Introduction
The ubiquitin-proteasome system is a major pathway
that contributes to intracellular proteostasis. Carboxy-terminus of
Hsc70 interacting protein (CHIP) has been identified as an E3
ubiquitin ligase and a potent regulator protein in maintaining
protein homeostasis in diverse cellular processes (1–3).
Structurally, 3 tandem repeats of the tetratricopeptide (TPR) motif
and a C-terminal U-box domain (U-box) contribute to the bioactivity
of CHIP as a chaperone-associated E3 ubiquitin ligase involved in
the ubiquitin-proteasome system (4,5). CHIP
post-translationally controls the turnover of its substrate
proteins and exerts regulatory roles in a myriad of biological
processes (6,7).
Until recently, accumulating evidence has suggested
a role for CHIP in the initiation and progression of cancers
(7–9). During the development of
carcinogenesis, the production of mutated or aberrant cellular
proteins must be gradually increased during malignant
transformation and in response to genetic instability processes
(10,11). As previously reported, CHIP not only
induces ubiquitylation and degradation of several oncogenic
proteins (7,12–14),
but also regulates tumor suppressor proteins (15,16),
which has increased interest in understanding the mechanisms of
CHIP in the context of carcinogenesis. Accordingly, the diversities
of CHIP substrate proteins present heterogeneity among different
tissue-derived cancers, and the biological functions of CHIP in
head and neck cancers (HNCs) remain unclear.
In the present study, we conducted a comprehensive
study on CHIP in HNCs from laboratory experiments to clinical data.
Our results showed that CHIP functions as a tumor suppressor in HNC
cell lines and a regular pattern of CHIP expression from well,
moderate, to poor differentiation state was illustrated. In
conclusion, we demonstrated that low expression of CHIP is a
potential risk factor for HNC patients.
Materials and methods
Cell culture and reagents
HNC cell lines (HN13, HN30, Cal27 and UMSCC12) were
cultured in Dulbecco's modified Eagle's medium (DMEM; Basal Media,
Shanghai, China) supplemented with 10% fetal bovine serum (FBS;
Gibco-Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100
µg/ml streptomycin, at 37°C in the presence of 5% carbon dioxide
(17,18). Thiazolyl blue tetrazolium bromide
(MTT) was purchased from Sigma (St. Louis, MO, USA).
Plasmid constructs and
transfections
The mammalian expression vector used was
pcDNA3.1(+)-myc-CHIP. Two loss-of-function CHIP mutants were
generated using the QuickChange XL Site-Directed Mutagenesis kit
(Life Technologies, Carlsbad, CA, USA) at the TPR domain (K30A),
and U-box domain (H260Q), respectively. The primer sequences used
for site-directed mutagenesis were: forward,
5′-GCGCGCAGGAGCTCGCGGAGCAGGGCAATC-3′ and reverse,
5′-GATTGCCCTGCTCCGCGAGCTCCTGCGCGC-3′ for CHIP (K30A); and forward,
5′-ACATCGAGGAGCAGCTGCAGCGTGTGG-3′ and reverse,
5′-CCACACGCTGCAGCTGCTCCTCGATGT-3′ for CHIP (H260Q) (6,16).
Plasmids were transfected into cells using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) diluted in Opti-MEM® I
(Life Technologies) according to the manufacturer's instructions.
UMSCC12-shCHIP stable cells were generated by transfecting UMSCC12
cells with the CHIP-specific short hairpin RNA
(5′-GGACGACATCCCCAGCGCTCT-3′) lentivirus and selected with 10 µg/ml
of puromycin (Calbiochem, La Jolla, CA, USA) for positive clones
(13), and UMSCC12-scrambled cells
were used as control.
Cell proliferation analysis
MTT assays were performed to examine the
proliferative ability of involved cells in the present study.
Absorbance measurements following dimethyl sulfoxide (DMSO)
resolution were performed at a wavelength of 490 nm (Bio-Rad
Laboratories, Hercules, CA, USA).
Colony formation assay
Cell suspension was counted, diluted and seeded at
1×103/well into a 6-well plate in triplicates. Cells
were kept culturing for 10 days, after which the cells were fixed
with paraformaldehyde and stained with Coomassie Brilliant Blue,
subsequently. The number of large colonies was counted and analyzed
(18).
Transwell assays
To determine cell migration, 5×104 cells
suspended in 200 µl of fresh DMEM were plated into Millicell
chambers (8 µm; Millipore Corp., Billerica, MA, USA) with 600 µl of
DMEM containing 10% FBS in the bottom chamber. Cells that migrated
through the filter at 24 h were fixed with paraformaldehyde and
stained with 10% crystal violet. To determine cell invasion, 50 µl
Matrigel (1:8 diluted; BD Biosciences, Franklin Lakes, NJ, USA) was
used to coat the chamber beforehand, and the invaded cells were
harvested at 48 h. The cells were photographed, and the number of
cells that migrated or invaded were counted based on 5 microscopic
×100 fields/insert.
Flow cytometric analysis
To analyze apoptotic cells, trypsinized adherent and
floating cells were harvested and prepared using the FITC/Annexin V
Apoptosis Detection Kit I (BD Pharmingen, San Jose, CA, USA)
according to the protocols recommended by the manufacturer. After
staining, the cells were analyzed using flow cytometry
(FACSCaliber; BD Biosciences).
