Introduction
Pituitary adenoma is one of the most common
intracranial tumors, with an increasing prevalence in recent years
(1). Of its multiple subtypes,
pituitary null cell adenoma was originally defined by electron
microscopy as a tumor containing cells with relatively few, poorly
developed organelles and with no sufficiently distinctive
structural characteristics to determine its cellular origin;
moreover, most cells in pituitary null cell adenomas do not yield
positive immunostaining results for anterior pituitary hormones
(2,3). Null cell adenomas, including their
oncocytic variants, account for approximately 25.1% of all
pituitary adenomas, and gonadotropinomas account for approximately
24.8% of all pituitary adenomas (3). Pituitary null cell adenomas are
clinically challenging because they often present as macroadenomas
and because there are no reliable biomarkers for monitoring and
effective targeted therapy for treating these tumors. Moreover, the
origin and pathogenesis of pituitary null cell adenomas remain to
be elucidated to date.
Long non-coding RNAs (lncRNAs) are transcribed from
at least 75% of the human genome (4). They play important roles in gene
regulation and maintenance of genomic stability and affect various
cellular process, including survival, proliferation, and migration
(5). Several lncRNAs function as
oncogenes, tumor suppressors, or prognostic markers (6–10).
Recent studies have reported that some novel tumor-associated
lncRNAs participate in the pathogenesis of different tumors, such
as TINCR, which is underexpressed and acts as a tumor suppressor in
colorectal cancer (11).
C5orf66-AS1 (also known as Epist and CTC-276P9.1) is underexpressed
in esophageal squamous cell carcinomas compared with that in normal
esophageal tissues (12). Results
of functional experiments have shown that C5orf66-AS1 functions as
a tumor suppressor (12). A more
recent study showed that expression of NORAD, which stabilizes the
genome and whose inactivation results in dramatic aneuploidy
(13), was elevated in invasive
breast cancer and was associated with decreased survival, and
showed that in vitro knockdown of NORAD inhibited tumor
growth (14). To date, only a few
studies have examined roles of lncRNAs in pituitary adenomas.
From RNA sequencing (RNA-seq) data of another cohort
with various types of pituitary adenomas, which is under study in
our laboratory, we carried out a manual review of the aberrantly
expressed long intergenic non-coding RNAs (lincRNAs) of the null
cell type. We found that the lncRNAs C5orf66-AS1, NORAD, and TINCR
were in the list of the 120 most significantly dysregulated
lincRNAs, and after the manual review we were specifically
interested in the functions of these three lincRNAs and their
targets. Since few studies have examined roles of lncRNAs in
pituitary null cell adenomas, we wondered if they play similar
roles in the development of pituitary null cell adenomas based on
previous studies on the participation of these three lncRNAs in
oncogenesis of other tumors.
Materials and methods
Examining gene expression abundance
across multiple tissues online
We first examined the expression of three lncRNAs,
C5orf66-AS1, NORAD, and TINCR, in different normal adult tissues by
using RNA-seq data from the Genotype-Tissue Expression (GTEx)
Project (http://www.gtexportal.org/home/). The data described
in this study were retrieved from the GTEx Portal 11/17/2016 (GTEx
Analysis Release V6p) and dbGaP (accession no. phs000424.v6.p1)
11/17/2016.
Patients and specimens
Patients who underwent endoscopic trans-sphenoidal
surgery from March 2013 to November 2014 at Beijing Tiantan
Hospital and who were diagnosed with pituitary null cell adenomas
through immunostaining and electron microscopy were enrolled in the
present study. Inclusion criteria were results of electron
microscopy showing cells with relatively few, poorly developed
organelles; absence of sufficiently distinctive structural
characteristics to determine the cellular origin of the tumor; and
negative results for the immunostaining of anterior pituitary
hormones, which was consistent with the definition of pituitary
null cell adenoma described above and according to the World Health
Organization classification (2004 version) (2,3,15).
Patients with a family history of pituitary adenoma or other tumors
of the endocrine system were excluded. Each patient underwent a
hormonal serum test and magnetic resonance imaging (MRI) before
undergoing surgery. The excised tissues were examined by performing
histopathological and immunohistochemical analysis for all anterior
pituitary hormones and by electron microscopy. Tumors with
Hardy-Wilson classification grade IV and/or Knosp classification
grades III and IV (16–18) were defined as invasive. Invasiveness
was determined by a group of neurosurgeons, radiologists, and
pathologists. Maximum tumor diameter was measured using gadolinium
(Gd)-enhanced T1WI MR images. Normal pituitary tissues were
obtained from donors who died from diseases other than neurologic
or endocrine diseases. All patients provided written informed
consent, and the study was approved by the Ethics Committee of the
Beijing Tiantan Hospital.
Total RNA extraction and cDNA
synthesis
Pituitary adenoma and normal anterior pituitary
tissues were stored in liquid nitrogen immediately after removal.
Total RNA was extracted from the frozen tissue samples by using
TRizol (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer's instructions. Integrity of the total RNA was
determined by performing agarose gel electrophoresis. Only samples
showing no RNA degradation (28S/18S ≥0.7) were used to generate
labeled targets. RNA purity and concentration were determined using
NAS-99 (Merinton, Beijing, China) and A260:A280 ratios yielded were
consistently close to 2.0. Total RNA was reverse transcribed to
cDNA by using HiFiScript gDNA Removal cDNA Synthesis kit (CWBio
Co., Ltd., Beijing, China), according to the manufacturer's
instructions.
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed using StepOnePlus™ Real-Time
PCR System (Applied Biosystems, Foster City, CA, USA) and Kapa SYBR
FAST qPCR Master Mix ABI Prism™ (2X, KK4601; Kapa Biosystems, Inc.,
Wilmington, MA, USA), according to the manufacturer's instructions.
SYBR Green assays were performed using 0.5 µl PCR forward primer
(10 µM), 0.5 µl PCR reverse primer (10 µM), 2 µl cDNA, 5 µl 2X
master mix, and 2 µl double distilled water in a total reaction
volume of 10 µl. The PCR protocol was initiated at 95°C for 3 min,
followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec.
GAPDH was used as a reference gene. Results were obtained
using three independent wells. Relative expression level was
calculated using 2−∆∆Ct method (19) after normalizing with GAPDH
expression level. Sequences of primers used for performing qRT-PCR
are presented in Table I.
 | Table I.Sequences of primers used in
qRT-PCR. |
Table I.
