Introduction
Gliomas, accounting for ~70% of human malignant
primary brain tumors, are the most lethal primary brain tumor which
include a heterogenous group of tumors. It is classified by the
World Health Organization (WHO) into pilocytic astrocytomas and
three groups of diffusely infiltrative astrocytomas, including
glioblastoma (1,2). Glioblastoma multiforme (GBM), the most
common form of malignant glioma, is characterized as presenting
with a highly heterogeneous composition of cells and exhibits
phenotypic heterogeneity (1,3).
Although a series of treatment protocol have been developed and
several potential drug targets have been discovered, the survival
rate of GBM patients has not been significantly increased (4,5). Thus,
it is still necessary to identify novel and effective biomarkers
which may help the development of therapeutic targeted drugs.
Long non-coding RNAs (lncRNAs), which were initially
argued to be spurious transcriptional noise, are now recognized as
a class of RNAs with transcripts longer than 200 nucleotides
without the function of encoding proteins (5–7).
Recent studies have found that lncRNAs: i) regulate downstream
target genes by multiple means via cis- and
trans-regulatory effects (8,9); and
ii) play a critical regulatory role in many human diseases,
including cancer (10,11). Other investogators have reported
that a growing number of lncRNAs can cooperate with neighbor genes
to form ‘lncRNA-mRNA’ pairs to affect their function (12–15).
Close relationships are often found between these lncRNAs and their
near-by mRNAs in expression or function. Based on microarray-based
data, Zhang et al previously demonstrated that specific
lncRNA expression patterns are associated with different
histological subtypes and malignant behaviors in glioma (16). Furthermore, certain lncRNAs were
found to be of prognostic significance, suggesting that lncRNAs may
have important roles in gliomagenesis and may serve as novel
therapeutic targets and biomarkers (17). Previously, downregulation of lncRNA
Prader Willi/Angelman region RNA 5 (PAR5; also known as PWAR5,
Homo sapiens, gene ID, 8123) was demonstrated to be
associated with GBM (17), yet the
functions of PAR5 have not been well investigated in this type of
tumor.
Histone modifications and global aberrations at the
histone level may result in distorted patterns of gene expression,
and malfunction of proteins that regulate chromatin modification
and remodeling (18,19). Reports have shown that histone
modifications play important roles in the pathogenesis of diffuse
gliomas in adults and children (20). Typical lncRNAs can coordinate
histone modifications by binding to various histone modification
enzymes. It is well known that polycomb repressive complex 2 (PRC2)
is a methyltransferase for histone H3 lysine 27 trimethylation
(H3K27me3), consisting of enhancer of zeste homolog 2 (EZH2),
suppressor of zeste 12 (SUZ12) and embryonic ectoderm development
(EED) (20). Lysine-specific
demethylase 1 (LSD1) is a demethylase that mediates the enzymatic
demethylation of histone H3 lysine 27 dimethylation (H3K4me2)
(21). It was revealed that the 5′
domain of lncRNA HOTAIR binds to PRC2, whereas the 3′ domain binds
to the LSD1/CoREST/REST complex, which acts as a modular scaffold
(22).
Previous studies have also shown that lncRNAs
influence the expression of downstream targets via recruiting PRC2
or LSD1 proteins (23,24). Therefore, the present study aimed to
explore the possible effects of PAR5 involved in the oncogenic
events of glioma and the interaction between PAR5 and PRC2 or LSD1
in glioma cells.
Materials and methods
Ethical approval
The present study protocol complied with the
Declaration of Helsinki and was approved by the Medical Ethics
Committee of Kunming Medical University. Glioma tumor specimens
were obtained from consenting patients at the Hospital of Yunnan
Province (Kunming, China).
Tissues preparation, molecular assay
and cell culture
Eighty-seven patients, attending (February 2013 to
December 2014) the clinic of the First Affiliated Hospital of
Kunming Medical University, provided consent to participate in the
present study. All of patients accepted to undergo surgery at our
hospital for the first time, without any antitumor treatment. The
specimens were histopathologically verified as primary glioma by
three independent senior pathologists according to the WHO
Classification of Tumors of the Central Nervous System (CNS)
(25,26). Subsequently, the primary carcinoma
tissues and the matched adjacent normal tissues (at least 3 cm away
from the tumor) (27) were
collected for analysis. The demographic and clinic characteristics
of the unselected 87 glioma population are summarized in Table I.
 | Table I.Summary of the patient cohort with
glioma in the present study. |
Table I.
Summary of the patient cohort with
glioma in the present study.
| Parameters | No. of cases
(%) |
|---|
| Total glioma
cases | 87 (100) |
| Age (≥50
years) | 41 (47.1) |
| Sex |
|
|
Male | 44 (50.6) |
|
Female | 43 (49.4) |
| Location of
tumor |
|
|
Frontal | 35 (40.2) |
|
Occipital | 12 (13.8) |
|
Temporal | 14 (16.1) |
|
Others | 26 (29.9) |
| WHO grade |
|
| I | 16 (18.4) |
| II | 18 (20.7) |
|
III | 25 (28.7) |
| IV | 28 (32.2) |
| IDH1
mut | 45 (51.7) |
| MGMT
methy | 51 (58.6) |
| 1p/19q
codel | 52 (59.8) |
| TERT
mut | 28 (32.2) |
DNA extraction from frozen tumor tissue was
performed using a DNeasy Blood and Tissue kit (Qiagen, Tokyo,
Japan). The presence of hotspot mutations in isocitrate
dehydrogenase (IDH) gene 1 (R132)/2 (R172), as well as the
two mutation hotspots in the TERT promoter (C228T and
C250T), were assessed mainly by pyrosequencing and partly by Sanger
sequencing, as previously reported (28,29).
The methylation status of the MGMT promoter was also
analyzed using a customized pyrosequencing assay, essentially as
previously described (30). A SALSA
MLPA kit probemix (P088-C1; MRC-Holland, Amsterdam, The
Netherlands) was employed for analysis of the copy number of 1p/19q
as previously described (31).
Human normal skin fibroblast HF cell line and glioma
cell lines U87 and U251 were purchased from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). The cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand
Island, NY, USA) supplemented with 10% fetal bovine serum, 100 U/ml
penicillin and 100 µg/ml streptomycin (BioWest, Nuaillé, France),
at 37°C in a humidified atmosphere containing 5% CO2.
All of the cells are negative for HIV-1, HBV, HCV, mycoplasma,
bacteria, yeast and fungi before experiment.
Cell transfection
RNA oligoribonucleotides targeting to PAR5 were
prepared for cell transfection. The small interfering RNAs (siRNAs)
that specifically target human PAR5 mRNA (si-PAR5) were designated
and purchased from RiboBio (Guangzhou, China). According to the
manufacturer's protocol, a final concentration of 2×105
U87 and U251 cells were seeded into each well of a 6-well plate and
transfected for 48 h using Lipofectamine 2000 reagent (Invitrogen,
Carlsbad, CA, USA). The negative control duplex containing siRNA
(si-NC; RiboBio), not homologous to any human genome sequences, was
employed as control. At the end of transfection, U87 and U251 cells
were collected for further analyses.
Full-length PAR5 cDNA was synthesized by Biomarker
Technologies (Beijing, China) and ligated into the pcDNA3.1(+)
vector (Invitrogen). PcDNA3.1-PAR5 (p-PAR5) and empty vector (p-NC)
were transfected into glioma cells, U87 and U251, cultured in
6-well plates using the Lipofectamine 2000 reagent (Invitrogen).
Cells were harvested for qRT-PCR analysis 48 h after
transfection.