Western blot analysis
Cellular extracts were acquired using whole cell
lysis buffer containing proteinase inhibitor cocktail. After
subjecting the lysates to SDS-PAGE electrophoresis, proteins were
transferred onto a polyvinylidene difluoride (PVDF) membrane by
electroblotting. The membranes were then blocked in 5% skimmed milk
for 1 h and incubated overnight at 4°C with specific primary
antibodies. Specific antibody-bound protein bands were detected
with fluorescent secondary antibody and visualized using an Odyssey
Infrared Imaging System (BD Biosciences, San Diego, CA, USA)
(17). In the present study, the
rabbit monoclonal antibody against CHIP (1:1,000; Abcam, Cambridge,
MA, USA) and rabbit polyclonal antibody against Myc Tag (1:1,000,
Abbkine, Redlands, CA, USA) were used. The mouse monoclonal against
β-tubulin (1:1,000, Boster, Wuhan, China) served as a loading
control.
Animal experiments
Male BALB/c nude mice (nu/nu, aged 4–5 weeks) were
purchased from Shanghai Laboratory Animal Center (Shanghai, China),
and were housed under specific pathogen-free conditions in the
Experimental Animal Care Center of Shanghai Ninth People's
Hospital. Animal welfare and experimental procedures were conducted
in compliance with the Guide for Care and Use of Laboratory Animals
(The Ministry of Science and Technology of China, 2006) and the
related ethical regulations of the hospital. The Animal Care and
Use Committees of the hospital approved all experimental
procedures.
Briefly, the nude mouse xenograft tumor models were
established by subcutaneous injection of 5×106
cells/site. To evaluate the effects of CHIP knockdown on UMSCC12
tumorigenesis, UMSCC12-scrambled cells were implanted on the right
flank, and UMSCC12-shCHIP cells were implanted on the left flank
(n=6). Tumor volumes (length × width2/2) were
monitored.
Immunohistochemistry staining
Fresh tissues were fixed in 4% formaldehyde,
embedded in paraffin and prepared into 5-µm sections. After
dewaxing, rehydration and antigen retrieval, endogenous peroxidase
activity was quenched. The slides were incubated with primary
antibody overnight at 4°C. Then, the samples were incubated with a
biotinylated secondary antibody for 50 min at 37°C, which was
followed by staining with a DAB kit (GTVision; China). The rabbit
polyclonal antibody against CHIP (1:250; Abcam) and mouse
monoclonal antibody against Ki67 (1:500; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA) were used as primary antibodies.
Scoring for tissue-array staining
A tissue microarray was constructed using primary
HNC samples from 101 patients who received radical surgery, and 10
out-patient biopsy samples for a potential cancer lesion, but
pathologically not (3 oral ulcer, 6 oral leukoplakia, and 1 normal
oral epithelial sample) from the Department of Oral
Maxillofacial-Head and Neck Oncology, Shanghai Ninth People's
Hospital (17). The patients
involved in the present study signed written informed consent, and
the study was approved by the Medical Ethics Committee of the Ninth
People's Hospital, Shanghai Jiaotong University School of
Medicine.
Three samples were excluded due to lack of tissue or
cancer cells in the array. The immunoreactivity score (IRS) for
CHIP immunohistochemistry (IHC) staining was recorded by two
independent observers, who scored based on staining intensity and
percentage of positive cancer cells. The staining intensity was
scored as follows: weak, scored 1; moderate, scored 2; and intense,
scored 3. Regarding the percentage of positive cancer cells, the
score was defined as follows: 0–25%, scored 1; >25–50%, scored
2; >50–75%, scored 3; >75%, scored 4. Finally, an overall
score (ranging from 1–12) was acquired by multiplying the above two
scores for each sample. A total score of 1–6 was considered low
expression; 7–12 was considered high expression.
Statistical analysis
The data were compiled using the software package
SPSS, version 12.0 (SPSS, Inc., Chicago, IL, USA). Chi-squared
tests were used to assess the statistical significance for
correlations between CHIP expression and clinicopathological
variables. Univariate survival analysis was performed using the
Kaplan-Meier method, and differences in survival curves were
assessed by the log-rank test. All experiments were performed in
triplicate; and representative results are displayed. The values
displayed correspond to the means ± SD. Significant differences
between two groups were determined based on Student's t-test and a
p-value <0.05 was considered statistically significant.