Sequences of primers used in
qRT-PCR.
| Gene | Primer
sequence |
|---|
| TINCR | F:
AAGGAGAGCCTACTTCCCTCAA |
|
| R:
TCTAGTTCCAAGCTGGGTGATC |
| C5orf66-AS1 | F:
GCTTCGCGTCAAGAGGGTAT |
|
| R:
GACCGACGTCTGCTGCTTTT |
| NORAD | F:
AGCTTTGGGATTTTGAATTGGT |
|
| R:
GATCCTGTGTGTAGGCACAACAT |
| SCGB3A1 | F:
ACAATGTTCGGTTGAGGGGAA |
|
| R:
AGGTGTGAGCAGCAGGGTTC |
| GAPDH | F:
ACAGCCTCAAGATCATCAGCAAT |
|
| R:
GATGGCATGGACTGTGGTCAT |
In silico target gene prediction
Two algorithms were used to predict possible
cis- and trans-acting target genes, as previously
described (20). Cis-acting
target genes were predicted using UCSC genome browser and genome
annotation to locate the nearest known protein-coding genes from
the lncRNAs (21). According to
distance stratification, protein-coding genes located within a
distance of 5 and 300 kb from the lncRNAs were captured as targets.
Trans-acting genes were predicted using RNAplex software
(http://www.bioinf.uni-leipzig.de/Software/RNAplex/).
RNAplex can determine possible hybridization sites for a query RNA
according to sequence complementarity and RNA duplex-binding energy
prediction (22,23). First, BLAST (e<1E-5) was
performed using the lncRNAs and known protein-coding genes; next,
RNAplex (parameter set as -e −70) was used to screen
trans-acting target genes.
Gene expression microarray
analysis
Expression of predicted target genes was determined
using microarray data of another cohort examined in our laboratory.
Microarray analysis was performed using Whole Human Genome Oligo
Microarray (4×44K) (Agilent Technologies, Santa Clara, CA, USA).
Total RNA was extracted and was purified using mirVana™ miRNA
Isolation kit (catalog no. AM1561; Ambion, Austin, TX, USA),
according to the manufacturer's instructions. RIN was determined
using Bioanalyzer 2100 (Agilent Technologies) to inspect RNA
integration. None of the samples showed RNA degradation (RIN ≥7.0
and 28S/18S ≥0.7). Total RNA was amplified and was labeled using
Low Input Quick Amp labeling kit, One-Color (catalog no. 5190-2305;
Agilent Technologies). Labeled cRNA was purified using RNeasy mini
kit (catalog no. 74106; Qiagen, GmBH, Hilden, Germany). Next, each
slide was hybridized with Cy3-labeled cRNA by using Gene Expression
Hybridization kit (catalog no. 5188-5242; Agilent Technologies),
according to the manufacturer's instructions. After hybridization,
the slides were scanned using a microarray scanner (catalog no.
G2565CA; Agilent Technologies). Data were extracted using Feature
Extraction software 10.7 (Agilent Technologies), and raw data were
normalized using Quantile algorithm, limma packages in R.
RNA sequencing
We previously performed RNA sequencing of a cohort
with various types of pituitary adenomas, which is under study in
our laboratory. After the total RNA extraction and DNase I
treatment, magnetic beads with Oligo (dT) were used to isolate
mRNAs. mRNAs were fragmented into short fragments by mixing with
fragmentation buffer. Then, cDNAs were synthesized using the mRNA
fragments as templates. Fragments were purified and resolved with
EB buffer for end reparation and single nucleotide adenine addition
and were connected with adapters. After agarose gel
electrophoresis, the suitable fragments were selected for PCR
amplification as templates. During the quality control steps, the
Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA)
and ABI StepOnePlus Real-time PCR System (Applied Biosystems) were
used in quantification and qualification of the sample library.
Finally, the library was sequenced using the Illumina HiSeq 2000
(Illumina, San Diego, CA, USA) at the BGI-Tech Bioinformatics
Institute (Shenzhen, China).
Cell culture and transfection
The mouse pituitary cell line GT1-1 was purchased
from the China Infrastructure of Cell Line Resources and cultured
in phenol red-free Dulbecco's modified Eagle's medium (DMEM;
Invitrogen) supplemented with 10% fetal bovine serum (Gibco,
Auckland, New Zealand) in a humidified incubator at 37°C with 5%
CO2. pUC57-C5orf66-AS1 was obtained from BGI (Shenzhen,
China). Transfection was performed using Lipofectamine 3000
(Invitrogen) according to the manufacturer's instructions. After 24
h, C5orf66-AS1 was examined using qRT-PCR in triplicates.
Assessment of cell viability
Log-phase GT1-1 cells were harvested and confirmed
to be >99% viable by trypan blue exclusion and adjusted to a
density of 1×105 cells/ml. A volume of 100 µl of cell
suspension was plated into 96-well plates and cultured for 24 h.
After transfection for 24, 48, and 72 h, 20 µl MTS solution was
added to each well and further incubated for 4 h. Absorbance of
each well at 490 nm was measured using an ELISA plate reader
(Thermo Labsystems, Franklin, MA, USA).
Transwell invasion assay
Tumor cell invasion was measured by examining cell
migration and invasion through fibronectin- and Matrigel-coated
polycarbonate filters, respectively, using modified Transwell
chambers (Corning Inc., Corning, NY, USA). GT1-1 cells
(2×104 cells) were added into the upper chambers.
Migrated cells adhered to the lower membrane were fixed in 4%
paraformaldehyde and stained with hematoxylin (Zhongshan Company,
Beijing, China). The average number of migrated cells was
quantified by counting five random high-power fields under a
fluorescent microscope (Carl Zeiss, Jena, Germany). Experiments
were performed in triplicates.
Statistical analyses
Statistical analysis of qRT-PCR results was
performed using SPSS software (v23.0; IBM Corp., Armonk, NY, USA).
Shapiro-Wilk test was used to test normality. Differences between
the groups were determined using Mann-Whitney U test. Correlation
between lncRNA expression level and maximum tumor diameter and
between lncRNA expression level and patient age was determined
using Spearman's rank correlation coefficient or Pearson's
correlation coefficient on the basis of data distribution.
Relationship between C5orf66-AS1 expression level and maximum tumor
diameter in male patients was determined by performing simple
linear regression analysis after calculating Pearson's correlation
coefficient. Co-expression analysis was performed with R based on
the Pearson correlation coefficients between C5orf66-AS1 and all
mRNAs with unique mapping from RNA-seq data after ruling out types
of pituitary adenomas other than the null cell type. P≤0.05 was
considered statistically significant, except in the Shapiro-Wilk
test, in which 0.1 was used as the level of significance to
decrease the probability of false-negative errors.
