Introduction
Glioma is one of the most common primary
intracranial malignant tumors in adults (1), and survival time after diagnosis is
approximately one year. The profuse infiltration of glioma cells
into healthy tissue surrounding the main tumor mass is one of the
major obstacles limiting the improvement of patient survival
(2). The incidence and mortality
rate of glioma still continues to increase (3). Although many genetic and epigenetic
changes were found to be related to glioma, the pathogenesis
remains poorly understood.
miRNAs are a class of small molecular non-coding
regulatory RNAs, which act as oncogenes or tumor suppressors in a
variety of tumors (4,5). Systematic and integrative analysis
could detect key miRNAs that contribute to cancer development
(6). The abnormal expression of
miRNAs involved in transcriptional regulation network leads to
tumor initiation and progression. Its main mechanism includes
one-to-many and many-to-one regulation between transcription factor
to miRNA and miRNA to target gene, which increases the complexity
of miRNA regulation, thus, affecting the biological behavior of the
tumor (7). Hence, there is
considerable interest in using deregulated miRNAs as prognosis
prediction markers and understanding their molecular targets for
cancer treatment.
During glioma occurrence and progression, abnormal
expression of miRNAs was found to exert important functions.
miR-182, miR-199b and miR-203 were upregulated (8–10),
while miR-451, miR-17 and miR-184 were downregulated in glioma
tissues (11,12). These miRNAs were also reported to be
involved in the development of glioma. However, miRNAs influencing
glioma occurrence and progression is far from completely
investigated, and no miRNA marker is used in clinic. Thus, there is
still a growing need for developing prognostic markers and
identifying therapeutic targets for improving the outcomes of
glioma patients.
In the present study, we found that miR-1908 was
significantly upregulated in glioma tissues. High level of miR-1908
was correlated with the shorter survival time of glioma, and
promoted glioma cell proliferation and invasion and suppressed
apoptosis. miR-1908 exerted these functions by regulating
SPRY4/RAF1 axis and changing the expression of apoptosis related
proteins Bcl-2/Bax and matrix metallopoteinase-2 (MMP-2). These
results help to elucidate the pathogenesis and predict prognosis of
glioma.
Materials and methods
Bioinformatics analysis
To identify the expression of miRNAs in primary
human glioma and normal cerebral tissue, data about miRNA
expression level in glioma patients were obtained from Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/). Clinical data such
as overall survival (OS), disease-free survival (DFS), lymph node
metastasis, TNM stage and miR-1908 expression level were extracted
from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). Target genes of
miR-1908 were analyzed by TargetScan (http://www.targetscan.org/) and miRTarBase database
(experimentally validated miRNA-target interactions database,
http://mirtarbase.mbc.nctu.edu.tw/).
Enrichment analysis was performed by cBioPortal online database
(http://www.cbioportal.org/). The
pathological images and IHC data of glioma were downloaded from the
Human Protein Atlas web portal (www.proteinatlas.org), the sum IOD was analyzed by
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.,
Rockville, MD, USA).
Gene Ontology and KEGG pathway
analyses
To explore the functional annotation enrichment of
the target genes of miR-1908 in glioma, GO and KEGG analyses were
conducted using the Database for Annotation, Visualization and
Integrated Discovery (DAVID) online analysis tool (https://david.ncifcrf.gov/). The database provides a
comprehensive set of functional annotation tools for investigators
to understand biological meaning behind large list of genes,
especially for identifying enriched biological themes.
Cell culture and miRNA
transfection
Human glioma cell line U251 was purchased from the
Cell Resource Center of Beijing Xiehe (Beijing, China) and
cultivated in an incubator at 37°C with 5% CO2. U251
cells were maintained in high-glucose Dulbeccos modified Eagles
medium (DMEM; Gibco, Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; HyClone Laboratories, Inc., Logan, UT, USA) as
well as penicillin (100 U/ml; Thermo Fisher Scientific, Waltham,
MA, USA).
miR-1908 mimics, miR-1908 inhibitor and their
negative control (NC) sequences were designed and synthesized by
Suzhou GenePharma Co., Ltd. (Shanghai, China). Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA) was used for miRNA oligo
transfection according to the manufacturers protocol. After 48 h of
transfection, cells were used for the following experiments.
RNA isolation and quantitative
real-time PCR assay (qRT-PCR)
Total RNA was extracted from U251 cells transfected
with miR-1908 mimics or inhibitor using the TRIzol reagent
(Invitrogen) according to the manufacturers protocol. RNA quantity
and purity were assessed using NanoDrop ND-1000 (Thermo Fisher
Scientific). Total RNA was reverse-transcribed to cDNA, and the
first-strand cDNA was used as a template for real-time PCR
(13). Reactions of qRT-PCR was
done on the ABI Prism 7500 (Applied Biosystems, Foster City, CA,
USA) using the commercially available gene expression assay for
miR-1908, SPRY4 and RAF1. All reactions were run in triplicate.
