Introduction
Colorectal cancer (CRC) is a leading cause of
cancer-related deaths worldwide (1). It is the second and third most
commonly diagnosed cancer in females and males, respectively, and
more than 1.2 million patients are diagnosed with CRC every year
(2,3). Currently, oxaliplatin-based
chemotherapy after surgical resection is one of the most widely
used therapeutic strategies (4).
However, a large proportion of patients receiving chemotherapy
finally become metastatic and chemoresistant, and this has been a
key barrier to the efficacy of CRC treatment (5). Hence, finding new therapeutic markers
is warranted to evade drug resistance mechanisms, and possibly find
a cure.
In recent years, a tremendous amount of effort has
been devoted to understanding the mechanisms of chemoresistance and
to elaborate the genes/pathways involved. Chemoresistance
mechanisms are extraordinarily complex, including inefficient
cellular drug uptake and accumulation (6), enhancement of DNA repair (7) and activation of the antioxidant
glutathione system for detoxification (8,9). One
major resistance mechanism utilized by tumor cells is to resist
drug-induced cell death through disruptions in apoptotic pathways
(10). Thus, it is also essential
to better understand the pathways related to chemoresistance in CRC
and discover novel strategies to further improve the effectiveness
of oxaliplatin.
Long non-coding RNAs (lncRNAs) are defined as
transcripts >200 nucleotides in length and are transcribed, but
are non-translated non-coding RNAs in the human genome (11). Recent studies have demonstrated that
lncRNAs play important roles in carcinogenesis and cancer
metastasis, and some lncRNAs function as oncogenes,
tumor-suppressor genes or both, depending on the circumstance
(12). However, aberrant lncRNA
expression has been detected and identified as a promising
biomarker for diagnosis and prognosis in breast and gastric cancer,
and CRC (13–15). The discovery and study of lncRNAs
are thus of major relevance to human biology and disease, as they
represent an extensive, largely unexplored and functional component
of the genome (16,17). To date, lncRNA expression has not
been extensively analyzed in CRC samples except in a few studies
that have used microarrays from TCGA (18), and the genome-wide screening of
relevant lncRNAs are essential for improving the prognosis of
cancer patients. Moreover, the association of the expression of
specific lncRNAs with drug resistance in CRC cells is not well
known.
In the present study, we conducted high-throughput
HiSeq sequencing followed by reverse transcription quantitative
real-time PCR (RT-qPCR) assays to test the hypothesis that specific
lncRNAs can be useful in predicting chemotherapeutic response with
the hope that such findings may guide the therapeutic choice. Our
data showed that MEG3 expression was significantly downregulated in
primary tissues and serum samples from CRC patients showing
resistance to oxaliplatin-based treatment. Moreover, the subsequent
functional assay revealed that MEG3 reverses oxaliplatin resistance
by promoting oxaliplatin-induced cell apoptosis.
Materials and methods
Patients and samples
A multiphase, case-control study was designed to
identify lncRNAs as potential biomarkers for differentiating the
chemoresponse to oxaliplatin therapy in CRC patients. Tumor
response status was evaluated according to the Response Evaluation
Criteria in Solid Tumors (RECIST) criteria and was assigned to
patients with complete or partial response (CR and PR,
respectively), and stable or progressive disease (SD and PD,
respectively) in tumor measurements confirmed by repeat studies
performed no less than 4 weeks after the criteria for response was
first met. Briefly, 316 patients diagnosed with CRC, but without
other diseases were recruited at Zhongnan Hospital of Wuhan
University between January 2009 and February 2013. All participants
were allocated to 3 phases. In the initial screening phase, tissue
samples pooled from 8 patients showing response and 8 patients
showing no response were subjected to HiSeq sequencing, to identify
lncRNAs that were significantly differentially expressed. In the
training phase, the candidate lncRNAs were tested with RT-qPCR in
an independent cohort of primary tissues from 80 CRC patients
responding to oxaliplatin treatment and 80 patients showing no
response to treatment. In the validation phase, another independent
group of 140 CRC patients who provided serum samples were enrolled.
Among these patients, there were 70 patients who showed response to
oxaliplatin treatment while the other 70 patients showed no
response.
All the patients were pathologically confirmed as
presenting with CRC and the clinical tissue samples were collected
before chemotherapy was started. They were classified according to
the tumor-node-metastasis (TNM) classification. Overall survival
(OS) was updated on February 1, 2012 and was defined as the time
from inclusion to death for any reason. Recurrence-free survival
(RFS) was defined as the time from inclusion to recurrence or
metastatic progression.
Sample preparation
Fresh tumor tissues were immediately cut from the
resected CRC tissues and kept at −80°C until RNA extraction. Venous
blood was collected and centrifuged at 4,000 rpm for 10 min, within
2 h. The supernatant fluids were then collected and further
centrifuged at 12,000 rpm for 15 min to completely remove the cell
debris. The whole process was strictly controlled to avoid
hemolysis, and the supernatant serum was stored at −80°C, until
analysis. Written informed consent was obtained from all patients
according to local ethical regulations of the Ethics Committee of
Zhongnan Hospital of Wuhan University.
HiSeq sequencing
Total tissue RNA was extracted by one-step
extraction using a TRIzol reagent kit (Life Technologies, Carlsbad,
CA, USA), and the purity and quantity of RNA were determined using
UV spectrophotometry. cDNA library construction and sequencing were
performed according to previously described methods (19). Briefly, after extraction of total
RNA, ribosomal RNA was separated to isolate as much ncRNA as
possible. RNA containing poly(A) was then removed. RNA fragments
were broken into short fragments randomly. The first chain of cDNA
was generated using RNA fragments as templates and 6-bp random
primers. Second chain of the cDNA was synthesized according to the
kit's instructions (Takara Bio Company, Dalian, China). After
purification, end repair, base A and sequencing joint adding, the
generated cDNA was fragmented using uracil-N-glycosylase (UNG).
cDNA fragments were chosen according to size, then PCR
amplification was performed to establish the complete sequencing
cDNA library. lncRNAs were sequenced using the high-throughput,
high-sensitivity HiSeq 2500 sequencing platform (Illumina Inc., San
Diego, CA, USA). Sequencing results were analyzed and treated using
Trim Galore software to dynamically remove joint sequence fragments
and low-quality segments from the 3′ end. FastQC software was used
for quality control of the pretreated data.
Cell culture
Human CRC cell lines HT29 and SW480 were obtained
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China) in 2014. All CRC cell lines were maintained in
RPMI-1640 medium (Thermo Fisher Scientific, Wilmington, DE, USA)
containing 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis,
MO, USA), 100 U/ml penicillin and 100 g/ml streptomycin (Life
Technologies, Grand Island, NY, USA) at 37°C in 5% CO2
and 95% air.
