Introduction
Gastric cancer (GC) represents one of the commonest
human malignancies characterized by an adverse clinical outcome
(1). Radical surgery may be the
only hope for curing GC in the stage of precursor lesions (2). However, even after surgical resection,
radiotherapy and chemotherapy provide little benefit (3). Previous studies revealed that a few
molecular markers were correlated with prognosis, but the mechanism
of GC remains to be elucidated (4).
Therefore, it is essential to recognize oncogenesis-associated
biomarkers, which are helpful for developing novel treatment in
view of GC.
Smad ubiquitin regulatory factor 1 (SMURF1), a
recently identified E3 ubiquitin ligase, targets substrate proteins
for ubiquitination and proteasomal degradation (5). Increasing evidence has demonstrated
that SMURF1 exerts a promoting role in carcinogenesis by targeting
downstream proteins for proteolysis. Epidermal growth factor
(EGF)-induced SMURF1 overexpression promoted breast cancer cell
migration and invasion by targeting RhoA (6). Tumor necrosis factor
receptor-associated factor 4 (TRAF4) was also reported to be a
substrate protein of SMURF1 and promoted the migration of breast
cancer cells (7). SMURF1 was
identified as a potential oncogene and a good candidate as a
therapeutic target of pancreatic cancer (PC) (8,9). The
overexpression of SMURF1 was observed in human colorectal cancer
(CRC), and contributed to tumor progression and poor prognosis
(10). Several studies have
reported that the upstream regulator of SMURF1, IQ motif containing
GTPase activating protein 1 (IQGAP1) promoted the ubiquitination
and degradation of TGF-β receptor II by facilitating the targeting
of SMURF1 to the plasma membrane in hepatic stellate cells
(11). Casein kinase-2 interacting
protein-1 (CKIP-1) suppressed colon cancer cell growth and
migration by inhibiting SMURF1 synthesis and facilitating SMURF1
autodegradation (12). Furthermore,
SMURF1 was recognized as a direct target of miR-497 in ovarian
cancer cells and exerted a pro-metastatic effect (13). TLX, a highly expressed nuclear
receptor, physically interacted with and stabilized SMURF1 in
glioblastoma (14). In addition,
SMURF1 facilitated T98G cell proliferation and migration by
regulation of DOC-2/DAB2 interactive protein (DAB2IP) (15). However, the clinical significance of
SMURF1 and its role in human GC remain poorly elucidated.
The present study revealed that SMURF1
overexpression predicted malignant clinical features and decreased
survival. We also demonstrated that SMURF1 promoted GC cell growth
and metastasis possibly by suppressing DAB2IP. In conclusion, the
present study revealed the first evidence that SMURF1 is a clinical
biomarker, and recognized as a potential therapeutic target for
GC.
Materials and methods
Patients
Sixty-eight cases of GC and corresponding
tumor-adjacent specimens were obtained from the Department of
Gastrointestinal Colorectal and Anal Surgery, China-Japan Union
Hospital, Jilin University. Tissue specimens were conserved in
liquid nitrogen for qRT-PCR or 10% formalin for IHC until use.
Informed consent from patients was obtained before the use of all
samples. All the clinicopathological information of patients is
presented in Table I. The Ethics
Committee of Jilin University approved the present study according
to the Declaration of Helsinki.
 | Table I.Correlation between SMURF1 expression
and clinicopathological features in gastric cancer. |
Table I.
Correlation between SMURF1 expression
and clinicopathological features in gastric cancer.
|
|
| SMURF1
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Total | Positive (48) | Negative (20) | P-value |
|---|
| Age (years) |
|
|
| 0.800 |
|
<65 | 29 | 20 | 9 |
|
| ≥65 | 39 | 28 | 11 |
|
| Sex |
|
|
| 0.485 |
| Male | 53 | 39 | 14 |
|
|
Female | 15 | 9 | 6 |
|
| Tumor
differentiation |
|
|
| 0.198 |
| I,
II | 32 | 25 | 7 |
|
| III,
IV | 36 | 23 | 13 |
|
| Size (cm) |
|
|
| 0.014a |
|
<5 | 32 | 18 | 14 |
|
| ≥5 | 36 | 30 | 6 |
|
| Invasive depth |
|
|
| 0.146 |
| Mucosa to
muscularis propria | 9 | 4 | 5 |
|
|
Adventitia to adjacent
structure | 59 | 44 | 15 |
|
| Lymph node
metastasis (regions) |
|
|
| 0.016a |
| ≤2 | 29 | 16 | 13 |
|
|
>2 | 39 | 32 | 7 |
|
| Distant
metastasis |
|
|
| 0.044a |
| No | 52 | 33 | 19 |
|
|
Yes | 16 | 15 | 1 |
|
| Venous
infiltration |
|
|
| 0.065 |
|
Absent | 51 | 33 | 18 |
|
|
Present | 17 | 15 | 2 |
|
| TNM stage |
|
|
| 0.042a |
| I,
II | 28 | 16 | 12 |
|
| III,
IV | 40 | 32 | 8 |
|
Cell culture and transfection
GC-derived cell lines (SGC-7901, MGC-803, MKN-28 and
BGC-823) and a normal gastric epithelium cell line (GES-1) were
obtained from the Cell Bank of the Shanghai Institute of Cell
Biology (Chinese Academy of Medical Sciences, Shanghai, China). The
cell lines were cultured in Dulbeccos modified Eagles medium (DMEM)
with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA)
with antibiotics (Sigma-Aldrich, St. Louis, MO, USA) in a incubator
containing a 5% CO2 humidified atmosphere at 37°C.
Small hairpin RNA (shRNA) targeting SMURF1 and
DAB2IP as well as non-targeting (NT) shRNA were obtained from Santa
Cruz Biotechnology (Santa Cruz, CA, USA). pcDNA3.1-SMURF1 and
pcDNA3.1-DAB2IP were synthesized and purchased from GeneChem
(Shanghai, China). All vectors were transferred into cells using
Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) on
the basis of the manufacturer's recommendations.
Immunohistochemistry (IHC)
The tissues that were previously formalin-fixed and
paraffin-embedded were sliced into 4 µm sections, and underwent
deparaffination and then rehydration. Antigen retrieval,
suppression of endogenous peroxidase activity and 10% skim milk
blocking were performed before primary antibody incubation. The
SMURF1 (Abcam, Cambridge, MA, USA) and DAB2IP (Santa Cruz
Biotechnology) antibodies were used as primary antibodies overnight
at 4°C. The slides were subsequently incubated with a
peroxidase-conjugated secondary antibody (ZSGB-BIO, Beijing, China)
for 90 min, and a peroxidase-labeled polymer, DAB solution was used
for signal development for 5 min. The sections were counterstained
with hematoxylin followed by dehydration and mounting. Staining
intensity was scored as: no staining, 0; weak staining, 1; moderate
staining, 2; and strong staining, 3. Staining quantity was graded
as: <25%, 1; 25–75%, 2; and >75%, 3. IHC score was manually
confirmed by two independent experienced pathologists using the
formula: IHC score = staining intensity × staining quantity.
Sections with an IHC score >1 were considered SMURF1- and
DAB2IP-positive expression.
