Introduction
Colorectal cancer (CRC) is a cause of high morbidity
and mortality worldwide (1).
Despite the use of multi-model treatment strategies, including
surgery, perioperative chemotherapy, radiotherapy and targeted
therapy, a subset of patients commonly develop local recurrence and
metachronous metastasis after resection of the primary tumor
(2,3). Therefore, the molecular and cellular
processes involved in CRC metastasis are urgently needed to be
detected in order to develop reliable biomarkers to predict poor
patient outcome.
A major pathway controlling protein degradation is
the ubiquitin-proteasome system (4). The attachment of ubiquitin to target
proteins is mediated by at least 3 enzymes: an ubiquitin-activating
enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin
ligase (E3) (5). Cullins are a
family of hydrophobic proteins providing a scaffold for ubiquitin
ligase E3. Cullin1 (Cul1) is the most representative member of the
Cullin protein family, which degrades many proteins by mediating
ubiquitination of proteins involved in cell cycle progression,
signal transduction and transcription (6–8). Cul1
regulates cell proliferation, cell cycle, migration, invasion,
metastasis and is associated with the patient prognosis in gastric
cancer and CRC (9,10). Recently, other investigators have
reported that Cul1 expression is associated with poor prognosis in
melanoma, lung and breast cancer (11–13).
The protoncogene c-Myc is a member of the Myc family
protein (14), which is involved in
many biological processes such as cell cycle, cell differentiation
and protein synthesis (15,16). Overexpression of c-Myc in CRC is a
deterioration index (17). The
c-Myc gene is known to regulate both oncogenes and tumor-suppressor
genes and therefore play an important role in the occurrence and
development of cancers. c-Myc directly governs cell mass and
progression through critical cell cycle transitions by promoting
G1 exit, and the regulation in part via Cul1-dependent
ubiquitination and degradation of the CDK inhibitor,
p27kip1 (18).
Herein, we aimed to elucidate the expression
patterns of Cul1 and c-Myc in a CRC patient cohort, and to examine
the possibility of Cul1 or c-Myc alone or in combination as a
prognostic and predictive biomarker.
Materials and methods
Patient specimens and tissue
samples
The present study was approved by the Institutional
Review Board of Yixing Hospital prior to the study. All subjects
provided written informed consent and were assured of their
anonymity and the confidentiality of the data obtained. The
10-paired fresh samples were frozen in liquid nitrogen immediately
after surgical removal and maintained at −80°C until use for
western blot analysis.
The cohort TMA consisting of 470 CRC surgical cases
was obtained from Yixing People's Hospital, Yixing City, in the
South of Jiangsu Province during January, 2006 and December, 2010.
The patients were followed up at least for 5 years. Overall
survival (OS) was the primary endpoint of this analysis, and
survival time was calculated from the date of surgery to the date
of death or to the last follow-up. The patient clinicopathologic
information including age at diagnosis, sex, differentiation stage,
depth of invasion, lymph node metastasis, tumor-node-metastasis
(TNM) stage and tumor diameter was collected. The median age of the
patients at tumor resection was 63 years; 281 (59.8%) were male and
189 (40.2%) were female cases (Table
I). TMA was constructed by taking tissue portions in the
identified and labeled area from the tumor samples which were fixed
in formalin and embedded in paraffin.
 | Table I.CRC patient clinicopathological
data. |
Table I.
CRC patient clinicopathological
data.
| Variables | n | % |
|---|
| All patients | 470 |
|
| Age (years) |
| ≤65 | 267 | 56.8 |
|
>65 | 203 | 43.2 |
| Sex |
| Male | 281 | 59.8 |
|
Female | 189 | 40.2 |
| Pathological
classificationa |
| I |
5 | 1.1 |
| II | 423 | 91.2 |
| III | 36 | 7.7 |
| Depth of
invasiona |
| T1 |
9 | 2.0 |
| T2 | 94 | 20.2 |
| T3 | 347 | 74.6 |
| T4 | 15 | 3.2 |
| Lymph node
metastasisa |
| N0 | 276 | 59.2 |
| N1 | 126 | 27.0 |
| N2 | 64 | 13.8 |
| TNM
stagea |
| I | 88 | 18.9 |
| II | 179 | 38.6 |
|
III | 180 | 38.8 |
| IV | 17 |
3.7 |
| Tumor diameter
(cm)a |
| ≤5 | 378 | 80.6 |
|
>5 | 91 | 19.4 |
| Distant
metastasis |
| M0 | 451 | 95.9 |
| M1 | 19 |
4.1 |
All procedures involving human participants were in
accordance with the ethical standards of the Institutional and/or
National Research Committee and with the 1964 Helsinki Declaration
and its later amendments or comparable ethical standards.
Western blotting
Western blot analyses were performed as previously
described (19). The rabbit
anti-Cul1 (1:1,000; Epitomics, Burlingame, CA, USA), anti-c-Myc
(1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA), and
monoclonal mouse anti-β-actin antibody (1:2,000; Beyotime
Biotechnology, Nantong, China) were used as the primary antibodies.
The intensity of the protein bands was analyzed using ImageJ
software (version 1.44; Wayne Rasband National Institutes of
Health, Bethesda, MD, USA), after normalization to the
corresponding β-actin level.
Construction of the tissue microarray
(TMA) and immunohistochemistry
Paraffin-embedded archived tissue material of tumor
and adjacent normal tissues was used for TMA construction. The CRC
TMA included 940 cores. Each sample was punched to a 1.5-mm
diameter. The standard protocol used for the immunostaining was
provided in a previous study (19).
The monoclonal rabbit anti-Cul1 (1:200; Epitomics) and anti-c-Myc
(1:200; Cell Signaling Technology, Inc.) were used for primary
antibody incubation at 4°C overnight. The omission of the primary
antibody served as the negative control. The staining scores of the
tissues controlled in each microarray slide were pre-evaluated as a
quality control of the immunostaining.