RNA extraction
Total RNA was isolated from primary CRC tissues and
cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Serum
RNA was isolated using acid phenol according to the manufacturer's
instructions. The extracted total RNA was eluted in 20 µl
nuclease-free water and the RNA concentration was measured by
NanoDrop 2000 (Thermo Fisher Scientific). The samples with
A260/A280 nm ratios between 1.8 and 2.0 were used for further
experiments.
Quantitative real-time PCR
(RT-qPCR)
For primary CRC tissues and cell lines, the cDNA was
synthesized from 200 ng extracted total RNA using the PrimeScript
RT reagent kit (Takara Bio Company) and amplified by RT-qPCR using
LightCycler 480 SYBR-Green I Master (Roche, Mannheim, Germany) and
GAPDH was used as the control gene. For serum cell-free MEG3
detection, we used a previously established RT-qPCR-D method
without RNA extraction (20).
Briefly, the 2X preparation buffer was prepared, which contained
2.5% Tween-20 (EMD Chemicals, Gibbstown, NJ, USA), 50 mmol/l Tris,
and 1 mmol/l EDTA (both from Sigma-Aldrich). First, 5 µl of serum
was mixed with an equal volume of 2X preparation buffer.
Subsequently, the above mixture was reverse transcribed (RT) in
triplicates in a 20 µl reaction volume. Finally, the RT product was
10-fold diluted and centrifuged at 16,000 × g for 5 min, and 5 µl
supernatant solution was used as a cDNA template for qPCR. The
reagents and reaction conditions were the same as those for
RT-qPCR. The 2−ΔΔCt method was used to determine the
relative quantification of gene expression levels. The primer
sequences were as follows: MEG3 forward, 5′-CTGCCCATCTACACCTCACG-3′
and reverse, 5′-CTCTCCGCCGTCTGCGCTAGGGGCT-3′; GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, ATGGTGGTGAAGACGCCAGT.
Development of oxaliplatin-resistant
(OxR) cell lines
Oxaliplatin (Sanofi-Synthelabo, Hangzhou, China) was
purchased from the pharmacy at Zhongnan Hospital of Wuhan
University. HT29 and SW480 cells were exposed to an initial
oxaliplatin concentration of 0.1 µmol/l in RPMI-1640 medium plus
10% FBS. The surviving population of cells was grown to 80%
confluency and passaged twice over 9 days to ensure viability. The
concentration of oxaliplatin that the surviving population was
exposed to was then sequentially increased in the same manner to
0.5 µmol/l (15 days), 1.0 µmol/l (30 days), and finally to the
clinically relevant plasma concentration of 2 µmol/l. For all in
vitro studies, OxR cells were used at no higher than 15
passages from creation and were maintained and exposed to 2 µmol/l
oxaliplatin unless otherwise indicated.
Cell transfection
The MEG3 overexpression plasmid (pMEG3) and control
vector (pVector) were purchased from Addgene (Cambridge, MA, USA).
CRC cells were plated in 24-well plates at 1×105/well.
Forty-eight hours after plating, 100 nM of si-MEG3 or pMEG3 as well
as the negative controls were transfected into the cells with
Lipofectamine 2000 (Invitrogen) according to the manufacturer's
instructions.
Cell viability assay
Cell viability was quantified using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT)
(Sigma-Aldrich) assay. Briefly, 100 µl of cells from the different
groups was seeded onto a 96-well plate at a concentration of 5,000
cells/well and were incubated at 37°C. At different time point, the
optical density was measured at 450 nm using a microtiter plate
reader, and the rate of cell survival was expressed as the