Results
Expression of the three lncRNAs across
multiple tissues in GTEx
First, we examined the expression of three lncRNAs,
C5orf66-AS1, NORAD, and TINCR, in different normal tissues by using
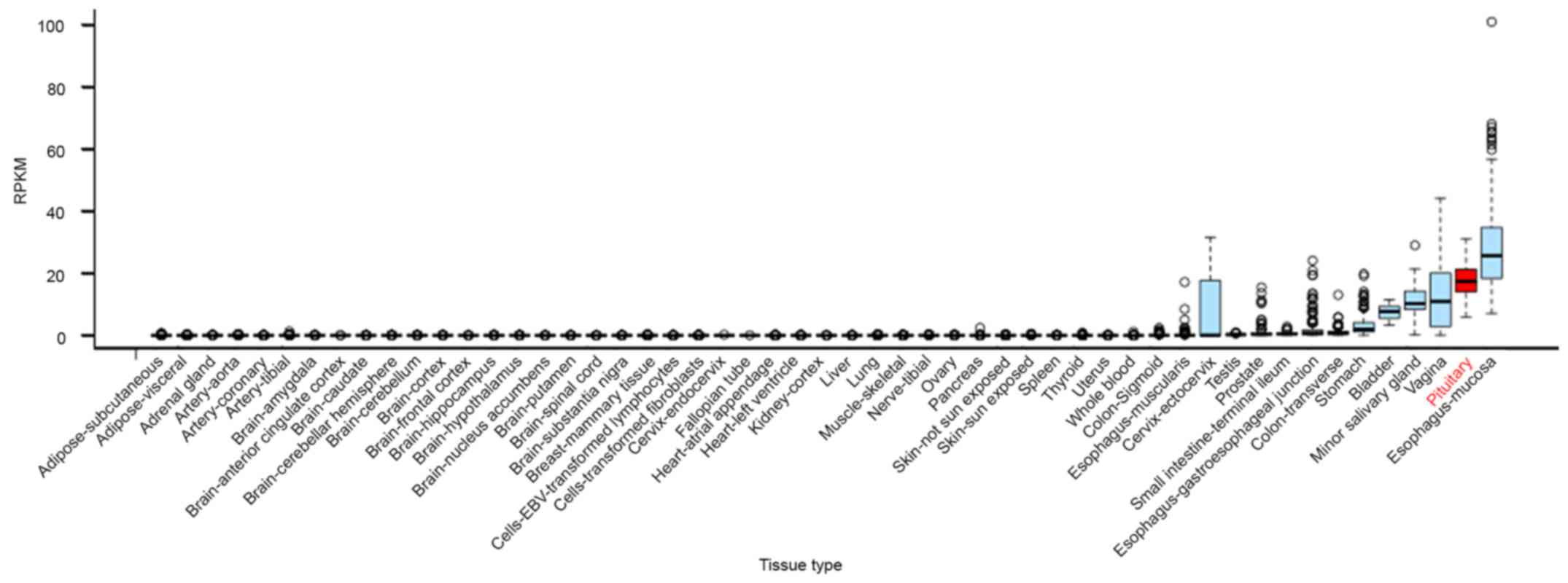
RNA-seq data from GTEx. We found that C5orf66-AS1 was overexpressed
specifically in the pituitary gland, with its expression level
being only lower than that in esophageal mucosa among the 53 tissue
types examined (Fig. 1). TINCR was
expressed at an average level, and no data are available on the
expression of NORAD. These results suggest that C5orf66-AS1
performs special functions in the pituitary gland.
Clinical characteristics of
patients
qRT-PCR analysis of TINCR, C5orf66-AS1, and NORAD
was performed using normal pituitary tissues from four donors and
tumor tissues from 11 patients, including six men and five women.
Characteristics of the patients are shown in Table II. Ages of the patients ranged from
36 to 74 years, with an average age of 53.3 years. The main
complaints of the patients were headache and visual disturbances;
in some patients, pituitary null cell adenoma was diagnosed by
performing incident MRI or CT scan. Time interval from the
appearance of the first symptoms to surgery varied from 20 days to
9 years. For each patient, serum test yielded negative results for
excessive pituitary hormone secretion, the sella turcica showed
enlargement, and radiologic performance was consistent with the
diagnosis of pituitary adenoma. Tumors of six patients were
invasive; of these, three patients had Hardy-Wilson grade III and
Knosp classification grade III tumors and the remaining three
patients had Hardy-Wilson grade IV and Knosp classification grade
IV tumors (Fig. 2). Maximum tumor
diameters ranged from 23.0 to 57.2 mm. No differences were observed
in maximum tumor diameters between male and female patients. All
the tumors were diagnosed as pituitary null cell adenomas, with no
major difficulty; some of these tumors showed oncocytic
changes.
 | Table II.Expression levels of lncRNAs in
pituitary adenomas grouped according to biological behavior, sex,
and other relative clinical characteristics. |
Table II.
Expression levels of lncRNAs in
pituitary adenomas grouped according to biological behavior, sex,
and other relative clinical characteristics.
|
|
|
| MD of tumor | C5orf66-AS1 | TINCR | NORAD |
|---|
|
|
|
|
|
|
|
|
|---|
| Group | n | m (sd) of age
(yrs.) | m (sd) (mm) | pa | m (sd) | pa | CCA | CCD | m (sd) | pa | CCA | CCD | m (sd) | pa | CCA | CCD |
|---|
| Behavior |
|
|
| 0.030 |
| 0.004 |
|
|
| 0.328 |
|
|
| 0.931 |
|
|
|
Inv | 5 | 52.6
(9.8)b | 41.0
(10.6)b |
| 0.0052
(0.0077)c |
| 0.400d | 0.700d | 0.0689
(0.0907)c |
| 0.600d | 0.300d | 0.2034
(0.2168)c |
| 0.200d | 0.100d |
|
N-inv | 6 | 53.8
(15.0)b | 29.1
(5.0)b |
| 0.0810
(0.0330)b |
| 0.922e,g | −0.323e | 0.0767
(0.0461)b |
| 0.653e | −0.441e | 0.1931
(0.1728)c |
| 0.371d | −0.943d,f |
| Sex |
|
|
| 0.329 |
| 0.126 |
|
|
| 0.361 |
|
|
| 0.247 |
|
|
|
Male | 6 | 57.0
(13.1)b | 31.1
(4.4)b |
| 0.0739
(0.0451)b |
| 0.515e | −0.884e,f | 0.0732
(0.0471)c |
| 0.543d | −0.486d | 0.1202
(0.0745)c |
| 0.943d,f | −0.886d,f |
|
Female | 5 | 48.8
(10.8)b | 38.6
(13.3)b |
| 0.0137
(0.0182)c |
| 0.400d | −0.100d | 0.0731
(0.0903)c |
| 0.100d | 0.100d | 0.2908
(0.2401)b |
| 0.105e | 0.109e |
| Total | 11 | 53.3
(12.3)b | 34.5
(9.8)b |
| 0.0465
(0.0462)c |
| 0.415d | −0.601d,f | 0.0732
(0.0661)c |
| −0.465d | −0.264d | 0.1978
(0.1838)c |
| 0.269d | −0.305d |
After predicting target genes, we examined some
target genes by using microarray data obtained from pituitary null
cell adenomas of 16 patients and normal pituitary tissues of six
donors from another study cohort. Patients in this cohort were
admitted during the same period and were diagnosed using the same
criteria as those described above. Of the 16 patients, 12 were men
and 4 were women. Moreover, six patients had invasive tumors, and
nine patients had non-invasive tumors (invasiveness of the tumor of
one patient could not be evaluated because of the absence of MRI
data). Other clinical characteristics of these patients were
similar to those of the 11 patients included in the present study.
