Introduction
Gastric cancer (GC), with nearly 1 million new cases
being diagnosed every year, is the second most common cause of
cancer-related deaths (1). Although
advancement in early diagnosis and therapeutic strategy has been
achieved, the prognosis of GC patients remains poor (2,3). The
poor prognosis of GC patients is partly due to the fact that the
molecular mechanisms underlying growth and metastasis of gastric
cancer remain unknown.
SIRT6, belong to a conserved family of NAD
positivity-dependent enzymes, playing an important role in
maintaining cellular and organismal homeostasis (4,5). It
was found to be involved in regulating mitochondrial oxidative
metabolism and cellular glucose uptake (6,7). It
was also found to suppress obesity (8), cardiac hypertrophy (9), inflammation (10) and cellular senescence (11). In human cancers, SIRT6 was found to
play different roles in different cancer types. SIRT6 has been
reported to have both tumor suppressor and oncogenic properties.
Reduced expression of SIRT6 has been found in colon cancer
(12), hepatocellular carcinoma
(13) and head and neck squamous
cell carcinoma (14). Its decreased
expression was correlated with advanced cancer stage and grade, and
with poorer survival of cancer patients. In contrast, high SIRT6
levels were demonstrated in breast cancer (15) and prostate cancer (16), and were associated with the drug
resistance and poor prognosis of cancer patients. However, the
expression and biological function of SIRT6 in GC remains
unknown.
In this study, we found that SIRT6 was significantly
decreased in GC tissues and cell lines. Decreased expression of
SIRT6 in GC patients was associated with poor clinicopathological
features and worse prognosis. Functionally, SIRT6 inhibited cell
viability, proliferation, and cell cycle progression while promoted
apoptosis of GC cells. In vivo experiments showed that SIRT6
inhibited the growth of SGC7901 cells in nude mice. Moreover, we
found that p-STAT3 expression was negatively correlated with SIRT6
expression in GC tissues. SIRT6 inhibited the JAK2/STAT3 pathway in
GC cells.
Materials and methods
Cell cultures
Gastric cancer cell lines SGC-7901, MKN-45, BGC-823,
and normal gastric epithelial GES-1 cells were obtained from ATCC
(Rockville, MD, USA) and the Cell Bank of Chinese Academy of
Sciences (Shanghai, China). These cells were cultured in RPMI-1640
medium (Life Technologies, Inc., Gaithersburg, MD, USA) along with
10% fetal bovine serum (Gibco Co., New York, NY, NY, USA),
penicillin (100 U/ml), and streptomycin (100 mg/ml). All cell
cultures were kept at 37°C in a humidified incubator with 5%
CO2.
Clinical tissues
GC tissues (68 pairs) and non-tumor tissues were
collected from Hangzhou Cancer Hospital, and were stored at liquid
nitrogen before using them for further experiments. Informed
consents were obtained from all enrolled patients in this study.
Demographic and clinicopathological information of the included
patients are presented in Table I.
Approval for carrying out the experiments in human tissue samples
were obtained from the Institutional Research Ethics Committee of
Hangzhou Cancer Hospital.
 | Table I.The correlation between SIRT6
expression and clinicopathological features in gastric cancer. |
Table I.
The correlation between SIRT6
expression and clinicopathological features in gastric cancer.
|
|
| SIRT6 expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Total | Low (52) | High (16) | P-value |
|---|
| Age (years) |
|
|
|
|
|
<65 | 30 | 21 | 9 | 0.264 |
| ≥65 | 38 | 31 | 7 |
|
| Sex |
|
|
|
|
| Male | 52 | 42 | 10 | 0.132 |
|
Female | 16 | 10 | 6 |
|
| Tumor
differentiation |
|
|
|
|
| I,
II | 31 | 19 | 12 | 0.007a |
| III,
IV | 37 | 33 | 4 |
|
| Size (cm) |
|
|
|
|
|
<5 | 30 | 19 | 11 | 0.023a |
| ≥5 | 38 | 33 | 5 |
|
| Invasive depth |
|
|
|
|
| Mucosa to
muscularis propria | 10 | 5 | 5 | 0.083 |
|
Adventitia to adjacent
structure | 58 | 47 | 11 |
|
| Lymph node
metastasis |
|
|
|
|
| ≤2
regions | 31 | 23 | 8 | 0.685 |
| >2
regions | 37 | 29 | 8 |
|
| Distant
metastasis |
|
|
|
|
| No | 54 | 44 | 10 | 0.119 |
| Yes | 14 | 8 | 6 |
|
| Venous
infiltration |
|
|
|
|
|
Absent | 50 | 39 | 11 | 0.864 |
|
Present | 18 | 13 | 5 |
|
| TNM stage |
|
|
|
|
| I,
II | 29 | 18 | 11 | 0.016a |
| III,
IV | 39 | 34 | 5 |
|
Cell transfection
Lentivirus encoding SIRT6 overexpressing vector and
control vector were obtained from Genechem (Shanghai, China).
Before virus transfection, GC cells were plated in 6-well plates,
and the viruses diluted in complete medium (2 ml) with polybrene (8
µg/ml) were added in the culture plates. Three days later, these
cells were collected and western blotting was used to confirm the
overexpression effect of the lentivirus.
