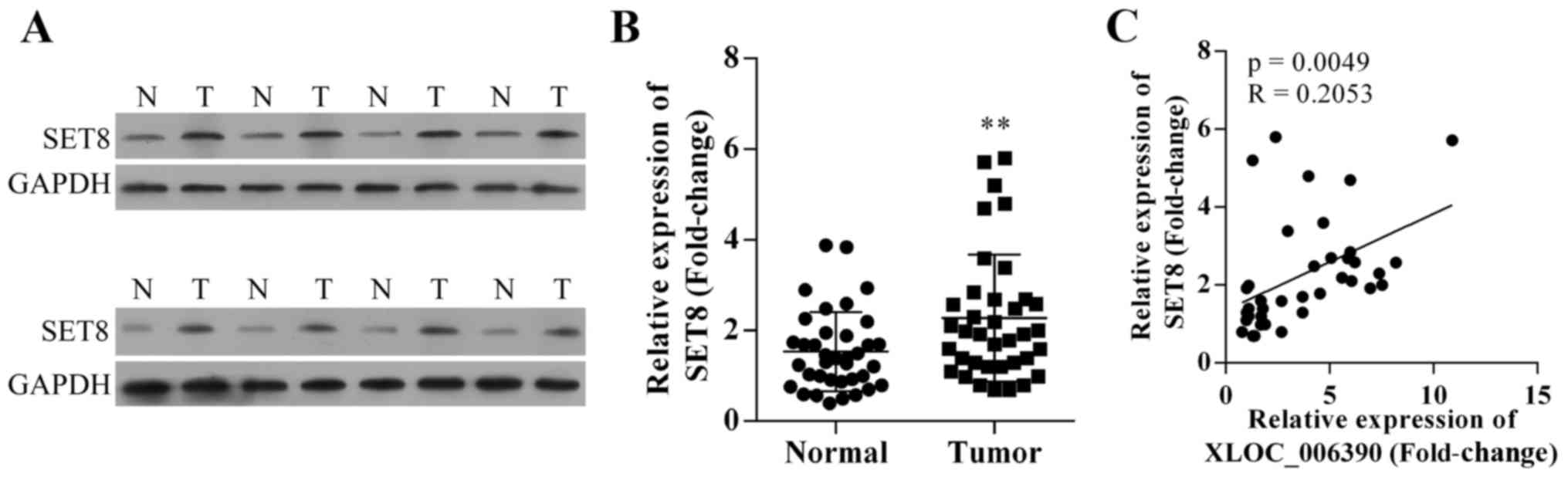
The protein and mRNA expression of SET8 in cervical
cancer tissues were detected using western blotting and qRT-PCR
methods, respectively. It was observed that both the protein and
mRNA expression level of SET8 were significantly increased in
cervical cancer tissues compared with matched adjacent
para-carcinoma tissues (Fig. 4A and
B). Due to the fact that both the expression of XLOC_006390 and
SET8 were significantly increased, we theorized a correlation
between XLOC_006390 expression and SET8 expression in cervical
cancer. The expression levels of SET8 in cervical cancer tissues
were positively associated with the expression levels of
XLOC_006390 (P=0.0049; R=0.2053; Fig.
4C).
Effects of SET8 on cervical cancer
cell proliferation
To evaluate the effects of SET8 on cervical cancer
cell proliferation, the expression of the SET8 protein in SiHa and
C4-1 cells after transfection with SET8-siRNA or SET8
overexpression vectors were examined by western blotting and IF.