Then, we performed a co-expression analysis of the RNA-seq data of
another cohort, including 12 pituitary null cell adenomas with
other types of pituitary adenomas and five normal pituitaries. Of
the 12 patients of pituitary null cell adenomas, six were men and
six were women; seven patients had invasive tumors, and 5 patients
had non-invasive tumors. Other clinical characteristics of these
patients were similar to those of the 11 patients included in the
present study.
Expression of C5orf66-AS1
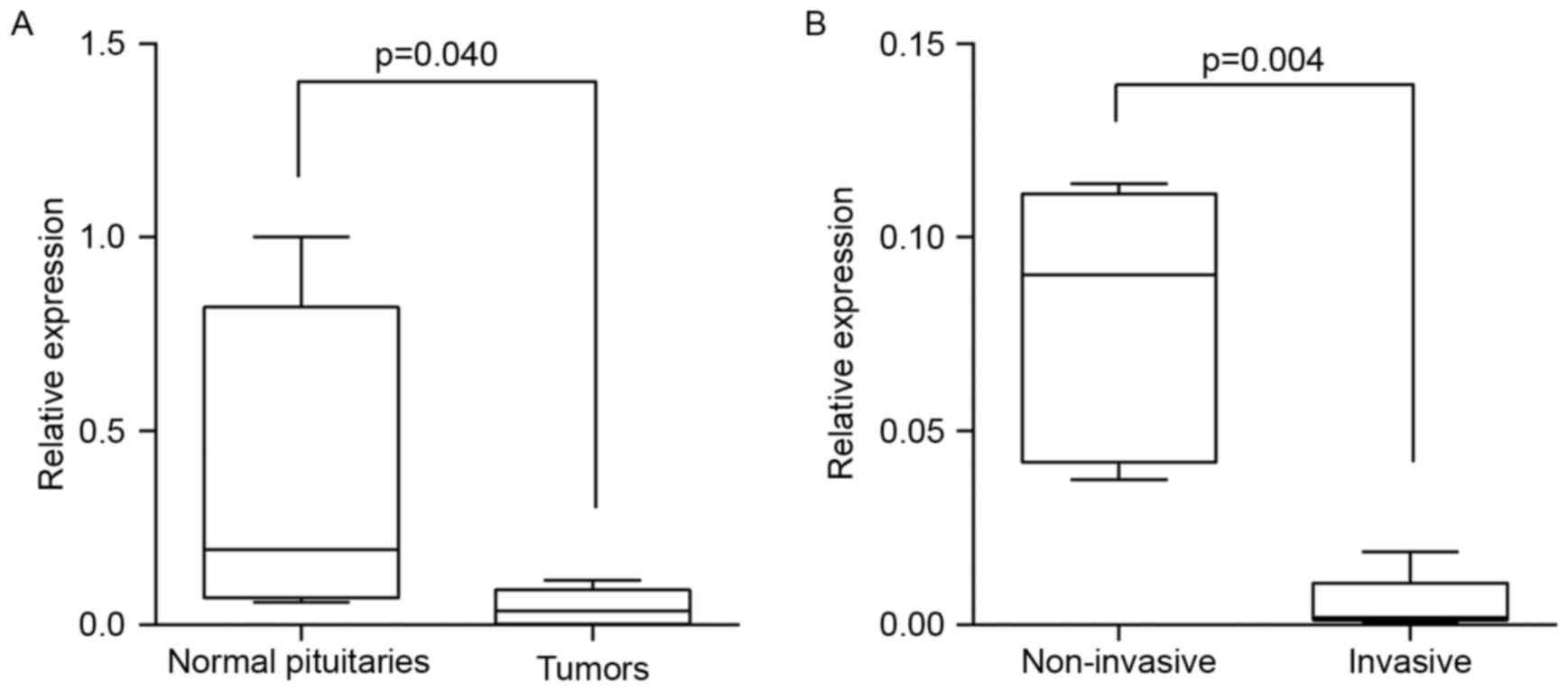
C5orf66-AS1 expression level was significantly lower
in pituitary null cell adenoma tissues than in normal pituitary
tissues (Mann-Whitney U test, Z= −2.089; two-tailed, P=0.040;
Table II, Fig. 3A). Moreover, C5orf66-AS1 expression
level was significantly lower in invasive tumors than in
noninvasive tumors (Mann-Whitney U test, Z= −2.739; two-tailed,
P=0.004; Table II, Fig. 3B).