Quantitative real-time PCR
(qRT-PCR)
The expression of PAR5 in the human glioma cells and
specimens from glioma patients was determined by qRT-PCR as
previously described (32). Total
RNA was isolated using TRIzol according to manufacturer's
instructions. qRT-PCR was performed using the iQ SYBR-Green
SuperMix (Bio-Rad, Hercules, CA, USA) as per the protocol of the
manufacturer. GAPDH served as an endogenous control. lncRNA
expression levels were normalized by calculating the lncRNAs/GAPDH
expression ratio (2−ΔCt). Amplification of qRT-PCR was
performed at 95°C for 3 min, followed by 40 cycles at 95°C for 15
sec and 60°C for 60 sec. The primer sequences were as follows:
GAPDH sense, 5′-CGAGATCCCTCCAAAATCAA-3′ and antisense,
5′-TTCACACCCATGACGAACAT-3′; PAR5 sense, 5′-TGATGTGGGTGTTGATAC-3′
and antisense, 5′-ACTCAAAGGCAAGAACTA-3′ (32).
Cell viability assay
Following various treatments, cell viability was
determined using the tetrazolium salt
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Briefly, 1×105 cells/ml cells were plated into
96-well culture plates in 200 µl of culture medium/well. After 48 h
of culture with treatment, 20 µl of 5 mg/ml MTT was added to each
well and incubated at 37°C for 4 h. The medium was then gently
aspirated and 150 µl of dimethyl sulfoxide (DMSO) was added to each
well to solubilize the formazan crystals. The absorbance of each
sample was immediately measured using a microplate reader
(Multiskan MK3; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
at 570 nm.
Migration and invasion assays
Transwell (Millipore, Billerica, MA, USA) and
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) chamber plates
were employed to evaluate the motility and invasiveness as
previously described (33). After
24 h of incubation, cells remaining in the upper chamber or on the
upper membrane were carefully removed. Cells adhering to the lower
membrane were stained, imaged and counted using an IBX3 inverted
microscope (Olympus, Tokyo, Japan) (34). The experiments were repeated three
times.
Scratch wound assay
Scratch wound assay was employed to detect the
migration of the glioma cell lines treated with various treatments,
as previously described (35).
Briefly, at ~80% confluency, the glioma cells transfected with
various chemicals were seeded onto 6-well plates and incubated at
37°C, respectively. Then, using a 10-µl pipette tip, a vertical
scratch wound was made through the center of each well plate. The
cells were then washed three times with phosphate-buffered saline
(PBS) to remove the scratched cells, and fresh serum-free medium
was transferred. After 48 h, the cells were examined by light
microscopy (Olympus) at a magnification of ×200 to determine the
resealing of the cell monolayer.
RNA immunoprecipitation (RIP)
assay
RIP was performed using the EZ-Magna RIP kit
(Millipore) according to the manufacturer's protocol. Briefly, at
80–90% confluency, U87 and U251 cells were scraped off the culture
plate and lysed in RIP lysis buffer. A total of 100 µl of whole
cell extract was incubated with RIP buffer containing magnetic
beads conjugated with antibodies against EZH2, LSD1, SUZ12 or
control IgG (Millipore) for 8 h at 4°C. The beads were treated with
wash buffer, then the complexes were incubated with 0.1% SDS/0.5
mg/ml proteinase K for 30 min at 55°C to remove proteins. Finally,
immunoprecipitated RNA was purified and analyzed by qRT-PCR.
RNA pull-down assays
The pCDNA3.1-PAR5 vector was cleaved by restriction
enzyme NruI and treated with RNase-free DNase I (Takara,
Dalian, China). PAR5 was transcribed from this vector using the
mMESSAGE mMACHINE T7® kit (Ambion, Carlsbad, CA, USA)
and purified using the RNeasy Mini kit (Qiagen, Valencia, CA, USA)
in vitro. The Pierce RNA 3′-End Desthiobiotinylation kit
(Thermo Fisher Scientific, Inc.) was employed to label the 3′ end
of PAR5, according to the instructions. The extensively expressed
messenger RNA (mRNA) stabilizing protein HuR (encoded by ELAVL1)
was employed and served as the positive control for RNA pull-down
assays. The nonspecific IgG antibody was used as a negative
control. One milligram of protein from the U87 cell extracts was
then mixed with 45 pmol of biotinylated RNA, and incubated with 50
µl of magnetic beads for 1 h at 4°C (Thermo Fisher Scientific,
Inc.). The RNA-protein complex was isolated from magnetic beads
using Biotin Elution Buffer and boiled in SDS buffer for 5 min. The
retrieved protein was detected using standard western blotting
techniques.
Western blotting
Using cell lysis buffer for western blotting
(Beyotime Biotechnology, Shanghai, China), total proteins were
isolated from the cells with various treatments. Subsequently, the
protein concentrations were detected using the BCA assay kit
(Beyotime Biotechnology) according to the manufacturer's protocol.
The protein expression of EZH2, epidermal growth factor receptor
(EGFR), VEGF-A, cyclin A, AKT and p-AKT (ser473) was detected as
previously described (14).
Briefly, protein samples were separated by 10% SDS-PAGE and
transferred onto nitrocellulose membranes (Beyotime Biotechnology).
The membranes were blocked with 5% non-fat milk for 2 h, and then
incubated overnight at 4°C with the primary antibody, including
EZH2 (1:1,000; Millipore), EGFR (1:1,000), VEGF-A (1:1,000), cyclin
A (1:500), AKT (1:500) and phosphorylated-AKT (p-AKT) (ser473;
1:1,000) (all from Cell Signaling Technology, Danvers, MA, USA).
Signals were detected using the enhanced chemiluminescence (ECL)
luminol reagent (Millipore).
Statistical analysis
Data are presented as means ± standard deviation
(SD). SPSS version 21.0 software (SPSS, Inc., Chicago, IL, USA) was
employed for analyses. Multivariate logistic regression analysis
was performed to evaluate the association between PAR5 expression
levels (low or high level) and clinicopathological/genetic
characteristics of the glioma patients. The strength of association
was measured using odds ratios (ORs) with 95% confidence interval
(CI). Logistic regression was used for ordinal data to estimate
adjusted ORs. The statistical significance of PAR5, EGFR, VEGF-A,
cyclin A, AKT and p-AKT (ser473) expression, MTT cell activity,
migration and invasion among groups following different treatments
were determined using one-way ANOVA. Significance level was
predetermined to be p≤0.05 unless otherwise indicated.