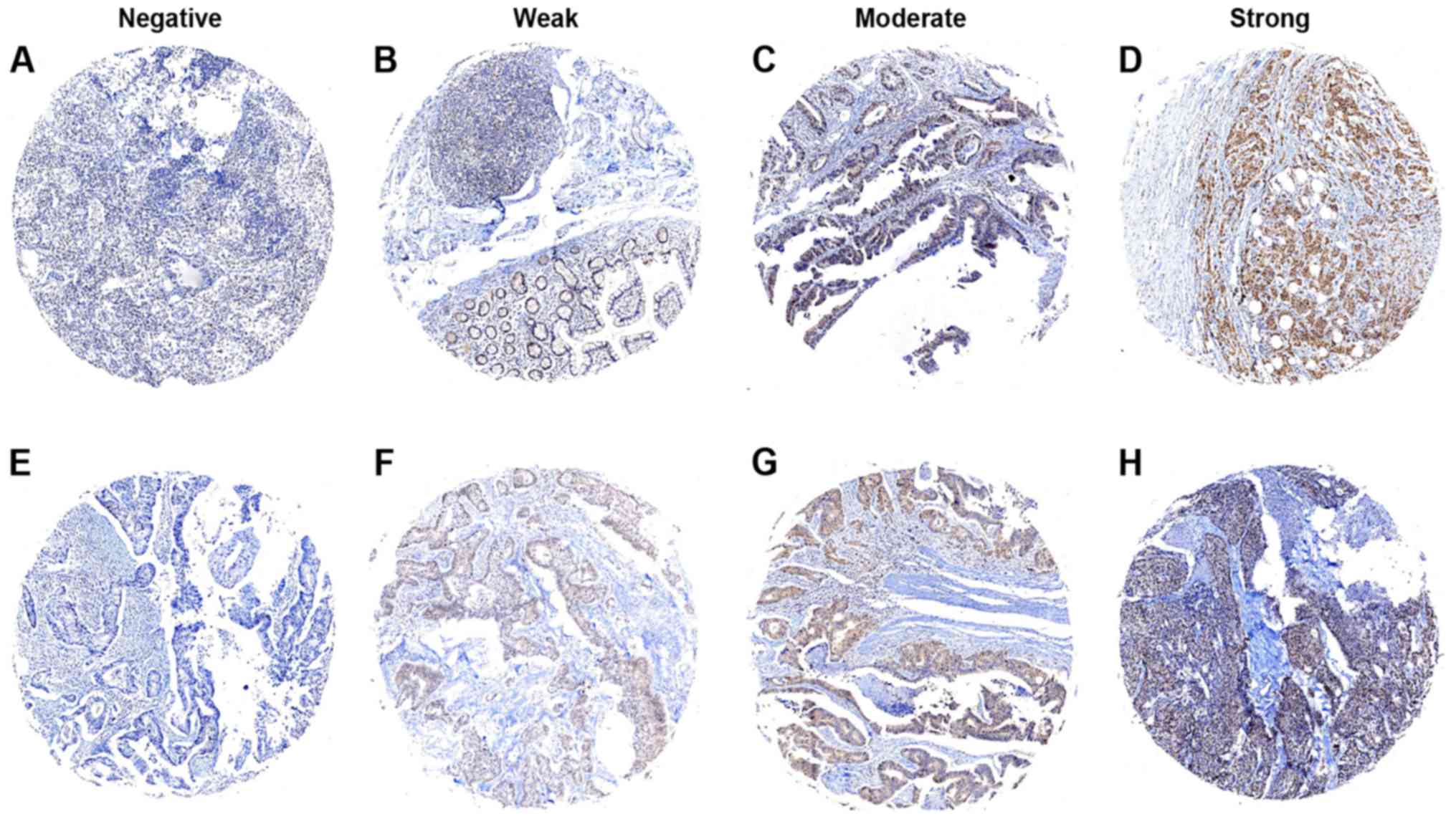
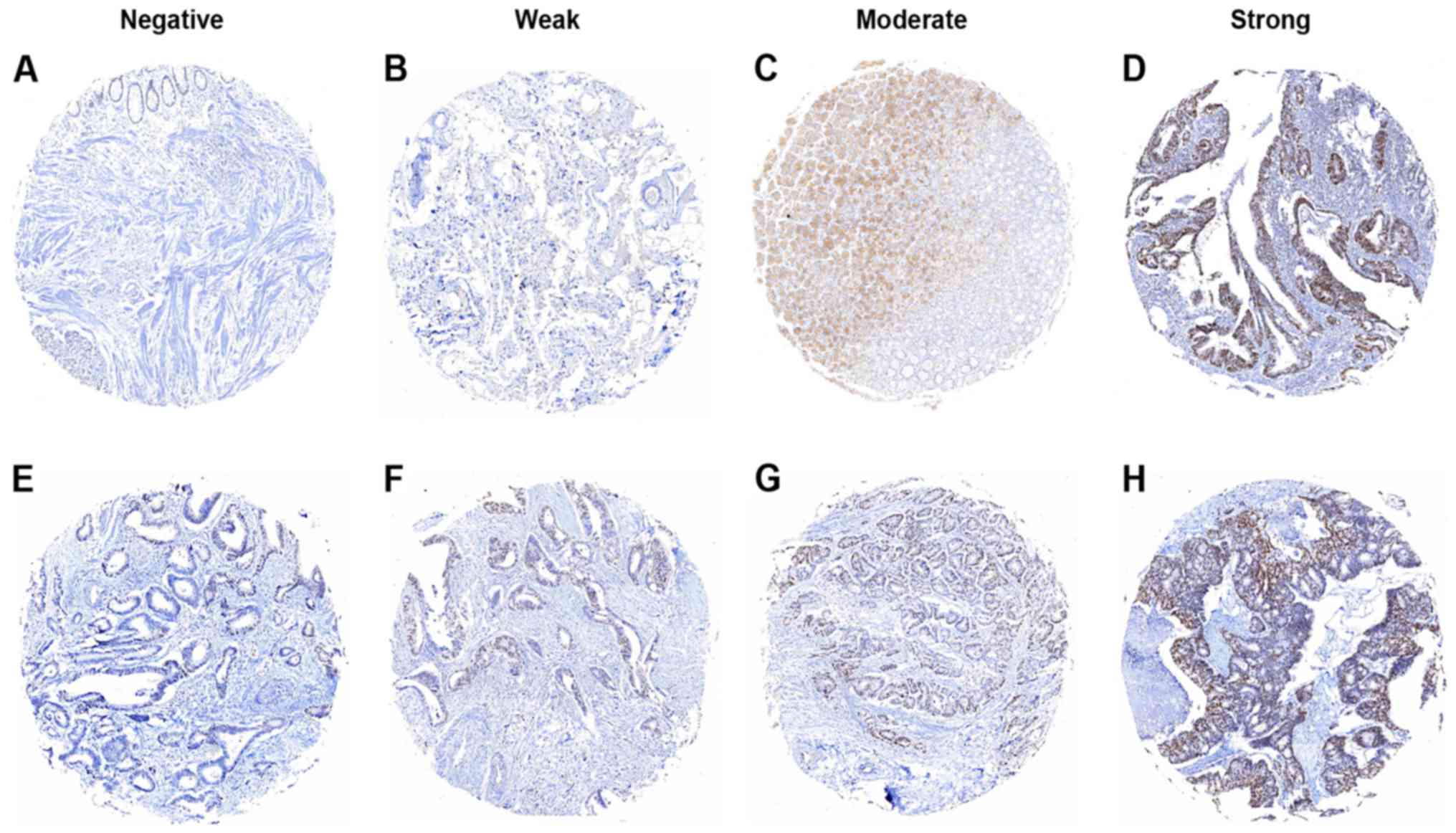
Evaluation of immunostaining
At first, staining of Cul1 or c-Myc in the tissues
was independently scored by two pathologists blinded to the
clinical data, by applying a semi-quantitative immunoreactivity
score (IRS) in the training cohort. The scoring criteria for IRS
were reported elsewhere (9,10,13).