No correlation was observed between C5orf66-AS1
expression level and patient age. Analysis of the relationship
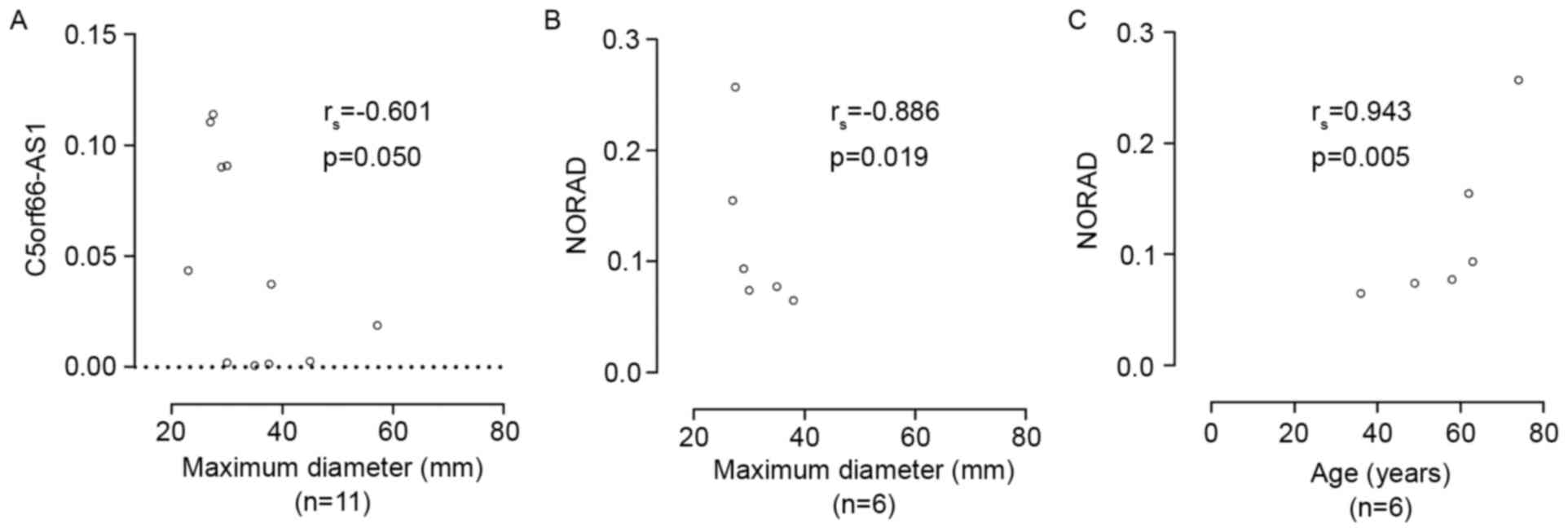
between C5orf66-AS1 expression and pituitary tumor growth showed
that C5orf66-AS1 lncRNA levels were negatively correlated with
maximum tumor diameter (Spearman's rank correlation coefficient
rs= −0.601, P=0.05; Table
II, Fig. 4A). Stratification of
tumors based on sex showed a negative linear correlation between
C5orf66-AS1 lncRNA levels and maximum tumor diameter in male
patients (Pearson correlation: r= −0.884, P=0.019; simple linear
regression analysis: R2=0.782, adjusted
R2=0.728, F=14.360, P=0.019, Y=0.354–0.009X, t= −3.790,
P=0.019; Table II). However, this
linear correlation was not observed in female patients. Moreover,
no significant difference was observed in C5orf66-AS1 expression
between male and female patients. Stratification of tumors based on
their invasive potential showed a significant positive linear
correlation between C5orf66-AS1 expression and patient age in
patients with non-invasive tumors (Pearson correlation, r=0.922,
P=0.009; Table II). However, no
correlation was observed between maximum tumor diameter and patient
age; and this linear correlation was not observed in patients with
invasive tumors.
Expression of NORAD and TINCR
NORAD and TINCR expression did not show normal
distribution in the tumor samples of the complete cohort examined
in the study. No difference was observed in NORAD and TINCR
expression levels between tumor and normal pituitary tissues and
between invasive and non-invasive tumors. Moreover, no correlation
was observed between NORAD and TINCR expression levels and maximum
tumor diameter or patient age. However, in non-invasive pituitary
null cell tumors, NORAD lncRNA level was significantly negatively
correlated with maximum tumor diameter (Spearman's rank
correlation, rs= −0.943, P=0.005; Table II). Furthermore, NORAD lncRNA level
was significantly negatively correlated with maximum tumor diameter
(Spearman's rank correlation, rs= −0.886, P=0.019;
Table II, Fig. 4B) but significantly positively
correlated with age (Spearman's rank correlation,
rs=0.943, P=0.005; Table
II, Fig. 4C) in male patients.
However, no correlation was observed between maximum tumor diameter
and age in male patients.
In silico prediction of target genes
of C5orf66-AS1 and their expression in microarrays
Nine cis-acting target genes of C5orf66-AS1
were detected in a distance of <300 kb from C5orf66-AS1
(Table III). Of these, the top
two genes, namely, C5orf66 and PITX1, were within a
distance of 5 kb from C5orf66-AS1. Location of C5orf66-AS1 is in
chromosome 5: 135,038,831-135,040,047 (reverse strand), while that
of C5orf66 is in chromosome 5: 135,033,280-135,344,680
(forward strand) and PITX1 is chromosome 5:
135,027,734-135,034,274 (reverse strand) (in assembly of
GRCh38.p7/GCF_000001405.33, Fig.
5). Furthermore, we predicted 347 trans-acting target
genes of C5orf66-AS1. The top 20 trans-acting genes with the
best interaction energy are listed in Table III.
 | Table III.Cis-acting and top 20 trans-acting
target genes of C5orf66-AS1 predicted in silico. |
Table III.
Cis-acting and top 20 trans-acting
target genes of C5orf66-AS1 predicted in silico.
| Type | Gene name |
|---|
|
Cisa | C5orf66,
PITX1d, CATSPER3,
PCBD2, TXNDC15d,
C5orf24d, DDX46,
CAMLG, H2AFY |
|
Transb | NME6,
VWA1c,
EVC2d, COL7A1,
GTF2IRD2B, GTF2IRD2d,
CYC1d,
RASSF1d, ANKRD33B,
LGI3d, DENND3,
ZNF621, QDPR, POLR1E, ADAM18, SCGB3A1d,e, AEBP1d, DBN1d, SH3TC1, SEPT8 |
Next, we examined the predicted target genes (nine
cis-acting genes and top 20 trans-acting genes) in
microarray data of another study cohort (16 patients with pituitary
null cell adenomas and six donors with normal pituitary gland)
examined in our laboratory. By using a stringency of P≤0.01 and
fold change of >1.5, we found that 3 of the 9 predicted
cis-acting genes and 8 of the 20 predicted
trans-acting genes were differentially expressed between
tumor tissues and normal pituitary tissues (Table III). Moreover, by using a
stringency of P≤0.05 and fold change of >1.5, we found that only
SCGB3A1 was significantly differentially expressed in
invasive and non-invasive pituitary null cell adenomas (P=0.046,
fold change=6.817). Then, we performed qRT-PCR to validate
expression of SCGB3A1 in pituitary null cell adenomas and in
normal pituitary tissues. The result showed that SCGB3A1 was
significantly underexpressed in pituitary adenomas compared with
normal pituitaries (P=0.030) and was significantly underexpressed
in invasive tumors compared with non-invasive ones (P=0.041).