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total RNA was drawn using TRIzol (Invitrogen,
Carlsbad, CA, USA). The first strand cDNA was compounded using a
TIANScript RT kit (Tiangen Biotech, Beijing, China). The expression
of SMURF1 mRNA was detected using the ABI 7300 system (Applied
Biosystems, Foster City, CA, USA). GAPDH was employed as the
internal control. The primers used for target genes were purchased
from Sangon Biotech (Shanghai, China).
Proliferation assay
For cell proliferation, GC cells that were treated
with corresponding vectors were seeded into 96-well plates
(1.5×103 cells/well). After transfection at 24, 48, 72
and 96 h, the cell proliferation assay was performed by addition of
10 µl Cell Counting Kit-8 (CCK-8) solution (Beyotime, Shanghai,
China) to each well, followed by incubation at 37°C for 2 h. The
absorbance was assessed at a wavelength of 490 nm using a
microplate reader (FlexStation III ROM V2.1.28; Molecular Devices,
Sunnyvale, CA, USA).
Wound healing assay
GC cells transduced with corresponding vectors were
seeded into 6-well plates to form a single confluent cell layer.
The wounds were made with 100 µl tips in the confluent cell layer.
After wound scratching at 0 and 24 h, the width of the wound was
photographed using a phase-contrast microscope.
Transwell invasion assay
We determined the invasion capacities of GC cells
using Transwell chambers of pore size 8-µm (Corning Costar,
Cambridge, MA, USA). Twenty-four hours after transduction,
5×104 cells were cultured in the 1:9 diluted
Matrigel-coated (BD Biosciences, Franklin Lakes, NJ, USA) upper
chamber with 250 µl of serum-free DMEM, while 700 µl DMEM with 10%
FBS were added in the lower chamber. After 24 h, we fixed the cells
with paraformaldehyde, and the cells in upper chamber were removed.
Cells in the lower chamber were subsequently stained using 0.1%
crystal violet solution and photographed.
Experimental mouse model
The in vivo growth ability of GC cells was
examined using the subcutaneous implantation nude mouse model.
MGC-803 cells transfected with NT-shRNA or SMURF1-shRNA were
subcutaneously injected into the left flank of nude mice. After 4
weeks, the subcutaneous tumors were finally resected and subjected
to weight assessments. A liver metastasis assay in nude mice was
performed using the subcapsular splenic injection model in which
the MGC-803 cells were injected to the spleen subcapsular. Nine
weeks after splenic injection, all mice were euthanized, and the
livers were obtained. Furthermore, analysis of micrometastasis was
assessed on the left lateral lobe of the liver, that was fixed and
cut sagittally into 4 parts, paraffin-embedded, sectioned and
stained for hematoxylin and eosin (H&E) (16). The protocol for these animal
experiments was approved by the Ethics Review Committee of Jilin
University.
Western blotting
Cancer cells were dissociated in RIPA lysis buffer
(P0013D) and PMSF (ST506) (both from Beyotime, Haimen, China).
Split products were centrifuged at 12,000 rpm, and then
supernatants were gathered. A Bradford protein assay kit (P0006)
(Beyotime) was used to analyze the protein concentration, and
proteins were loaded onto 10% SDS-PAGE. Then proteins after
separation were transferred onto polyvinylidene fluoride (PVDF)
membranes (Sigma-Aldrich). Then, the PVDF membranes were obstructed
with 5% skim milk (GuangMing, Shanghai, China) and incubated with
SMURF1 (Abcam), DAB2IP (Santa Cruz Biotechnology), p-Akt (Ser473),
Akt, c-Myc or ZEB1 (all from Cell Signaling Technology, Beverly,
MA, USA) primary antibodies at 4°C overnight. Then, the specimens
were incubated with a secondary antibody conjugated with HRP (Cell
Signaling Technology). Signals were detected using an HRP
chemiluminescent kit (Thermo Fisher Scientific) and an optional CCD
camera as well as an image processing system (Bio-Rad, Hercules,
CA, USA). GAPDH (G8140; US Biological, Swampscott, MA, USA) was
used as a loading control.
Statistical analysis
Data were presented as the mean ± SEM and analyzed
by GraphPad Prism 5 software (GraphPad Software, Inc., San Diego,
CA, USA). Chi-squared test was employed to explore the association
between two variables. The Student's t-test and ANOVA were carried
out to analyze continuous variables. Survival analysis was
performed using Kaplan-Meier's method and the log-rank test. A
P-value <0.05 was considered to have statistical
significance.
Results
SMURF1 overexpression is a clinical
biomarker of GCs
Sixty-eight samples of GC and corresponding
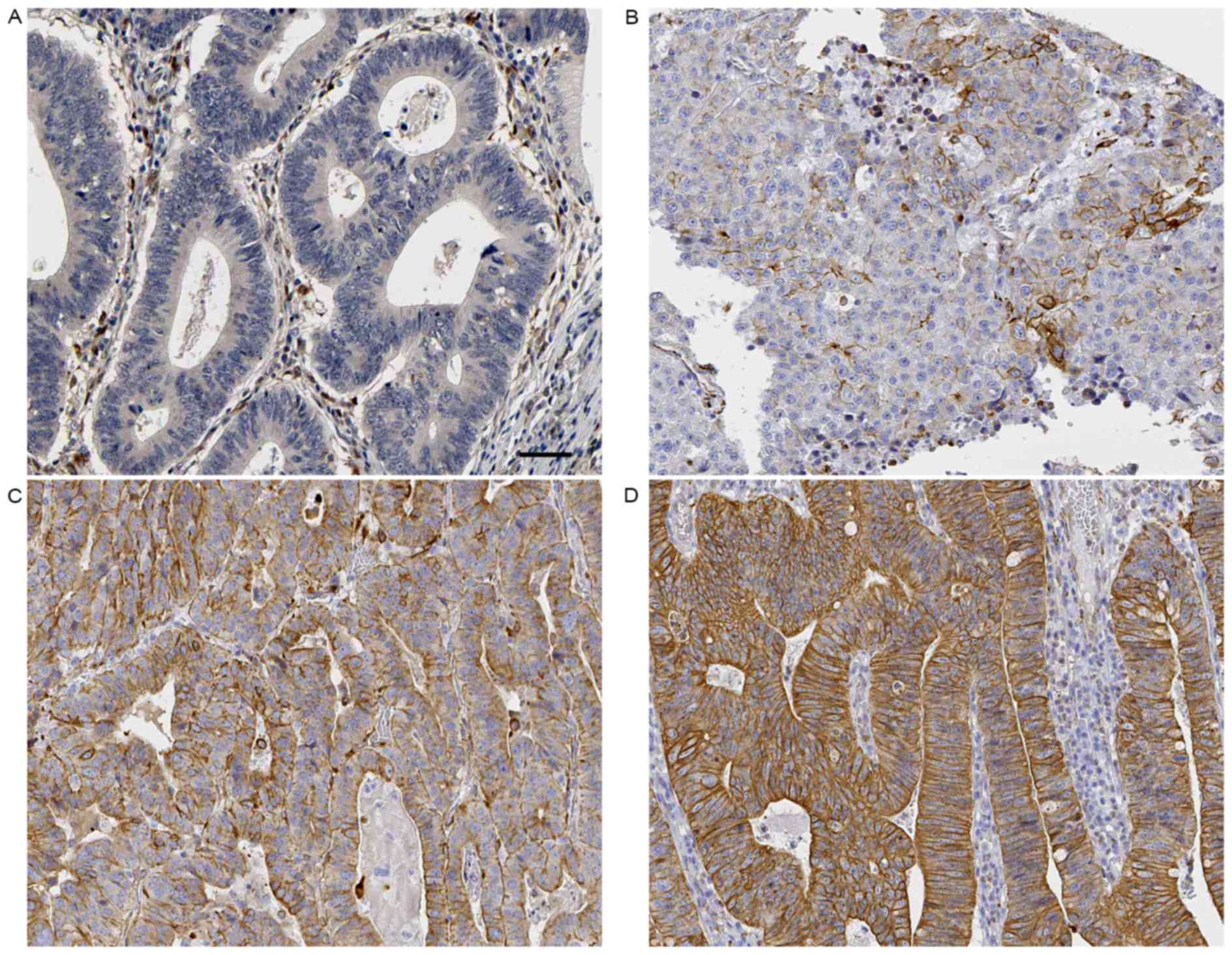
tumor-adjacent specimens were detected by IHC for SMURF1 staining.