Results
Abnormal expression of PAR5 in glioma
patients and cells
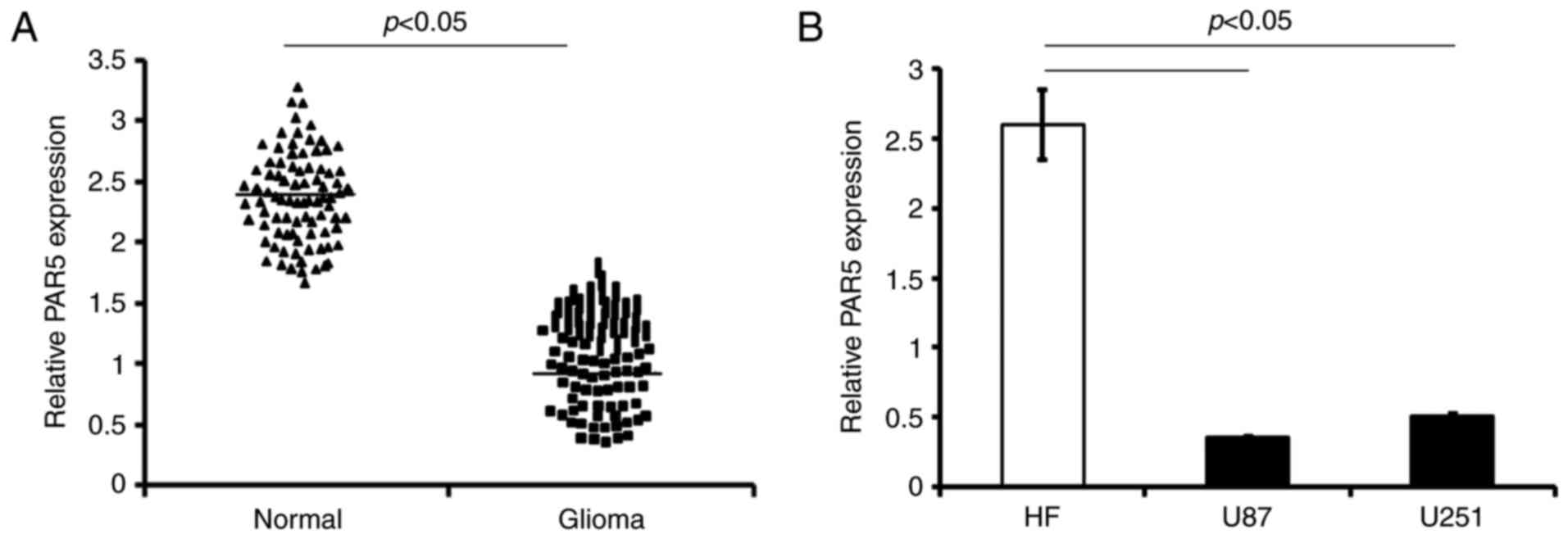
The levels of PAR5 were examined in the tumor
tissues of glioma patients by qRT-PCR analysis. The PAR5 expression
was significantly decreased in the glioma tissue compared with that
noted in the adjacent normal brain tissues (p<0.05; Fig. 1A).
Consistently, the PAR5 level was lower in both human
glioma cell lines, U87 and U251 (U87 vs. HF cells, and U251 vs. HF
cells respectively, p<0.05; Fig.
1B).
PAR5 expression is associated with
glioma progression
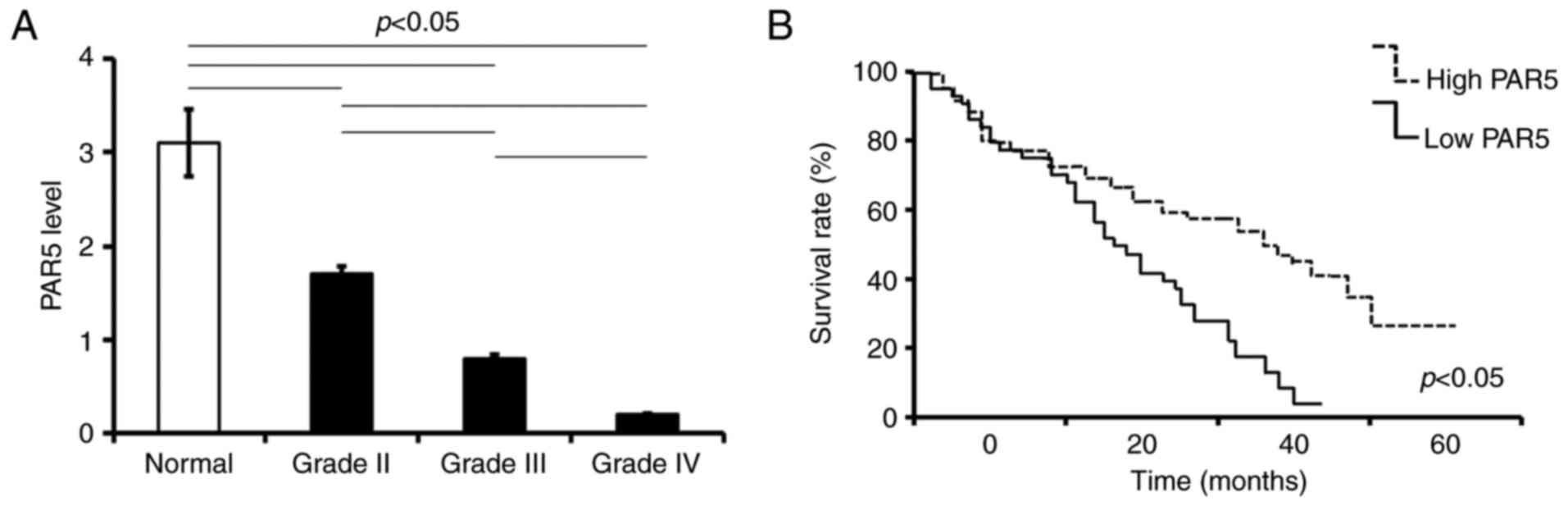
The expression of PAR5 in GBM tumors collected from
patients was determined by qRT-PCR. According to the PAR5 level,
the enrolled patients were divided into two cohorts: those with
less than or equal to the median expression level of PAR5 (low
level) and those with more than the median expression level of PAR5
(high level). The clinicopathologic association of PAR5 mRNA levels
in the glioma tumors were analyzed. The level of PAR5 expression in
glioma tissues was lower when compared to that in the adjacent
normal brain tissues, and decreased with increasing WHO grade
(p<0.05; Fig. 2A). Kaplan-Meier
survival analysis showed that patients with high PAR5 expression
had significantly increased overall survival compared with patients
with low PAR5 expression (p<0.05; Fig. 2B).
Analysis showed that low PAR5 expression was
significantly correlated with tumor size (≥5 cm, r=0.469, p=0.0019,
<0.05), WHO grade (r=0.376, p=0.0035, <0.05) and Karnofsky
performance score (KPS) score (r=0.297, p=0.005, <0.05)
(Table II). Notably, there were 25
patients with low PAR5 expression classified as having highly
malignant grade IV glioblastomas (GBMs). Age at diagnosis, sex and
resection status were not correlated with PAR5 expression (Table II).
 | Table II.PAR5 level is associated with
clinicopathological features of the glioma cases. |
Table II.
PAR5 level is associated with
clinicopathological features of the glioma cases.
|
| PAR5 levels
(n=87) |
|
|
|---|
|
|
|
|
|
|---|
|
| Low level | High level |
|
|
|---|
|
| n=47 | n=40 | rs | P-value |
|---|
| Ages (years) |
|
| 0.005 | 0.65 |
|
<50 | 22 | 24 |
|
|
|
≥50 | 25 | 16 |
|
|
| Sex |
|
| 0.069 | 0.32 |
|
Male | 25 | 19 |
|
|
|
Female | 22 | 21 |
|
|
| Resection
status |
|
| 0.128 | 0.19 |
|
Total | 23 | 22 |
|
|
|
Subtotal | 24 | 18 |
|
|
| Tumor size
(cm) |
|
| 0.469 | 0.0019 |
|
<5 | 19 | 31 |
|
|
| ≥5 | 28 | 9 |
|
|
| Location of
tumor |
|
| 0.024 | 0.59 |
|
Frontal | 20 | 15 |
|
|
|
Occipital | 7 | 5 |
|
|
|
Temporal | 8 | 6 |
|
|
|
Others | 12 | 14 |
|
|
| Histopathological
diagnosis (WHO) |
|
| 0.376 | 0.0035 |
| Grade
I | 2 | 14 |
|
|
|
Astrocytoma II | 6 | 12 |
|
|
|
Astrocytoma III | 11 | 9 |
|
|
|
Oligodendro-gliomas
II/III | 3 | 2 |
|
|
| IV,
glioblastomas | 25 | 3 |
|
|
| KPS score |
|
| 0.297 | 0.005 |
|
<80 | 33 | 8 |
|
|
|
≥80 | 14 | 32 |
|
|
Assessment of the molecular features of the
unselected glioma patients revealed that PAR5 expression was
significantly associated with IDH1 mutation (r=0.809,
p=0.000, <0.05), MGMT methylated status (r=0.201,
p=0.013, <0.05) and TERT promoter mutation (r=0.721,
p=0.000, <0.05) (Table
III).
 | Table III.Correlation between PAR5 level and
molecular features of the glioma cases. |
Table III.