Western blotting
Cellular proteins from GC cells were extracted with
the RIPA lysis buffer and were quantified with a BCA protein assay
kit (Pierce, Rockford, IL, USA), and 30 µg cellular proteins were
separated on 10% SDS-PAGE and transferred to NC membrane. Primary
antibodies were incubated with the membranes overnight at 4°C. The
following antibodies were used in this study SIRT6 (1:1,000, Cell
Signaling Technologies, Danvers, MA, USA), p-JAK2 (1:1,000, Cell
Signaling Technologies), JAK (1:1,000, Cell Signaling
Technologies), p-STAT3 (1:1000, Cell Signaling Technologies), STAT3
(1:1,000, Cell Signaling Technologies), cyclin D1 (1:1,500, Cell
Signaling Technologies), Bcl-2 (1:500, Cell Signaling Technologies)
and GAPDH (1:2,000, Santa Cruz, CA, USA). The membranes were
incubated with secondary antibodies (1:3,000, Santa Cruz) at room
temperature for 1 h. The protein signals were visualized with ECL
reagents (Amersham Biosciences Corp., USA). Western blots were
semi-quantified by ImageJ software (1.46; National Institutes of
Health, Bethesda, MD, USA).
Cell viability and proliferation
assay
MTT assay was used for measuring cell viability. In
brief, 5,000 GC cells were plated into 96-well plates. These cells
were stained with MTT (Sigma, St. Louis, MO, USA) for 4 h at 37°C
at corresponding time points (0, 24, 48 and 72 h), and the
absorbance at 490 nm was measured to reflect the cell viability.
For cell proliferation, colony formation assay was performed. GC
cells transfected with control vector or SIRT6 overexpressing
vector were seeded in 6-well plates and maintained in cell
incubators for 10 days. The formed cell colonies were stained with
crystal violet solution. The number of cell colonies was counted to
represent the cell proliferation ability of GC cells.
IHC staining
GC tissues and the adjacent non-tumor tissues were
subjected to formalin fixation and were embedded with paraffin, 5
μm thick tissue sections were used for IHC staining following the
standard IHC protocol. Staining intensity was scored as no 0,
staining; 1, weak staining; 2, moderate staining; and 3, strong
staining. Staining quantity was graded as 1, <25%; 2, 25–75%;
and 3, >75%. IHC score was manually confirmed by two independent
experienced pathologists using the formula: IHC score = staining
intensity × staining quantity.
Cell cycle and apoptosis analysis
For cell cycle assay, GC cells were collected 72 h
after virus transfection. These cells were fixed with 80% ethanol
overnight, and then were stained with propidium iodide (50 µg/ml,
BD Biosciences, Franklin Lakes, NJ, USA) at room temperature for 30
min. The percentage of cells in each cell cycle was measured with
FACSCalibur system (BD Biosciences). For apoptosis assay, GC cells