The intensity of immunostaining is shown in Figs. 1 and 2. The concordance for IRS staining score
of Cul1 between the two pathologists was 423 in 470 tumors (90%),
and the discrepancies were resolved by consensus using a multihead
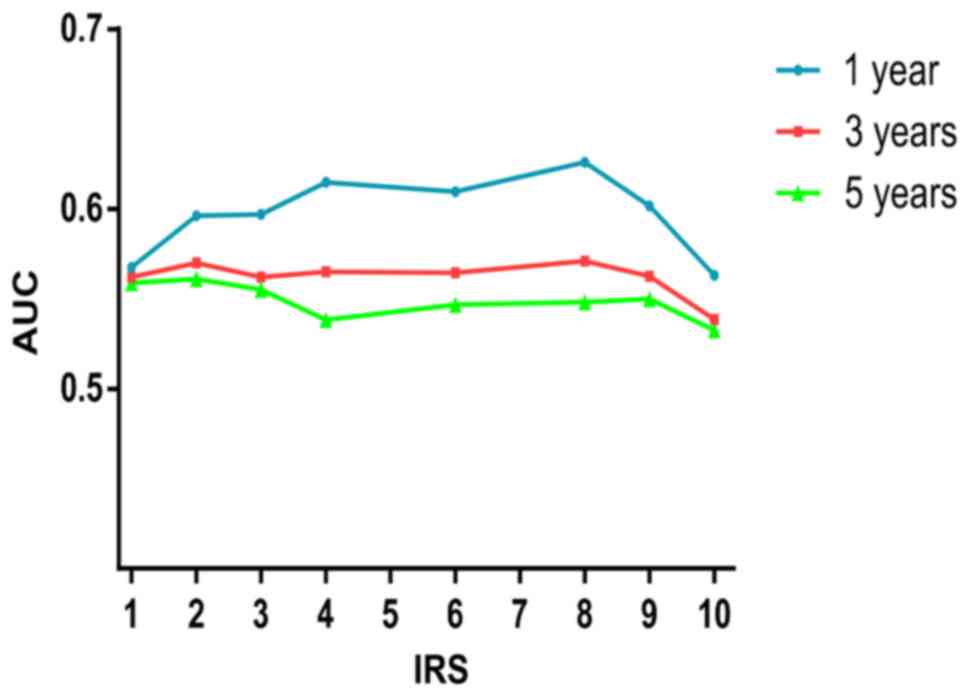
microscope. The optimum cut-off value of IRS was obtained by
receiver operator characteristic (ROC) analysis; the area under the
curve (AUC) at different cut-off values for Cul1 IRS for 1, 3 and 5
years of OS time was calculated. The optimum value of cut-off
points for Cul1 IRS was shown to be 4 since it had the best
predictive value for survival (Fig.
3). Under these conditions, samples with IRS 0–3 and IRS 4–12
were classified as low or high expression of Cul1, respectively. By
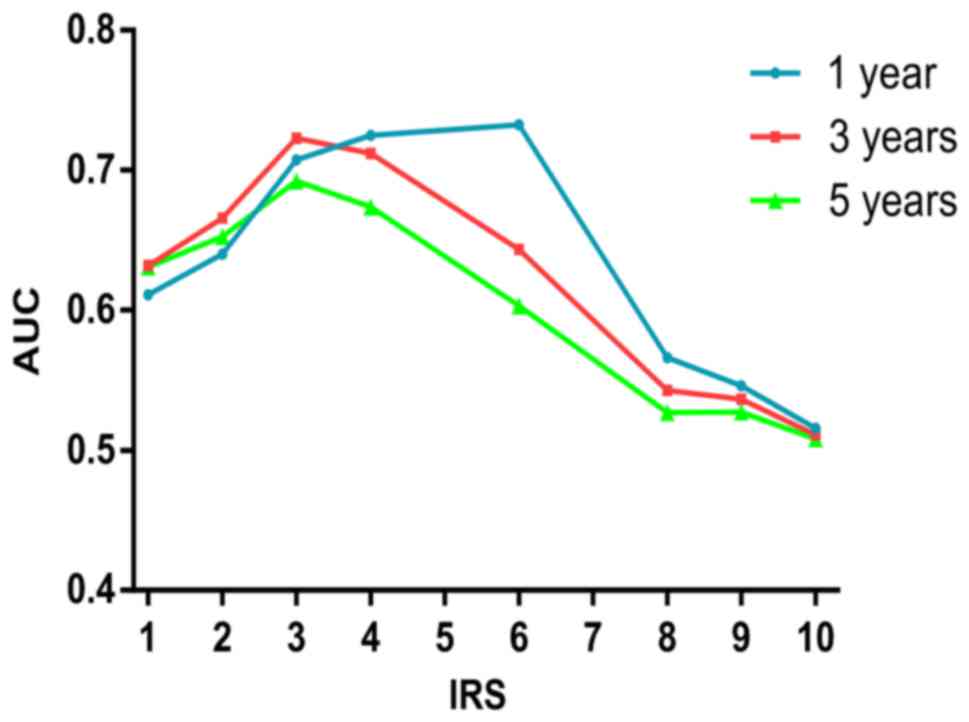
use of the same method, the optimum value of cut-off points of
c-Myc IRS was shown to be 3 (Fig.
4), and samples with IRS 0–2 and IRS 3–12 were classified as
low or high expression of c-Myc, respectively.
Statistical analysis
For TMA, statistical processing was performed using
SPSS 20.0 software (SPSS, Inc, Chicago, IL, USA). Fisher's exact
test was used to evaluate the association between Cul1 and c-Myc
expression and clinicopathological parameters. Differences in IRS
for Cul1 or c-Myc staining in primary tumors, and their paired
adjacent normal tissues were assessed by the paired Wilcoxon test
(raw scores). The correlation between the expression of Cul1 and
c-Myc was established by Spearman rank-order correlation (raw
scores) and Fisher's exact test (grouped). Probability of
differences in OS as a function of time was ascertained by use of
the Kaplan-Meier method, with a log-rank test probe for
significance. Univariate and multivariate Cox proportional hazards
regression analyses were performed to estimate the crude hazard
ratios (HRs), adjusted HRs and 95% confidence interval (CI) of HRs.