Correlation between PAR5 level and
molecular features of the glioma cases.
|
| PAR5 (n=87) |
|
|
|---|
|
|
|
|
|
|---|
|
| Low level n=47
(%) | High level n=40
(%) | rs | P-value |
|---|
| IDH1/2
status |
|
| 0.809 | 0.000 |
| WT | 37 (78.7) | 5 (12.5) |
|
|
|
mut | 10 (21.3) | 35 (87.5) |
|
|
| MGMT
status |
|
| 0.201 | 0.013 |
|
Methylation | 30 (63.8) | 6 (15) |
|
|
|
Unmethylation | 17 (36.2) | 34 (85) |
|
|
| 1p/19q
status |
|
| 0.137 | 0.087 |
|
Non-codel | 20 (42.6) | 17 (42.5) |
|
|
|
Codel | 27 (57.4) | 23 (57.5) |
|
|
| TERT
status |
|
| 0.721 | 0.000 |
| WT | 15 (31.9) | 27 (67.5) |
|
|
|
mut | 32 (68.1) | 13 (32.5) |
|
|
Evaluation of the effectiveness of
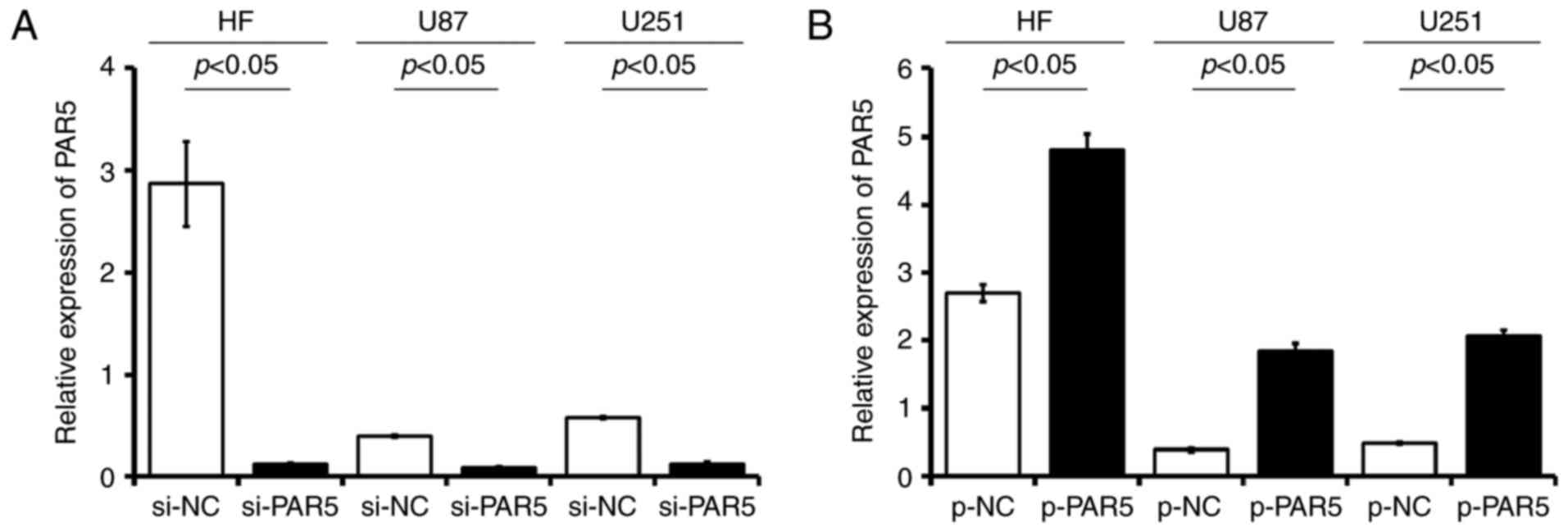
PAR5 siRNA and overexpression plasmid transfection
The glioma cell lines, U87 and U251, as well as the
HF cells were stably transfected with si-PAR5, p-PAR5 or the
matched control si-NC, p-NC. The data showed that stable expression
of PAR5 siRNA (si-PAR5) in U87, U251 and HF cells resulted in
>70% decrease in PAR5 RNA (Fig.
3A). In the p-PAR5-transfected glioma cells, the PAR5 levels as
detected were significantly higher than levels in the matched p-NC
controls, respectively (p<0.05; Fig.
3B). The PAR5 level was upregulated at least 1.7-fold in the HF
cells, while the level was increased significantly in the glioma
cells (4.7-fold in U87 cells; 4.2-fold in U251 cells), compared
with the matched p-NC cell lines, respectively.
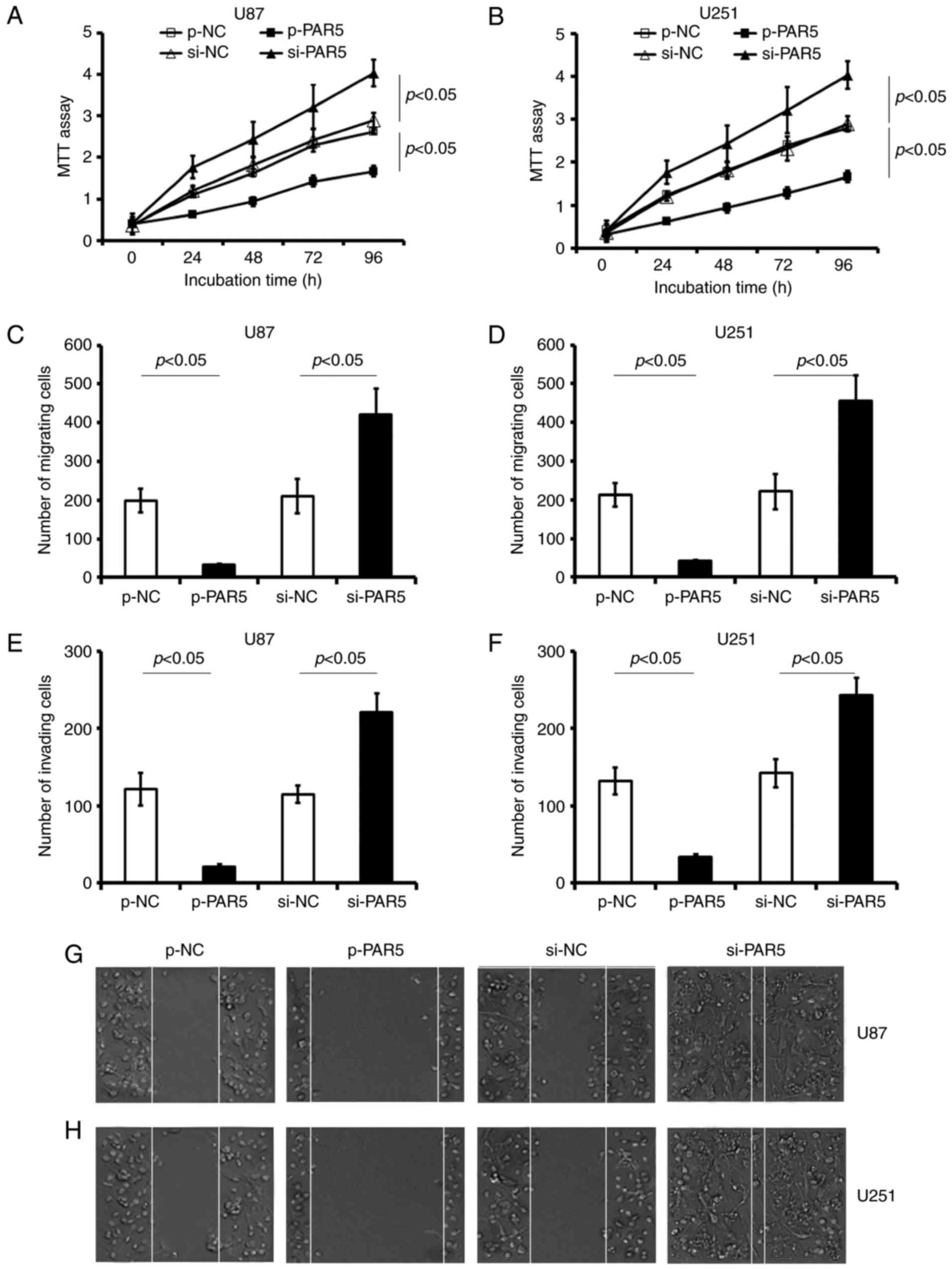
PAR5 inhibits proliferation, migration
and invasion of glioma cells
Transfection with p-PAR5 significantly decreased
cell viability, and suppressed migration and invasion in both U87
and U251 cell lines, compared with these parameters in the matched
p-NC treated cell lines, respectively. Functional inhibition of
endogenous PAR5 by special si-PAR5 induced a significant promotion
in viability in the U87 (Fig. 4A;
p<0.05) and U251 cell lines (Fig.