after transfection were subjected to an apoptosis assay. The
percentage of apoptotic GC cells were measured using Annexin
V/propidium iodide kit (BD Pharmingen, San Diego, CA, USA)
according to the manufacturer's instructions.
In vivo tumor growth assay
For tumor growth studies, nude mice were injected
subcutaneously with 1×106 SGC7901 cells transfected with
control vector or SIRT6 vector. Tumor sizes were measured every 3
days after subcutaneous injection. Three weeks later, the mice were
sacrificed by cervical dislocation under anesthesia, and tumors
were removed for the volume measurement. The protocols for animal
experiments were approved by the Animal Care Committee of Hangzhou
Cancer Hospital.
Statistical analysis
All quantitative data in this study are presented as
mean ± standard error of the mean (SEM). Statistical analysis
including Student's t-test, ANOVA analysis, Chi-square test,
Correlation analysis, and Kaplan-Meier analysis was performed with
Graphpad software. P<0.05 was considered as statistically
significant.
Results
SIRT6 expression is decreased in GC
tissues and cells
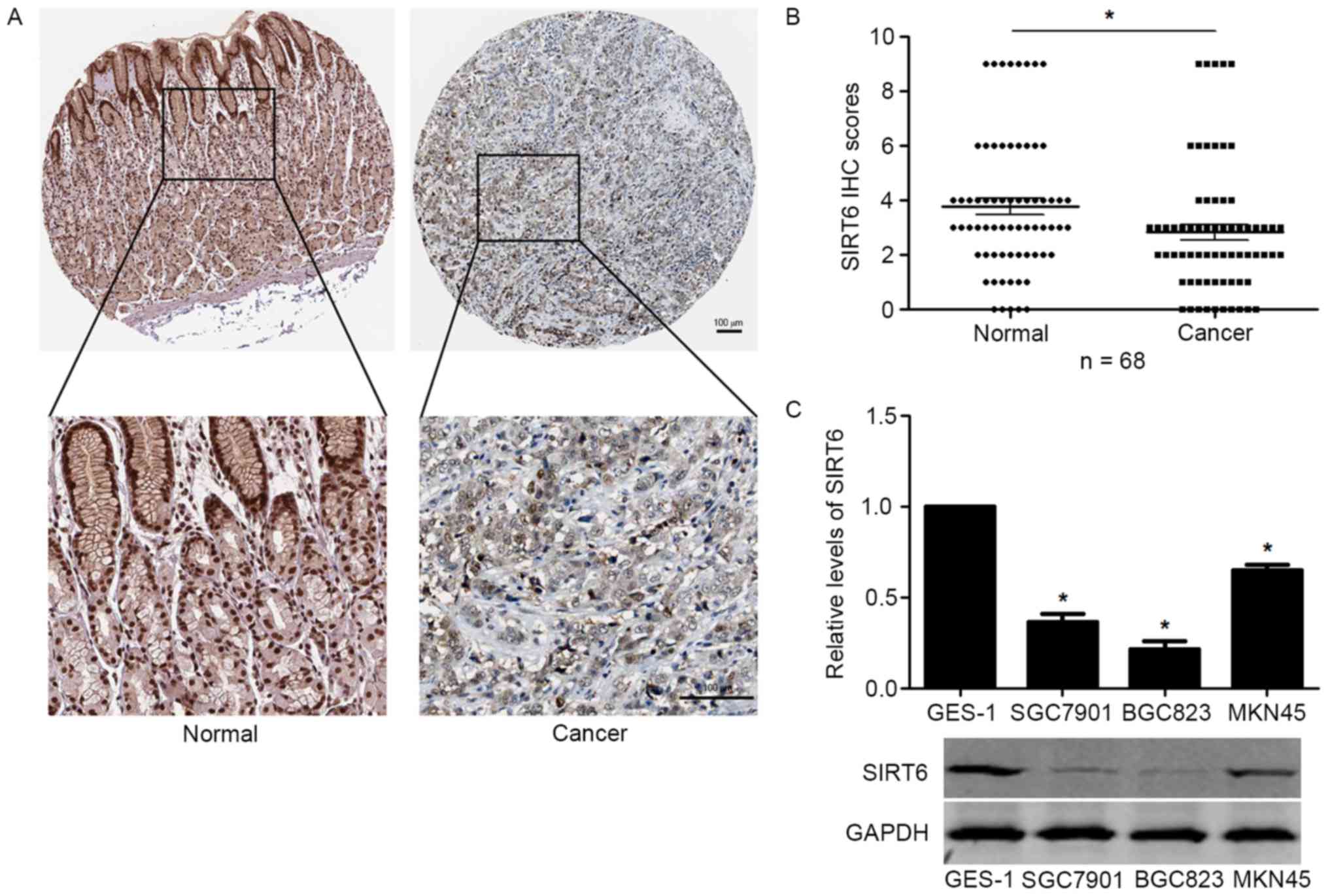
To investigate the expression level of SIRT6 in GC,
we performed IHC staining of SIRT6 in GC tissues. IHC staining
showed that SIRT6 expression was significantly lower in GC tissues
compared with that in adjacent non-tumor tissues (Fig. 1A) compared with that in adjacent
non-tumor tissues. IHC scoring for SIRT6 in GC confirmed that the
expression level of SIRT6 in GC tissues was significantly increased
(P<0.05, Fig. 1B). Moreover, we
investigated the expression level of SIRT6 in GC cells lines and
GES-1 cells. Compared with that in GES-1 cells, the protein level
of SIRT6 was significantly decreased in GC cell lines including
SGC7901, BGC823 and MKN45 cells (P<0.05, Fig. 1C).
Decreased SIRT6 expression in GC
tissues is associated with poor clinicopathological features and
prognosis of GC patients
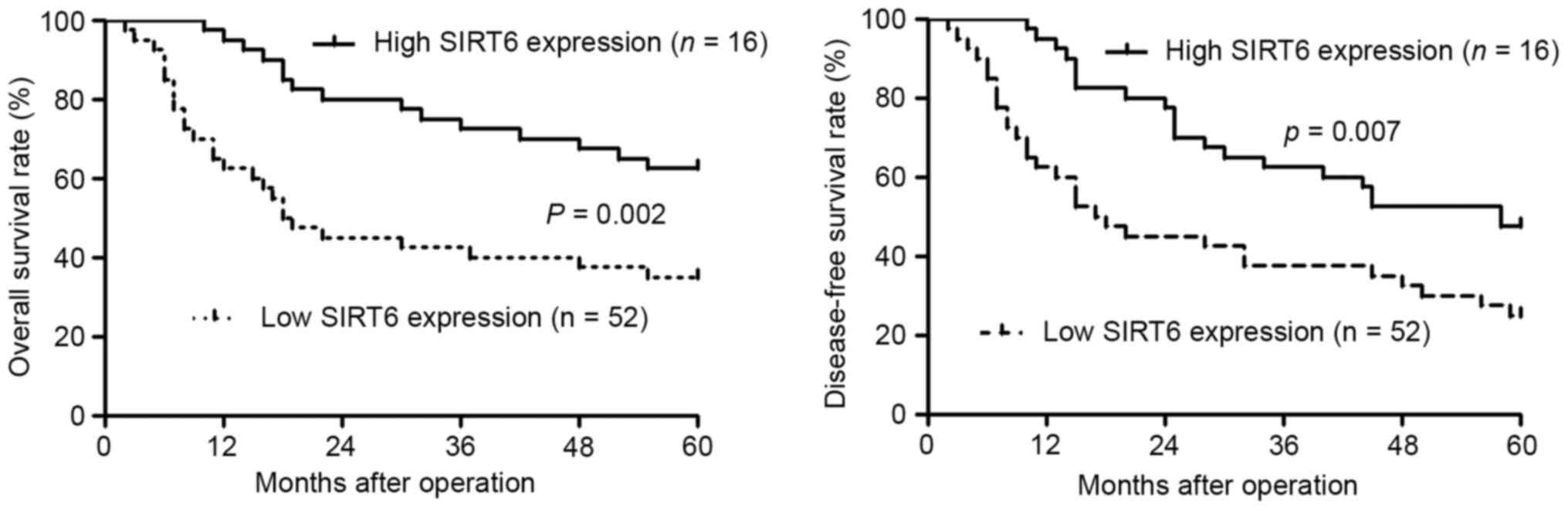
After confirming the decreased expression of SIRT6
expression in GC, we further determined whether decreased SIRT6
expression level was correlated with the clinical features and the
prognosis of GC patients. GC patients were divided into two groups
according to the IHC scores: SIRT6 low expression group (IHC score
<4) and SIRT6 high expression group (IHC score ≥4). Clinical
association analysis showed that patients with lower SIRT6 level
had lower grade of tumor differentiation, large tumor size and
advanced TNM stage (P<0.05, respectively). More importantly,
decreased level of SIRT6 was associated with decreased rate of
overall survival (P=0.002, Fig. 2)
and disease-free survival (P=0.007, Fig. 2).