We evaluated the performances of different scores by plotting [t,
AUC (t)] for different values of follow-up time (t). All the
statistical analyses were performed by STATA statistical software
(version 10.1; StataCorp, College Station, TX, USA). A P-value of
<0.05 was considered statistically significant, and all tests
were two-sided.
Results
Expression of Cul1 and c-Myc is
increased in CRC vs. adjacent normal tissues
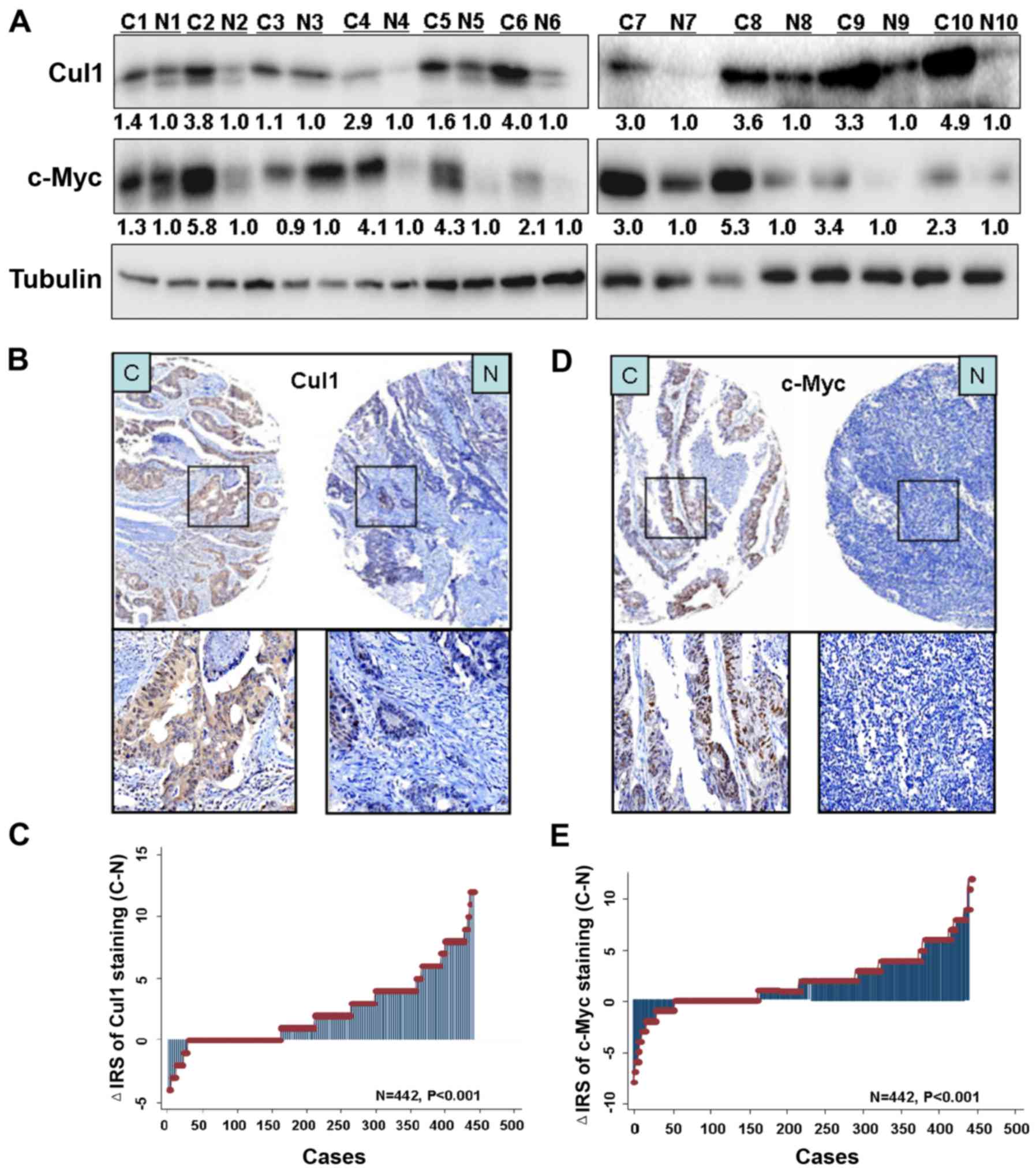
Ten pairs of human CRC samples, including primary
CRC and matched normal colorectal tissues were collected to test
the expression of Cul1 and c-Myc protein by western blotting,
respectively. Data showed increased expression levels of Cul1 and
c-Myc in all tumor tissues compared with the levels in the matched
normal tissues (Fig. 5A).
Immunohistochemical staining further confirmed higher Cul1 and
c-Myc expression levels in the CRC tissues than levels in the
paired adjacent normal tissues (Fig. 5B
and D). There were 442 cases of CRC tissues, and with paired
adjacent non-cancer tissues available for evaluating the score. As
a result, Cul1 and c-Myc expression was upregulated in tumor
tissues compared with that noted in the paired adjacent non-tumor
tissues (P<0.001; Fig. 5C and
E).
Cul1 and c-Myc expression correlates
with clinicopathological parameters
In the CRC cohort, Fisher's exact analysis revealed
that there was s significant positive association between high Cul1
expression in cancer tissues and depth of invasion (P=0.005), lymph
node metastasis (P=0.001) and TNM stage (P=0.015). However, there
was no association between Cul1 expression and age, sex,
pathological classification and tumor diameter (Table II).
 | Table II.Relationship between the expression
level of Cul1 and clinicopathological features of the CRC
patients. |
Table II.