4B; p<0.05).
Restoration of the PAR5 level in glioma cells by
transfection with p-PAR5 significantly reduced cellular motility
compared with that noted in the p-NC-treated groups. The migration
assay revealed that the number of crystal violet-stained cells was
significantly decreased in the p-PAR5-treated cells, compared with
that in the matched p-NC control groups (p<0.05; Fig. 4C and D). p-PAR5 transfection
decreased the number of U87 and U251 cells that migrated through
the Transwell membrane by at least 80% compared with the
p-NC-treated cells. Scratch wound assay showed that transfection
with p-PAR5 significantly impaired the invasiveness of U87
(Fig. 4E and G) and U251 (Fig. 4F and H) cells, in comparison with
that of the control p-NC-treated cells, respectively (p<0.05).
In contrast, si-PAR5 transfection significantly promoted the
migration and invasion in both glioma cell lines (vs. the matched
si-NC control, p<0.05, respectively; Fig. 4).
PAR5 interacts directly with EZH2 in
glioma cells
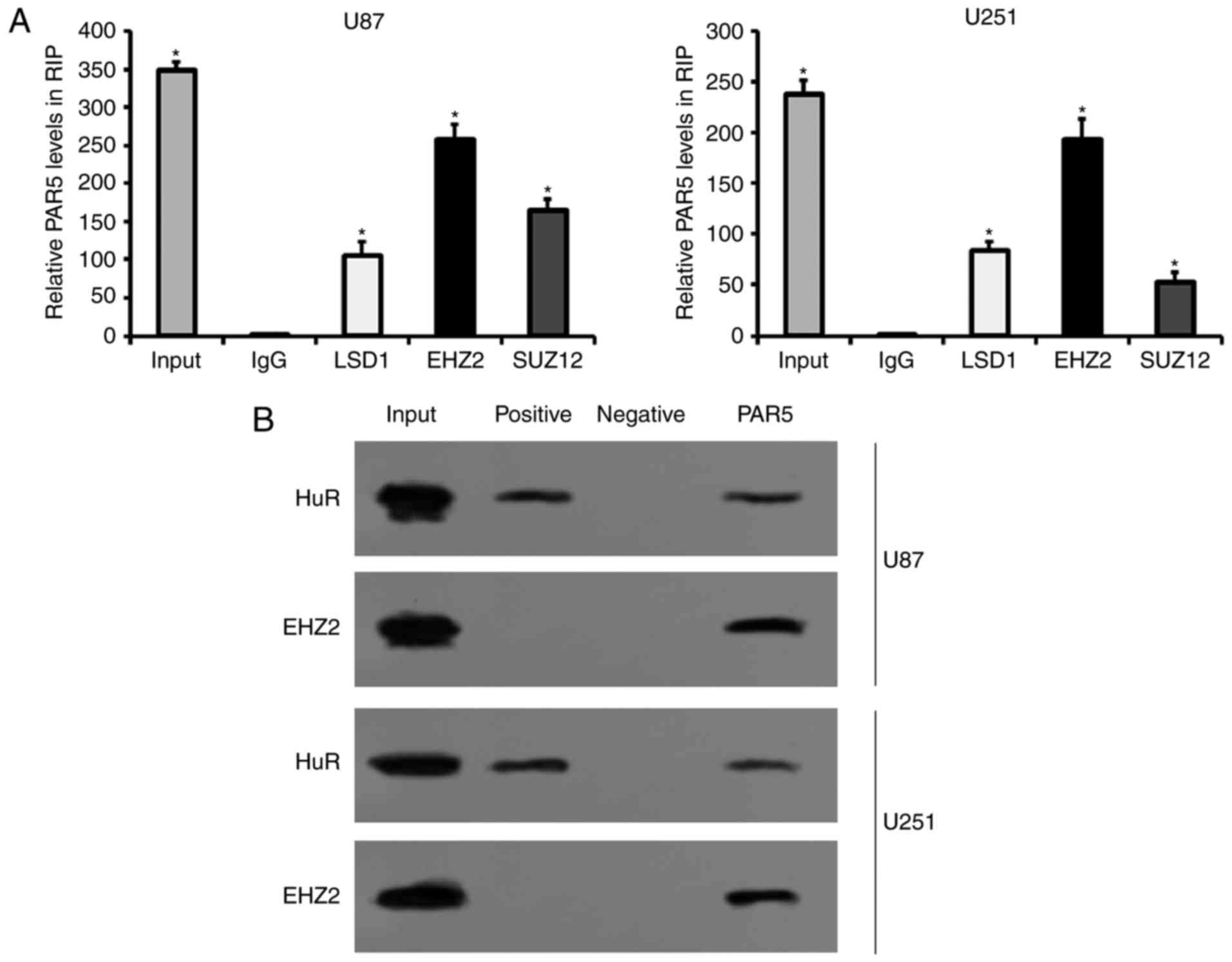
The binding of PAR5 with EZH2 was confirmed by RIP
and qRT-PCR in U87 and U251 cells. RIP assays and subsequent
qRT-PCR revealed that PAR5 was abundant in the RIP samples using
LSD1, EZH2 and SUZ12 antibodies as compared with the samples using
nonspecific antibodies IgG, which confirmed the specificity of RIP
assays and qRT-PCR performed in the present study (Fig. 5A). The relative expression of PAR5
was significantly enriched in the EZH2 antibody-treated glioma U87
and U251 cells, compared with that in the LSD1 and SUZ12
antibody-treated ones, respectively (Fig. 5A). The results showed that PAR5
bound directly to EZH2 in both U87 and U251 cells. The following
RNA pull-down assays and western blotting also confirmed the
interaction between PAR5 and EZH2 (Fig.
5B). Additionally, HuR served as the positive control,
indicating the sensitivity of RNA pull-down performed in the
present study (Fig. 5B). These data
suggest that EZH2 specifically binds to PAR5 in U87 and U251
cells.