SIRT6 inhibits cell viability and
proliferation of GC cells
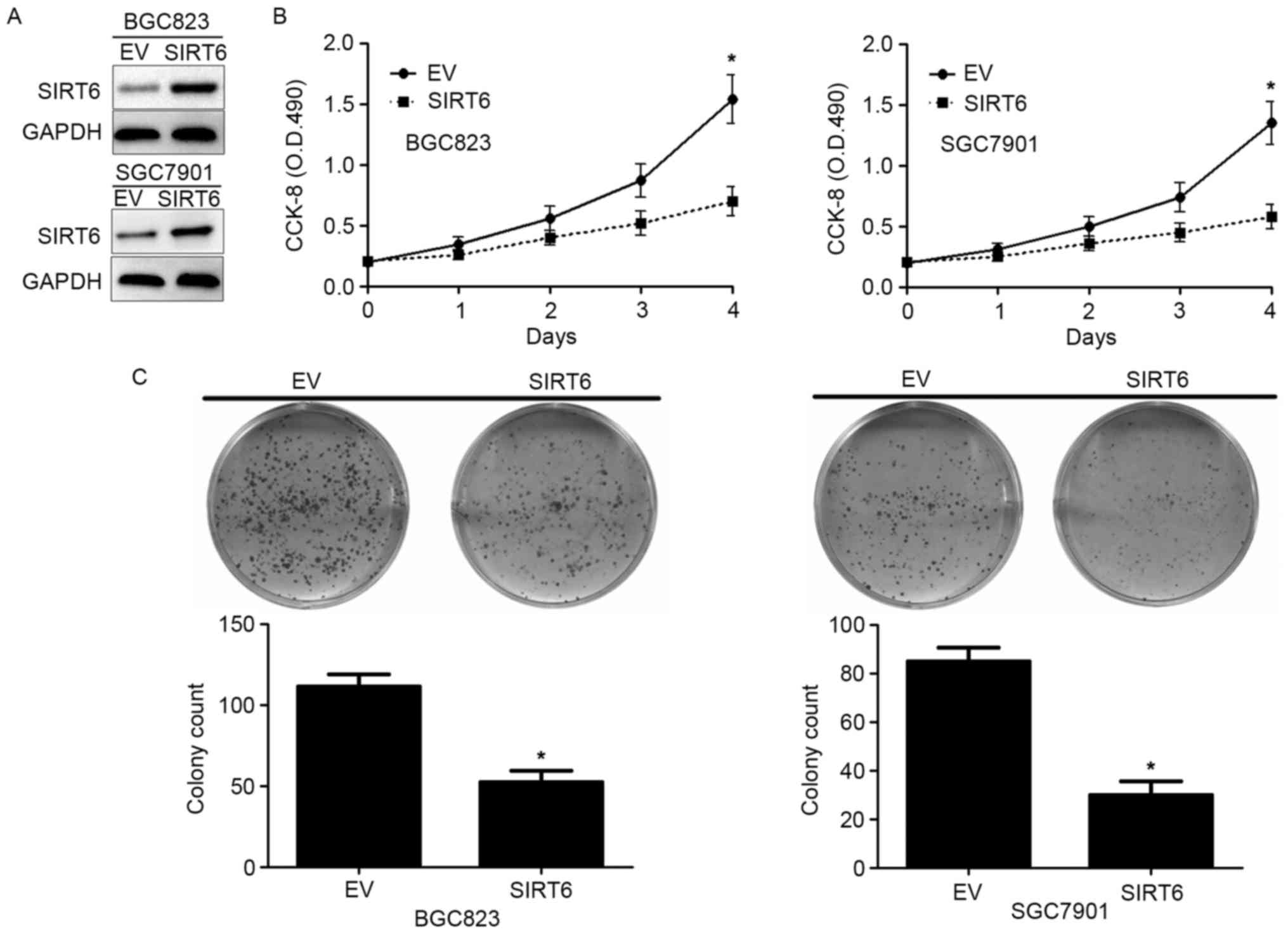
Then, we investigated the biological function of
SIRT6 in GC cells. Lentivirus encoding SIRT6 overexpressing vector
significantly increased SIRT6 expression in BGC823 and SGC7901
cells as suggested by western blotting (Fig. 3A). MMT assay showed that forced
expression of SIRT6 inhibited the cell viability of BGC823 and
SGC7901 cells (P<0.05, Fig. 3B).
Moreover, colony formation assay showed that overexpressing SIRT6
significantly inhibited the colony formation of BGC823 and SGC7901
cells (P<0.05, Fig. 3C). These
data indicate SIRT6 inhibited the cell viability and proliferation
of GC cells.