Relationship between the expression
level of Cul1 and clinicopathological features of the CRC
patients.
| Variables | Low n (%) | High n (%) |
P-valuea |
|---|
| All patients
(N=464) | 266 (57.3) | 198 (42.7) |
|
| Age (years) |
|
| 0.139 |
|
≤65 | 157 (59.7) | 106 (40.3) |
|
|
>65 | 109 (54.2) | 92 (45.8) |
|
| Sex |
|
| 0.175 |
|
Male | 154 (55.4) | 124 (44.6) |
|
|
Female | 112 (60.2) | 74 (39.8) |
|
| Pathological
classificationb |
|
| 0.302 |
| I | 4 (80.0) | 1 (20.0) |
|
| II | 242 (57.9) | 176 (42.1) |
|
|
III | 17 (47.2) | 19 (52.8) |
|
| Depth of
invasionb |
|
| 0.005 |
|
T1/T2 | 71 (68.9) | 32 (31.1) |
|
|
T3/T4 | 193 (54.1) | 164 (45.9) |
|
| Lymph node
metastasisb |
|
| 0.001 |
| N0 | 173 (63.4) | 100 (36.6) |
|
|
N1/N2 | 91 (48.4) | 97 (51.6) |
|
| TNM
stageb |
|
| 0.015 |
| I | 60 (68.2) | 28 (31.8) |
|
| II | 107 (60.8) | 69 (39.2) |
|
|
III | 88 (49.4) | 90 (50.6) |
|
| IV | 8 (47.1) | 9 (52.9) |
|
| Tumor diameter
(cm)b |
|
| 0.543 |
| ≤5 | 214 (57.2) | 160 (42.8) |
|
|
>5 | 51 (57.3) | 38 (42.7) |
|
| Distant
metastasis |
|
| 0.423 |
| M0 | 256 (57.5) | 189 (42.5) |
|
| M1 | 10 (52.6) | 9 (47.4) |
|
| c-Myc
expression |
|
Low | 200 (72.2) | 77 (27.8) | 0.001 |
|
High | 65 (34.9) | 121 (65.1) |
|
We also analyzed the relationship between c-Myc
expression and clinicopathological parameters. Data showed that
high c-Myc expression in cancer tissues was significantly
associated with age (P=0.004), depth of invasion (P<0.001),
lymph node metastasis (P<0.001) and TNM stage (P<0.001).
There was no association between c-Myc expression and sex,
pathological classification and tumor diameter (Table III).
 | Table III.Relationship between the expression
level of c-Myc and clinicopathological features of the CRC
patients. |
Table III.
Relationship between the expression
level of c-Myc and clinicopathological features of the CRC
patients.
| Variables | Low n (%) | High n (%) |
P-valuea |
|---|
| All patients
(N=464) | 278 (59.9) | 186 (40.1) |
|
| Age (years) |
|
≤65 | 172 (65.4) | 91 (34.6) | 0.004 |
|
>65 | 106 (52.7) | 95 (47.3) |
|
| Sex |
|
Male | 167 (60.3) | 110 (39.7) | 0.458 |
|
Female | 111 (59.4) | 76 (40.6) |
|
| Pathological
classificationb |
|
| 0.091 |
| I | 5 (100.0) | 0 (0.0) |
|
| II | 251 (60.1) | 167 (39.9) |
|
|
III | 18 (50.0) | 18 (50.0) |
|
| Depth of
invasionb |
|
|
<0.001 |
|
T1/T2 | 77 (74.8) | 26 (25.2) |
|
|
T3/T4 | 198 (55.5) | 159 (44.5) |
|
| Lymph node
metastasisb |
|
|
<0.001 |
| N0 | 192 (70.3) | 81 (29.7) |
|
|
N1/N2 | 84 (44.7) | 104 (55.3) |
|
| TNM
stageb |
|
|
<0.001 |
| I | 66 (75.0) | 22 (25.0) |
|
| II | 121 (68.7) | 55 (31.3) |
|
|
III | 84 (47.2) | 94 (52.8) |
|
| IV | 3 (17.7) | 14 (82.3) |
|
| Tumor diameter
(cm)b |
|
| 0.191 |
| ≤5 | 219 (58.7) | 154 (41.3) |
|
|
>5 | 58 (64.4) | 32 (35.6) |
|
| Distant
metastasis |
|
| 0.001 |
| M0 | 274 (61.6) | 171 (38.4) |
|
| M1 | 4 (21.1) | 15 (78.9). |
|
Increased Cul1 or c-Myc expression
correlates with the poor survival of CRC patients
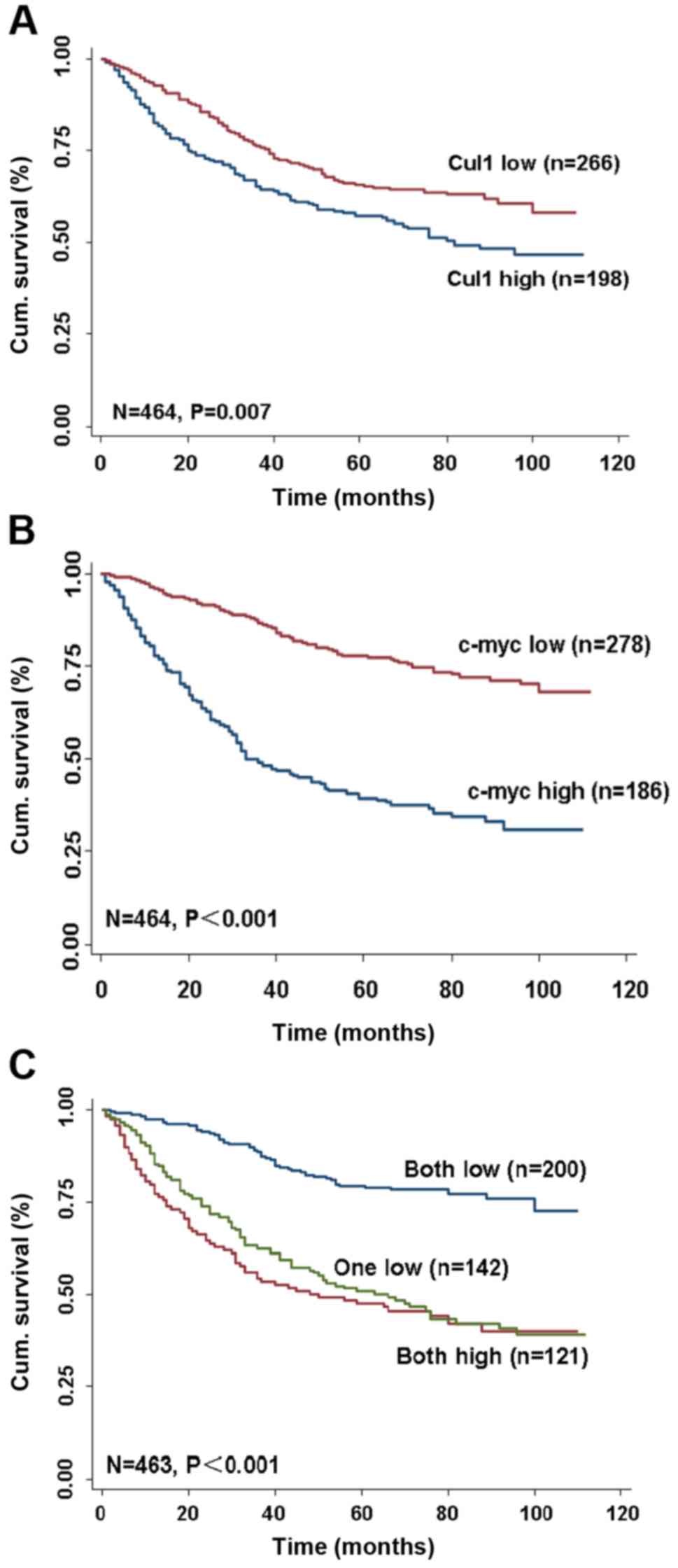
Kaplan-Meier survival assay was conducted and the
data revealed that higher Cul1 and c-Myc expression in cancer
tissues was correlated with a worse OS in CRC patients (P<0.001
and P<0.001, respectively, log-rank test; Fig. 6A and B). Cul1 and c-Myc expression
in cancer tissues was an independent marker for the prognosis of
CRC patients by univariate and multivariate Cox regression
analysis. The univariate Cox regression analysis also showed that
age, pathological classification, depth of invasion, lymph node
metastasis, TNM stage and Cul1 or c-Myc expression were associated
with OS of the CRC patients (Table
IV). The multivariate Cox regression analysis revealed that
Cul1 expression was an independent and unfavorable prognostic
factor for CRC patients (HR, 0.749, 95% CI, 0.563–0.996, P<0.05;
Table V). Similarly, c-Myc
expression was also an independent and unfavorable prognostic
factor for CRC patients (HR, 0.384, 95% CI 0.257–0.472, P<0.001;
Table V).