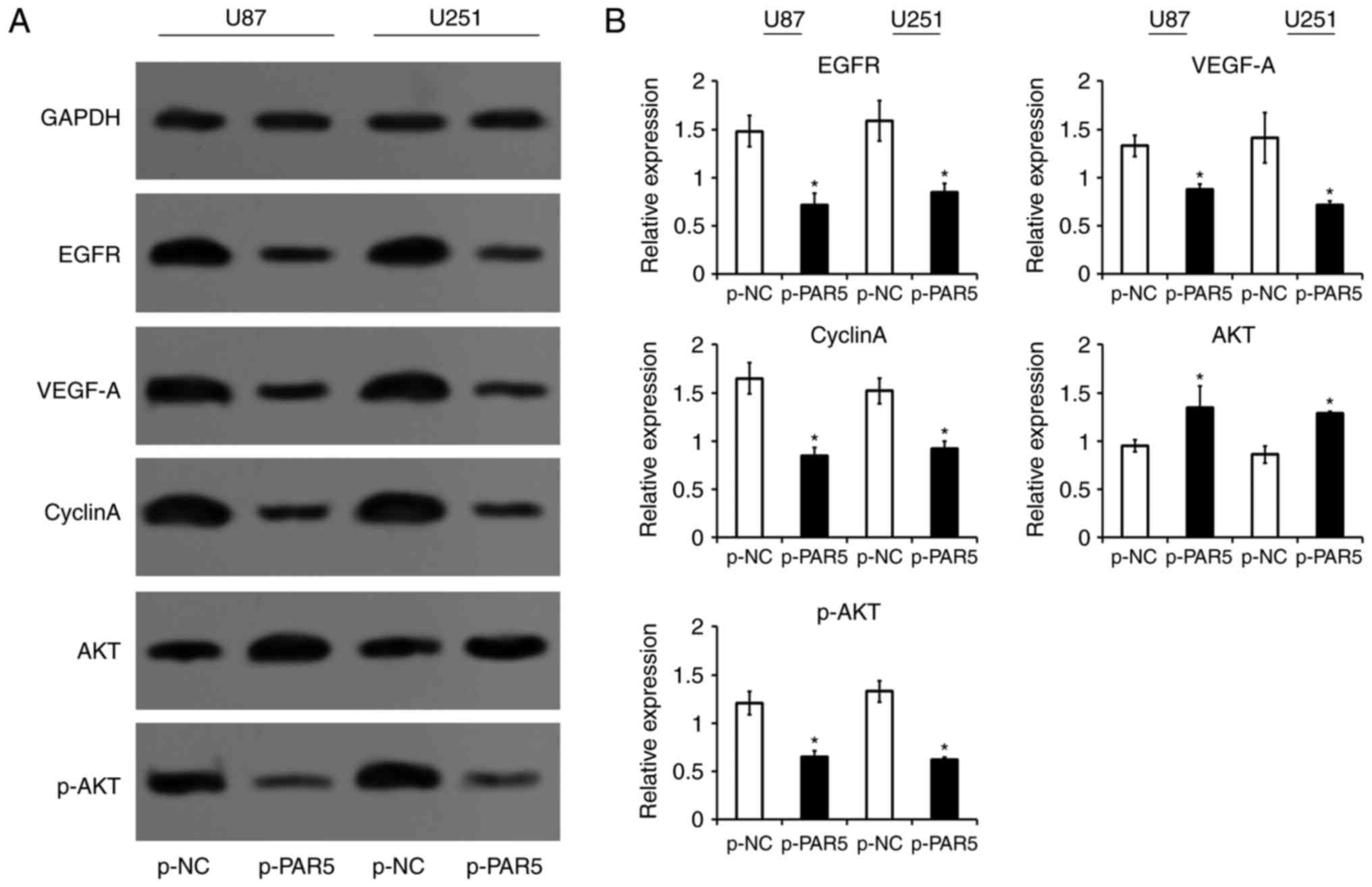
PAR5 inhibits expression of oncogenes
in glioma cells
Given the significant inhibition of cell growth and
motility by PAR5 in the glioma cells, the involvements of several
genes related with proliferation, cell cycle and tube formation of
tumor cells were investigated. The data showed that PAR5 reduced
the expression of EGFR, VEGF-A and cyclin A (Fig. 6). Additionally, the expression of
p-AKT was decreased by PAR5 overexpressed plasmid transfection
(Fig. 6).
Discussion
In the present study, we found that: i)
significantly downregulated PAR5 was negatively correlated with
clinicopathological and genetic features; ii) a cohort with PAR5
low expression in tumors had a worse overall survival rate compared
to those with PAR5 high expression; iii) PAR5 inhibited human
glioma cell proliferation, invasion and migration; iv) PAR5
interacted with EZH2 and suppressed expression of various oncogenes
including EFGR, VEGF-A, cyclin A and p-AKT in glioma cells.
PAR5 was previously reported to be: i) significantly
downregulated predominantly in one specific hepatitis C
virus-related hepatocellular carcinoma (HCC) type (32); ii) correlated with poor prognostic
outcomes in human glioblastoma multiforme (17). However, there is limited information
concerning PAR5. Recently, the functions of PAR5 have not been well
investigated. Thus, additional studies may be needed to elucidate
the mechanistic connection between PAR5 and the molecular
pathogenesis of tumors, particularly glioma. We found that
endogenous PAR5 expression was significantly decreased in glioma
tissues and cell lines, while a low level of PAR5 was associated
with pathological features and the survival rate of glioma
patients. Low PAR5 expression was correlated with tumor size (≥5
cm), WHO grade (III–IV) and KPS score (<80), which indicated the
deterioration of glioma patients. Evaluation of the molecular
genetic features of this unselected glioma cohort revealed that
PAR5 expression was also associated with IDH1/2 mutation,
MGMT methylated status and TERT promoter
mutation.
The presence of the IDH1/2 mutation in
astrocytoma patients was found to have a positive effect on overall
survival (36,37). They are rare in primary GBM and
absent in pilocytic astrocytomas and are often associated with
MGMT promoter hypermethylation (‘MGMT methylation’),
a well-established prognostic marker for primary GBM and a
predictive marker for the response to temozolomide in elderly GBM
patients (38–40). In the present study, presence of the
IDH1/2 mutation was associated with significantly enhanced
overall survival in glioma patients with a high PAR5 level, while
the presence of IDH-wild-type in patients with a low level
of PAR5 suffered from poor overall survival. In the present
unselected cohort, the presence of MGMT methylation was
frequently detected in those with low PAR5 expression (63.8%), in
which more patients were diagnosed with glioblastomas (GBMs) (25
vs. 3 cases). PAR5 expression was highly consistent with
IDH1/2 mutation and MGMT methylation. Our data also
revealed that the TERE promoter mutations and
IDH-wild-type are the common genotypes observed in the
cohort with a PAR5 low expression level, which tend to be
associated with poor prognosis. Research has demonstrated that
TERT promoter mutations almost always coincide with
IDH mutations and 1p/19q codeletion in oligodendrogliomas,
whereas a combination of TERT mutation and
IDH-wild-type is the most common genotype observed in GBM
(41). Given these findings, the
combination of PAR5 expression, IDH and TERT
mutations may be useful to define glioma subclasses and predict
outcome in GBM. Assessment of expression of PAR5 in glioma patients
may be valuable for application as a molecular classification of
IDH-wild-type gliomas and a prognostic indicator of GBM.
However, further investigation between PAR5 and the
well-established molecular markers is necessary.