Co-expression analysis of C5orf66-AS1
in RNA-seq data
Co-expression analysis of C5orf66-AS1 in the RNA-seq
data showed that among the total 19,378 mRNAs which were uniquely
mapped to, there were 32 mRNAs with r>0.90 or r< −0.90
(P<0.001) and 3,002 mRNAs with r>0.70 or <-0.70
(P<0.001). The most correlated gene was PAQR7 (r=0.938,
P<0.001). In the top 20 trans-acting genes we predicted
in silico, there were eight genes with r>0.70
(P<0.001). They were NME6 (r=0.851), RASSF1
(r=0.822), EVC2 (r=0.821), COL7A1 (r=0.746),
POLR1E (r=0.730), SCGB3A1 (r=0.730), LGI3
(r=0.728), and GTF2IRD2 (r=0.723). Out of these eight genes,
seven genes (NME6, RASSF1, EVC2, COL7A1, SCGB3A1, LGI3, and
GTF2IRD2) were differentially expressed between tumor
tissues and normal pituitary tissues.
Cell viability and invasion assay
after overexpressing C5orf66-AS1
To confirm the tumor suppressor role of C5orf66-AS1,
we first overexpressed it in mouse pituitary cell line GT1-1 by
transfection. We performed qRT-PCR to examine expression of
C5orf66-AS1 before transfection and at 24 h after transfection. The
results showed that C5orf66-AS1 was significantly overexpressed
after transfection. A cell viability assay was performed at 24, 48,
and 72 h after transfection. The result showed that after
C5orf66-AS1 transfection, the viability of tumor cells was
significantly inhibited compared with that of the control group in
a time-dependent manner. After that, we performed the Transwell
invasion assay. The result showed that overexpression of
C5orf66-AS1 markedly reduced the effect of invasion.
Discussion
Null cell adenoma is a special type of pituitary
adenoma, which has no structural characteristics sufficient to
determine its cellular origin and which does not yield positive
immunostaining results for adenohypophyseal hormones, except for a
few scattered cells (2). Results of
electron microscopy show that pituitary null cell adenoma cells
have special features such as electron-lucent cytoplasm, with
relatively few, poorly developed organelles and small, spherical,
and usually sparse secretory granules (2,3).
Although pituitary null cell adenomas are presumed to originate
from gonadotropic cells (3,24), some studies argue that null cell
adenomas are different from gonadotropic adenomas (25,26).
However, the actual origin and detailed mechanism underlying the
pathogenesis of pituitary null cell adenoma have not been
elucidated to date.
From RNA-seq data of another cohort with various
types of pituitary adenomas, we found that lncRNAs C5orf66-AS1,
NORAD, and TINCR were significantly dysregulated in the null cell
type. Since recent studies indicate that lncRNAs C5orf66-AS1
(12), NORAD (13,14),
and TINCR (11) are involved in the
pathogenesis of other tumors, we examined whether these lncRNAs
played similar roles in pituitary adenomas. We performed qRT-PCR
analysis of the three lncRNAs. We found that C5orf66-AS1 expression
was significantly lower in pituitary null cell adenoma tissues than
in normal pituitary tissues and in invasive adenomas than in
non-invasive adenomas. Moreover, we observed a negative correlation
between C5orf66-AS1 expression and maximum tumor diameter in the
complete cohort as well as in male patients (this was not observed
in female patients possibly because of small sample size). After
transfection into GT1-1 cells, assessment of cell viability and
invasion suggested that overexpressed C5orf66-AS1 inhibited cell
viability and cell invasion. C5orf66-AS1 acts as a tumor suppressor
in different tumor types (12,27).
The pattern of C5orf66-AS1 expression in pituitary null cell
adenomas and results of in vitro experiments were consistent
with its role as a suppressor of tumor development and invasion. A
significant positive linear correlation was observed between
C5orf66-AS1 expression and age in patients with non-invasive
pituitary null cell adenomas. This finding might be of interest;
however, further studies must be conducted to validate this finding
because of the small sample size used in the present study.
Next, we performed in silico prediction of
target genes of C5orf66-AS1. We found that two cis-acting
target genes, including PITX1 (28–32),
which is important for pituitary development and tumorigenesis,
were located in a distance of <5 kb from C5orf66-AS1. In
addition, we determined 347 trans-acting target genes by
setting RNAplex parameter as -e −70. Among the top 20
trans-acting target genes with the most stable interaction
(Table III), several genes such
as RASSF1, SCGB3A1, and AEBP1 were found to be
associated with transcription regulation or tumor development
(33–38). Especially RASSF1 was
indicated to play a role in pituitary tumorigenesis and suppression
of tumor progression (33,39). Next, we explored the 29 target genes
in microarray data obtained from another study cohort. By using a
stringency of P≤0.01, we found that three of the nine predicted
cis-acting target genes and eight of the 20 predicted
trans-acting target genes, including PITX1, RASSF1,
SCGB3A1, and AEBP1, were significantly differentially
expressed between tumor and normal pituitary tissues.
Notably, a considerable proportion of
trans-targeting genes (8 of 20) were differentially
expressed in pituitary tumor tissues compared to normal pituitary
tissues; this implicates the role of a trans-acting
mechanism in the effect exerted by C5orf66-AS1. Furthermore, by
using a stringency of P≤0.05, we found that SCGB3A1 was
differentially expressed between invasive and non-invasive
pituitary null cell adenomas, which was then confirmed by qRT-PCR.
Since SCGB3A1 was the only gene that was differentially
expressed in both comparisons, which was consistent with the
expression pattern of C5orf66-AS1, it may be the most probable
target gene of C5orf66-AS1. Next, we performed co-expression
analysis of C5orf66-AS1 in the RNA-seq data. We found that the most
correlated gene with C5orf66-AS1 was PAQR7 (r=0.938), a
membrane progesterone receptor that may mediate a reduction in GnRH
in the progesterone negative feedback action in a PR
(A/B)-independent way (40). This
is interesting because pituitary is pivotal in the
hypothalamic-pituitary-gonadal axis for progesterone feedback
action, and several researchers have presumed that pituitary null
cell adenomas originate from gonadotropic cells (3,24). In
the top 20 trans-acting genes we predicted in silico,
there were eight genes with r>0.70, and seven genes out of those
are differentially expressed in tumor tissues and normal pituitary
tissues, including RASSF1 and SCGB3A1.