Results
Involvement of CHIP expression in the
proliferative and colony forming ability of HNCs
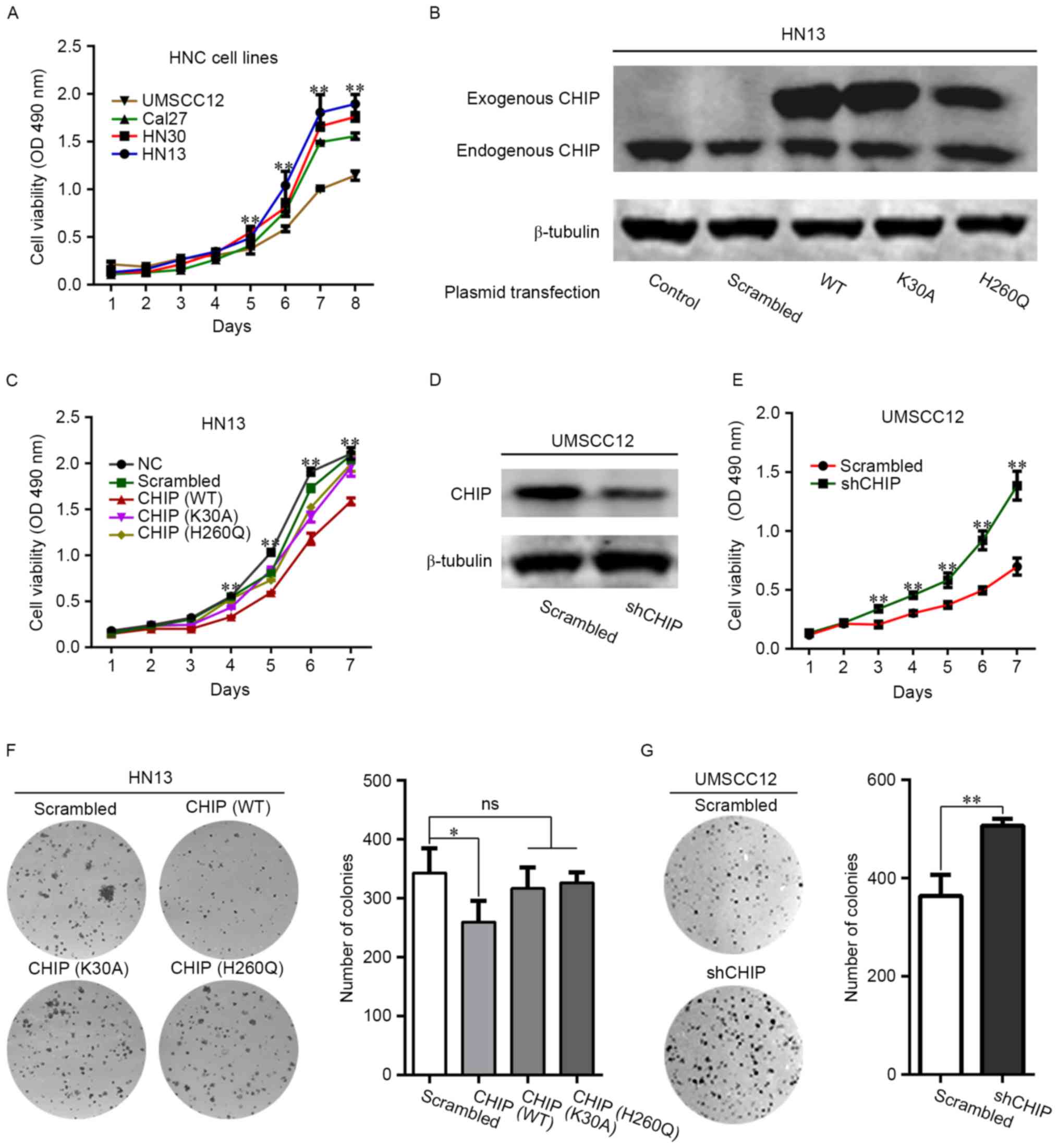
By evaluating the cellular proliferation rate of 4
HNC cell lines, we found that HN13 displayed the greatest
proliferative capacity, whereas UMSCC12 displayed the least
proliferative capacity (Fig. 1A).
Next, we managed to overexpress CHIP (CHIPOE) and its
two loss-of-function mutants in the HN13 cells (Fig. 1B). CHIPOE significantly
suppressed cellular proliferation in HN13 cells, whereas
overexpression of loss-of-function CHIP mutants (K30A and H260Q)
abolished such effects (Fig. 1C).
The expression of CHIP in UMSCC12 cells was suppressed by stable
transfection with CHIP-specific short hairpin RNA lentivirus
(Fig. 1D). Likewise, CHIP knockdown
promoted the proliferation of UMSCC12 cells (Fig. 1E). The results of colony formation
assays revealed that CHIPOE significantly decreased the
colony formation number (Fig. 1F),
which was not observed upon overexpression of the mutant constructs
CHIP (K30A) or CHIP (H260Q). The rate of colony formation was
significantly enhanced in the UMSCC12-shCHIP cells (Fig. 1G).
The biological effects of CHIP on the