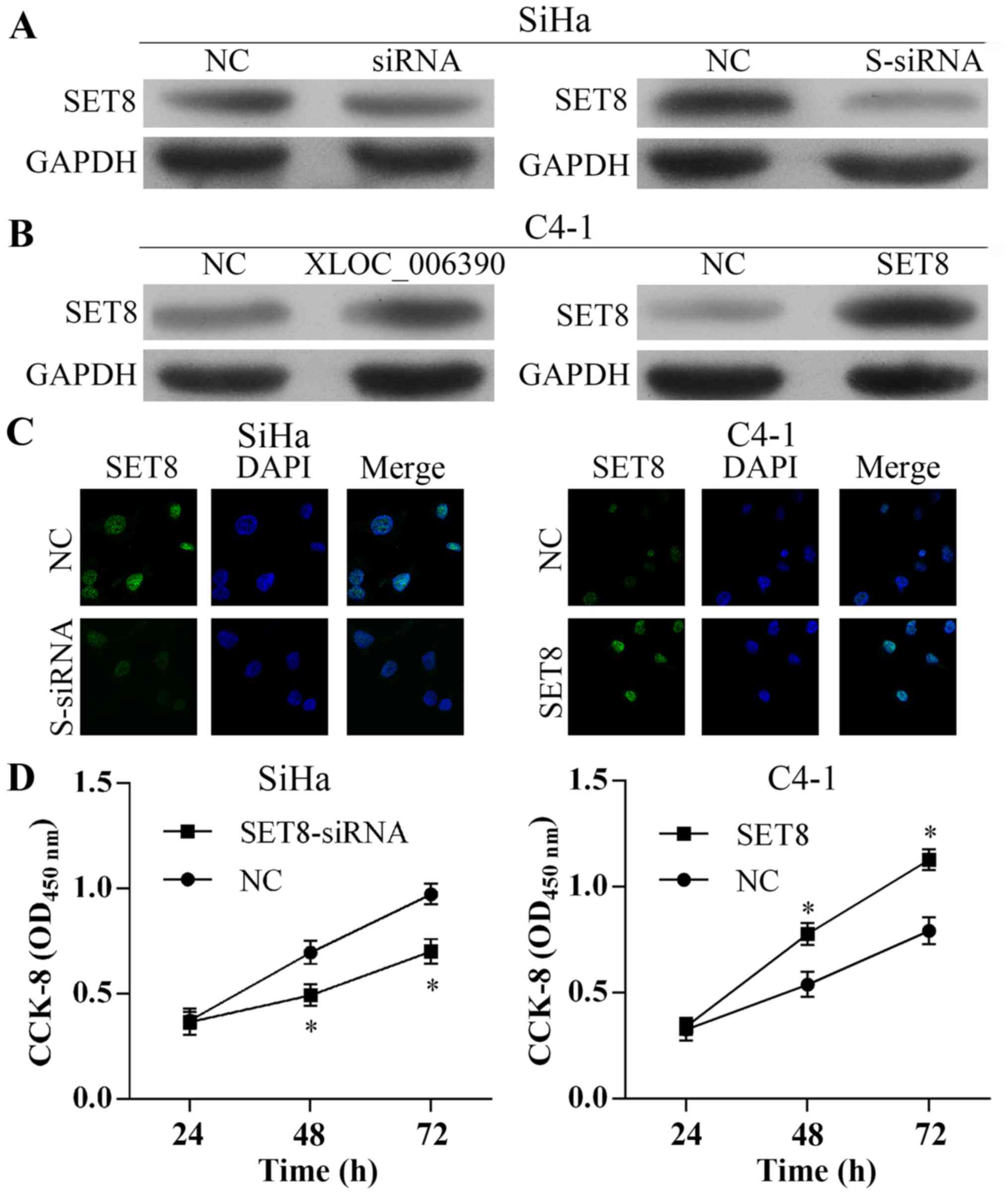
Firstly, as shown in Fig. 4C, we
found that the expression level of SET8 was positively associated
with XLOC_006390 expression. In the present experiment we found
that SET8 was decreased or increased after transfection with
XLOC_006390-siRNA or XLOC_006390 overexpression vectors in SiHa or
C4-1 cells, respectively (Fig. 5A and
B). Furthermore, western blotting and IF results revealed that
the expression of SET8 was suppressed or increased in SiHa or C4-1
cells after transfection with SET8-siRNA or SET8 overexpression
vectors, respectively (Fig. 5A-C).
In addition, CCK-8 assay results revealed that suppressed SET8
expression inhibited the proliferation of SiHa cells and
overexpression of XLOC_006390 promoted C4-1 cell proliferation
(Fig. 5D).