absorbance. The concentration-dependent curves were generated based
on the cell viability after the cells were cultured for 72 h at
different concentrations of oxaliplatin. The results represent the
mean of 3 replicates under the same conditions.
Cell apoptosis assay
Cells (1×105/well) were collected 48 h
after transfection and were stained with Annexin V-FITC and
propidium iodide (PI) according to the manufacturer's instructions
(BD Biosciences, Erembodegem, Belgium). Apoptosis was assessed
using flow cytometry (BD FACSCalibur; BD Biosciences).
Statistical analysis
Statistical analyses were performed using SPSS 19.0
software (SPSS, Inc., Chicago, IL, USA). Kolmogorov-Smirnov test
was used to determine the normality of the distribution of data in
each group. Data are presented as median (interquartile range).
Difference in lncRNA levels among multiple groups in HiSeq
sequencing was determined using Bonferroni adjustment method.
Mann-Whitney U test or Kruskal-Wallis test was employed to compare
differences of lncRNAs among clinical cohort groups. A log-rank
test was used to analyze the statistical differences in survival as
deduced from Kaplan-Meier curves. Cox proportional-hazard
regression analysis was performed to calculate hazard ratio (HR)
and 95% confidence interval (CI) for each covariable. All
differences were regarded as statistical significant when
P<0.05.
Results
Identification of candidate lncRNAs by
high-throughput HiSeq sequencing
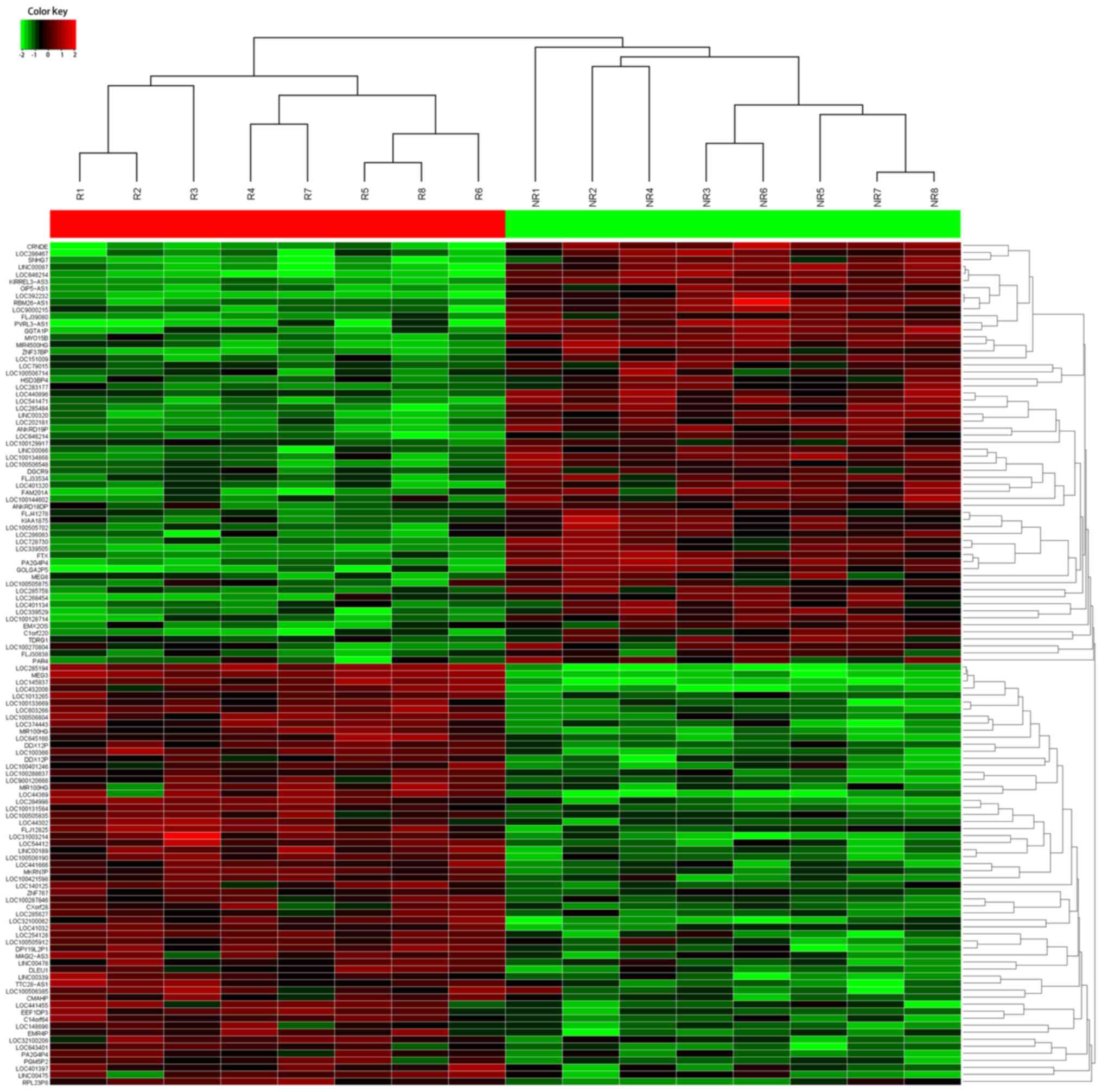
The HiSeq sequencing with 8 tissue samples pooled
from CRC patients showing response and 6 from patients showing no
response to oxaliplatin treatment was conducted. In total, 735
lncRNAs were identified with significant differential expression
(fold-change, ≥2.0). To identify the lncRNAs that are potential
biomarkers, we concentrated on the top 60 most upregulated and 60
downregulated lncRNAs that were differentially expressed between
CRC patients showing response or no response (Fig. 1). Starting from those lncRNAs with
the greatest fold-change, we filtered appropriate candidate lncRNAs
in descending order. Candidates should be plausible for primer
designing, and only those having steady expression in tissue
samples were selected. Finally, we chose 3 candidate lncRNAs from
the upregulated group and 3 from the downregulated group as well
(Table I). Another 4 lncRNAs were
also tested by RT-qPCR since they were peviously shown to be
dysregulated in CRC chemoresistance (21–24).
Thus, 10 lncRNAs were selected as candidates for further testing
via RT-qPCR.
 | Table I.Candidate lncRNAs selected on a basis
of the HiSeq analysis. |
Table I.
Candidate lncRNAs selected on a basis
of the HiSeq analysis.
| Seqname | Location | Regulation (NR vs.
R) | Fold-change | P-value |
|---|
| LOC286467 | ChrXq26.2 | Up | 73.6341872 | 0.00006546 |
| CRNDE | Chr16q12.2 | Up | 42.6729041 | 0.00109347 |
| SNHG7 | Chr9q34.3 | Up | 27.8730264 | 0.00960371 |
| LOC145837 | Chr15q23 | Down | 48.8710538 | 0.00054087 |
| MEG3 | Chr14q32.2 | Down | 46.4837692 | 0.00079283 |
| LOC285194 | Chr3q13.13 | Down | 22.7845924 | 0.01947853 |
MEG3 is downregulated in CRC patients
showing response to oxaliplatin treatment by RT-qPCR
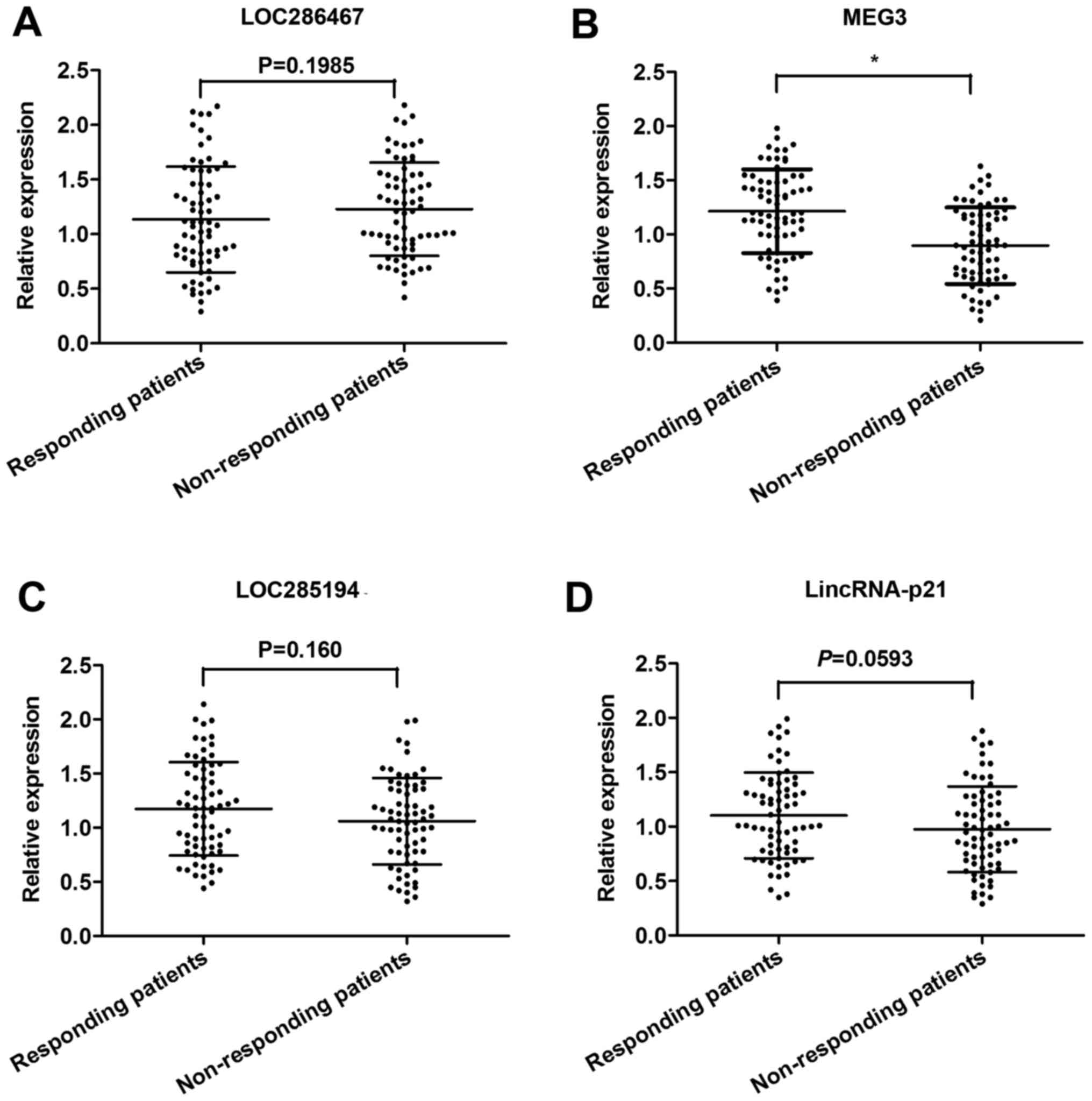
The expression of all 10 candidate lncRNAs was
evaluated by RT-qPCR, using 80 CRC tissues from patients showing
response to oxaliplatin and 80 from patients showing no response.