SIRT6 inhibits cell cycle progression
and increases apoptosis of GC cells
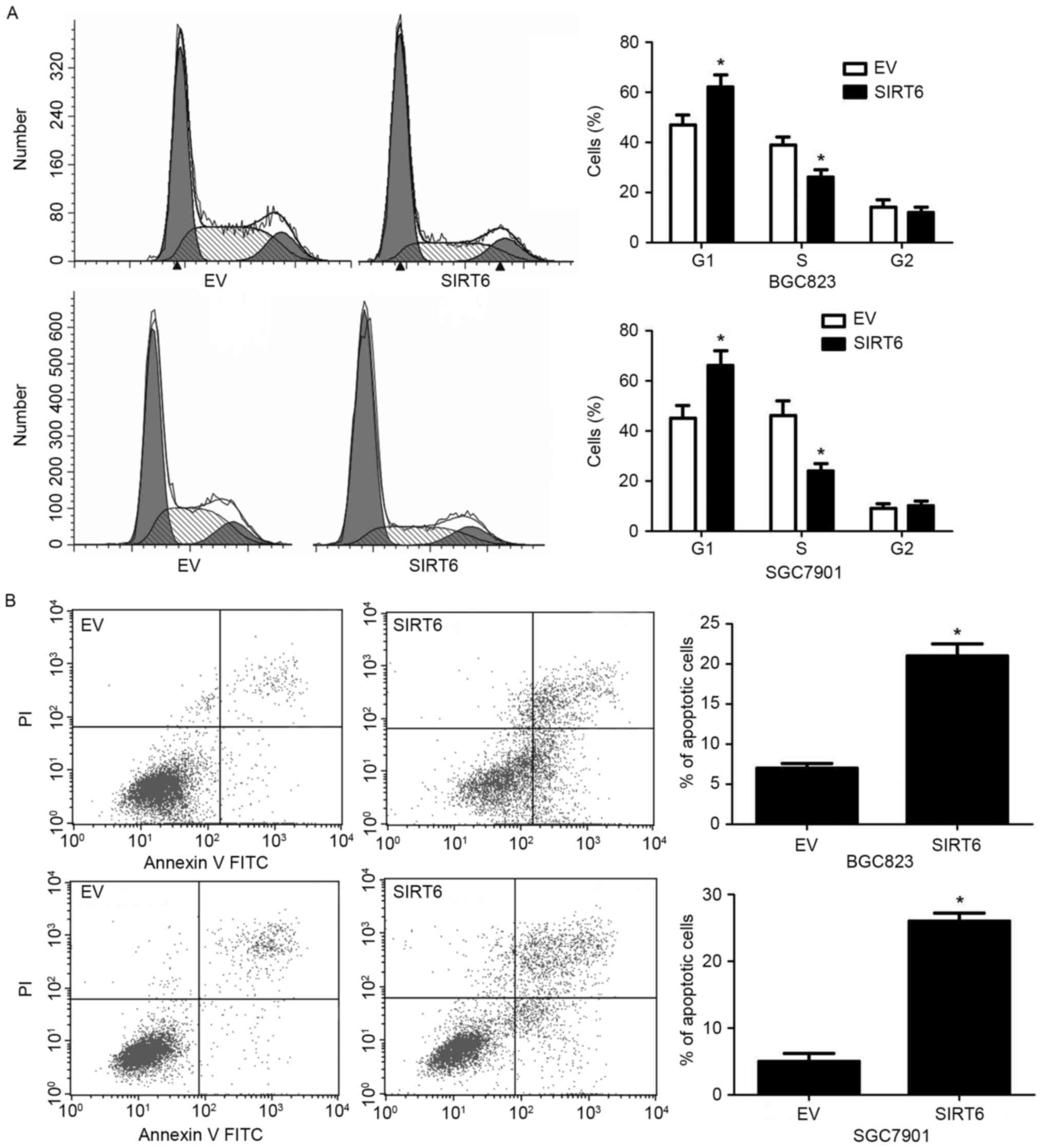
Then, we investigated whether SIRT6 could influence
cell cycle progression and apoptosis of GC cells. SIRT6
overexpression in both BGC823 and SGC7901 cells increased the
percentage of GC cells in G1 stage while decreased the percentage
of GC cells in S phase (P<0.05, Fig.
4A). On the other hand, overexpression of SIRT6 increased the
percentage of apoptotic BGC823 and SGC7901 cells (P<0.05,
Fig. 4B).
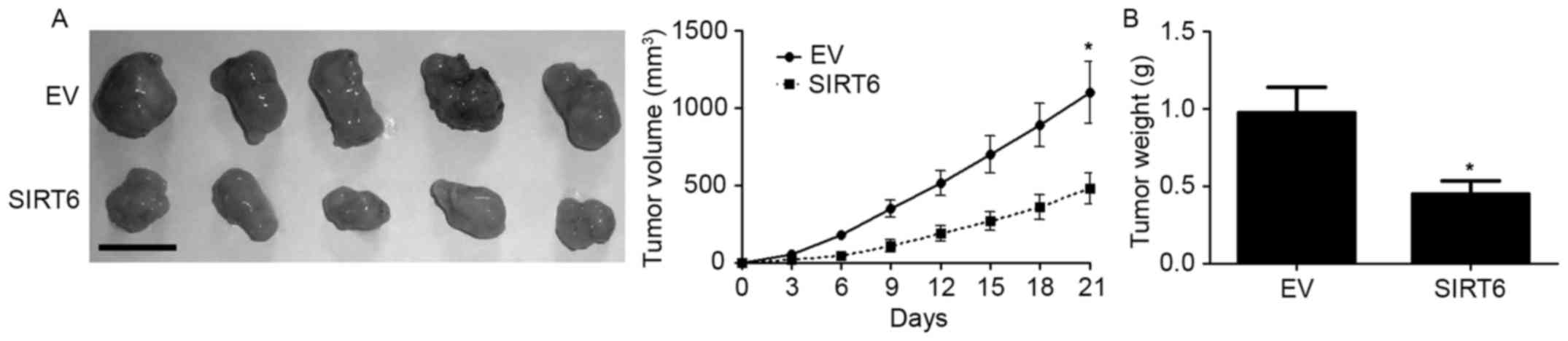
SIRT6 inhibits the growth of SGC7901
cells in nude mice
To further investigate whether SIRT6 affects the
in vivo growth of GC cells, we carried out subcutaneous
injection experiment with SGC7901. The result of subcutaneous tumor
formation showed that the in vivo growth of SGC7901 cells
was significantly inhibited after overexpression of SURT6 (Fig.5, P<0.05). These data suggest that
SIRT6 inhibited the growth of GC cells in vivo.