 | Table IV.Univariate Cox regression analysis of
Cul1 or c-Myc expression and clinicopathological variables
predicting survival in CRC patients. |
Table IV.
Univariate Cox regression analysis of
Cul1 or c-Myc expression and clinicopathological variables
predicting survival in CRC patients.
|
| n=470 cases |
|---|
|
|
|
|---|
| Variables | HR (95% CI) | P-value |
|---|
| Age (≤65 vs. >65
years) | 1.607
(1.215–2.126) |
0.001 |
| Sex (male vs.
female) | 1.013
(0.762–1.347) |
0.927 |
| Pathological
classification (I/II vs. III) | 2.475
(1.587–3.860) | <0.001 |
| Depth of invasion
(T1/T2 vs. T3/T4) | 3.687
(2.270–5.990) | <0.001 |
| Lymph node
metastasis (N0 vs. N1/N2) | 2.807
(2.112–3.731) | <0.001 |
| TNM stage (I/II vs.
III/IV) | 3.214
(2.407–4.291) | <0.001 |
| Distant metastasis
(M0 vs. M1) | 8.150
(4.849–13.699) | <0.001 |
| Tumor diameter (≤5
vs. >5 cm) | 1.196
(0.848–1.688) |
0.307 |
| Cul1 expression
(low vs. high) | 0.683
(0.515–0.905) |
0.008 |
| c-Myc expression
(low vs. high) | 0.280
(0.209–0.374) |
<0.001 |
| Cul1/c-Myc
expression |
| Both
low vs. one low | 1.762
(1.472–2.109) |
<0.001 |
| Both
low vs. both high | 3.422
(2.348–4.987) |
<0.001 |
 | Table V.Multivariate Cox regression analysis
of Cul1, c-Myc, Cul1/c-Myc expression and clinicopathological
variables predicting survival in patients with CRC. |
Table V.
Multivariate Cox regression analysis
of Cul1, c-Myc, Cul1/c-Myc expression and clinicopathological
variables predicting survival in patients with CRC.
| Variables | HR (95% CI) |
P-valuea |
|---|
| Cul1 |
| Age
(≤65 vs. >65 years) | 1.834
(1.376–2.443) | <0.001 |
| Sex
(male vs. female) | 0.925
(0.691–1.237) |
0.597 |
|
Pathological classification
(I/II vs. III) | 1.993
(1.252–3.174) |
0.004 |
| TNM
stage (I/II vs. III/IV) | 3.443
(2.554–4.642) | <0.001 |
| Tumor
diameter (≤5 vs. >5 cm) | 1.157
(0.805–1.662) |
0.432 |
| Cul1
expression (low vs. high) | 0.757
(0.569–1.007) |
0.046 |
| c-Myc |
| Age
(≤65 vs. >65 years) | 1.723
(1.289–2.302) | <0.001 |
| Sex
(male vs. female) | 0.913
(0.682–1.223) |
0.542 |
|
Pathological classification
(I/II vs. III) | 1.959
(1.227–3.129) |
0.005 |
| TNM
stage (I/II vs. III/IV) | 2.868
(2.117–3.887) | <0.001 |
| Tumor
diameter (≤5 vs. >5 cm) | 1.309
(0.908–1.888) |
0.149 |
| c-Myc
expression (low vs. high) | 0.337
(0.249–0.455) |
<0.001 |
| Cul1/c-Myc |
| Age
(≤65 vs. >65 years) | 1.903
(1.432–2.529) | <0.001 |
| Sex
(male vs. female) | 0.896
(0.672–1.196) |
0.458 |
|
Pathological classification
(I/II vs. III) | 1.961
(1.234–3.167) |
0.004 |
| TNM
stage (I/II vs. III/IV) | 3.386
(2.522–4.546) | <0.001 |
| Tumor
diameter (≤5 vs. >5 cm) | 1.159
(0.810–1.658) |
0.420 |
| Cul1/c-Myc
expression |
| Both
low vs. one low | 2.704
(1.862–3.927) |
<0.001 |
| Both
low vs. both high | 0.073
(0.247–0.540) |
<0.001 |
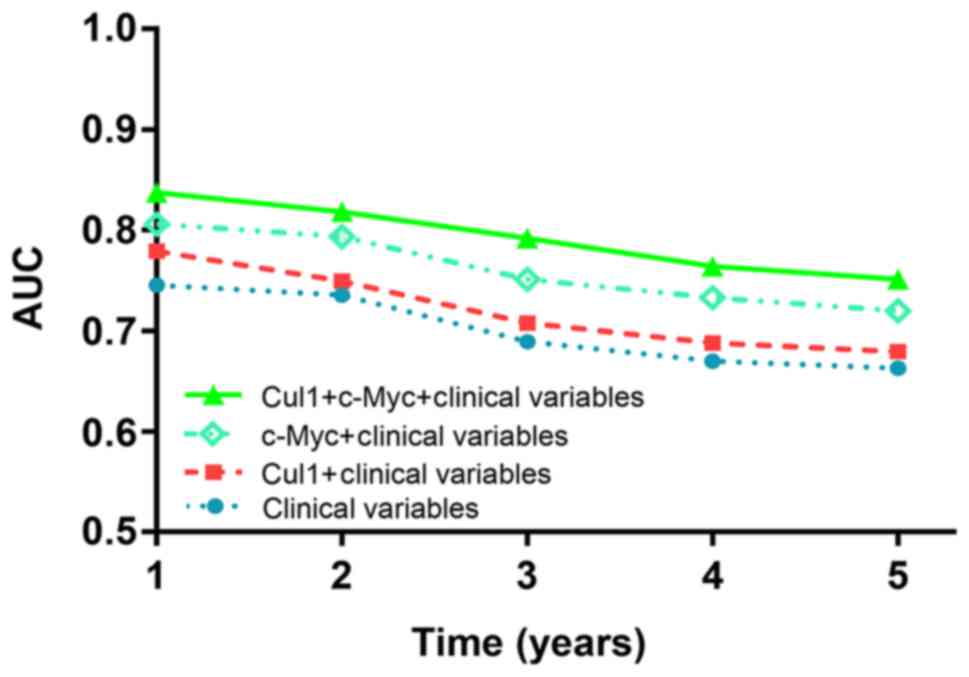
Synergisic effect of Cul1 and c-Myc
expression on OS in CRC patients
To further evaluate whether Cul1 combined with c-Myc
has a synergetic effect on the prognosis of CRC patients, we
conducted a time-dependent ROC analysis for the censored data. The
data indicated that the combination of the clinical risk score (TNM
stage, histologic type and tumor diameter) and Cul1 or c-Myc or
Cul1 plus c-Myc expression contributed much more than any one of
these markers alone in CRC patients (Fig. 7). For instance, in the TMA cohort,
the AUC at year 5 was 0.663 (95% CI, 0.476–0.703) for only clinical
risk score, whereas it was increased to 0.751 (95% CI, 0.499–0.748)
when combined with the clinical risk score and with Cul1 plus c-Myc
risk score.
The stratified analysis indicated that patients with
both low expression of Cul1 and c-Myc had a more favorable outcome
of survival (P<0.001; log-rank test; Fig. 6C) when compared with one low or both
high expression groups. The multivariate Cox regression analysis
indicated that low Cul1 and c-Myc expression alone was a favorable
independent prognostic factor for CRC patients (P<0.05 for all;
Table V).
Discussion
Prognostic studies of tumor biomarkers are valuable
as they facilitate early diagnosis, treatment efficacy and
prevention of malignancies (20).
During the occurrence and development of CRC, abnormal expression