Sections with an IHC score >1 were considered as positive
expression of SMURF1. GC tissue samples, 70.59% (48/68) exhibited
positive staining of SMURF1, while SMURF1 signal were detected in
only 47.06% (32/68) samples of tumor-adjacent specimens (P=0.005;
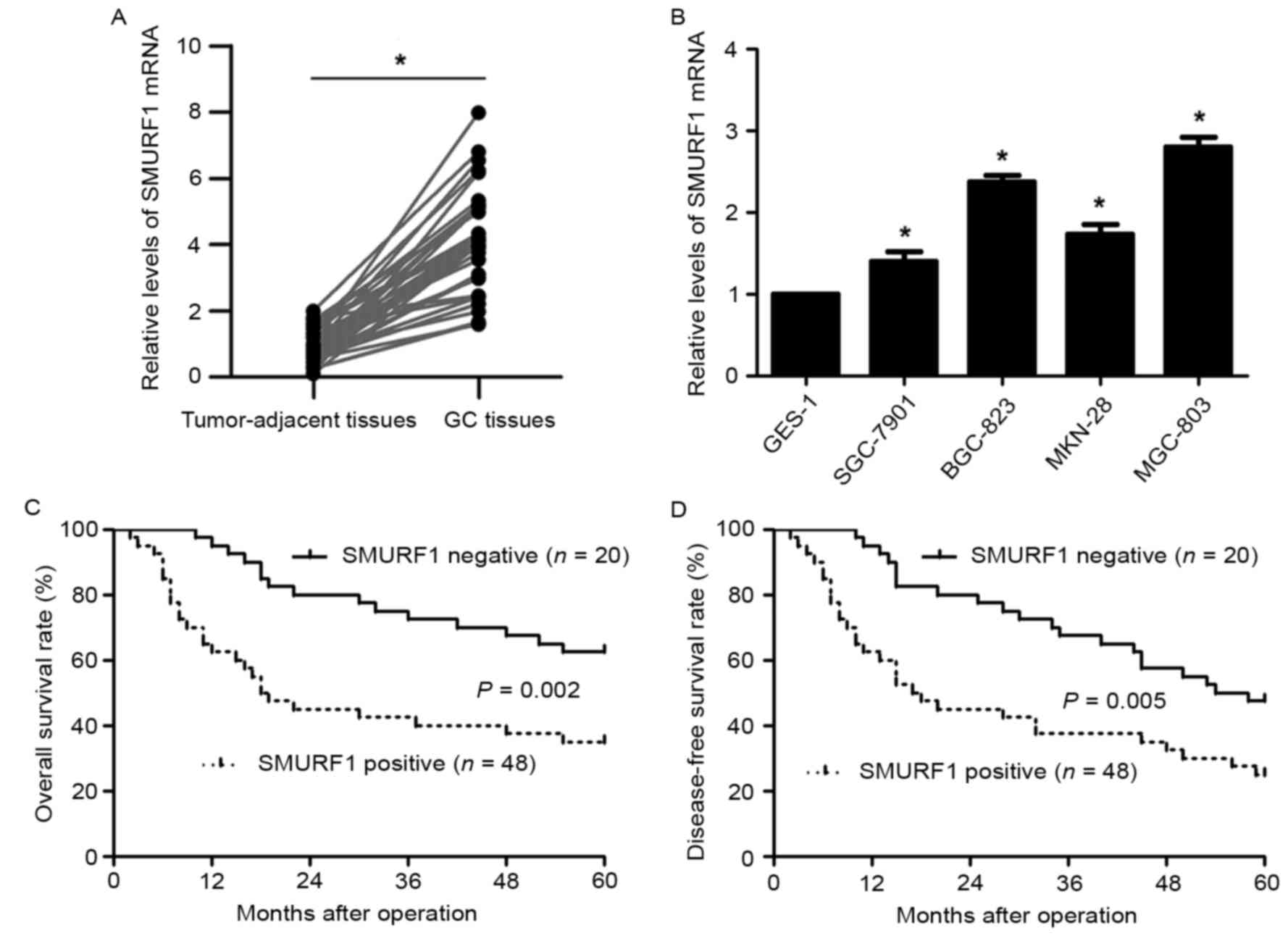
Fig. 1). Next, qRT-PCR further
demonstrated that the levels of SMURF1 mRNA in GC tissues was
upregulated compared with corresponding tumor-adjacent tissues
(P<0.05; Fig. 2A). Moreover, the
levels of SMURF1 mRNA expression in GC cell lines (SGC-7901,
MGC-803, MKN-28 and BGC-823) were significantly increased when
compared to the normal gastric epithelium cell line (GES-1)
(P<0.05, respectively; Fig. 2B).
In addition, positive expression of SMURF1 in GC patients was
correlated with large-sized tumors, more lymph node metastasis and
distant metastasis as well as tumor-node-metastasis (TNM) grade
(P<0.05, respectively; Table I).
Survival analysis revealed that GC patients with SMURF1-positive
expression exhibited a prominent decreased 5-year overall and
disease-free survival (P<0.05, respectively; Fig. 2C and D). Thus, SMURF1 may be a
potential prognostic biomarker in GC.