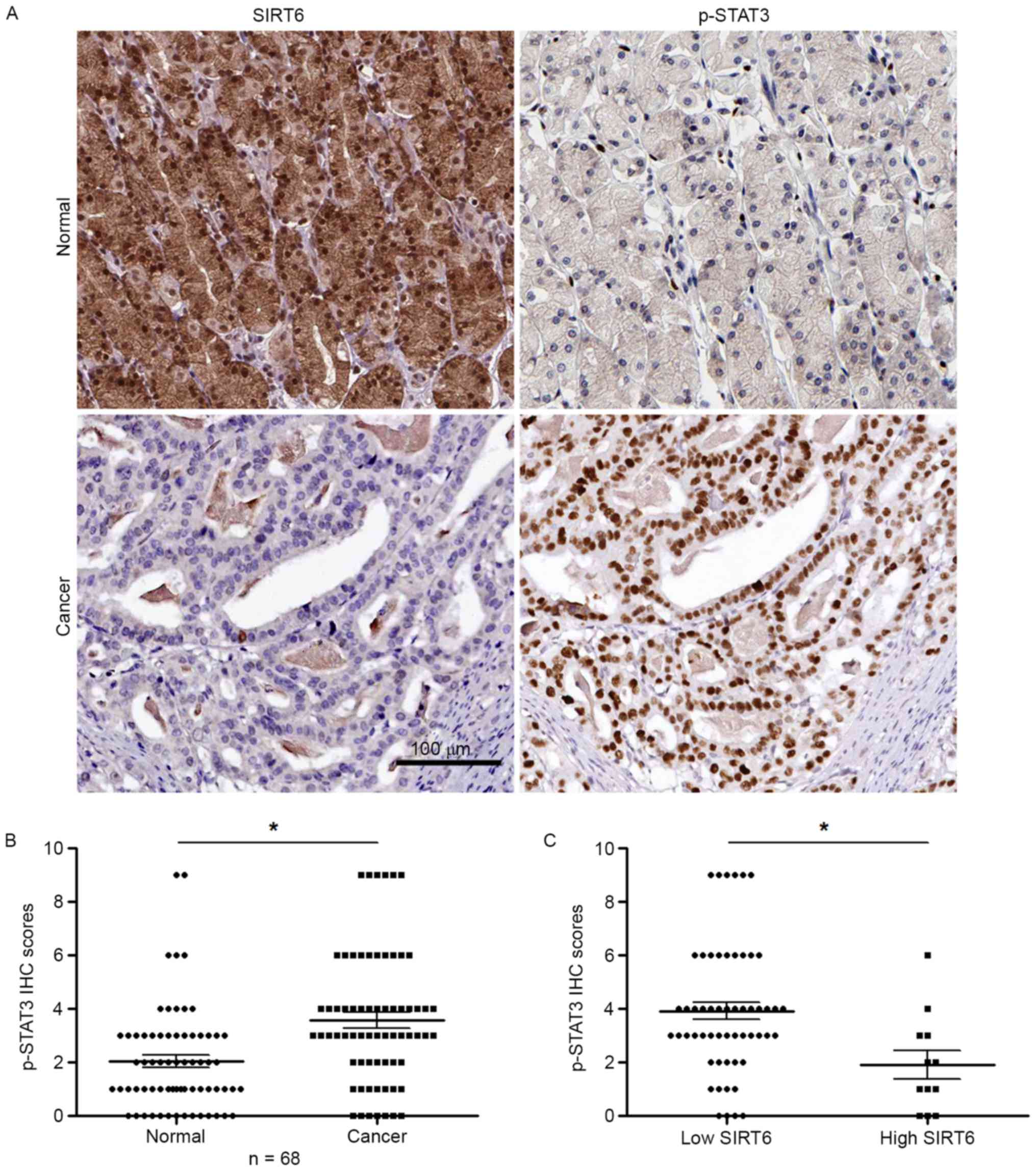
SIRT6 inhibits JAK2/STAT3 signaling
pathway in GC cells
Since JAK2/STAT3 signaling pathway has been
confirmed to be a critical pathway in regulating the growth of GC
cells, we investigated whether SIRT6 could inhibit JAK2/STAT3
pathway in GC cells. To answer this question, we used IHC staining
to determine whether the expression of p-STAT3, which was marker of
JAK2/STAT3 pathway, was negatively correlated with SIRT6 expression
in GC tissues. Serial sectioning of GC tissues showed that in
adjacent non-tumor tissue with high SIRT6 expression, the
expression of p-STAT3 was low (Fig.