Relative gene expression was quantified using U6 or GAPDH as an
internal control.
CCK-8 assay
Cells transfected with miR-1908 mimics or inhibitor
sequence were plated at a density of 5×103 cells/well
onto 96-well plates at 37°C in an incubator with 5% CO2.
Cell proliferation was then assessed every 24 h using Cell Counting
kit-8 (CCK-8; Sigma Aldrich St. Louis, MO, USA) according to
standard protocol. For each sample at each time-point, six wells
were analyzed. The experiment was repeated three times.
Cell invasion
Cell invasion abilities were determined in Transwell
assays. Briefly, to determine the invasion potential, DMEM
containing 10% FBS was added to the lower chambers, and
1×105 cells suspended in 200 µl serum-free medium were
added to the upper chambers with Matrigel matrix gel and cultured
for 24 h. Finally, the cells that traversed the membrane were
stained with crystal violet and counted.
Gelatin zymography assay
Activities of MMP-2 and MMP-9 were assessed by
gelatin zymography assay. U251 cells were cultured for 24 h after
transfecting the miR-1908 mimics or inhibitor. At the end of
incubation, 40 µl of culture supernatant was mixed with sample
buffer and resolved on a 10% SDS-PAGE under non-reducing
conditions. The gel was co-polymerized containing 0.5 mg/ml of
gelatin (Sigma-Aldrich). Gel was washed twice for 30 min with
renaturation buffer (2.5% Triton X-100) at room temperature before
incubation in the incubation buffer (50 mM Tris-HCl pH 7.5, 200 mM
NaCl, 10 mM CaCl2, 1 µM ZnCl2) at 37°C for 36
h. Thereafter, gel was stained for 2 h in 0.25% Coomassie brilliant
blue R-250 and then de-stained. White bands were observed against a
blue background after de-staining, indicating gelatinolytic
activities of MMP-2 and MMP-9.
Western blotting
Cells were collected and lysed in lysis buffer
(Beijing CoWin Biotech Co., Ltd., Beijing, China) in the presence
of protease inhibitors for 30 min to extract total protein from
cells transfected with miR-1908 mimics or inhibitor, and protein
levels were quantified using bicinchoninic acid assays (Beijing
CoWin Biotech). Subsequently, 30 µg protein from each sample was
loaded onto 10% sodium dodecyl sulfate (SDS) polyacrylamide gels
and subjected to SDS-polyacrylamide gel electrophoresis (PAGE;
Beijing CoWin Biotech). Protein was then transferred to
nitrocellulose membranes (Sigma-Aldrich), which were then blocked
with bovine serum albumin buffer (BSA; Invitrogen) for 1 h.
Membranes were then incubated with primary antibodies targeting
SPRY4 (1:1,000 dilution, cat. no. ab59785; Abcam), RAF1 (1:1,000
dilution, cat. no. ab154754; Abcam), Bax (1:2,000 dilution, cat.
no. ab182733; Abcam), Bcl-2 (1:1,000 dilution, cat. no. ab194583;
Abcam) or GAPDH (1:5,000 dilution, cat. no. ab70699; Abcam)
overnight at 4°C, followed by incubation with goat anti-rabbit
horseradish peroxidase (HRP)-conjugated secondary antibody (1:3,000
dilution, cat. no. CW0103; Beijing CoWin Biotech) for 1 h at room
temperature. Detection was facilitated using an enhanced
chemiluminescence kit and images were analyzed using ImageJ
software (version 1.62; National Institutes of Health, Bethesda,
MD, USA).
Annexin V-FITC/PI apoptosis
detection
Annexin V-FITC/PI (BD Biosciences, Bedford, MA, USA)
was used to detect cell apoptosis rate. In brief, U251 cells
transfected with miR-1908 mimics or inhibitor were washed twice
with cold phosphate-buffered saline (PBS), and cells were added in
400 µl binding buffer (provided by the manufacturer) at a
concentration of 1×106 cells/ml. Annexin V-FITC (5 µl)
and PI (5 µl) were then added, and the cells were analyzed with a
flow cytometer (BD Accuri C6; BD Biosciences) within 1 h (13).
Protein-protein interaction (PPI)
network construction
Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING; Search Tool for the Retrieval of
Interacting Genes, http://string-db.org/) is a database of known and
predicted protein interactions that may aid in the comprehensive
description of cellular mechanisms and functions. The PPI network
of the selected target genes of miR-1908 in glioma was constructed
using the STRING database.
Statistical analysis
Statistical analyses were performed using the SPSS
21.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism (GraphPad
Software, Inc., San Diego, CA, USA). GEO microarray data and TCGA
data were analyzed by paired sample t-test. The log-rank test for
the generated Kaplan-Meier (K-M) curve was conducted to evaluate
the association between the expression level of miR-1908 and the
survival rate. OS was defined as the time between the first surgery
for primary glioma and death for any reason. DFS was defined as the
time between the first surgical resection and disease recurrence.