SMURF1 regulates proliferation and
mobility of GC cells
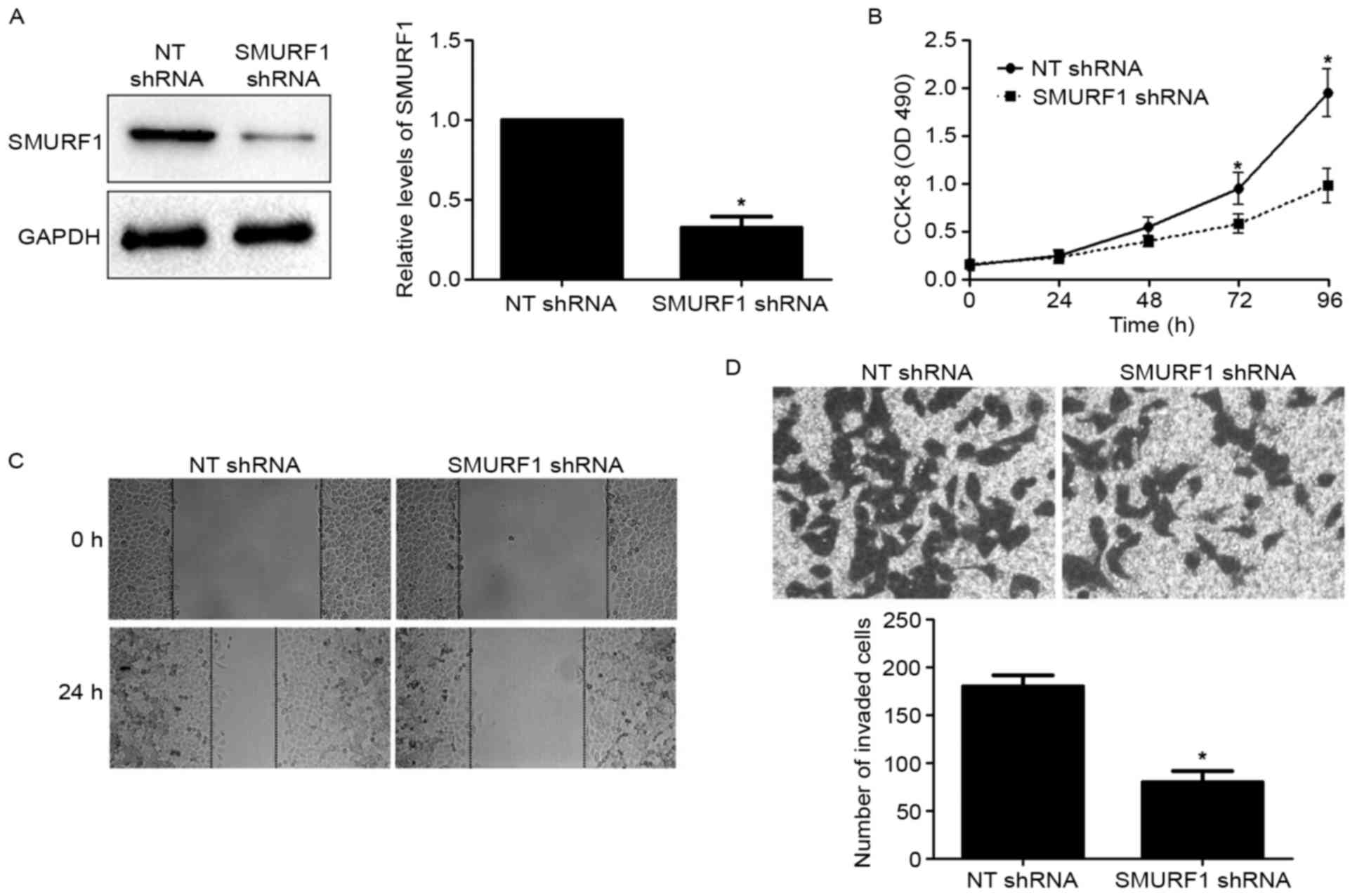
Since SMURF1 was overexpressed in GC, we speculated
that the biological functions of SMURF1 may participate in
controlling cell proliferation and metastasis. To verify this
hypothesis, SMURF1 expression was silenced by a specific shRNA in
MGC-803 cells (P<0.05; Fig. 3A).
CCK-8 assays were used to analyze the effect of SMURF1 silencing on
proliferation of GC cells. We found that silencing of SMURF1
suppressed MGC-803 cell proliferation as compared with control
cells (P<0.05; Fig. 3B). To
disclose the potential role of SMURF1 in the metastasis of GC, we
analyzed the migration and invasion of GC cells using wound healing
and Transwell invasion assays. The results revealed that SMURF1
knockdown caused a prominent decrease in the migratory and invasive
abilities of MGC-803 cells compared to the control cells
(P<0.05, respectively; Fig. 3C and
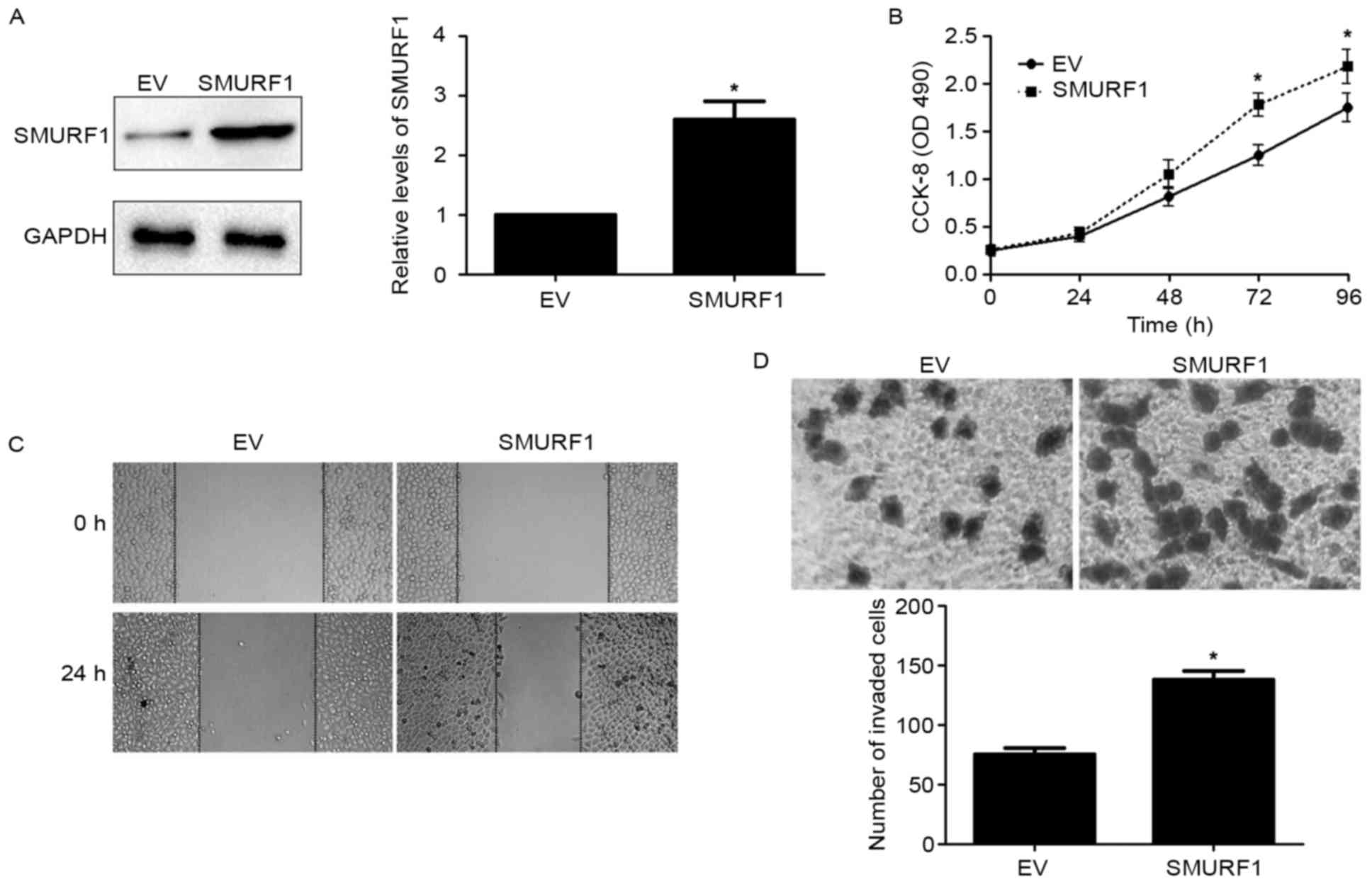
D). Next, SGC-7901 cells were transduced with an empty vector
(EV) and pcDNA3.1-SMURF1, respectively. SMURF1 overexpression was
confirmed by western blotting in SGC-7901 cells (P<0.05;
Fig. 4A). Overexpression of SMURF1
notably enhanced the proliferation, migration and invasion of
SGC-7901 cells (P<0.05; respectively; Fig. 4B-D). Collectively, all the results
demonstrated that SMURF1 can markedly inhibit GC cell proliferation
and mobility in vitro.