6A, upper part). In tumor tissues with low SIRT6 expression,
the expression of p-STAT3 was high (Fig. 6A, lower part). The result of p-STAT3
IHC score showed that the expression of p-STAT3 was significantly
increased in GC tissues (P<0.05, Fig. 6B). Importantly, in tissues with low
SIRT6 level, the expression of p-STAT3 was significantly higher
than that in tissues with high SIRT6 level (P<0.05, Fig. 6C). To further confirm SIRT6 could
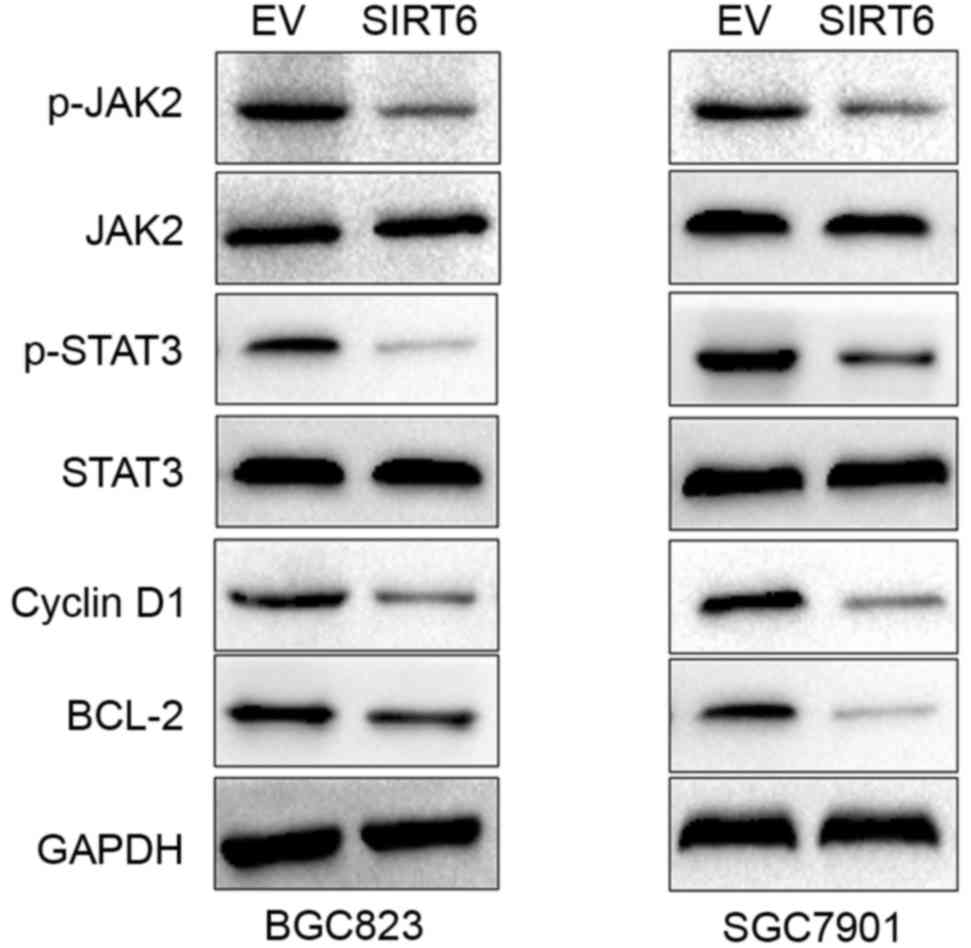
affect the JAK2/STAT3 pathway in GC cells, we used western blotting
to investigate the expression of p-JAK2, JAK2, p-STAT3 and STAT3,
and Bcl-2 and cyclin D1 which are downstream markers of the
activation of JAK2/STAT3 pathway, after SIRT6 overexpression.
Overexpression of SIRT6 in BGC823 and SGC7901 cells significantly
reduced the level of p-JAK2, p-STAT3, cyclin D1 and Bcl-2 (Fig. 7). These data indicate that SIRT6
inhibited JAK2/STAT3 pathway in GC cells.
Discussion
SIRT6 was found to be a versatile protein which was
involved in various biological processes including mitochondrial
oxidative metabolism (6), cellular
glucose uptake (7), inflammation
(10), cardiac hypertrophy
(9), DNA repair (17), and resistance to heat stress
(18). Studies on cancer showed
that SIRT6 could either act as tumor suppressors or as oncogenic
protein in different types of human cancers. However, the
expression and function of SIRT6 in GC were not previously
investigated. In this study, we showed that the expression of SIRT6
was significantly decreased in GC tissues and cell lines.
Association analysis further confirmed that decreased expression of
SIRT6 was associated with unfavorable clinical features and poor
prognosis of GC patients indicating that SIRT6 played tumor
suppressive role in GC.
Previously, SIRT6 expression was also found to be
decreased in colon (12), liver
(13), and head and neck squamous
cell carcinomas (14).
Functionally, SIRT6 was found to play tumor suppressive roles by
inhibiting the cell proliferation, lipogenesis and glycosis and
enhancing apoptosis (19). In this
study, in vitro assays showed that SIRT6 could inhibit cell
viability, proliferation and cell cycle progression while promoted
the apoptosis of GC cells. In vivo experiments further
confirmed that SIRT6 suppressed the in vivo growth of
SGC7901 cells in nude mice. These date indicate SIRT6 played tumor
suppressive role in GC cells by inhibiting tumor growth.
Previous studies showed that SIRT6 could regulate
p53 and p73 apoptosis pathways (20), HIF-1α pathway (21) and insulin-IGF-1-like signaling
pathway (22). JAK2/STAT3 signaling
pathway is a well-characterized pathway which promotes the growth
of GC cells (23). Since we found
SIRT6 inhibits the growth of GC cells, we further investigated
whether SIRT6 could inhibit the growth of GC cells by inhibiting
JAK2/STAT3 pathway. The results of IHC staining in GC tissues and
western blotting in GC cells demonstrated that SIRT6 inhibited
JAK2/STAT2 pathway activation in GC. The above suggest that SIRT6
inhibited the growth of GC cells by inhibiting the JAK2/STAT3
pathway.