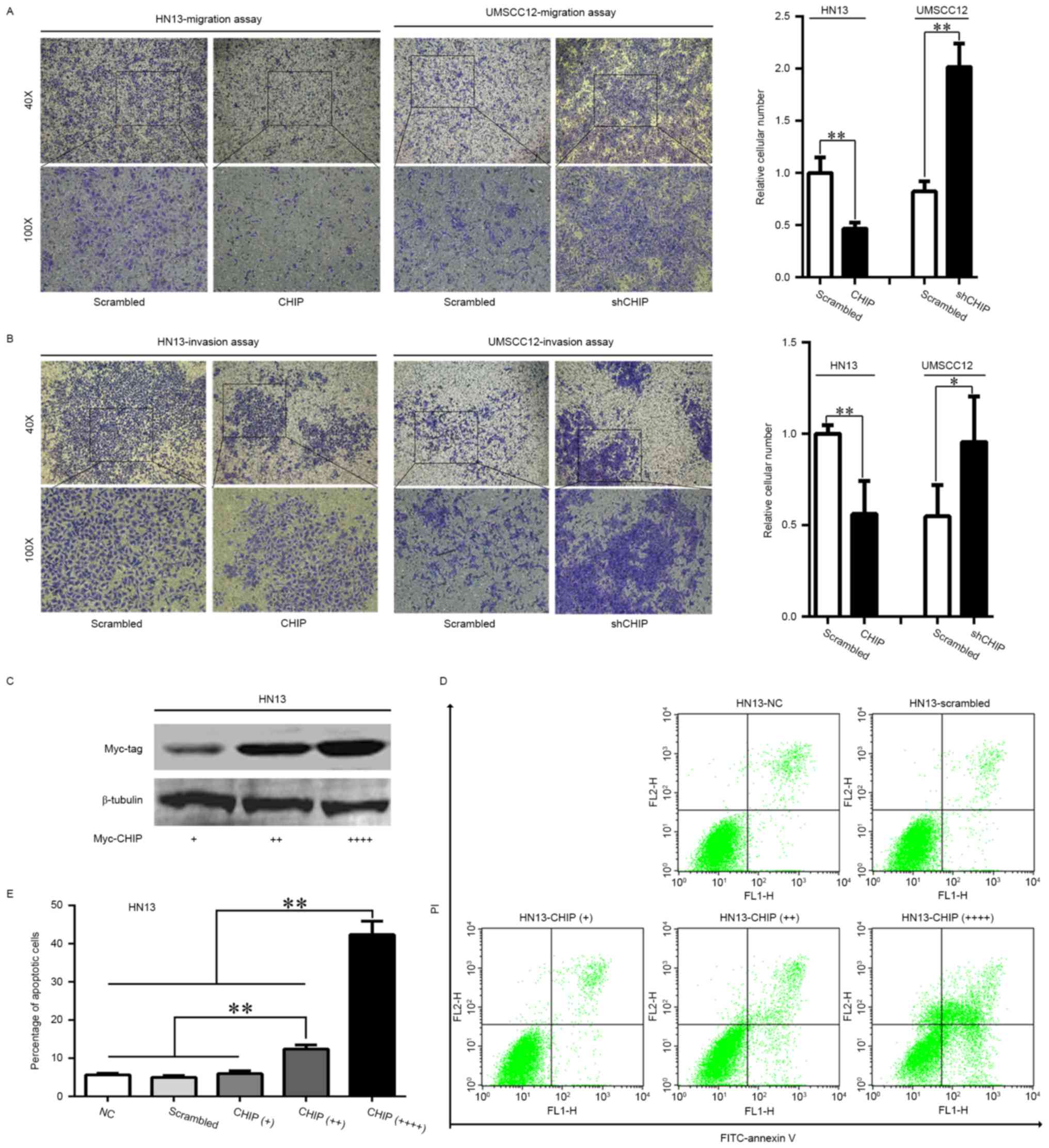
migration and invasion abilities of HNCs
The migration and invasion abilities of the involved
cells were detected by Transwell assays. The results indicated that
CHIPOE caused a dramatic decrease in the migration and
invasion abilities of the HN13 cells, whereas the migration and
invasion abilities of the UMSCC12-shCHIP cells were significantly
increased compared with the UMSCC12-scrambled cells (Fig. 2A and B).
Apoptotic induction of CHIP
overexpression in HNCs
In HN13 cells, we managed to increase the expression
of CHIP to 2- and 4-fold of the regular dose used in the previous
assays (1.5 µg myc-CHIP plasmid in 5×105 cells; Fig. 2C). We observed an increased rate of
cellular apoptosis in a dose-dependent manner for CHIP
overexpression 48 h after transfection (Fig. 2D and E; p<0.01), suggesting that
CHIP overexpression may serve as an apoptotic inducible factor in
HNCs.
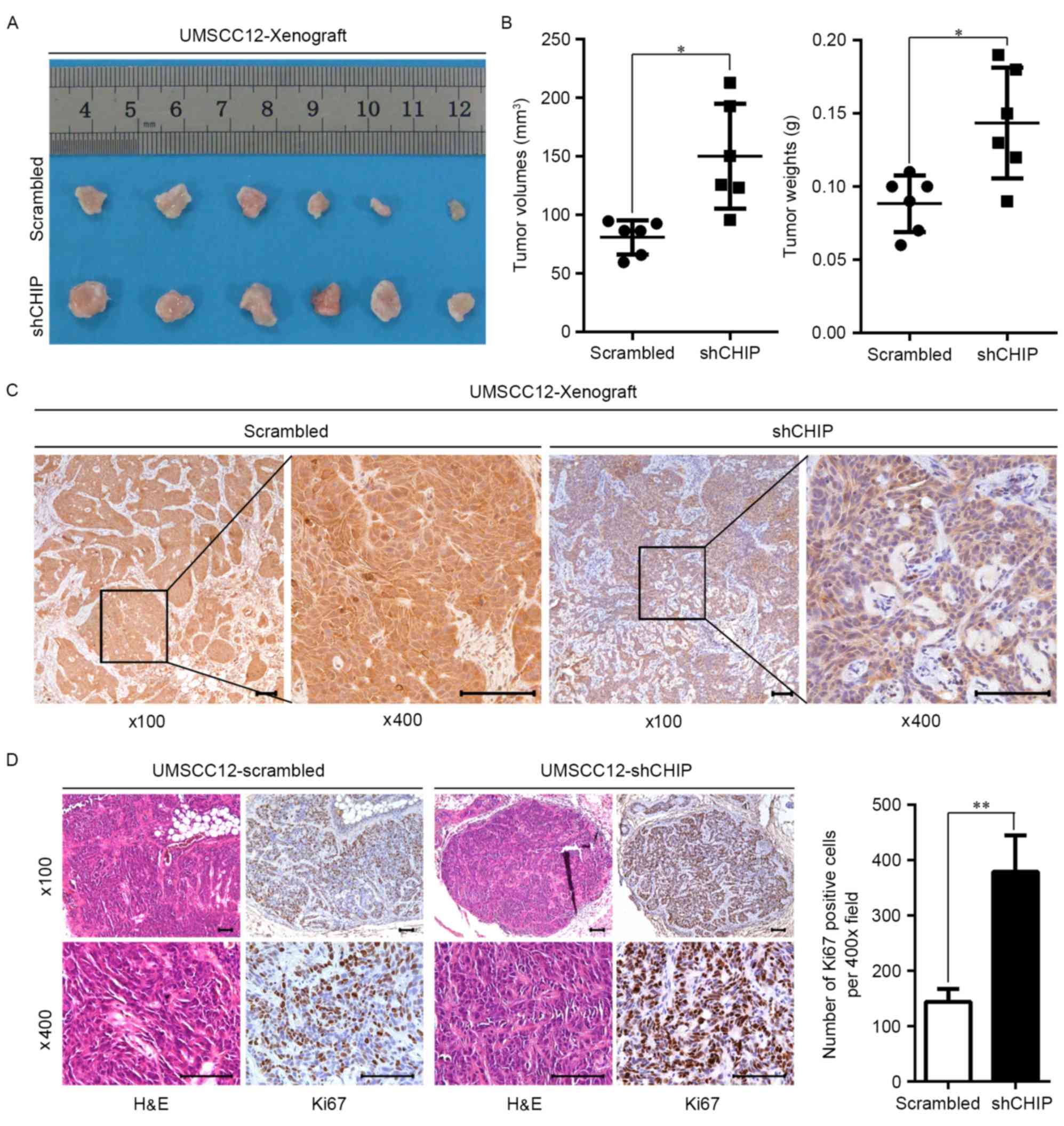
Involvement of CHIP knockdown in
tumorigenesis of HNCs
To investigate the biological effect of CHIP in the
tumorigenesis of HNCs, UMSCC12-scrambled and UMSCC12-shCHIP stable
transfected cells were subcutaneously inoculated in the nude mouse.