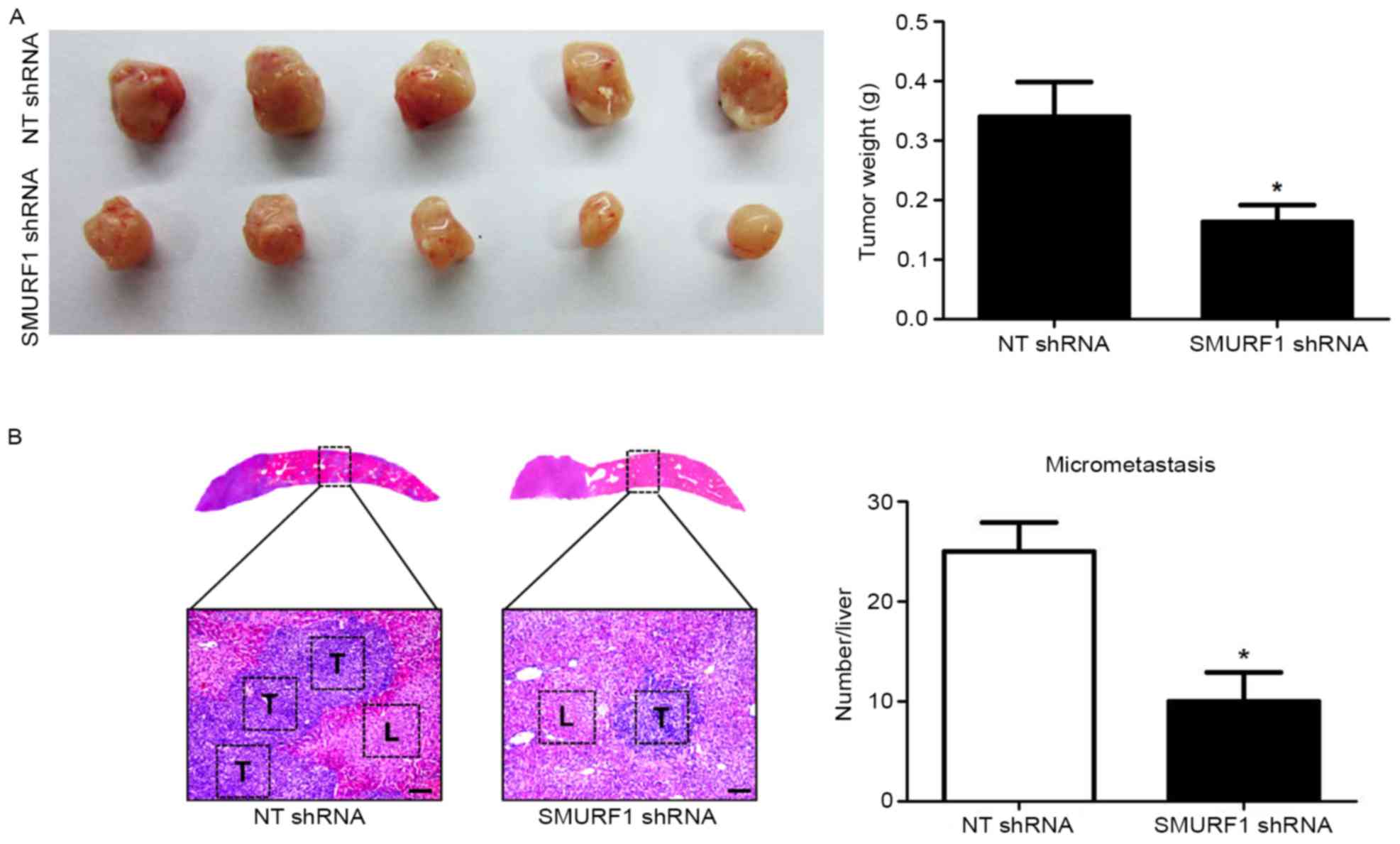
SMURF1 knockdown decreases the growth
and metastasis of GC cells in nude mice
To further confirm the effects of SMURF1 on the
growth and metastasis of GC cells, a mouse experimental growth or
liver metastasis model was constructed using MGC-803 cells via
subcutaneous or subcapsular splenic injection. Silencing of SMURF1
markedly decreased the subcutaneous growth of GC in nude mice as
determined by the tumor weights (P<0.05; Fig. 5A). In addition, liver metastasis
experiments revealed that SMURF1 knockdown notably decreased the
number of metastatic nodules in the livers of nude mice (P<0.05;
Fig. 5B). Altogether, our data
revealed that SMURF1 prominently prohibited GC cell growth and
metastasis in vivo.