Moreover, restoration of PAR5 expression using a
PAR5-overexpressing plasmid resulted in retardation of
proliferation, decreased invasion and migration in the human glioma
cells, U87 and U251. Reversely, si-RNA targeting PAR5 promoted
proliferation and increased motility in the glioma cells. These
results suggest that PAR5 may act as a tumor suppressor in glioma,
and may serve as a novel therapeutic target and biomarker. The
abnormal expressional loss of PAR5 in glioma tissues may contribute
to tumor progression and the poor prognosis of patients with
glioma.
There is evidence to show that lncRNAs regulate
target expression through distinct mechanisms in different cancer
cells. Using RIP and RNA pull-down analysis, we showed that PAR5
could bind directly to several RNA binding proteins, including
EZH2, LSD1 and SUZ12, particularly EZH2. EZH2 is the core
structural catalytic subunit of PRC2, which can silence target
genes through induction of methylating lysine 27 of histone 3
(H3K27) and mediation of DNA methylation (42). EZH2 was identified as an RNA-binding
component protein of PRC2 and may bind cooperatively to target RNAs
(43). Moreover, EZH2 is pivotal to
maintain the undifferentiated status of neuroblastoma by epigenetic
repression of various tumor-suppressor genes (44). Reseach investigating the prognostic
role of EZH2 in glioma patients, suggested that EZH2 may be applied
as a potential biomarker for glioma (45–47).
Recently, researchers have proposed that lncRNAs may interact with
EZH2, thereby repressing target oncogene expression (48–50).
To determine whether PAR5 regulates targets using a similar
mechanism, we carried out western blotting to evaluate the relative
expression of various oncogene-coding proteins involved in
proliferation, tube formation and motility of glioma cells.
Cyclin A is a key molecular regulator of cell cycle
progression from S to G2 phase by activating CDK2. This regulatory
pathway is involved in cell proliferation and tube formation and is
closely associated with tumor development and morbidity (51,52).
Glioma cells commonly share abnormalities in pathway signaling
through the EGFR (53,54) and its downstream phosphoinositide
3-kinase/protein kinase B (PI3K/AKT) pathways (55). EGFR and VEGF-A are demonstrated to
play key regulatory roles in tumor tube formation (56–58).
Reports have revealed that EGFR overexpression in general carries
worse prognosis (59). Furthermore,
p-AKT and EGFR are well-known oncogenes that play essential roles
in the control of glioma cell proliferation (60). The present data found that the
expression levels of EGFR, VEGF-A, cyclin A and p-AKT are
downregulated after PAR5 restoration in glioma cells, suggesting
that these genes are involved in the carcinogenic progression of
glioma by PAR5-EZH2.
Given these findings, the present data indicated
that PAR5 may elicit decreased proliferation and motility in glioma
cells by interacting with EZH2 and altering expression levels of
various oncogenes, including EGFR, VEGF-A, cyclin A, AKT and p-AKT.
However, further high-throughput RNA sequencing, novel targets of
PAR5 and correlation analysis in glioma cells are needed.
In conclusion, the present study revealed decreased
PAR5 in glioma tissues of patients and glioma cells. The reduced
PAR5 expression was correlated with clinicopathological features
and the 5-year survival rate of glioma patients. Restoration of
PAR5 expression by pcDNA3.1-PAR5 transfection inhibited the
proliferation and motility of human glioma cells. RIP and RNA
pull-down assay showed that PAR5 directly interacted with EZH2.
Furthermore, restoration of PAR5 also suppressed expression levels
of EFGR, VEGF-A, cyclin A and p-AKT in glioma cells. These data
demonstrated that PAR5, as a tumor suppressor, reduced
proliferation and motility of glioma cells by interacting with EZH2
and regulating oncogene expression. This finding may provide a
therapeutic approach for the future treatment of glioma.
Acknowledgements
The present study was supported by a fund of the
Applied Basic Research of Yunnan Province, joint special project of
Kunming Medical University [grant no. 2017FE467 (−174)]; and
Research institutes in Yunnan medical and health units project
(grant no. 2017NS058).
References
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van den Bent MJ: Interobserver variation
of the histopathological diagnosis in clinical trials on glioma: A
clinician's perspective. Acta Neuropathol. 120:297–304. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Joseph JV, Balasubramaniyan V, Walenkamp A
and Kruyt FA: TGF-β as a therapeutic target in high grade gliomas -
promises and challenges. Biochem Pharmacol. 85:478–485. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nicolaidis S: Biomarkers of glioblastoma
multiforme. Metabolism. 64 Suppl 1:S22–S27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long non-coding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Caley DP, Pink RC, Trujillano D and Carter
DR: Long non-coding RNAs, chromatin, and development. Sci World J.
10:90–102. 2010. View Article : Google Scholar
|
|
8
|
Wapinski O and Chang HY: Long non-coding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JT: Epigenetic regulation by long
non-coding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takayama K, Horie-Inoue K, Katayama S,
Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K,
Takahashi S, et al: Androgen-responsive long noncoding RNA CTBP1-AS
promotes prostate cancer. EMBO J. 32:1665–1680. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang X, Song JH, Cheng Y, Wu W, Bhagat T,
Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al: Long
non-coding RNA HNF1A-AS1 regulates proliferation and migration in
oesophageal adenocarcinoma cells. Gut. 63:881–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo H, Wu L, Yang Q, Ye M and Zhu X:
Functional linc-POU3F3 is overexpressed and contributes to
tumorigenesis in glioma. Gene. 554:114–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Sun S, Pu JK, Tsang AC, Lee D,
Man VO, Lui WM, Wong ST and Leung GK: Long non-coding RNA
expression profiles predict clinical phenotypes in glioma.
Neurobiol Dis. 48:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and Leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maleszewska M and Kaminska B: Deregulation
of histone-modifying enzymes and chromatin structure modifiers
contributes to glioma development. Future Oncol. 11:2587–2601.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee J, Solomon DA and Tihan T: The role of
histone modifications and telomere alterations in the pathogenesis
of diffuse gliomas in adults and children. J Neurooncol. 132:1–11.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Conway E, Healy E and Bracken AP: PRC2
mediated H3K27 methylations in cellular identity and cancer. Curr
Opin Cell Biol. 37:42–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J, Hu L, Du Y, Kong F and Pan Y:
Prognostic role of LSD1 in various cancers: Evidence from a
meta-analysis. Onco Targets Ther. 8:2565–2570. 2015.PubMed/NCBI
|
|
22
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long non-coding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nie F, Yu X, Huang M, Wang Y, Ma H, Wang
Z, De W and Sun M: Long noncoding RNA ZFAS1 promotes gastric cancer
cells proliferation by epigenetically repressing KLF2 and NKD2
expression. Oncotarget. 8:38227–38238. 2017.PubMed/NCBI
|
|
24
|
Li W, Sun M, Zang C, Ma P, He J, Zhang M,
Huang Z, Ding Y and Shu Y: Upregulated long non-coding RNA
AGAP2-AS1 represses LATS2 and KLF2 expression through interacting
with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death
Dis. 7:e22252016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the Central Nervous System: a summary.
Acta Neuropathol. 131:803–820. 2016.doi: 10.1007/s00401-016-1545-1.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO Classification of Tumours of the Central Nervous System. Acta
Neuropathol. 114:97–109. 2007.doi: 10.1007/s00401-007-0243-4.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song S, Fajol A, Tu X, Ren B and Shi S:
miR-204 suppresses the development and progression of human
glioblastoma by targeting ATF2. Oncotarget. 7:70058–70065.