Among these, 4 lncRNAs (LOC286467, MEG3, LOC285194 and LincRNA-p21)
were found to be significantly dysregulated in responding tissues
compared with non-responding tissues (Table II). Subsequently, these 4 lncRNAs
were further validated in an independent cohort of 140 serum
samples from 70 CRC patients showing response and 70 showing no
response. Among the 4 candidate lncRNAs, only MEG3 was
significantly desregulated with a markedly suppressed expression in
non-responding patients compared to responding patients (Fig. 2). Thus, we focused on the role of
MEG3 in CRC chemoresistance.
 | Table II.Expression of 10 candidate lncRNAs in
CRC patients showing response or non-response to oxaliplatin
treatment [median (interquartile range)]. |
Table II.
Expression of 10 candidate lncRNAs in
CRC patients showing response or non-response to oxaliplatin
treatment [median (interquartile range)].
| lncRNA | Response | Non-response | P-value |
|---|
| LOC286467 | 1.39
(0.47–2.02) | 1.86
(1.13–2.45) | <0.05 |
| CRNDE | 0.94
(0.43–1.95) | 1.18
(0.49–2.03) | 0.37 |
| SNHG7 | 1.14
(0.33–2.51) | 1.57
(0.52–3.08) | 0.24 |
| H19 | 0.87
(0.35–1.89) | 1.27
(0.44–1.97) | 0.06 |
| MALAT1 | 1.03
(0.42–2.07) | 1.21
(0.66–2.49) | 0.18 |
| LOC145837 | 0.99
(0.38–2.15) | 0.77
(0.23–1.85) | 0.09 |
| MEG3 | 1.32
(0.40–2.31) | 0.74
(0.35–1.62) | <0.01 |
| LOC285194 | 1.47
(0.46–2.28) | 0.92
(0.37–1.89) | <0.01 |
| LincRNA-p21 | 1.22
(0.54–2.18) | 0.88
(0.33–1.76) | <0.05 |
| SLC25A25-AS1 | 1.15
(0.60–2.22) | 1.02
(0.47–1.59) | 0.44 |
Decreased serum MEG3 expression
predicts poor response to oxaliplatin treatment in CRC
patients
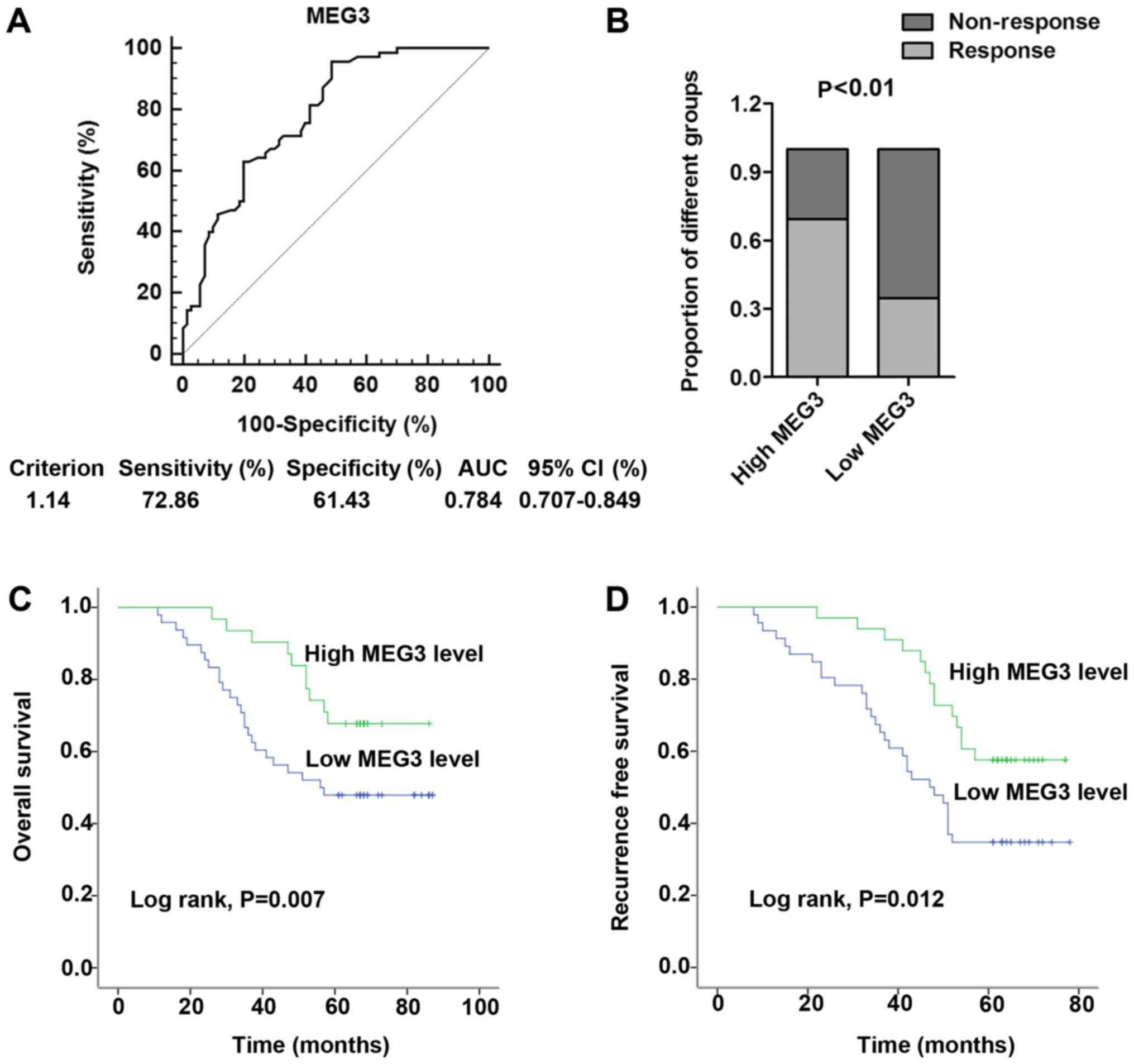
Predictive biomarkers are better when blood-based,
as blood is easily available and provides the chance to monitor
cancer progression. Thus, receiver operator characteristic (ROC)
curve analysis was firstly performed to investigate the potential
value of serum MEG3 in distinguishing the chemotherapeutic
responding and non-responding CRC patients. Our data showed that
the area under the curve (AUC) was 0.784, providing a diagnostic
sensitivity of 72.86% and a specificity of 61.43% (Fig. 3A). Under the stratification criteria
(1.14) established by the ROC curve, patients were stratified into
a high (n=62) and a low (n=78) MEG3-expressing group. The
proportion of patients that responded to oxaliplatin treatment was
significantly higher in the high MEG3-expressing group than in the
low MEG3-expressing group (P<0.01; Fig. 3B). More importantly, Kaplan-Meier
survival analysis was performed to further investigate the effect
of serum MEG3 on oxaliplatin treatment for CRC patients. The
results indicated that low MEG3 expression was associated with poor
OS (P=0.007; Fig. 3C) and RFS
(P=0.012; Fig. 3D). Furthermore, we
performed Cox regression univariate/mutivariate analyses to
identify whether MEG3 or another clinical parameter was an
independent indicator for OS of CRC patients who received
oxaliplatin chemotherapy. The results indicated that the serum MEG3
expression level and distant metastasis maintained their
significance as independent prognostic factors for OS of CRC
patients receiving oxaliplatin treatment (Table III).