Effects of SET8 on cervical cancer
cell migration and invasion
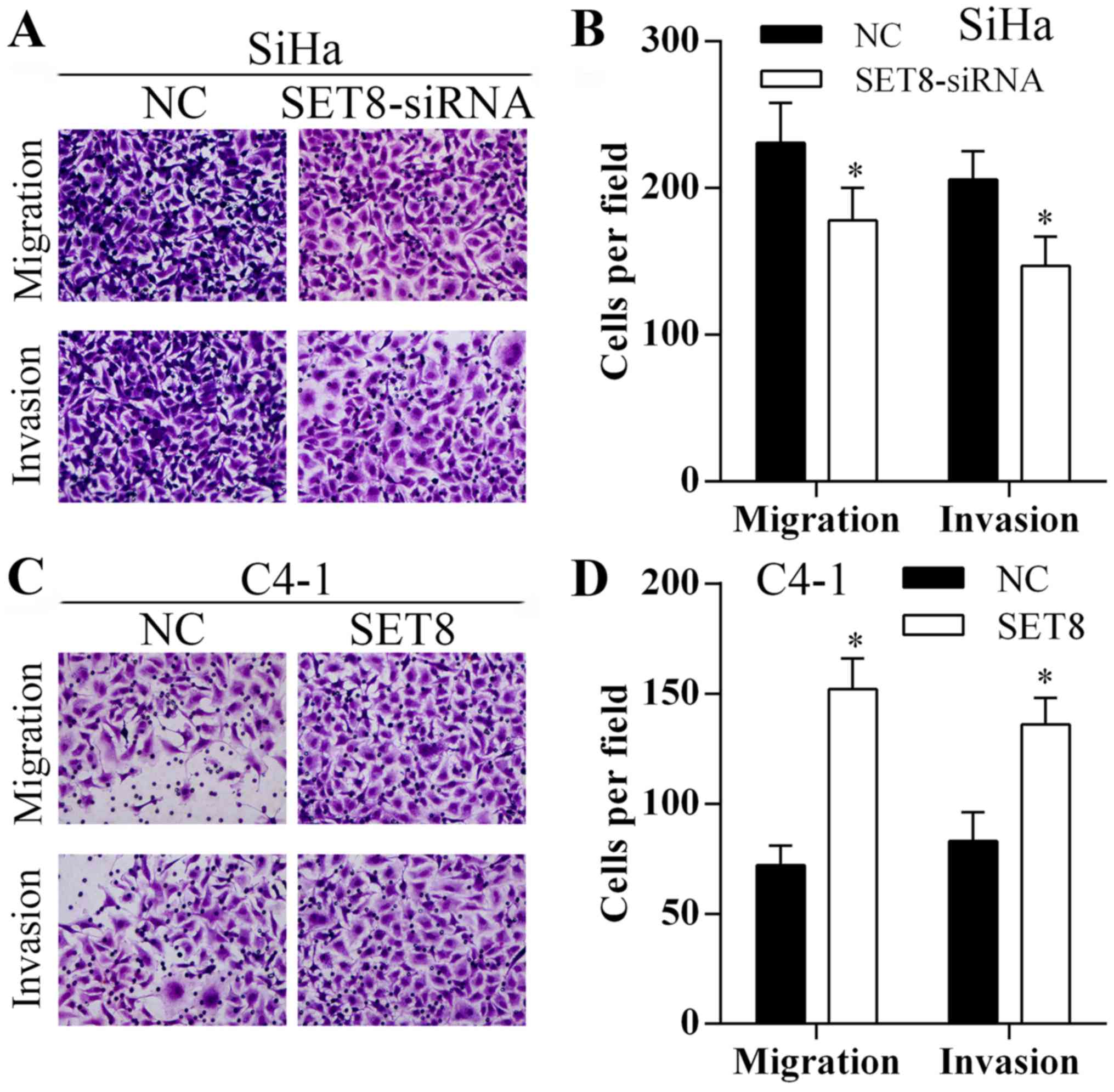
To investigate the role of SET8 on cervical cancer
cell migration and invasion, Transwell assays were performed. As
shown in Fig. 6, knockdown or
overexpression of SET8 inhibited or promoted the migration and
invasion abilities of the SiHa and C4-1 cells, respectively.
Discussion
Cervical cancer is still one of the leading causes
of cancerrelated deaths in females worldwide (26). Although, the treatment methods
including surgery, radiotherapy and chemotherapy have been greatly
improved, the survival rate of cervical cancer patients is still
unsatisfactory due to the high recurrence and metastasis rate
(3,4). Therefore, searching for a useful
molecular biomarker may provide a new strategy to improve the
survival of cervical cancer patients.
Numerous studies have indicated that lncRNAs play
critical roles in various physiological processes, including cell
proliferation, cell differentiation and epigenetic information
(27–29). Increased research has revealed that
dysregulated expression of lncRNAs is involved in pathological
processes, such as malignant tumors (30,31).
Effective control of cell growth, survival and aggressiveness plays
a critical role in preventing and treating tumors, successfully.