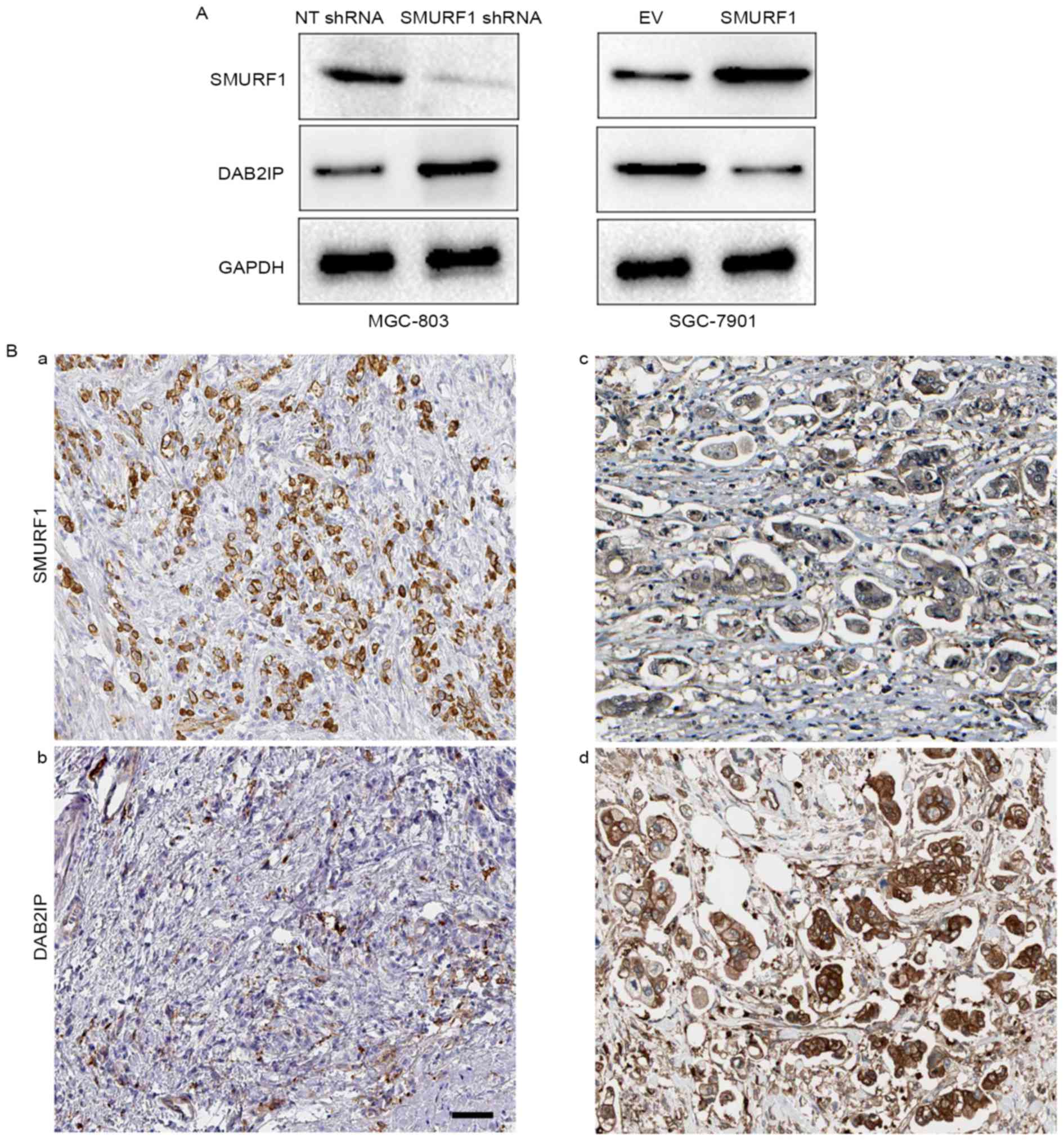
DAB2IP may be involved in the role of
SMURF1
Previous research revealed that SMURF1 promotes
cancer cell proliferation and migration by negatively regulating
DAB2IP (15). Therefore, we
investigated whether DAB2IP was involved in the role of SMURF1 in
GC. Notably, we found that the expression of DAB2IP was increased
after SMURF1 knockdown in MGC-803 cells (Fig. 6A), while, SMURF1 overexpression led
to a decreased level of DAB2IP in SGC-7901 cells (Fig. 6A). Furthermore, DAB2IP staining was
performed in GC tissues using IHC. Our data revealed that the
positive expression of DAB2IP was observed in 36.76% (25/68) GC
tissue samples. Notably, highly-expressing SMURF1 in GC tissues
exhibited weak staining of DAB2IP, while strong signals of DAB2IP
were observed in tissues with low expression of SMURF1 (Fig. 6B). Next, DAB2IP was markedly
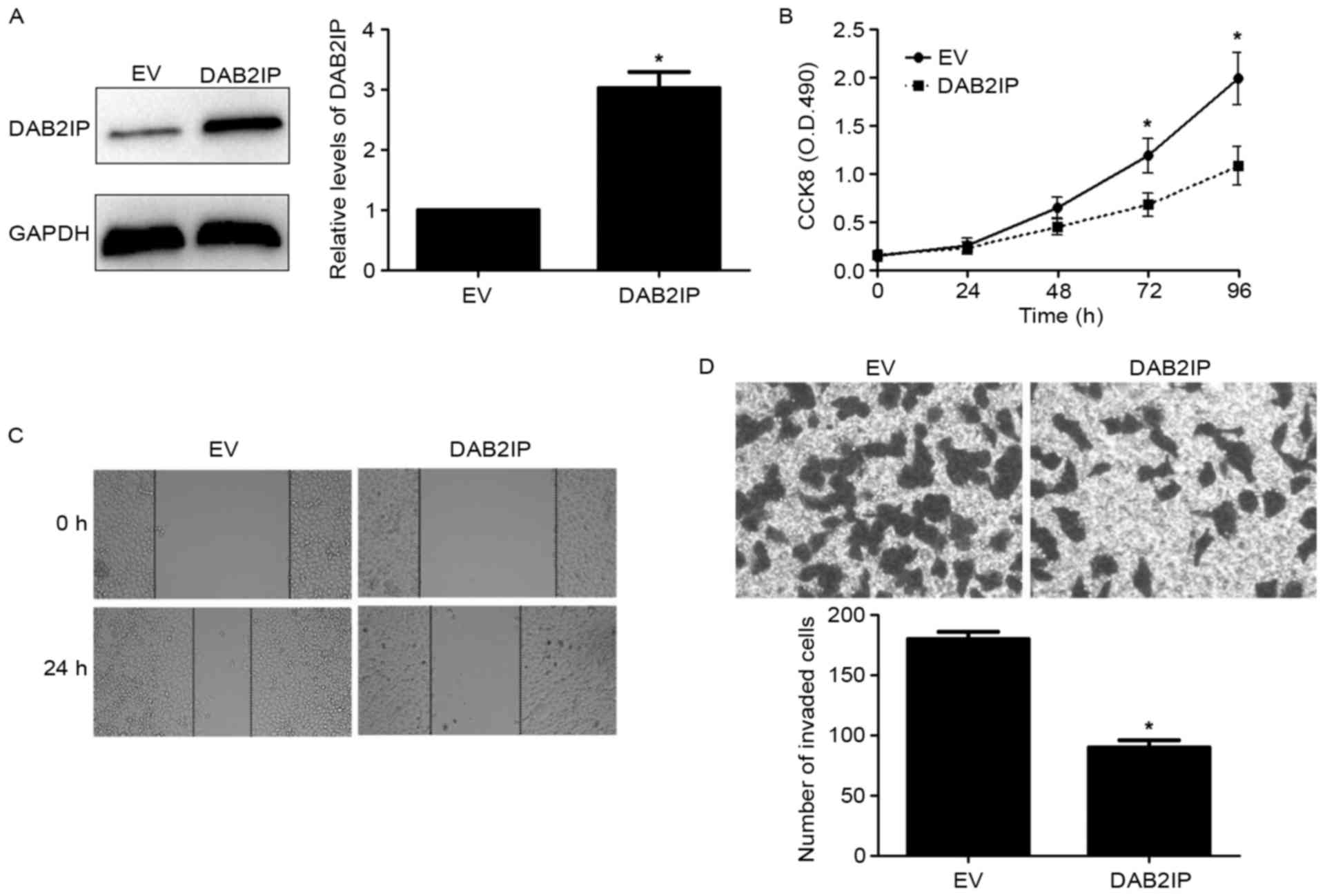
overexpressed by a pcDNA3.1-mediated expression plasmid in MGC-803
cells (P<0.05; Fig. 7A).