As showed in Fig. 5,
C5orf66-AS1 is located within C5orf66 and is <5 kb
upstream of PITX1. It is transcribed in the antisense
direction of C5orf66 and in the same direction as
PITX1. According to the definitions provided by Luo et
al (7), for PITX1,
C5orf66-AS1 belongs to the biotype of sense lncRNAs, which are
transcribed in the same direction as the nearby protein-coding gene
and are included in the class of genic lncRNAs. Luo et al
found that lncRNAs showed non-random genomic distribution within a
5-kb distance from neighboring coding gene, which defines genic
lncRNAs. The authors provided multiple solid proofs to identify the
genuine cis-regulation of some divergent genic lncRNAs,
including binding to and acting on chromatin and interacting with
mediators. Their results suggest that the sense biotype of genic
lncRNAs may regulate neighboring protein-coding genes in cis
as well as by adjusting transcription or through another yet
unknown mechanism. This function was partly validated by Wei et
al (12) who determined the
suppressive role of C5orf66-AS1 in esophageal squamous cell
carcinoma. They determined high coexpression of three genes,
namely, C5orf66-AS1, neighboring PITX1, and RASAL1,
which is a major target of PITX1 (29). Knockdown of C5orf66-AS1
significantly altered the expression of two downstream targets of
PITX1, namely RASAL1 and TERT. Since PITX1 is
a pan-pituitary and pituitary-specific regulator of transcription
involved in the early development and differentiation of the
pituitary gland (28) and because
it suppresses the tumorigenicity in multiple tumors (29) especially prolactinomas (32), we hypothesized that C5orf66-AS1
exerted a tumor suppressor effect partly through PITX1.
However, further studies are needed to confirm this hypothesis.
Although NORAD and TINCR showed non-normal
distribution most probably because of the small sample size, we
obtained a few interesting results after analyzing the data
further. Stratification of tumors according to their invasive
potential showed that NORAD expression level was significantly
negatively correlated with maximum tumor diameter in patients with
non-invasive pituitary null cell adenomas. Stratification of tumors
according to sex showed that NORAD expression level was
significantly negatively correlated with maximum tumor diameter and
was significantly positively correlated with patient age in male
patients. Although these results should be validated by performing
further studies involving a large sample size, the negative
correlation between NORAD expression level and tumor size in the
male and non-invasive subgroups suggests a tumor suppressive role
of NORAD in pituitary null cell adenomas.
In summary, we found that C5orf66-AS1 expression was
lower in null cell adenomas than in normal pituitary tissues and in
invasive adenomas than in non-invasive adenomas. These results
along with the negative correlation between C5orf66-AS1 expression
and maximum tumor diameter and the results of in vitro
experiments on pituitary adenoma cells indicate that C5orf66-AS1
suppresses the development and invasion of pituitary null cell
adenomas. However, sufficient statistical evidence is not available
to support the roles of NORAD and TINCR in the development and
invasion of pituitary null cell adenomas, although our results
suggest that NORAD may also play a tumor suppressive role. To our
knowledge, the present study is the first to investigate the roles
of these three lncRNAs in pituitary adenomas. However, further
studies are needed to validate the findings of the present study
and to determine detailed mechanisms underlying the functions of
these lncRNAs.
Acknowledgements
We used some data from the GTEx Project, which was
supported by the Common Fund of the Office of the Director of the
National Institutes of Health, NCI, NHGRI, NHLBI, NIDA, NIMH, and
NINDS. This study was supported by the Research Special Fund for
Public Welfare Industry of Health (201402008); the National High
Technology Research and Development Program of China (863 Program,
2015AA020504). An English Language Service from Elsevier's WebShop
was used for language editing.
Glossary
Abbreviations
Abbreviations:
|
Gd
|
gadolinium
|
|
GTEx
|
Genotype-Tissue Expression
|
|
lincRNAs
|
long intergenic non-coding RNAs
|
|
lncRNAs
|
long non-coding RNAs
|
|
MRI
|
magnetic resonance imaging
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reaction
|
|
RNA-seq
|
RNA sequencing
|
References
|
1
|
Aflorei ED and Korbonits M: Epidemiology
and etiopathogenesis of pituitary adenomas. J Neurooncol.
117:379–394. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kovacs K, Horvath E, Ryan N and Ezrin C:
Null cell adenoma of the human pituitary. Virchows Arch A Pathol
Anat Histol. 387:165–174. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saeger W, Lüdecke DK, Buchfelder M,
Fahlbusch R, Quabbe HJ and Petersenn S: Pathohistological
classification of pituitary tumors: 10 years of experience with the
German Pituitary Tumor Registry. Eur J Endocrinol. 156:203–216.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng GXY, Do BT, Webster DE, Khavari PA
and Chang HY: Dicer-microRNA-Myc circuit promotes transcription of
hundreds of long noncoding RNAs. Nat Struct Mol Biol. 21:585–590.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X,
Wu B, Xu R, Liu W, Yan P, et al: Divergent lncRNAs regulate gene
expression and lineage differentiation in pluripotent cells. Cell
Stem Cell. 18:637–652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li L, Liu B, Wapinski OL, Tsai MC, Qu K,
Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, et al: Targeted
disruption of Hotair leads to homeotic transformation and gene
derepression. Cell Rep. 5:3–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chunharojrith P, Nakayama Y, Jiang X, Kery
RE, Ma J, De La Hoz Ulloa CS, Zhang X, Zhou Y and Klibanski A:
Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived
cell line. Mol Cell Endocrinol. 416:27–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Z, Li C, Liu C, Yu S and Zhang Y:
Expression of the long non-coding RNAs MEG3, HOTAIR, and MALAT-1 in
non-functioning pituitary adenomas and their relationship to tumor
behavior. Pituitary. 18:42–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang ZY, Lu YX, Zhang ZY, Chang YY, Zheng
L, Yuan L, Zhang F, Hu YH, Zhang WJ and Li XN: Loss of TINCR
expression promotes proliferation, metastasis through activating
EpCAM cleavage in colorectal cancer. Oncotarget. 7:22639–22649.
2016.PubMed/NCBI
|
|
12
|
Wei G, Luo H, Sun Y, Li J, Tian L, Liu W,
Liu L, Luo J, He J and Chen R: Transcriptome profiling of
esophageal squamous cell carcinoma reveals a long noncoding RNA
acting as a tumor suppressor. Oncotarget. 6:17065–17080. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee S, Kopp F, Chang T-C, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu H, Li J, Koirala P, Ding X, Chen B,
Wang Y, Wang Z, Wang C, Zhang X and Mo YY: Long non-coding RNAs as
prognostic markers in human breast cancer. Oncotarget.
7:20584–20596. 2016.PubMed/NCBI
|
|
15
|
Lloyd R, Kovacs K, Young WJ, et al: Tumour
of the pituitary gland. World Health Organization Classification of
Tumours: Pathology and Genetics of Tumours of Endocrine Organs.