of oncogenes and tumor-suppressor genes play an important role.
These genes may be potentially valuable as biomarkers to determine
the prognosis of CRC.
Cullin1 (Cul1) is the most representative member of
the Cullin protein family, and its complex has been identified to
exert ubiquitin ligase activity involved in the degradation of
proteins associated with the cell cycle and cancer-associated
processes. We and other investigators previously provided evidence
that Cul1 overexpression is associated with the poor prognosis of
gastric (10), breast (13) and non-small cell lung cancer
(21). In the present study, we
provided new evidence that Cul1 expression is higher in CRC tumor
tissues compared with that observed in matched adjacent normal
tissues. We also demonstrated that high Cul1 expression in CRC
tumor tissues was significantly correlated with depth of invasion,
lymph node metastasis and TNM stage. In addition, Kaplan-Meier
survival analysis revealed that high Cul1 expression in tumor
tissues was correlated with poor OS in CRC patients. Furthermore,
univariate and multivariate Cox proportional hazards regression
analyses showed that Cul1 expression is an independent negative
prognostic factor of CRC.
c-Myc is a well known oncogene and plays a vital
role in the developmental process of many types of cancers
(22). c-Myc was found to promote
cell proliferation, accelerate the cell cycle, inhibit cell
differentiation and induce apoptosis by activating the
PTEN/PI3K/AKT pathway (23). c-Myc
is highly expressed in CRC and promotes the malignant proliferation
of tumor cells (17); c-Myc gene
copy-number was identified as an independent factor for poor
prognosis in CRC (24,25). c-Myc overexpression is associated
with a worse prognosis in prostate cancer (26), lymphoma (27,28),
lung (29) and breast cancer
(30). Nevertheless, few studies
have examined the clinicopathological and prognostic implications
of c-Myc status in CRC. In the present study, c-Myc expression was
upregulated in tumor tissues compared with that noted in the paired
adjacent non-tumor tissues in both CRC fresh tissues and a TMA
cohort. We demonstrated that high c-Myc expression in CRC cancer
tissures was significantly correlated with age, depth of invasion,
lymph node metastasis and TNM stage. High c-Myc expression was also
correlated with worse OS and was found to be an independent
negative prognostic factor in CRC patients. Our cohort indicated
that increased expression of Cul1 and c-Myc was significantly
associated with unfavorable clinicopathologic parameters and worse
OS for CRC. c-Myc enhances expression of Cul1 and promotes
ubiquitin-dependent proteolysis and cell cycle progression
(18). On the contrary, Cul1
assembled SCF (Skp1/Cul1/F-box)FBXO28 plays an important
role in the regulation of MYC-driven cancers (31). In the present study, we also
demonstrated that Cul1 combined with c-Myc has synergistic
potential and may be more effective than Cul1 or cMyc alone in
predicting the prognosis of CRC patients.
In conclusion, our findings indicate that Cul1 and
c-Myc are unfavorable prognostic factors for CRC patients. In
addition, to the best of our knowledge, we first revealed the
combined value of Cul1 and c-Myc as efficient prognostic factors.
Although the co-action of these two proteins can be used to predict
the prognosis of CRC, further investigation is warranted to
elucidate their role in the occurrence and development of CRC.
Acknowledgements
The present study was supported in part by the
Jiangsu Key Laboratory of Cancer Biomarkers, Prevention and
Treatment.
Glossary
Abbreviations
Abbreviations:
|
Cul1
|
Cullin1
|
|
CRC
|
colorectal cancer
|
References
|
1
|
Wan DS: Epidemiologic trend of and
strategies for colorectal cancer. Ai Zheng. 28:897–902. 2009.(In
Chinese). PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weitz J, Koch M, Debus J, Höhler T, Galle
PR and Büchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paul S: Dysfunction of the
ubiquitin-proteasome system in multiple disease conditions:
Therapeutic approaches. BioEssays. 30:1172–1184. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nakayama KI and Nakayama K: Regulation of
the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol.