Morphologically, xenograft tumors derived from the UMSCC12-shCHIP
cells were larger than the xenograft tumors from the
UMSCC12-scrambled cells (Fig. 3A).
Quantitatively, UMSCC12-shCHIP cells formed xenografts with
significantly larger tumor volume (p=0.011) and greater tumor mass
(Fig. 3B; p=0.010). In addition,
the subsequent IHC staining further validated the suppressed
expression of CHIP in the UMSCC12-shCHIP xenograft specimens
(Fig. 3C). Moreover, we observed
significantly increased Ki67-positive cells, a marker for
proliferation, in the UMSCC12-shCHIP xenografts compared with this
number in the UMSCC12-scrambled xenografts (Fig. 3D). These results suggest that
expression of CHIP negatively affects the malignant characteristics
of HNC cell lines.
Suppression of CHIP expression acts as
a risk factor for HNC patients
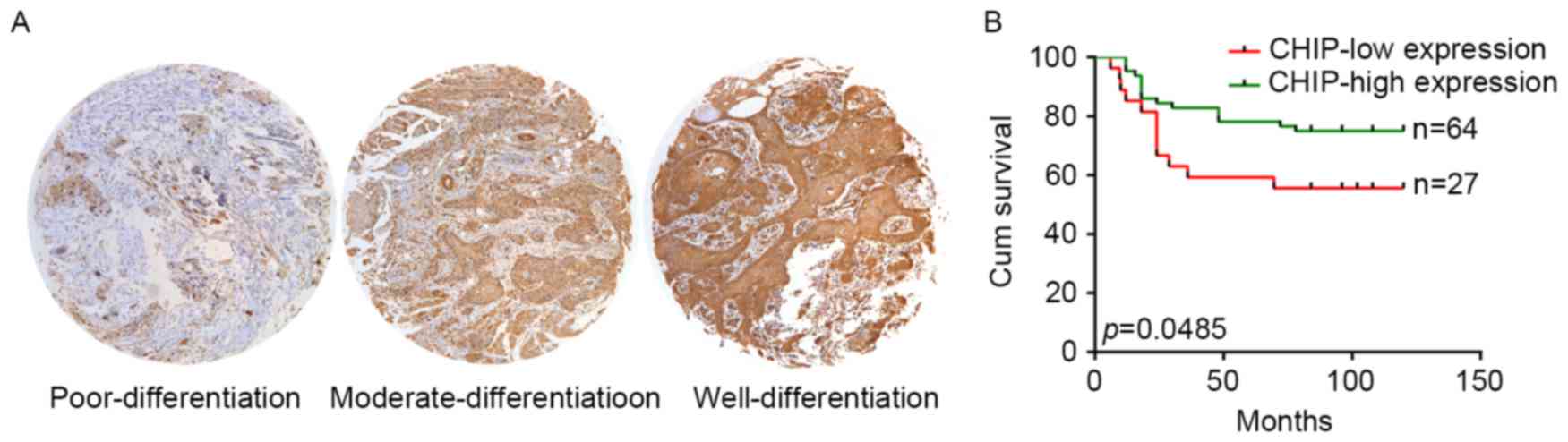
In order to illustrate the clinical significance of
variable CHIP expression in HNCs, a retrospective cohort was
included and analyzed with matched tissue array. The IHC analysis
of clinical samples revealed that high CHIP expression occurred in
70 of 98 HNC samples. In the included cohort, the CHIP expression
level was significantly associated with the differentiation status
of cancer cells, whereas no significance was observed for the other
parameters (Table I). As shown in
Fig. 4A, we observed a changing
expression pattern of CHIP from poor, moderate, to well
differentiation pathological status in the HNC specimens. Since 7
samples lacked follow-up data, the effect of CHIP expression on the
overall survival (OS) was analyzed in 91 cases. Patients with HNC
cancers that expressed lower CHIP showed poorer OS (p=0.0485;
Fig. 4B).
 | Table I.Demographic characteristics of the
patient population according to CHIP expression. |
Table I.