Notably, DAB2IP restoration revealed similar effects to SMURF1
knockdown in MGC-803 cells with decreased cell proliferation,
migration and invasion (P<0.05, respectively; Fig. 7B-D). Therefore, these results
indicate that SMURF1 plays an oncogenic role possibly by
suppressing DAB2IP in GC cells.
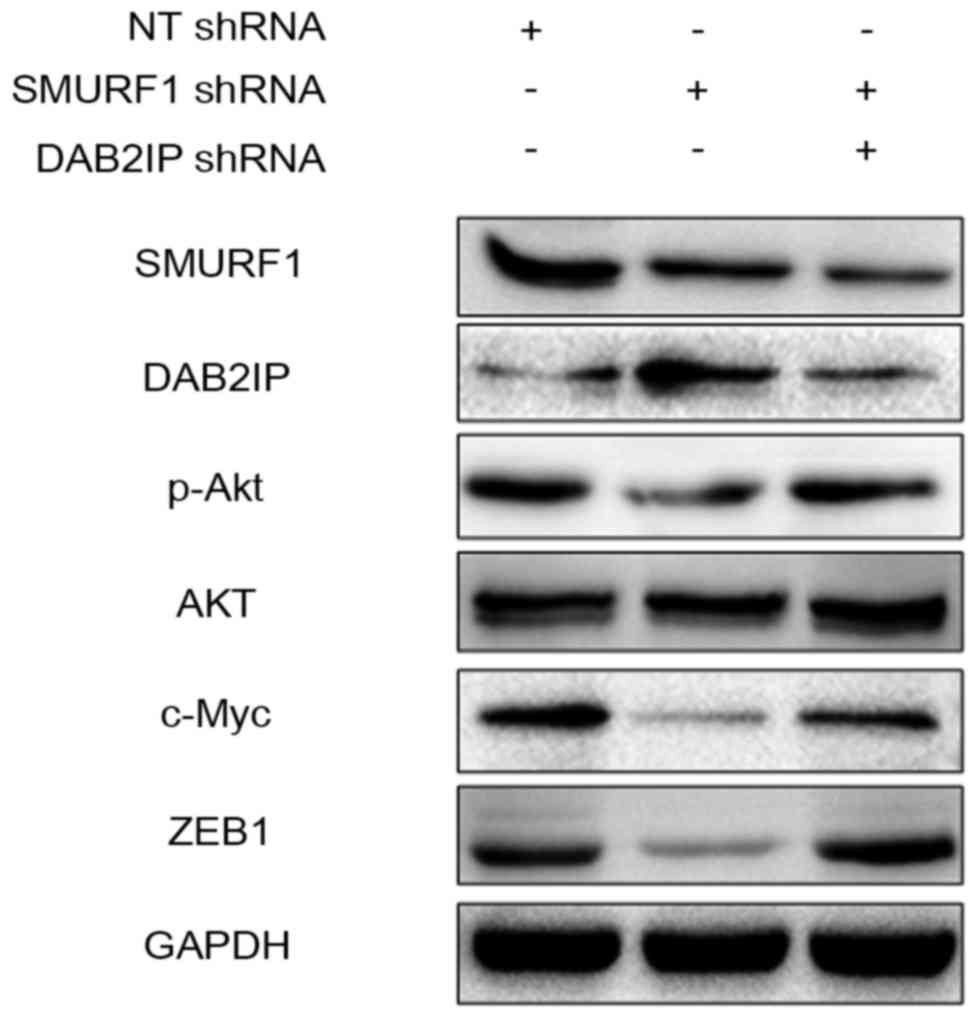
The PI3K/Akt pathway is potentially
involved in the oncogenic role of the SMURF1/DAB2IP axis
DAB2IP has been reported to regulate various
signaling pathways including the PI3K/Akt pathway in human cancer
(17). Furthermore, the downstream
targets of PI3K/Akt pathway such as c-Myc and ZEB1 regulate
proliferation, migration and invasion in GC cells (18,19).
The results from western blot analysis revealed that SMURF1
knockdown upregulated the level of DAB2IP and subsequently
downregulated the expression of phosphorylated Akt as well as c-Myc
and ZEB1 in MGC-803 cells (Fig. 8).
However, DAB2IP-silencing abrogated the effects of SMURF1 knockdown
on the activation of the PI3K/Akt pathway with increased levels of
poshorylated Akt, c-Myc and ZEB1 (Fig.
8). Thus, SMURF1 probably suppresses DAB2IP and subsequently
enhances the activation of the PI3K/Akt pathway in GC cells.
Discussion
Increasing evidence has demonstrated that SMURF1
participates in the initiation and progression of numerous
malignancies (6,8,9,12,13,15).
Dysregulation of SMURF1 is a common event in various types of
cancer including PC (8,9), breast (6,7) and
ovarian cancer (13), and CRC
(12). To the best of our
knowledge, few studies have focused on the clinical values and the
role of SMURF1 in GC to date. Thus, further studies concerning
SMURF1 in GC may be helpful in the treatment of this deadly
disease. The present study demonstrated the pattern of SMURF1
expression in the carcinogenesis of GC and its potential clinical
significance in GC patients, and further investigated its effect
and potential mechanisms in GC cells. The present study identified
SMURF1 as a promising clinical biomarker of GC patients. The
expression of SMURF1 in the GC tissues was notably increased
compared to the tumor-adjacent specimens, and its increase was
correlated with large tumor size, more lymph node metastasis and
distant metastasis, as well as advanced TNM stage and decreased
survival. Thus, SMURF1 potentially functions as a clinical marker
in GC.
Then, we explored the biology of SMURF1 in GC, and
demonstrated that silencing of SMURF1 expression inhibited cell
proliferation and mobility in vitro and in vivo. In
turn, SMURF1 overexpression significantly promoted cell
proliferation as well as migration and invasion in GC cells. The
prominent ability of SMURF1 to promote tumorigenesis reveals that
it plays an oncogenic role in GC. Therefore, targeting SMURF1 may
represent a favorable therapeutic strategy for GC treatment. Next,
we explored a potential target gene that may be involved in the
role of SMURF1. DAB2IP, a tumor-suppressor, plays a critical role
in cancer cell growth and metastasis as well as other aspects
during tumor progression (17).
Epigenetic silencing is the main cause of dysregulated expression
of DAB2IP in human cancers (20).
The expression of DAB2IP is silenced by promoter methylation and
histone modification in prostate cancer (21,22).
In bladder cancer, DAB2IP expression was suppressed by
post-transcriptional regulation of miR-92b (23). In the present study, we revealed
that silencing of SMURF1 upregulated DAB2IP expression while SMURF1
overexpression decreased DAB2IP expression in GC cells, suggesting
that SMURF1 is a novel negative regulator of DAB2IP as previously
reported (15). Notably, DAB2IP
restoration resulted in similar effects to SMURF1 silencing in GC
cells with weakened proliferation, migration and invasion. Thus, we
support a preliminary theory that SMURF1 inversely regulates DAP2IP
expression, resulting in the induction of growth and metastasis in
GC. The PI3K/Akt pathway has been implicated in the growth and
metastasis of GC (24,25). In addition, the downstream targets
of the PI3K/Akt pathway such as c-Myc and ZEB1 regulate
proliferation, migration and invasion in GC cells (18,19).