Delellis RA, Lloyd RV, Heitz PU and Eng C: Lyon: International
Agency for Research and Cancer (IARC) Press; pp. 9–48. 2004
|
|
16
|
Wilson CB: A decade of pituitary
microsurgery. The Herbert Olivecrona lecture. J Neurosurg.
61:814–833. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Knosp E, Steiner E, Kitz K and Matula C:
Pituitary adenomas with invasion of the cavernous sinus space: A
magnetic resonance imaging classification compared with surgical
findings. Neurosurgery. 33:610–617; discussion 617–618. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Micko ASG, Wöhrer A, Wolfsberger S and
Knosp E: Invasion of the cavernous sinus space in pituitary
adenomas: Endoscopic verification and its correlation with an
MRI-based classification. J Neurosurg. 122:803–811. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin XC, Zhu Y, Chen WB, Lin LW, Chen DH,
Huang JR, Pan K, Lin Y, Wu BT, Dai Y, et al: Integrated analysis of
long non-coding RNAs and mRNA expression profiles reveals the
potential role of lncRNAs in gastric cancer pathogenesis. Int J
Oncol. 45:619–628. 2014.PubMed/NCBI
|
|
21
|
Jia H, Osak M, Bogu GK, Stanton LW,
Johnson R and Lipovich L: Genome-wide computational identification
and manual annotation of human long noncoding RNA genes. RNA.
16:1478–1487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tafer H and Hofacker IL: RNAplex: A fast
tool for RNA-RNA interaction search. Bioinformatics. 24:2657–2663.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tafer H, Amman F, Eggenhofer F, Stadler PF
and Hofacker IL: Fast accessibility-based prediction of RNA-RNA
interactions. Bioinformatics. 27:1934–1940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kontogeorgos G and Thodou E: The
gonadotroph origin of null cell adenomas. Hormones (Athens).
15:243–247. 2016.PubMed/NCBI
|
|
25
|
Ishii Y, Suzuki M, Takekoshi S, Egashira
N, Yamazaki M, Miyai S, Sanno N, Teramoto A and Osamura RY:
Immunonegative ‘null cell’ adenomas and gonadotropin (Gn) subunit
(SUs) immunopositive adenomas share frequent expression of multiple
transcription factors. Endocr Pathol. 17:35–43. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Balogun JA, Monsalves E, Juraschka K,
Parvez K, Kucharczyk W, Mete O, Gentili F and Zadeh G: Null cell
adenomas of the pituitary gland: An institutional review of their
clinical imaging and behavioral characteristics. Endocr Pathol.
26:63–70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu YP, Bian XJ, Ye DW, Yao XD, Zhang SL,
Dai B, Zhang HL and Shen YJ: Long noncoding RNA expression
signatures of bladder cancer revealed by microarray. Oncol Lett.
7:1197–1202. 2014.PubMed/NCBI
|
|
28
|
Tremblay JJ, Lanctôt C and Drouin J: The
pan-pituitary activator of transcription, Ptx1 (pituitary homeobox
1), acts in synergy with SF-1 and Pit1 and is an upstream regulator
of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 12:428–441.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kolfschoten IGM, van Leeuwen B, Berns K,
Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM and Agami
R: A genetic screen identifies PITX1 as a suppressor of RAS
activity and tumorigenicity. Cell. 121:849–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lamolet B, Pulichino AM, Lamonerie T,
Gauthier Y, Brue T, Enjalbert A and Drouin J: A pituitary
cell-restricted T box factor, Tpit, activates POMC transcription in
cooperation with Pitx homeoproteins. Cell. 104:849–859. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pellegrini-Bouiller I, Manrique C, Gunz G,
Grino M, Zamora AJ, Figarella-Branger D, Grisoli F, Jaquet P and
Enjalbert A: Expression of the members of the Ptx family of
transcription factors in human pituitary adenomas. J Clin
Endocrinol Metab. 84:2212–2220. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wierinckx A, Auger C, Devauchelle P,
Reynaud A, Chevallier P, Jan M, Perrin G, Fèvre-Montange M, Rey C,
Figarella-Branger D, et al: A diagnostic marker set for invasion,
proliferation, and aggressiveness of prolactin pituitary tumors.
Endocr Relat Cancer. 14:887–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qian ZR, Sano T, Yoshimoto K, Yamada S,
Ishizuka A, Mizusawa N, Horiguchi H, Hirokawa M and Asa SL:
Inactivation of RASSF1A tumor suppressor gene by aberrant promoter
hypermethylation in human pituitary adenomas. Lab Invest.
85:464–473. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pefani D-E, Latusek R, Pires I, Grawenda
AM, Yee KS, Hamilton G, van der Weyden L, Esashi F, Hammond EM and
O'Neill E: RASSF1A-LATS1 signalling stabilizes replication forks by
restricting CDK2-mediated phosphorylation of BRCA2. Nat Cell Biol.
16:962–971, 1–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang KH, Huang SF, Chen I-H, Liao CT,
Wang HM and Hsieh LL: Methylation of RASSF1A, RASSF2A, and HIN-1 is
associated with poor outcome after radiotherapy, but not surgery,
in oral squamous cell carcinoma. Clin Cancer Res. 15:4174–4180.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Krop I, Parker MT, Bloushtain-Qimron N,
Porter D, Gelman R, Sasaki H, Maurer M, Terry MB, Parsons R and
Polyak K: HIN-1, an inhibitor of cell growth, invasion, and AKT
activation. Cancer Res. 65:9659–9669. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shigematsu H, Suzuki M, Takahashi T,
Miyajima K, Toyooka S, Shivapurkar N, Tomlinson GE, Mastrangelo D,
Pass HI, Brambilla E, et al: Aberrant methylation of HIN-1 (high in
normal-1) is a frequent event in many human malignancies. Int J
Cancer. 113:600–604. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ladha J, Sinha S, Bhat V, Donakonda S and
Rao SM: Identification of genomic targets of transcription factor
AEBP1 and its role in survival of glioma cells. Mol Cancer Res.
10:1039–1051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Y, Zhang X and Klibanski A: Genetic
and epigenetic mutations of tumor suppressive genes in sporadic
pituitary adenoma. Mol Cell Endocrinol. 386:16–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sleiter N, Pang Y, Park C, Horton TH, Dong
J, Thomas P and Levine JE: Progesterone receptor A (PRA) and
PRB-independent effects of progesterone on gonadotropin-releasing
hormone release. Endocrinology. 150:3833–3844. 2009. View Article : Google Scholar : PubMed/NCBI
|