Demographic characteristics of the
patient population according to CHIP expression.
| Variable | N | CHIP expression
(IRS=1-6) n (%) | CHIP expression
(IRS=7-12) n (%) | P-value |
|---|
| All cases | 98 | 28 | 70 |
|
| Age, years |
|
|
| 0.602 |
|
<60 | 59 | 18 (64.3) | 41 (58.6) |
|
|
≥60 | 39 | 10 (35.7) | 29 (41.4) |
|
| Sex |
|
|
| 0.337 |
|
Male | 52 | 17 (60.7) | 35 (50) |
|
|
Female | 46 | 11 (39.3) | 35 (50) |
|
| Smoking |
|
|
| 0.752 |
|
Yes | 32 | 10 (41.7) | 22 (37.9) |
|
| No | 50 | 14 (58.3) | 36 (62.1) |
|
| Alcohol |
|
|
| 0.418 |
|
Yes | 22 | 8 (33.3) | 14 (24.6) |
|
| No | 59 | 16 (66.7) | 43 (75.4) |
|
| Histological grade
(differentiation) |
|
|
| 0.000 |
|
Well/moderate | 87 | 18 (64.3) | 69 (98.6) |
|
|
Poor | 11 | 10 (35.7) | 1 (1.4) |
|
| Tumor size |
|
|
| 0.808 |
|
T1+T2 | 59 | 17 (65.4) | 42 (62.7) |
|
|
T3+T4 | 34 | 9 (34.6) | 25 (37.3) |
|
| Nodal status |
|
|
| 0.373 |
| N0 | 67 | 17 (65.4) | 50 (74.6) |
|
|
N1+ | 26 | 9 (34.6) | 17 (25.4) |
|
| Metastasis |
|
|
| 1.000 |
| M0 | 92 | 26 (100) | 66 (98.5) |
|
| M1 | 1 | 0 (0) | 1 (1.5) |
|
| Local
recurrence |
|
|
| 1.000 |
|
Yes | 5 | 1 (3.7) | 4 (5.8) |
|
| No | 91 | 26 (96.3) | 65 (94.2) |
|
Discussion
In the present study, we illustrated the inhibition
of cancer cell growth by the E3 ligase CHIP in HNCs with a series
of in vitro and in vivo assays. In HNC clinical
samples, CHIP expression was shown to be significantly related to
the differentiation status of the HNCs, and low expression of CHIP
indicated poorer OS in HNC patients.
Increasing evidence strongly suggests that
ubiquitin-dependent proteolysis, mediated by the E3 ligase CHIP,
plays an essential role in regulating various biological processes
during carcinogenesis. Previously, we reviewed the reported studies
on the biological effects of CHIP on cancers by searching PubMed
database up to December, 2016. In summary, both oncogene and tumor
suppressor functions of CHIP have been reported in variant cancers
by regulating underlying targets (2,7,8,19).
In addition, the E3 ligase CHIP has been illustrated to exert its
biological functions in cancer by targeting more than 30 types of
proteins (data not shown). In breast cancer, the oncogene effects
of CHIP were reported by regulating PTEN and Pfn1 (15,20,21).
However, the tumor suppressor effects of CHIP were reported by
targeting TRAF2, ErbB2, ERα, MIF, PTK6, SRC-3, CtBP2 (7,22–29).
However, in prostate cancer, CHIP was reported as an oncogene by
targeting PTEN and Mst1, and as a tumor suppressor by targeting AR
(16,19,30).
The production of mutated proteins or abnormally expressed proteins
seems to be unavoidable during carcinogenesis. Undoubtedly, the
biological functions of CHIP in each cancer type may not only
target just one specific protein. Thus, it seems to be more
meaningful to investigate the biological effect of E3 ligase CHIP
on certain behaviors of cancer cells. In our previous studies, we
illustrated that CHIP regulated the cancer stem-like properties of
HNCs by targeting CD166 protein (31).
Previous studies have reported that CHIP functions
as a tumor-suppressor in pancreatic, colorectal and gastric cancer
(12–14,23).
In pancreatic cancer, CHIP was reported to impair cell
proliferation, migration and invasion by targeting EGFR (12). In colorectal cancer, CHIP impaired
tumor growth, migration and invasion by repressing NF-κB-mediated
signaling (13). In breast cancer
cells, anti-apoptotic protein Bcl-2 was downregulated by CHIP
(32). Furthermore, CHIP was
reported to modulate mitotic arrest by degradation of the androgen
receptor and c-Myc (30,33). In the present study, we identified
the function of CHIP as a candidate tumor suppressor using a series
of in vitro and in vivo assays. We found that altered
CHIP expression regulated the cellular proliferation,
migration/invasion abilities and tumor growth in HNCs. However,
overexpression of CHIP induced increased apoptotic levels in HNC
cells. In HNC samples, low expression of CHIP indicated a poor
differentiation status and a higher risk of reduced OS.
Accordingly, we conclude that suppressed expression of CHIP
participates in the progression of HNCs. Ηerein, the underlying
targets of CHIP were not further investigated and further studies
are warranted to investigate the underlying mechanisms.
CHIP is a cochaperone E3 ligase containing 3 TPR
motifs and a U-box domain (34). In
the present study, we managed to construct two loss-of-function
mutants of CHIP at the TPR motifs and U-box domain, respectively.
Expectedly, these two mutants abolished the suppressive function of
wild-type CHIP protein in regards to the proliferative and colony
forming abilities of HNCs, indicating that the biological effects
of CHIP in HNCs were TPR motif- and U-box domain-dependent.
Functionally, CHIP has been identified as being associated with
Hsc70 and Hsp90 to achieve the subsequent ubiquitination of
targeting protein via the U-box domain (35). Accordingly, the inhibition of Hsp90
by 17-AAG or 17-DMAG was found to increase the expression levels of
Hsp70 and Hsp90, resulting in enhanced CHIP-mediated ubiquitination
and the following proteasomal degradation (10,36,37).