In the present study, we reported that SMURF1 promoted the
activation of PI3K/Akt and its downstream targets including c-Myc
and ZEB1 via inhibition of DAB2IP in GC cells.
In conclusion, we revealed that SMURF1 as an
oncogene is important in GC. Firstly, our results demonstrated that
SMURF1 expression was increased in GC cell lines and tissues. Then,
our clinical data revealed that SMURF1 may be used as a novel
biomarker for GC. Moreover, SMURF1 overexpression resulted in
enhanced proliferation and mobility possibly by DAB2IP inhibition
in GC cells. Collectively, our results ascertained that SMURF1 may
serve as a potential target in cancer therapeutics of GC.
Acknowledgements
The authors thank all the patients who participated
in the present study.
References
|
1
|
Wang J, Yu JC, Kang WM and Ma ZQ:
Treatment strategy for early gastric cancer. Surg Oncol.
21:119–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marqués-Lespier JM, González-Pons M and
Cruz-Correa M: Current perspectives on gastric cancer.
Gastroenterol Clin North Am. 45:413–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thrumurthy SG, Chaudry MA, Chau I and
Allum W: Does surgery have a role in managing incurable gastric
cancer? Nat Rev Clin Oncol. 12:676–682. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McLean MH and El-Omar EM: Genetics of
gastric cancer. Nat Rev Gastroenterol Hepatol. 11:664–674. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cao Y and Zhang L: A Smurf1 tale: Function
and regulation of an ubiquitin ligase in multiple cellular
networks. Cell Mol Life Sci. 70:2305–2317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kwon A, Lee HL, Woo KM, Ryoo HM and Baek
JH: SMURF1 plays a role in EGF-induced breast cancer cell migration
and invasion. Mol Cells. 36:548–555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Jin C, Tang Y, Tang LY and Zhang
YE: Ubiquitination of tumor necrosis factor receptor-associated
factor 4 (TRAF4) by Smad ubiquitination regulatory factor 1
(Smurf1) regulates motility of breast epithelial and cancer cells.
J Biol Chem. 288:21784–21792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki A, Shibata T, Shimada Y, Murakami
Y, Horii A, Shiratori K, Hirohashi S, Inazawa J and Imoto I:
Identification of SMURF1 as a possible target for 7q21.3–22.1
amplification detected in a pancreatic cancer cell line by in-house
array-based comparative genomic hybridization. Cancer Sci.
99:986–994. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwei KA, Shain AH, Bair R, Montgomery K,
Karikari CA, van de Rijn M, Hidalgo M, Maitra A, Bashyam MD and
Pollack JR: SMURF1 amplification promotes invasiveness in
pancreatic cancer. PLoS One. 6:e239242011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie P, Zhang M, He S, Lu K, Chen Y, Xing
G, Lu Y, Liu P, Li Y, Wang S, et al: The covalent modifier Nedd8 is
critical for the activation of Smurf1 ubiquitin ligase in
tumorigenesis. Nat Commun. 5:37332014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Billadeau DD, Abdelhakim H, Leof E,
Kaibuchi K, Bernabeu C, Bloom GS, Yang L, Boardman L, Shah VH, et
al: IQGAP1 suppresses TβRII-mediated myofibroblastic activation and
metastatic growth in liver. J Clin Invest. 123:1138–1156. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nie J, Liu L, Xing G, Zhang M, Wei R, Guo
M, Li X, Xie P, Li L, He F, et al: CKIP-1 acts as a colonic tumor
suppressor by repressing oncogenic Smurf1 synthesis and promoting
Smurf1 autodegradation. Oncogene. 33:3677–3687. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Ren F, Wu Q, Jiang D, Li H, Peng
Z, Wang J and Shi H: MicroRNA-497 inhibition of ovarian cancer cell
migration and invasion through targeting of SMAD specific E3
ubiquitin protein ligase 1. Biochem Biophys Res Commun.
449:432–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johansson E, Zhai Q, Zeng ZJ, Yoshida T
and Funa K: Nuclear receptor TLX inhibits TGF-β signaling in
glioblastoma. Exp Cell Res. 343:118–125. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Dai X, Wan L, Inuzuka H, Sun L and
North BJ: Smurf1 regulation of DAB2IP controls cell proliferation
and migration. Oncotarget. 7:26057–26069. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mendonsa AM, VanSaun MN, Ustione A, Piston
DW, Fingleton BM and Gorden DL: Host and tumor derived MMP13
regulate extravasation and establishment of colorectal metastases
in the liver. Mol Cancer. 14:492015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu L, Xu C, Hsieh JT, Gong J and Xie D:
DAB2IP in cancer. Oncotarget. 7:3766–3776. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu SQ, Yu JP, Yu HG, Lv P and Chen HL:
Activation of Akt and ERK signalling pathways induced by etoposide
confer chemoresistance in gastric cancer cells. Dig Liver Dis.
38:310–318. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuan D, Xia H, Zhang Y, Chen L, Leng W,
Chen T, Chen Q, Tang Q, Mo X, Liu M, et al: P-Akt/miR-200 signaling
regulates epithelial-mesenchymal transition, migration and invasion
in circulating gastric tumor cells. Int J Oncol. 45:2430–2438.
2014.PubMed/NCBI
|
|
20
|
Tsai YS, Lai CL, Lai CH, Chang KH, Wu K,
Tseng SF, Fazli L, Gleave M, Xiao G, Gandee L, et al: The role of
homeostatic regulation between tumor suppressor DAB2IP and
oncogenic Skp2 in prostate cancer growth. Oncotarget. 5:6425–6436.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen H, Toyooka S, Gazdar AF and Hsieh JT:
Epigenetic regulation of a novel tumor suppressor gene
(hDAB2IP) in prostate cancer cell lines. J Biol Chem.
278:3121–3130. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen H, Tu SW and Hsieh JT:
Down-regulation of human DAB2IP gene expression mediated by
polycomb Ezh2 complex and histone deacetylase in prostate cancer. J
Biol Chem. 280:22437–22444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang J, Wang B, Hui K, Zeng J, Fan J,
Wang X, Hsieh JT, He D and Wu K: miR-92b targets DAB2IP to promote
EMT in bladder cancer migration and invasion. Oncol Rep.
36:1693–1701. 2016.PubMed/NCBI
|
|
24
|
Xiong J, Li Z, Zhang Y, Li D, Zhang G, Luo
X, Jie Z, Liu Y, Cao Y, Le Z, et al: PRL-3 promotes the peritoneal
metastasis of gastric cancer through the PI3K/Akt signaling pathway
by regulating PTEN. Oncol Rep. 36:1819–1828. 2016.PubMed/NCBI
|
|
25
|
Shen X, Si Y, Wang Z, Wang J, Guo Y and
Zhang X: Quercetin inhibits the growth of human gastric cancer stem
cells by inducing mitochondrial-dependent apoptosis through the
inhibition of PI3K/Akt signaling. Int J Mol Med. 38:619–626.
2016.PubMed/NCBI
|