Therefore, increased understanding of the biological functions of
lncRNAs may provide potential useful approaches for cervical cancer
diagnosis and treatment. The expression of lncRNA GAS5 was
decreased in cervical cancer tissues and a lower expression of
lncRNA GAS5 was associated with a poorer survival rate of patients
(32). Cervical cancer cell
proliferation was promoted by lncNRA CCHE1 through regulation of
PCNA expression (33). lncRNA
HOTAIR was increased in cervical cancer tissues and a higher
expression of HOTAIR was associated with poorer prognosis. HOTAIR
could promote cervical cancer cell aggressiveness through the
promotion of VEGF and MMP-9 expression in vitro (34). However, there are no studies
concerning lncRNA XLOC_006390 in the progression and metastasis of
cervical cancer. In the present study, we found that lncRNA
XLOC_006390 was increased in cervical cancer tissues and a higher
expression of XLOC_006390 was associated with advanced FIGO stage,
lymphatic metastasis and distant metastasis. Furthermore, knockdown
of XLOC_006390 suppressed SiHa proliferation, migration and
invasion in vitro, and overexpression of XLOC_006390
promoted C4-1 cell proliferation, migration and invasion. Moreover,
SET domain containing 8 (SET8) was found to be overexpressed in
cervical cancer tissues and its expression level was positively
associated with XLOC_006390 expression. In addition, knockdown of
XLOC_006390 suppressed SET8 expression, and overexpression of
XLOC_006390 induced SET8 expression. Our results revealed that
XLOC_006390 may play a critical role in the proliferation and
metastatic potential of cervical cancer through the regulation of
SET8 expression. Nevertheless, only 37 patients were involved in
the present study, this may be a limitation for analyzing the
correlation of lncRNA XLOC_006390 expression with
clinicopathological factors in cervical cancer patients. More
volunteers are needed for the investigation of the role of lncRNA
XLOC_006390 in the progression and metastasis of cervical cancer,
and to confirm the potential role of lncRNA XLOC_006390 as a
diagnostic and treatment target for cervical cancer.
SET8 also called PR-Set7/9, KMT5A or SETD8, is one
of the SET domain-containing methyltransferases that specifically
targets histone H4 lysine 20 (H4K20) for monomethylation, and which
exerts diverse biological processes, including activation or
silencing of gene transcription (35,36),
maintaining chromosome structure and stability (37,38),
regulating the cell cycle and preventing premature chromatin
compaction in the S phase (39,40),
and rescuing and degrading DNA damage (41,42).
Yu et al found that miR-7 suppressed the invasive potential
of breast cancer cells by targeting SET8 (43). However, the role of SET8 in cervical
cancer metastasis and how the expression of SET8 is regulated are
poorly understood. In the present study, we found that SET8 was
increased in cervical cancer tissues. Knockdown of SET8 suppressed
SiHa proliferation, migration and invasion in vitro, and
overexpression of SET8 promoted C4-1 cell growth and metastasis.
Furthermore, the expression level of SET8 was positively associated
with the expression of XLOC_006390. All the results indicated that
upregulation of lncRNA XLOC_006390 may promote cervical cancer cell
proliferation and metastasis through the regulation of SET8.
Nevertheless, our results only revealed that knockdown of
XLOC_006390 suppressed the expression SET8 and overexpression of
XLOC_006390 induced the expression of SET8. How XLOC_006390
regulates SET8 expression and its regulatory mechanism need to be
confirmed in the future.