 | Table III.Univariate and multivariate Cox
proportional hazards regression model analysis of factors for OS in
patients with CRC in a validation cohort. |
Table III.
Univariate and multivariate Cox
proportional hazards regression model analysis of factors for OS in
patients with CRC in a validation cohort.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
Characteristics | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender | 1.018 | 0.617–2.012 | 0.639 |
|
|
|
| Age (years) | 1.533 | 0.741–2.882 | 0.202 |
|
|
|
| Tumor size | 1.679 | 0.537–2.729 | 0.418 |
|
|
|
|
Differentiation | 1.885 | 1.029–3.352 | 0.087 |
|
|
|
| Local invasion | 1.661 | 0.902–2.798 | 0.152 |
|
|
|
| Distant
metastasis | 2.771 | 1.580–3.998 | 0.008 | 2.768 | 1.445–4.473 | 0.007 |
| TNM stage | 2.257 | 1.141–3.505 | 0.043 | 2.285 | 1.027–3.555 | 0.058 |
| Serum MEG3
level | 1.353 | 0.321–2.107 | 0.007 | 1.390 | 0.324–2.089 | 0.007 |
MEG3 is downregulated in OxR CRC cell
lines
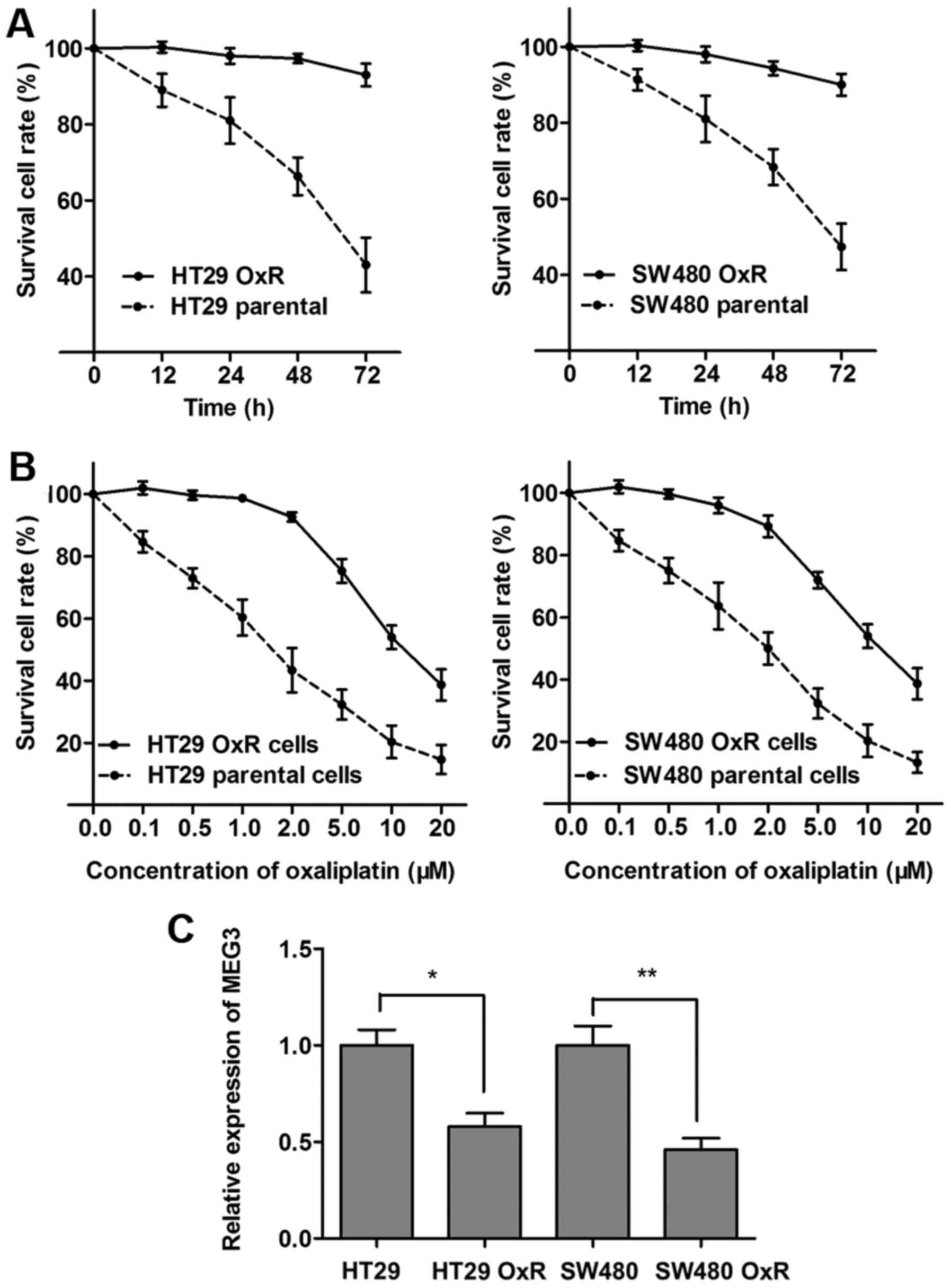
As a follow-up to our patient data that revealed a
lower expression of MEG3 in the OxR population, we further assessed
the expression of MEG3 in the oxaliplatin-resistant CRC cell lines.
HT29 and SW480 cell lines were constantly exposed to a high
concentration of oxaliplatin (2 µM) to establish the HT29 OxR (OxR)
cell line and SW480 OxR cell line. The established HT29-resistant
cells were maintained and exposed to 2 µM oxaliplatin unless
otherwise indicated. As shown in Fig.