Hsp90-targeted therapy has been advanced and well documented and
has been proposed as a prospective treatment method in various
types of cancers (38,39). Above all, to enhance the tumor
suppressor functions of CHIP in HNCs using an Hsp90 inhibitor may
be a new treatment strategy for HNC patients.
In conclusion, the present study demonstrated that
CHIP functions as a candidate tumor suppressor in HNCs. Meanwhile,
we demonstrated that suppression of the expression of CHIP may
participate in the development and progression of HNCs. Thus,
identifying a strategy by which to increase the expression or to
enhance the biological function of CHIP may benefit HNC patients in
clinical practice.
Acknowledgements
We thank Professor Zeguang Han, Professor Li Mao and
Wenyi Wei for providing insightful comments for the present study.
We also thank members of the Chenping Zhang's Department of Oral
and Maxillofacial Head and Neck Oncology for providing the clinical
information of the involved HNC specimens in the present study. The
present study was supported by the National Program on Key Research
Project of China (2016YFC0902700), the National Natural Science
Foundation of China (81272978 and 91229103), the Project of the
Shanghai Science and Technology Committee (15DZ22992200) and the
Innovation Fund for Doctoral Program of Shanghai Jiaotong
University, School of Medicine (BXJ201628).
References
|
1
|
Kumar P, Pradhan K, Karunya R, Ambasta RK
and Querfurth HW: Cross-functional E3 ligases Parkin and C-terminus
Hsp70-interacting protein in neurodegenerative disorders. J
Neurochem. 120:350–370. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yan S, Sun X, Xiang B, Cang H, Kang X,
Chen Y, Li H, Shi G, Yeh ET, Wang B, et al: Redox regulation of the
stability of the SUMO protease SENP3 via interactions with CHIP and
Hsp90. EMBO J. 29:3773–3786. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seo J, Lee EW, Sung H, Seong D,
Dondelinger Y, Shin J, Jeong M, Lee HK, Kim JH, Han SY, et al: CHIP
controls necroptosis through ubiquitylation- and lysosome-dependent
degradation of RIPK3. Nat Cell Biol. 18:291–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paul I and Ghosh MK: The E3 ligase CHIP:
Insights into its structure and regulation. Biomed Res Int.
2014:9181832014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Edkins AL: CHIP: A co-chaperone for
degradation by the proteasome. Subcell Biochem. 78:219–242. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei Q, Sha Y, Bhattacharya A, Abdel Fattah
E, Bonilla D, Jyothula SS, Pandit L, Hershey GK Khurana and Eissa
NT: Regulation of IL-4 receptor signaling by STUB1 in lung
inflammation. Am J Respir Crit Care Med. 189:16–29. 2014.PubMed/NCBI
|
|
7
|
Kajiro M, Hirota R, Nakajima Y, Kawanowa
K, So-ma K, Ito I, Yamaguchi Y, Ohie SH, Kobayashi Y, Seino Y, et
al: The ubiquitin ligase CHIP acts as an upstream regulator of
oncogenic pathways. Nat Cell Biol. 11:312–319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gaude H, Aznar N, Delay A, Bres A,
Buchet-Poyau K, Caillat C, Vigouroux A, Rogon C, Woods A, Vanacker
JM, et al: Molecular chaperone complexes with antagonizing
activities regulate stability and activity of the tumor suppressor
LKB1. Oncogene. 31:1582–1591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hatakeyama S, Watanabe M, Fujii Y and
Nakayama KI: Targeted destruction of c-Myc by an engineered
ubiquitin ligase suppresses cell transformation and tumor
formation. Cancer Res. 65:7874–7879. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruckova E, Muller P, Nenutil R and
Vojtesek B: Alterations of the Hsp70/Hsp90 chaperone and the
HOP/CHIP co-chaperone system in cancer. Cell Mol Biol Lett.
17:446–458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: Diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang T, Yang J, Xu J, Li J, Cao Z, Zhou L,
You L, Shu H, Lu Z, Li H, et al: CHIP is a novel tumor suppressor
in pancreatic cancer through targeting EGFR. Oncotarget.
5:1969–1986. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Ren F, Wang Y, Feng Y, Wang D, Jia
B, Qiu Y, Wang S, Yu J, Sung JJ, et al: CHIP/Stub1 functions as a
tumor suppressor and represses NF-κB-mediated signaling in
colorectal cancer. Carcinogenesis. 35:983–991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang S, Wu X, Zhang J, Chen Y, Xu J, Xia
X, He S, Qiang F, Li A, Shu Y, et al: CHIP functions as a novel
suppressor of tumour angiogenesis with prognostic significance in
human gastric cancer. Gut. 62:496–508. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ying Z, Haiyan G and Haidong G: BAG5
regulates PTEN stability in MCF-7 cell line. BMB Rep. 46:490–494.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahmed SF, Deb S, Paul I, Chatterjee A,
Mandal T, Chatterjee U and Ghosh MK: The chaperone-assisted E3
ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN
for proteasomal degradation. J Biol Chem. 287:15996–16006. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan M, Yang X, Wang L, Clark D, Zuo H, Ye
D, Chen W and Zhang P: Plasma membrane proteomics of tumor spheres
identify CD166 as a novel marker for cancer stem-like cells in head
and neck squamous cell carcinoma. Mol Cell Proteomics.
12:3271–3284. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv Z, Wu X, Cao W, Shen Z, Wang L, Xie F,
Zhang J, Ji T, Yan M and Chen W: Parathyroid hormone-related
protein serves as a prognostic indicator in oral squamous cell
carcinoma. J Exp Clin Cancer Res. 33:1002014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ren A, Yan G, You B and Sun J:
Down-regulation of mammalian sterile 20-like kinase 1 by heat shock
protein 70 mediates cisplatin resistance in prostate cancer cells.