16:323–333. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Salon C, Brambilla E, Brambilla C,
Lantuejoul S, Gazzeri S and Eymin B: Altered pattern of Cul-1
protein expression and neddylation in human lung tumours:
Relationships with CAND1 and cyclin E protein levels. J Pathol.
213:303–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen BB, Glasser JR, Coon TA and
Mallampalli RK: F-box protein FBXL2 exerts human lung tumor
suppressor-like activity by ubiquitin-mediated degradation of
cyclin D3 resulting in cell cycle arrest. Oncogene. 31:2566–2579.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen BB, Glasser JR, Coon TA, Zou C,
Miller HL, Fenton M, McDyer JF, Boyiadzis M and Mallampalli RK:
F-box protein FBXL2 targets cyclin D2 for ubiquitination and
degradation to inhibit leukemic cell proliferation. Blood.
119:3132–3141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Chen Y, Deng J, Zhou J, Gu X, Tang
Y, Zhang G, Tan Y, Ge Z, Huang Y, et al: Cullin1 is a novel
prognostic marker and regulates the cell proliferation and
metastasis in colorectal cancer. J Cancer Res Clin Oncol.
141:1603–1612. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan
Y, Zhou J and Li G: Overexpression of Cullin1 is associated with
poor prognosis of patients with gastric cancer. Hum Pathol.
42:375–383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen G and Li G: Increased Cul1 expression
promotes melanoma cell proliferation through regulating p27
expression. Int J Oncol. 37:1339–1344. 2010.PubMed/NCBI
|
|
12
|
Chen G, Cheng Y, Martinka M and Li G: Cul1
expression is increased in early stages of human melanoma. Pigment
Cell Melanoma Res. 23:572–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bai J, Yong HM, Chen FF, Mei PJ, Liu H, Li
C, Pan ZQ, Wu YP and Zheng JN: Cullin1 is a novel marker of poor
prognosis and a potential therapeutic target in human breast
cancer. Ann Oncol. 24:2016–2022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Riggelen J, Yetil A and Felsher DW:
MYC as a regulator of ribosome biogenesis and protein synthesis.
Nat Rev Cancer. 10:301–309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meyer KD, Donner AJ, Knuesel MT, York AG,
Espinosa JM and Taatjes DJ: Cooperative activity of cdk8 and GCN5L
within Mediator directs tandem phosphoacetylation of histone H3.
EMBO J. 27:1447–1457. 2008.PubMed/NCBI
|
|
17
|
Böckelman C, Koskensalo S, Hagström J,
Lundin M, Ristimäki A and Haglund C: CIP2A overexpression is
associated with c-Myc expression in colorectal cancer. Cancer Biol
Ther. 13:289–295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O'Hagan RC, Ohh M, David G, De Alboran IM,
Alt FW, Kaelin WG Jr and DePinho RA: Myc-enhanced expression of
Cul1 promotes ubiquitin-dependent proteolysis and cell cycle
progression. Genes Dev. 14:2185–2191. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang S, Wu X, Chen Y, Zhang J, Ding J,
Zhou Y, He S, Tan Y, Qiang F, Bai J, et al: Prognostic and
predictive role of JWA and XRCC1 expressions in gastric cancer.
Clin Cancer Res. 18:2987–2996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Takano Y and Zheng HC: The
pathobiological features of gastrointestinal cancers (Review).
Oncol Lett. 3:961–969. 2012.PubMed/NCBI
|
|
21
|
Xu M, Yang X, Zhao J, Zhang J, Zhang S,
Huang H, Liu Y and Liu J: High expression of Cullin1 indicates poor
prognosis for NSCLC patients. Pathol Res Pract. 210:397–401. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hatakeyama S, Watanabe M, Fujii Y and
Nakayama KI: Targeted destruction of c-Myc by an engineered
ubiquitin ligase suppresses cell transformation and tumor
formation. Cancer Res. 65:7874–7879. 2005.PubMed/NCBI
|
|
23
|
Blackburn JS, Liu S, Wilder JL, Dobrinski
KP, Lobbardi R, Moore FE, Martinez SA, Chen EY, Lee C and Langenau
DM: Clonal evolution enhances leukemia-propagating cell frequency
in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway
activation. Cancer Cell. 25:366–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB,
Choe G, Kim WH and Lee HS: c-MYC copy-number gain is an independent
prognostic factor in patients with colorectal cancer. PLoS One.
10:e01397272015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khaleghian M, Shakoori A, Razavi AE and
Azimi C: Relationship of amplification and expression of the C-MYC
gene with survival among gastric cancer patients. Asian Pac J
Cancer Prev. 16:7061–7069. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zeng W, Sun H, Meng F and Liu Z, Xiong J,
Zhou S, Li F, Hu J, Hu Z and Liu Z: Nuclear C-MYC expression level
is associated with disease progression and potentially predictive
of two year overall survival in prostate cancer. Int J Clin Exp
Pathol. 8:1878–1888. 2015.PubMed/NCBI
|
|
27
|
Huang W, Guo L, Liu H, Zheng B, Ying J and
Lv N: C-MYC overexpression predicts aggressive transformation and a
poor outcome in mucosa-associated lymphoid tissue lymphomas. Int J
Clin Exp Pathol. 7:5634–5644. 2014.PubMed/NCBI
|
|
28
|
Zhang Y, Li J, Xi Y, Bai W, Bai W and Sun
R: Significance of C-myc expression in T-lymphoblastic
lymphoma/leukemia and its relation with prognosis. Zhonghua Bing Li
Xue Za Zhi. 44:571–577. 2015.PubMed/NCBI
|
|
29
|
Seo AN, Yang JM, Kim H, Jheon S, Kim K,
Lee CT, Jin Y, Yun S, Chung JH and Paik JH: Clinicopathologic and
prognostic significance of c-MYC copy number gain in lung
adenocarcinomas. Br J Cancer. 110:2688–2699. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deming SL, Nass SJ, Dickson RB and Trock
BJ: C-myc amplification in breast cancer: A meta-analysis of its
occurrence and prognostic relevance. Br J Cancer. 83:1688–1695.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cepeda D, Ng HF, Sharifi HR, Mahmoudi S,
Cerrato VS, Fredlund E, Magnusson K, Nilsson H, Malyukova A,
Rantala J, et al: CDK-mediated activation of the SCFFBXO28
ubiquitin ligase promotes MYC-driven transcription and
tumourigenesis and predicts poor survival in breast cancer. EMBO
Mol Med. 5:1067–1086. 2013. View Article : Google Scholar : PubMed/NCBI
|