4A, both HT29 OxR and SW480 OxR cells showed elevated cell
viability compared with the HT29 and SW480 parental cells when
incubated with culture medium containing 2 µM concentration of
oxaliplatin. In contrast, the concentration-effect curve indicated
that the IC50 value of oxaliplatin for HT29 OxR cells
was 11.6 µM, while the IC50 value of oxaliplatin for the
HT29 parental cells was 1.5 µM, which means that the HT29 OxR cells
exhibited a 7.7 times higher ability of oxaliplatin resistance than
the HT29 cells. Similarly, the SW480 OxR cells exhibited a 5.4
times higher ability of oxaliplatin resistance than the SW480
parental cells (10.3/1.9 µM; Fig.
4B). After the OxR CRC sub-lines were established, we
determined the MEG3 expression level, and found that MEG3 was
significantly downregulated in the HT29 OxR and SW480 OxR cells
compared with the parental HT29 and SW480 cells, respectively
(Fig. 4C).
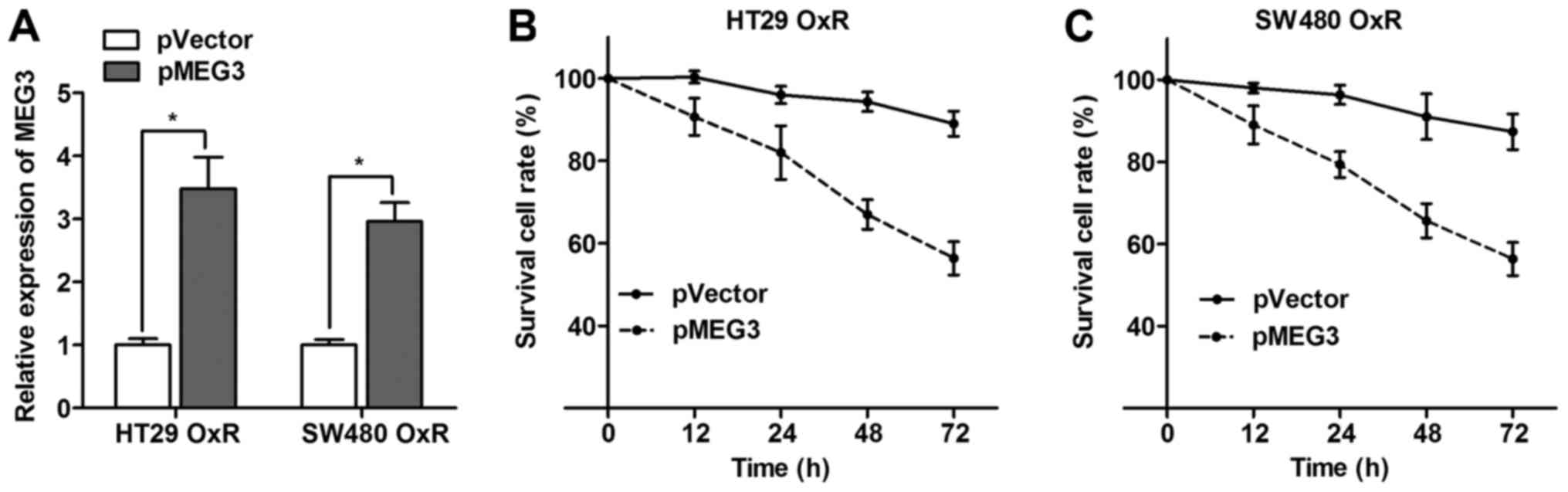
Overexpression of MEG3 partially
reverses the chemoresistance status of OxR cells
After having validating the disruption of MEG3 in
OxR cells, we further investigated whether MEG3 plays a role during
the formation of oxaliplatin resistance in CRC cells. MEG3
overexpression plasmid (pMEG3) was transfected into HT29 OxR and
SW480 OxR cells, and RT-qPCR assay showed that the MEG3 expression
level was significantly increased in cells transfected with pMEG3
(Fig. 5A). Moreover, the survival
cell rate was significantly impaired when MEG3 was overexpressed in
the HT29 OxR and SW480 OxR cells incubated with 2 µM concentration
of oxaliplatin (Fig. 5B and C).
This suggests that MEG3 overexpression partially reverses the
oxaliplatin resistance in CRC.
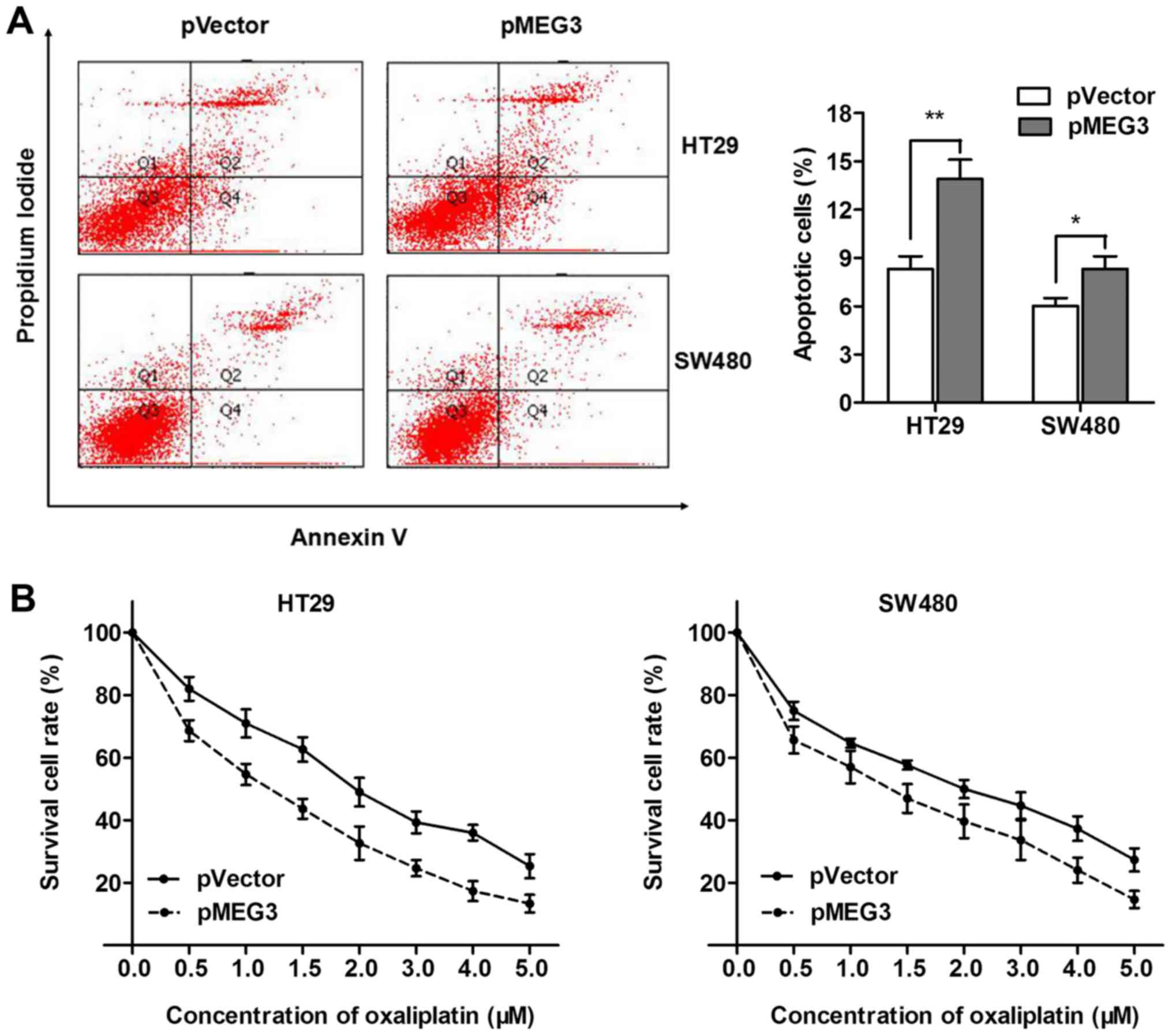
MEG3 promotes oxaliplatin-induced
apoptosis in CRC cells
Previous studies have demonstrated that MEG3 may
have an effect on cell apoptosis during cancer angiogenesis
(25). Thus, flow cytometric
apoptosis analysis was performed to explore whether MEG3 reversed
oxaliplatin resistance through promotion of cell apoptosis. Our
results showed that pMEG3 significantly enhanced cell apoptosis in
the HT29 and SW480 cells (Fig. 6A).