In conclusion, we demonstrated that SIRT6 expression
was decreased in GC tissues and cells. Decreased expression level
of SIRT6 was associated with poor clinical features and prognosis
of GC patients. SIRT6 inhibited the cell viability, proliferation
and cell cycle progression and increased apoptosis of GC cells.
In vivo experiments confirmed that SIRT6 suppressed the
growth of SGC7901 cells in nude mice. Mechanically, we demonstrated
that SIRT6 inhibited JAK2/STAT3 pathway activation in GC cells.
Therefore, this study indicates that SIRT6 is a promising biomarker
and therapeutic target for GC patients.
References
|
1
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saragoni L, Morgagni P, Gardini A, Marfisi
C, Vittimberga G, Garcea D and Scarpi E: Early gastric cancer:
Diagnosis, staging, and clinical impact. Evaluation of 530
patients. New elements for an updated definition and
classification. Gastric Cancer. 16:549–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carcas LP: Gastric cancer review. J
Carcinog. 13:142014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gertler AA and Cohen HY: SIRT6, a protein
with many faces. Biogerontology. 14:629–639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan PW, Feldman JL, Devries MK, Dong A,
Edwards AM and Denu JM: Structure and biochemical functions of
SIRT6. J Biol Chem. 286:14575–14587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Houtkooper RH, Pirinen E and Auwerx J:
Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol
Cell Biol. 13:225–238. 2012.PubMed/NCBI
|
|
7
|
Xiao C, Kim H-S, Lahusen T, Wang RH, Xu X,
Gavrilova O, Jou W, Gius D and Deng CX: SIRT6 deficiency results in
severe hypoglycemia by enhancing both basal and insulin-stimulated
glucose uptake in mice. J Biol Chem. 285:36776–36784. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kanfi Y, Peshti V, Gil R, Naiman S, Nahum
L, Levin E, Kronfeld-Schor N and Cohen HY: SIRT6 protects against
pathological damage caused by diet-induced obesity. Aging Cell.
9:162–173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sundaresan NR, Vasudevan P, Zhong L, Kim
G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam
V, et al: The sirtuin SIRT6 blocks IGF-Akt signaling and
development of cardiac hypertrophy by targeting c-Jun. Nat Med.
18:1643–1650. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao C, Wang R-H, Lahusen TJ, Park O,
Bertola A, Maruyama T, Reynolds D, Chen Q, Xu X, Young HA, et al:
Progression of chronic liver inflammation and fibrosis driven by
activation of c-JUN signaling in Sirt6 mutant mice. J Biol Chem.
287:41903–41913. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mao Z, Tian X, Van Meter M, Ke Z,
Gorbunova V and Seluanov A: Sirtuin 6 (SIRT6) rescues the decline
of homologous recombination repair during replicative senescence.
Proc Natl Acad Sci USA. 109:11800–11805. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Z-G and Qin C-Y: Sirt6 suppresses
hepatocellular carcinoma cell growth via inhibiting the
extracellular signal-regulated kinase signaling pathway. Mol Med
Rep. 9:882–888. 2014.PubMed/NCBI
|
|
14
|
Lu C-T, Hsu C-M, Lin P-M, Lai CC, Lin HC,
Yang CH, Hsiao HH, Liu YC, Lin HY, Lin SF, et al: The potential of
SIRT6 and SIRT7 as circulating markers for head and neck squamous
cell carcinoma. Anticancer Res. 34:7137–7143. 2014.PubMed/NCBI
|
|
15
|
Khongkow M, Olmos Y, Gong C, Gomes AR,
Monteiro LJ, Yagüe E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong
S, et al: SIRT6 modulates paclitaxel and epirubicin resistance and
survival in breast cancer. Carcinogenesis. 34:1476–1486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Xie QR, Wang B, Shao J, Zhang T,
Liu T, Huang G and Xia W: Inhibition of SIRT6 in prostate cancer
reduces cell viability and increases sensitivity to
chemotherapeutics. Protein Cell. 4:702–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao Z, Hine C, Tian X, Van Meter M, Au M,
Vaidya A, Seluanov A and Gorbunova V: SIRT6 promotes DNA repair
under stress by activating PARP1. Science. 332:1443–1446. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiang W-C, Tishkoff DX, Yang B,
Wilson-Grady J, Yu X, Mazer T, Eckersdorff M, Gygi SP, Lombard DB
and Hsu AL: C. elegans SIRT6/7 homolog SIR-2.4 promotes
DAF-16 relocalization and function during stress. PLoS Genet.
8:e10029482012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roth M and Chen WY: Sorting out functions
of sirtuins in cancer. Oncogene. 33:1609–1620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van Meter M, Mao Z, Gorbunova V and
Seluanov A: SIRT6 overexpression induces massive apoptosis in
cancer cells but not in normal cells. Cell Cycle. 10:3153–3158.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barneda-Zahonero B and Parra M: Histone
deacetylases and cancer. Mol Oncol. 6:579–589. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Leiser SF, Fletcher M, Begun A and
Kaeberlein M: Life-span extension from hypoxia in Caenorhabditis
elegans requires both HIF-1 and DAF-16 and is antagonized by
SKN-1. J Gerontol A Biol Sci Med Sci. 68:1135–1144. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Judd LM, Menheniott TR, Ling H, Jackson
CB, Howlett M, Kalantzis A, Priebe W and Giraud AS: Inhibition of
the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and
in vivo. PLoS One. 9:e959932014. View Article : Google Scholar : PubMed/NCBI
|