Cancer Res. 68:2266–2274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lv Y, Song S, Zhang K, Gao H and Ma R:
CHIP regulates AKT/FoxO/Bim signaling in MCF7 and MCF10A cells.
PLoS One. 8:e833122013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Choi YN, Lee SK, Seo TW, Lee JS and Yoo
SJ: C-Terminus of Hsc70-interacting protein regulates profilin1 and
breast cancer cell migration. Biochem Biophys Res Commun.
446:1060–1066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jang KW, Lee KH, Kim SH, Jin T, Choi EY,
Jeon HJ, Kim E, Han YS and Chung JH: Ubiquitin ligase CHIP induces
TRAF2 proteasomal degradation and NF-κB inactivation to regulate
breast cancer cell invasion. J Cell Biochem. 112:3612–3620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jan CI, Yu CC, Hung MC, Harn HJ, Nieh S,
Lee HS, Lou MA, Wu YC, Chen CY, Huang CY, et al: Tid1, CHIP and
ErbB2 interactions and their prognostic implications for breast
cancer patients. J Pathol. 225:424–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yi X, Wei W, Wang SY, Du ZY, Xu YJ and Yu
XD: Histone deacetylase inhibitor SAHA induces ERalpha degradation
in breast cancer MCF-7 cells by CHIP-mediated ubiquitin pathway and
inhibits survival signaling. Biochem Pharmacol. 75:1697–1705. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raja SM, Clubb RJ, Bhattacharyya M, Dimri
M, Cheng H, Pan W, Ortega-Cava C, Lakku-Reddi A, Naramura M, Band
V, et al: A combination of Trastuzumab and 17-AAG induces enhanced
ubiquitinylation and lysosomal pathway-dependent ErbB2 degradation
and cytotoxicity in ErbB2-overexpressing breast cancer cells.
Cancer Biol Ther. 7:1630–1640. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang SA, Cho HS, Yoon JB, Chung IK and Lee
ST: Hsp90 rescues PTK6 from proteasomal degradation in breast
cancer cells. Biochem J. 447:313–320. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jeong JH, An JY, Kwon YT, Li LY and Lee
YJ: Quercetin-induced ubiquitination and down-regulation of
Her-2/neu. J Cell Biochem. 105:585–595. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee JS and Yoo SJ: C-terminus of
Hsc70-interacting protein regulates C-terminal binding protein 2
and the expression of its target genes. Biochem Biophys Res Commun.
432:418–424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schulz R, Marchenko ND, Holembowski L,
Fingerle-Rowson G, Pesic M, Zender L, Dobbelstein M and Moll UM:
Inhibiting the HSP90 chaperone destabilizes macrophage migration
inhibitory factor and thereby inhibits breast tumor progression. J
Exp Med. 209:275–289. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sarkar S, Brautigan DL, Parsons SJ and
Larner JM: Androgen receptor degradation by the E3 ligase CHIP
modulates mitotic arrest in prostate cancer cells. Oncogene.
33:26–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao M, Yan M, Zhang J, Xu Q, Qi S, Wang X
and Chen W: Cancer stem-like cell related protein CD166 degrades
through E3 ubiquitin ligase CHIP in head and neck cancer. Exp Cell
Res. 353:46–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsuchiya M, Nakajima Y, Waku T, Hiyoshi H,
Morishita T, Furumai R, Hayashi Y, Kishimoto H, Kimura K and
Yanagisawa J: CHIP buffers heterogeneous Bcl-2 expression levels to
prevent augmentation of anticancer drug-resistant cell population.
Oncogene. 34:4656–4663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paul I, Ahmed SF, Bhowmik A, Deb S and
Ghosh MK: The ubiquitin ligase CHIP regulates c-Myc stability and
transcriptional activity. Oncogene. 32:1284–1295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
McDonough H and Patterson C: CHIP: A link
between the chaperone and proteasome systems. Cell Stress
Chaperones. 8:303–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murata S, Chiba T and Tanaka K: CHIP: A
quality-control E3 ligase collaborating with molecular chaperones.
Int J Biochem Cell Biol. 35:572–578. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tsuchiya M, Nakajima Y, Hirata N,
Morishita T, Kishimoto H, Kanda Y and Kimura K: Ubiquitin ligase
CHIP suppresses cancer stem cell properties in a population of
breast cancer cells. Biochem Biophys Res Commun. 452:928–932. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Muller P, Ruckova E, Halada P, Coates PJ,
Hrstka R, Lane DP and Vojtesek B: C-terminal phosphorylation of
Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP
and HOP to determine cellular protein folding/degradation balances.
Oncogene. 32:3101–3110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ghadban T, Jessen A, Reeh M, Dibbern JL,
Mahner S, Mueller V, Wellner UF, Güngör C, Izbicki JR and Vashist
YK: In vitro study comparing the efficacy of the
water-soluble HSP90 inhibitors, 17-AEPGA and 17-DMAG, with that of
the nonwater-soluble HSP90 inhibitor, 17-AAG, in breast cancer cell
lines. Int J Mol Med. 38:1296–1302. 2016.PubMed/NCBI
|
|
39
|
Haque A, Alam Q, Alam MZ, Azhar EI, Sait
KH, Anfinan N, Mushtaq G, Kamal MA and Rasool M: Current
understanding of HSP90 as a novel therapeutic target: An emerging
approach for the treatment of cancer. Curr Pharm Des. 22:2947–2959.
2016. View Article : Google Scholar : PubMed/NCBI
|