2016.PubMed/NCBI
|
|
28
|
Arita H, Narita Y, Matsushita Y, Fukushima
S, Yoshida A, Takami H, Miyakita Y, Ohno M, Shibui S and Ichimura
K: Development of a robust and sensitive pyrosequencing assay for
the detection of IDH1/2 mutations in gliomas. Brain Tumor Pathol.
32:22–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arita H, Narita Y, Fukushima S, Tateishi
K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP,
Kawahara N, et al: Upregulating mutations in the TERT promoter
commonly occur in adult malignant gliomas and are strongly
associated with total 1p19q loss. Acta Neuropathol. 126:267–276.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mulholland S, Pearson DM, Hamoudi RA,
Malley DS, Smith CM, Weaver JM, Jones DT, Kocialkowski S, Bäcklund
LM, Collins VP, et al: MGMT CpG island is invariably methylated in
adult astrocytic and oligodendroglial tumors with IDH1 or IDH2
mutations. Int J Cancer. 131:1104–1113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okita Y, Narita Y, Miyakita Y, Ohno M,
Matsushita Y, Fukushima S, Sumi M, Ichimura K, Kayama T and Shibui
S: IDH1/2 mutation is a prognostic marker for survival and predicts
response to chemotherapy for grade II gliomas concomitantly treated
with radiation therapy. Int J Oncol. 41:1325–1336. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Q, Matsuura K, Kleiner DE, Zamboni
F, Alter HJ and Farci P: Analysis of long non-coding RNA expression
in hepatocellular carcinoma of different viral etiology. J Transl
Med. 14:3282016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen ZH, Hu HK, Zhang CR, Lu CY, Bao Y,
Cai Z, Zou YX, Hu GH and Jiang L: Down-regulation of long
non-coding RNA FOXD3 antisense RNA 1 (FOXD3-AS1) inhibits cell
proliferation, migration, and invasion in malignant glioma cells.
Am J Transl Res. 8:4106–4119. 2016.PubMed/NCBI
|
|
34
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b
suppressesed human liver cancer cell proliferation and metastasis
by directly targeting oncogene LIN28B2. Hepatology. 52:1731–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He Z, Huang C, Lin G and Ye Y:
siRNA-induced TRAF6 knockdown promotes the apoptosis and inhibits
the invasion of human lung cancer SPC-A1 cells. Oncol Rep.
35:1933–1940. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Turkalp Z, Karamchandani J and Das S: IDH
mutation in glioma: New insights and promises for the future. JAMA
Neurol. 71:1319–1325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang YA, Ma X, Sathe A, Fujimoto J,
Wistuba I, Lam S, Yatabe Y, Wang YW, Stastny V, Gao B, et al:
Validation of SCT methylation as a hallmark biomarker for lung
cancers. J Thorac Oncol. 11:346–360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Malmström A, Grønberg BH, Marosi C, Stupp
R, Frappaz D, Schultz H, Abacioglu U, Tavelin B, Lhermitte B, Hegi
ME, et al Nordic Clinical Brain Tumour Study Group (NCBTSG), :
Temozolomide versus standard 6-week radiotherapy versus
hypofractionated radiotherapy in patients older than 60 years with
glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol.
13:916–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wick W, Platten M, Meisner C, Felsberg J,
Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M,
et al NOA-08 Study Group of Neuro-oncology Working Group (NOA) of
German Cancer Society, : Temozolomide chemotherapy alone versus
radiotherapy alone for malignant astrocytoma in the elderly: The
NOA-08 randomised, phase 3 trial. Lancet Oncol. 13:707–715. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arita H, Yamasaki K, Matsushita Y,
Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M,
Shimizu S, et al: A combination of TERT promoter mutation and MGMT
methylation status predicts clinically relevant subgroups of newly
diagnosed glioblastomas. Acta Neuropathol Commun. 4:792016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chinaranagari S, Sharma P and Chaudhary J:
EZH2 dependent H3K27me3 is involved in epigenetic silencing of ID4
in prostate cancer. Oncotarget. 5:7172–7182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Betancur JG and Tomari Y: Cryptic
RNA-binding by PRC2 components EZH2 and SUZ12. RNA Biol.
12:959–965. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei
JS, Marquez VE, Bates SE, Jin Q, Khan J, et al: EZH2 mediates
epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU,
RUNX3, and NGFR. Cancer Res. 72:315–324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wu Z, Wang Q, Wang L, Li G, Liu H, Fan F,
Li Z, Li Y and Tu Y: Combined aberrant expression of Bmi1 and EZH2
is predictive of poor prognosis in glioma patients. J Neurol Sci.
335:191–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Chen L, Han L, Shi Z, Zhang J, Pu
P and Kang C: EZH2 is a negative prognostic factor and exhibits
pro-oncogenic activity in glioblastoma. Cancer Lett. 356:929–936.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Y, Yu X, Chen L, Zhang Z and Feng S:
EZH2 overexpression is associated with poor prognosis in patients
with glioma. Oncotarget. 8:565–573. 2017.PubMed/NCBI
|
|
48
|
Huang M, Hou J, Wang Y, Xie M, Wei C, Nie
F, Wang Z and Sun M: Long noncoding RNA LINC00673 is activated by
SP1 and exerts oncogenic properties by interacting with LSD1 and
EZH2 in gastric cancer. Mol Ther. 25:1014–1026. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun CC, Li SJ, Li G, Hua RX, Zhou XH and
Li DJ: Long intergenic non-coding RNA 00511 acts as an oncogene in
non-small-cell lung cancer by binding to EZH2 and suppressing p57.
Mol Ther Nucleic Acids. 5:e3852016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ding J, Xie M, Lian Y, Zhu Y, Peng P, Wang
J, Wang L and Wang K: Long non-coding RNA HOXA-AS2 represses P21
and KLF2 expression transcription by binding with EZH2, LSD1 in
colorectal cancer. Oncogenesis. 6:e2882017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu F, Tong D, Li H, Liu M, Li J, Wang Z
and Cheng X: Bufalin enhances antitumor effect of paclitaxel on
cervical tumorigenesis via inhibiting the integrin α2/β5/FAK
signaling pathway. Oncotarget. 7:8896–8907. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ivkovic S, Canoll P and Goldman JE:
Constitutive EGFR signaling in oligodendrocyte progenitors leads to
diffuse hyperplasia in postnatal white matter. J Neurosci.
28:914–922. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Galvez-Contreras AY, Quiñones-Hinojosa A
and Gonzalez-Perez O: The role of EGFR and ErbB family related
proteins in the oligodendrocyte specification in germinal niches of
the adult mammalian brain. Front Cell Neurosci. 7:2582013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huse JT and Holland EC: Targeting brain
cancer: Advances in the molecular pathology of malignant glioma and
medulloblastoma. Nat Rev Cancer. 10:319–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan
G and Jiang Y: HULC long non-coding RNA silencing suppresses
angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling
pathway in human gliomas. Oncotarget. 7:14429–14440. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang Q, Cheng F, Ma TT, Xiong HY, Li ZW,
Xie CL, Liu CY and Tu ZG: Interleukin-12 inhibits the
hepatocellular carcinoma growth by inducing macrophage polarization
to the M1-like phenotype through downregulation of Stat-3. Mol Cell
Biochem. 415:157–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sun L, Zhang Q, Li Y, Tang N and Qiu X:
CCL21/CCR7 up-regulate vascular endothelial growth factor-D
expression via ERK pathway in human non-small cell lung cancer
cells. Int J Clin Exp Pathol. 8:15729–15738. 2015.PubMed/NCBI
|
|
59
|
Jansen M, Yip S and Louis DN: Molecular
pathology in adult gliomas: Diagnostic, prognostic, and predictive
markers. Lancet Neurol. 9:717–726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|