To directly validate that MEG3 enhanced oxaliplatin-induced cell
apoptosis in CRC, we treated HT29 and SW480 cells with a
concentration gradient of 0–5 µM oxaliplatin after being
transfected with pMEG3 or the negative control. The dose-effect
curve showed that pMEG3 transfection was followed by significantly
increased cell apoptosis compared with the pVector control
(Fig. 6B). However, the
IC50 values for oxaliplatin were 1.8 and 2 µM in the
non-transfected HT29 and SW480 cells, and were markedly decreased
to 1.2 and 1.4 after transfection with pMEG3, respectively. To
conclude, MEG3 enhanced the chemosensitivity of oxaliplatin by
promoting oxaliplatin-induced apoptosis in CRC cells.
Discussion
Despite recent chemotherapeutic regimens that have
significantly increased the survival of patients with metastatic
disease, invariably, nearly all CRC patients finally become
chemoresistant accompanied by distant metastasis (5). Identification of new therapeutic
markers and better understanding of the pathways related to
chemoresistance are essential to improving the prognosis of CRC
patients. In the present study, high-throughput HiSeq sequencing
was firstly employed to provide basic information concerning the
lncRNAs significantly dysregulated in CRC tissues. Candidate
lncRNAs were selected and then evaluated by RT-qPCR in tissues and
serum samples to validate their consistent pattern of dysregulation
in these clinical materials. MEG3 was finally identified to show
considerable discriminating potential to identify responding
patients from non-responding patients with high AUC value. Serum
MEG3 expression was associated with chemoresponse and predicted the
prognosis of CRC patients receiving oxaliplatin treatment. More
importantly, we demonstrated the molecular mechanism by which MEG3
exerted its function in oxaliplatin chemoresistance in CRC cells.
We found that MEG3 promoted chemosensitivity by enhancing
oxaliplatin-induced cell cytotoxicity in CRC cells.
In recent years, surgical resection with subsequent
first-line chemotherapy regimens are common clinical therapeutic
strategies. A major challenge, however, is that only approximately
half of the patients obtain an objective response to the regimens,
and that partial cross-resistance exists between different drugs
(26). For this reason, it is of
utmost importance to identify molecular bio-markers that have
effective diagnostic and prognostic meaning. In the present study,
we systematically investigated the expression of specific lncRNAs
using a 3-phase study. lncRNAs dysregulated in both tissue and
serum with a consistent pattern could effectively represent the
lncRNA expression alteration in CRC and simultaneously satisfy the
demand of noninvasive biomarkers. This stringent analysis led to
identification of only one significantly altered lncRNA, MEG3.
MEG3 is located on chromosome 14q32 and is widely
expressed in many types of normal tissues (27,28).
MEG3 was first identified as the ortholog of gene traplocus 2
(Gtl2) in mice (29). It belongs to
the DLK1-MEG3 imprinting locus which contains some maternally and
paternally imprinted genes (30).
Accumulating studies have demonstrated that MEG3 expression is
downregulated in various types of cancers (27). Thus, MEG3 is a tumor-suppressor and
overexpression of MEG3 could inhibit tumor cell proliferation and
promote tumor cell apoptosis in different cancers (13,31–33).
In our research, we confirmed the downregulation of MEG3 and found
that serum MEG3 expression considerably distinguished responding
patients from non-responding ones with high diagnostic efficiency.
However, the proportion of patients that responded to oxaliplatin
treatment was significantly higher in the high MEG3-expressing
group than in the low MEG3-expressing group. Most importantly, low
serum MEG3 expression was associated with poor OS and RFS in
patients receiving oxaliplatin treatment. Collectively, we revealed
that MEG3 is a lncRNA lowly expressed in non-responding CRC
patients and closely correlated to the chemoresponse to oxaliplatin
treatments.
Taking one step further, we aimed to verify the
underlying regulatory mechanism that could account for these
findings. By establishing two oxaliplatin-resistant cell lines, we
revealed that the MEG3 expression level was significantly
suppressed in oxaliplatin-resistant cells compared with that noted
in the parental cells. However, MEG3 overexpression promoted while
MEG3 knockdown inhibited CRC cell apoptosis. More importantly,
enhanced MEG3 expression significantly promoted oxaliplatin-induced
cell cytotoxicity in the CRC cell lines. This indicates that MEG3
overexpression can reverse oxaliplatin resistance in CRC cells;
thus, suppression of expression of MEG3 may be an important
contributor to oxaliplatin resistance of CRC. Our results are
partly consistent with previous studies. Yin et al found
that MEG3 suppressed cell proliferation, caused cell cycle arrest
and promoted cell apoptosis in CRC (13). Chak et al demonstrated that
MEG3 is also downregulated in nasopharyngeal carcinoma (NPC) and
predicted better survival by promoting NPC cell apoptosis (33). Luo et al also found that MEG3
inhibited cell proliferation and induced apoptosis in prostate
cancer (34). Thus, our data
demonstrated that MEG3 promoted oxaliplatin-induced cell apoptosis
in CRC, which may explain why MEG3 is downregulated in
non-responding CRC patients and oxaliplatin-resistant cells.
In conclusion, our integrated approach demonstrated
that MEG3 expression is downregulated in CRC patients who are
resistant to oxaliplatin treatment, and is closely associated with
the chemoresponse status to oxaliplatin treatment. We also revealed
that MEG3 promoted the chemosensitivity to oxaliplatin of CRC by
enhancing oxaliplatin-induced cell cytotoxicity. These findings
indicate that downregulation of MEG3 confers a potent poor
therapeutic efficacy. Thus, MEG3 may be a potential prognostic
marker and therapeutic target in CRC patients.