In conclusion, we demonstrated that lncRNA
XLOC_006390 and SET8 were increased in cervical cancer tissues, and
the expression level of SET8 was positively associated with the
expression of XLOC_006390. Furthermore, lncRNA XLCO_006390
regulated SET8 expression. Moreover, knockdown or overexpression of
XLOC_006390 and SET8 expression suppressed or promoted cervical
cancer cell proliferation and metastasis in vitro,
respectively. In conclusion, our data demonstrated that lncRNA
XLOC_006390 promoted cervical cancer cell growth and metastasis
through the regulation of SET8, at least partly, which indicated
the critical roles of XLOC_006390 and SET8 in cervical cancer
progression and metastasis.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang W, Jia HL, Huang JM, Liang YC, Tan H,
Geng HZ, Guo LY and Yao SZ: Identification of biomarkers for lymph
node metastasis in early-stage cervical cancer by tissue-based
proteomics. Br J Cancer. 110:1748–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fleming ND, Frumovitz M, Schmeler KM, dos
Reis R, Munsell MF, Eifel PJ, Soliman PT, Nick AM, Westin SN and
Ramirez PT: Significance of lymph node ratio in defining risk
category in node-positive early stage cervical cancer. Gynecol
Oncol. 136:48–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ying TH, Lee CH, Chiou HL, Yang SF, Lin
CL, Hung CH, Tsai JP and Hsieh YH: Knockdown of Pentraxin 3
suppresses tumorigenicity and metastasis of human cervical cancer
cells. Sci Rep. 6:293852016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xin M, Qiao Z, Li J, Liu J, Song S, Zhao
X, Miao P, Tang T, Wang L, Liu W, et al: miR-22 inhibits tumor
growth and metastasis by targeting ATP citrate lyase: Evidence in
osteosarcoma, prostate cancer, cervical cancer and lung cancer.
Oncotarget. 7:44252–44265. 2016.PubMed/NCBI
|
|
8
|
Zhou Q, Han LR, Zhou YX and Li Y: MiR-195
suppresses cervical cancer migration and invasion through targeting
Smad3. Int J Gynecol Cancer. 26:817–824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mattick JS: The genetic signatures of
noncoding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Zhong H, Xie X, Chen CY, Huang D,
Shen L, Zhang H, Chen ZW and Zeng G: Long noncoding RNA derived
from CD244 signaling epigenetically controls CD8+ T-cell immune
responses in tuberculosis infection. Proc Natl Acad Sci USA.
112:E3883–E3892. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carlson HL, Quinn JJ, Yang YW, Thornburg
CK, Chang HY and Stadler HS: LncRNA-HIT functions as an epigenetic
regulator of chondrogenesis through its recruitment of p100/CBP
complexes. PLoS Genet. 11:e10056802015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou C, York SR, Chen JY, Pondick JV,
Motola DL, Chung RT and Mullen AC: Long noncoding RNAs expressed in
human hepatic stellate cells form networks with extracellular
matrix proteins. Genome Med. 8:312016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi K, Yan I, Haga H and Patel T:
Long noncoding RNA in liver diseases. Hepatology. 60:744–753. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Z, Luo Y, Yang W, Ding L, Wang J, Tu
J, Geng B, Cui Q and Yang J: Comparison analysis of dysregulated
LncRNA profile in mouse plasma and liver after hepatic
ischemia/reperfusion injury. PLoS One. 10:e01334622015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li P, Zhang G, Li J, Yang R, Chen S, Wu S,
Zhang F, Bai Y, Zhao H, Wang Y, et al: Long noncoding RNA RGMB-AS1
indicates a poor prognosis and modulates cell proliferation,
migration and invasion in lung adenocarcinoma. PLoS One.
11:e01507902016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li G, Zhang H, Wan X, Yang X, Zhu C, Wang
A, He L, Miao R, Chen S and Zhao H: Long noncoding RNA plays a key
role in metastasis and prognosis of hepatocellular carcinoma.
Biomed Res Int. 2014:7805212014.PubMed/NCBI
|
|
19
|
Liu T, Zhang X, Yang YM, Du LT and Wang
CX: Increased expression of the long noncoding RNA CRNDE-h
indicates a poor prognosis in colorectal cancer, and is positively
correlated with IRX5 mRNA expression. Onco Targets Ther.
9:1437–1448. 2016.PubMed/NCBI
|
|
20
|
Kong J, Sun W, Li C, Wan L, Wang S, Wu Y,
Xu E, Zhang H and Lai M: Long non-coding RNA LINC01133 inhibits
epithelial-mesenchymal transition and metastasis in colorectal
cancer by interacting with SRSF6. Cancer Lett. 380:476–484. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen S, Wu H, Lv N, Wang H, Wang Y, Tang
Q, Shao H and Sun C: LncRNA CCAT2 predicts poor prognosis and
regulates growth and metastasis in small cell lung cancer. Biomed
Pharmacother. 82:583–588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong R, Liu GB, Liu BH, Chen G, Li K,
Zheng S and Dong KR: Targeting long non-coding RNA-TUG1 inhibits
tumor growth and angiogenesis in hepatoblastoma. Cell Death Dis.