Acknowledgements
The authors would like to thank Professor Bing Xia
for his great contribution to their study. The present study was
supported by research grants from the National Natural Science
Foundation of China (no. 81270467).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li PL, Zhang X, Wang LL, Du LT, Yang YM,
Li J and Wang CX: MicroRNA-218 is a prognostic indicator in
colorectal cancer and enhances 5-fluorouracil-induced apoptosis by
targeting BIRC5. Carcinogenesis. 36:1484–1493.
2015.PubMed/NCBI
|
|
3
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long non-coding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang AD, Fan F, Camp ER, van Buren G, Liu
W, Somcio R, Gray MJ, Cheng H, Hoff PM and Ellis LM: Chronic
oxaliplatin resistance induces epithelial-to-mesenchymal transition
in colorectal cancer cell lines. Clin Cancer Res. 12:4147–4153.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldberg RM, Sargent DJ, Morton RF, Fuchs
CS, Ramanathan RK, Williamson SK, Findlay BP, Pitot HC and Alberts
SR: A randomized controlled trial of fluorouracil plus leucovorin,
irinotecan, and oxaliplatin combinations in patients with
previously untreated metastatic colorectal cancer. J Clin Oncol.
22:23–30. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hector S, Bolanowska-Higdon W, Zdanowicz
J, Hitt S and Pendyala L: In vitro studies on the mechanisms of
oxaliplatin resistance. Cancer Chemother Pharmacol. 48:398–406.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ahmad S: Platinum-DNA interactions and
subsequent cellular processes controlling sensitivity to anticancer
platinum complexes. Chem Biodivers. 7:543–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sau A, Tregno F Pellizzari, Valentino F,
Federici G and Caccuri AM: Glutathione transferases and development
of new principles to overcome drug resistance. Arch Biochem
Biophys. 500:116–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Landriscina M, Maddalena F, Laudiero G and
Esposito F: Adaptation to oxidative stress, chemoresistance, and
cell survival. Antioxid Redox Signal. 11:2701–2716. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes
bladder cancer metastasis by associating with suz12. Clin Cancer
Res. 20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long non-coding RNA MEG3
affects cell proliferation and predicts a poor prognosis in
patients with colorectal cancer. Tumour Biol. 36:4851–4859. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai Y, Xu P, Li Q, Ren D, Wang J, Xu K and
Gao W: Downregulation of long non-coding RNA ZMAT1 transcript
variant 2 predicts a poor prognosis in patients with gastric
cancer. Int J Clin Exp Pathol. 8:5556–5562. 2015.PubMed/NCBI
|
|
15
|
Yang F, Liu YH, Dong SY, Ma RM, Bhandari
A, Zhang XH and Wang OC: A novel long non-coding RNA FGF14-AS2 is
correlated with progression and prognosis in breast cancer. Biochem
Biophys Res Commun. 470:479–483. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long non-coding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mattick JS: The genetic signatures of
non-coding RNAs. PLoS Genet. 5:e10004592009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang R, Du L, Yang X, Jiang X, Duan W, Yan
S, Xie Y, Zhu Y, Wang Q, Wang L, et al: Identification of long
non-coding RNAs as potential novel diagnosis and prognosis
biomarkers in colorectal cancer. J Cancer Res Clin Oncol.
142:2291–2301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long non-coding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Yang X, Zhang Y, Liu X, Zheng G,
Yang Y, Wang L, Du L and Wang C: Direct serum assay for cell-free
bmi-1 mRNA and its potential diagnostic and prognostic value
for colorectal cancer. Clin Cancer Res. 21:1225–1233. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Huang S, Li Y, Zhang W, He K, Zhao
M, Lin H, Li D, Zhang H, Zheng Z, et al: Decreased expression of
LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and
EMT in colorectal cancer cells. Tumour Biol. 37:14205–14215. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Q, Dai Y, Wang F and Hou S:
Differentially expressed long non-coding RNAs and the prognostic
potential in colorectal cancer. Neoplasma. 63:977–983. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu KF, Liang WC, Feng L, Pang JX, Waye MM,
Zhang JF and Fu WM: H19 mediates methotrexate resistance in
colorectal cancer through activating Wnt/β-catenin pathway. Exp
Cell Res. 350:312–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Lei ZJ, Guo Y, Wang T, Qin ZY,
Xiao HL, Fan LL, Chen DF, Bian XW, Liu J, et al: miRNA-regulated
delivery of lincRNA-p21 suppresses β-catenin signaling and
tumorigenicity of colorectal cancer stem cells. Oncotarget.
6:37852–37870. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar MM and Goyal R: LncRNA as a
therapeutic target for angiogenesis. Curr Top Med Chem.
17:1750–1757. 2017.doi: 10.2174/1568026617666161116144744.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pfeiffer P, Qvortrup C and Eriksen JG:
Current role of antibody therapy in patients with metastatic
colorectal cancer. Oncogene. 26:3661–3678. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benetatos L, Vartholomatos G and
Hatzimichael E: MEG3 imprinted gene contribution in tumorigenesis.
Int J Cancer. 129:773–779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y and Klibanski A Zhangxand: MEG3
non-coding RNA: A tumor suppressor. J Mol Endocrinol. 48:R45–R53.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schuster-Gossler K, Bilinski P, Sado T,
Ferguson-Smith A and Gossler A: The mouse Gtl2 gene is
differentially expressed during embryonic development, encodes
multiple alternatively spliced transcripts, and may act as an RNA.
Dev Dyn. 212:214–228. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miyoshi N, Wagatsuma H, Wakana S,
Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino
T and Ishino F: Identification of an imprinted gene,
Meg3/Gtl2 and its human homologue MEG3, first mapped
on mouse distal chromosome 12 and human chromosome 14q. Genes
Cells. 5:211–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Balik V, Srovnal J, Sulla I, Kalita O,
Foltanova T, Vaverka M, Hrabalek L and Hajduch M: MEG3: A novel
long non-coding potentially tumour-suppressing RNA in meningiomas.
J Neurooncol. 112:1–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang P, Ren Z and Sun P: Overexpression of
the long non-coding RNA MEG3 impairs in vitro glioma cell
proliferation. J Cell Biochem. 113:1868–1874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chak WP, Lung RW, Tong JH, Chan SY, Lun
SW, Tsao SW, Lo KW and To KF: Downregulation of long non-coding RNA
MEG3 in nasopharyngeal carcinoma. Mol Carcinog. 56:1041–1054. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo G, Wang M, Wu X, Tao D, Xiao X, Wang
L, Min F, Zeng F and Jiang G: Long non-coding RNA MEG3 inhibits
cell proliferation and induces apoptosis in prostate cancer. Cell
Physiol Biochem. 37:2209–2220. 2015. View Article : Google Scholar : PubMed/NCBI
|