7:e22782016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Li Z, Zheng S, Zhou Y, Zhao L, Ye
H, Zhao X, Gao W, Fu Z, Zhou Q, et al: Expression profile of long
non-coding RNAs in pancreatic cancer and their clinical
significance as biomarkers. Oncotarget. 6:35684–35698.
2015.PubMed/NCBI
|
|
24
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Huang H, Li Y, Li L, Hou W and You
Z: Decreased expression of long non-coding RNA GAS5 promotes cell
proliferation, migration and invasion, and indicates a poor
prognosis in ovarian cancer. Oncol Rep. 36:3241–3250.
2016.PubMed/NCBI
|
|
26
|
Arbyn M, Castellsagué X, de Sanjosé S,
Bruni L, Saraiya M, Bray F and Ferlay J: Worldwide burden of
cervical cancer in 2008. Ann Oncol. 22:2675–2686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15INK4B
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang
G and Zhang Y: lncRNA GAS5 enhances G1 cell cycle arrest via
binding to YBX1 to regulate p21 expression in stomach cancer. Sci
Rep. 5:101592015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan Y, Li G, Zhong H, Chen M, Chen T, Gao
L, Wu H and Guo J: RIG-I inhibits pancreatic β cell proliferation
through competitive binding of activated Src. Sci Rep. 6:289142016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hrdlickova B, De Almeida RC, Borek Z and
Withoff S: Genetic variation in the non-coding genome: Involvement
of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys
Acta. 1842:1910–1922. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao S, Liu W, Li F, Zhao W and Qin C:
Decreased expression of lncRNA GAS5 predicts a poor prognosis in
cervical cancer. Int J Clin Exp Pathol. 7:6776–6783.
2014.PubMed/NCBI
|
|
33
|
Yang M, Zhai X, Xia B, Wang Y and Lou G:
Long noncoding RNA CCHE1 promotes cervical cancer cell
proliferation via upregulating PCNA. Tumour Biol. 36:7615–7622.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim
SW and Kim YT: Long non-coding RNA HOTAIR is associated with human
cervical cancer progression. Int J Oncol. 46:521–530.
2015.PubMed/NCBI
|
|
35
|
Congdon LM, Houston SI, Veerappan CS,
Spektor TM and Rice JC: PR-Set7-mediated monomethylation of histone
H4 lysine 20 at specific genomic regions induces transcriptional
repression. J Cell Biochem. 110:609–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Z, Nie F, Wang S and Li L: Histone H4
Lys 20 monomethylation by histone methylase SET8 mediates Wnt
target gene activation. Proc Natl Acad Sci USA. 108:3116–3123.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Houston SI, McManus KJ, Adams MM, Sims JK,
Carpenter PB, Hendzel MJ and Rice JC: Catalytic function of the
PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for
mitotic entry and genomic stability. J Biol Chem. 283:19478–19488.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oda H, Okamoto I, Murphy N, Chu J, Price
SM, Shen MM, Torres-Padilla ME, Heard E and Reinberg D:
Monomethylation of histone H4-lysine 20 is involved in chromosome
structure and stability and is essential for mouse development. Mol
Cell Biol. 29:2278–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jørgensen S, Elvers I, Trelle MB, Menzel
T, Eskildsen M, Jensen ON, Helleday T, Helin K and Sørensen CS: The
histone methyltransferase SET8 is required for S-phase progression.
J Cell Biol. 179:1337–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Centore RC, Havens CG, Manning AL, Li JM,
Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC and Zou L:
CRL4Cdt2-mediated destruction of the histone methyltransferase Set8
prevents premature chromatin compaction in S phase. Mol Cell.
40:22–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sanders SL, Portoso M, Mata J, Bähler J,
Allshire RC and Kouzarides T: Methylation of histone H4 lysine 20
controls recruitment of Crb2 to sites of DNA damage. Cell.
119:603–614. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Oda H, Hübner MR, Beck DB, Vermeulen M,
Hurwitz J, Spector DL and Reinberg D: Regulation of the histone H4
monomethylase PR-Set7 by CRL4Cdt2-mediated PCNA-dependent
degradation during DNA damage. Mol Cell. 40:364–376. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu N, Huangyang P, Yang X, Han X, Yan R,
Jia H, Shang Y and Sun L: microRNA-7 suppresses the invasive
potential of breast cancer cells and sensitizes cells to DNA
damages by targeting histone methyltransferase SET8. J Biol Chem.
288:19633–19642. 2013. View Article : Google Scholar : PubMed/NCBI
|