Growth curves were analyzed using a repeated measures ANOVA
followed by Bonferroni post hoc analysis. Cell invasion data,
apoptosis data and IHC data were analyzed by independent samples
t-test. The correlation of target genes of miR-1908 expression
level was analyzed by the Pearson correlation analysis. P<0.05
was considered to indicate a statistically significant
difference.
Results
Increased expression of miR-1908 in
glioma and its association with shorter survival time in the
patients
We used GEO microarray and TCGA miRNA dataset to
screen miRNAs which related to glioma occurrence and development.
Three pairs of glioma tissues and adjacent normal cerebral tissues
were compared by miRNA microarray, 53 differentially expressed
miRNAs (P<0.05; FC>1 and FC<-1) were identified. Of which,
miR-1908 was one of rarely reported miRNAs in glioma and attracted
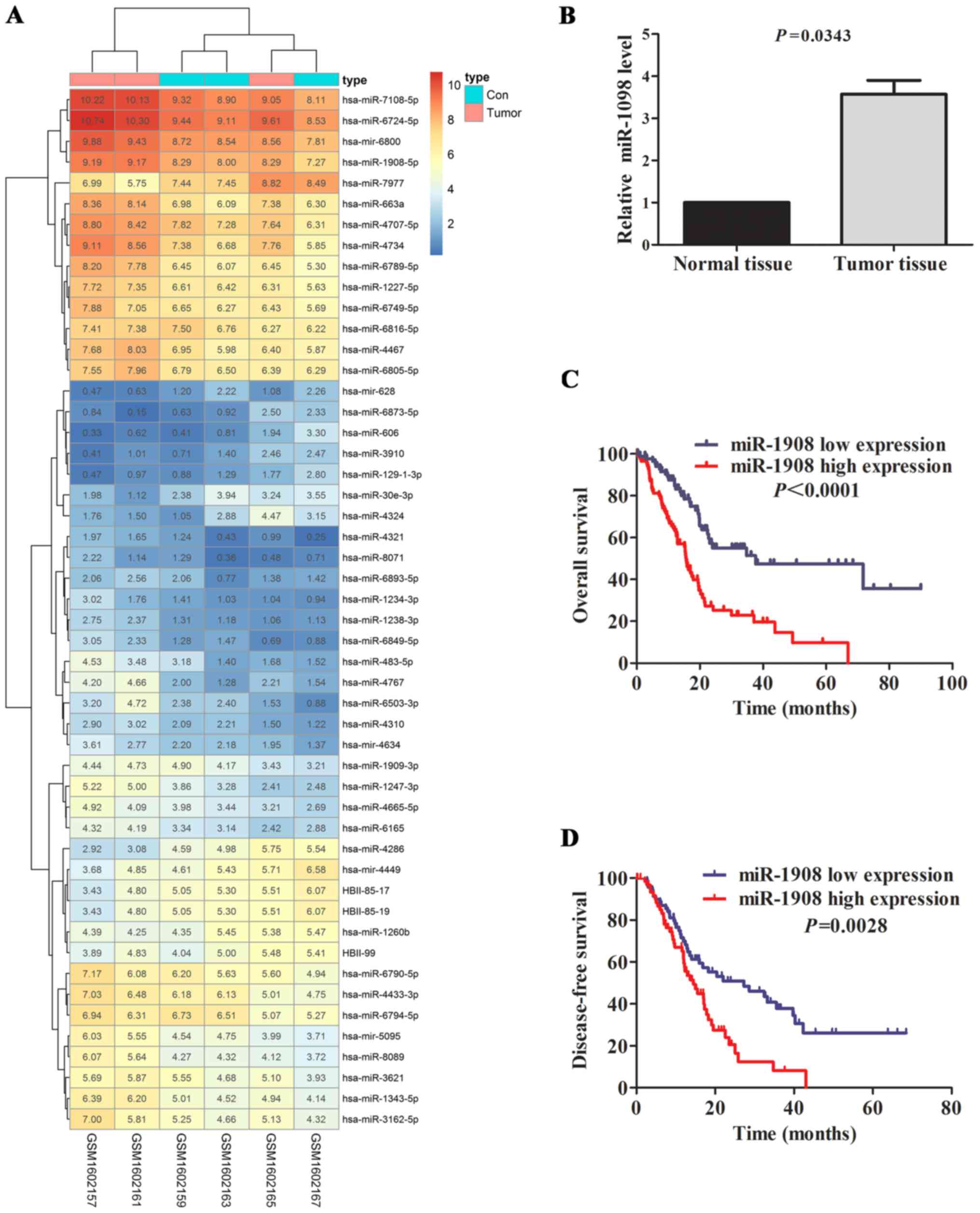
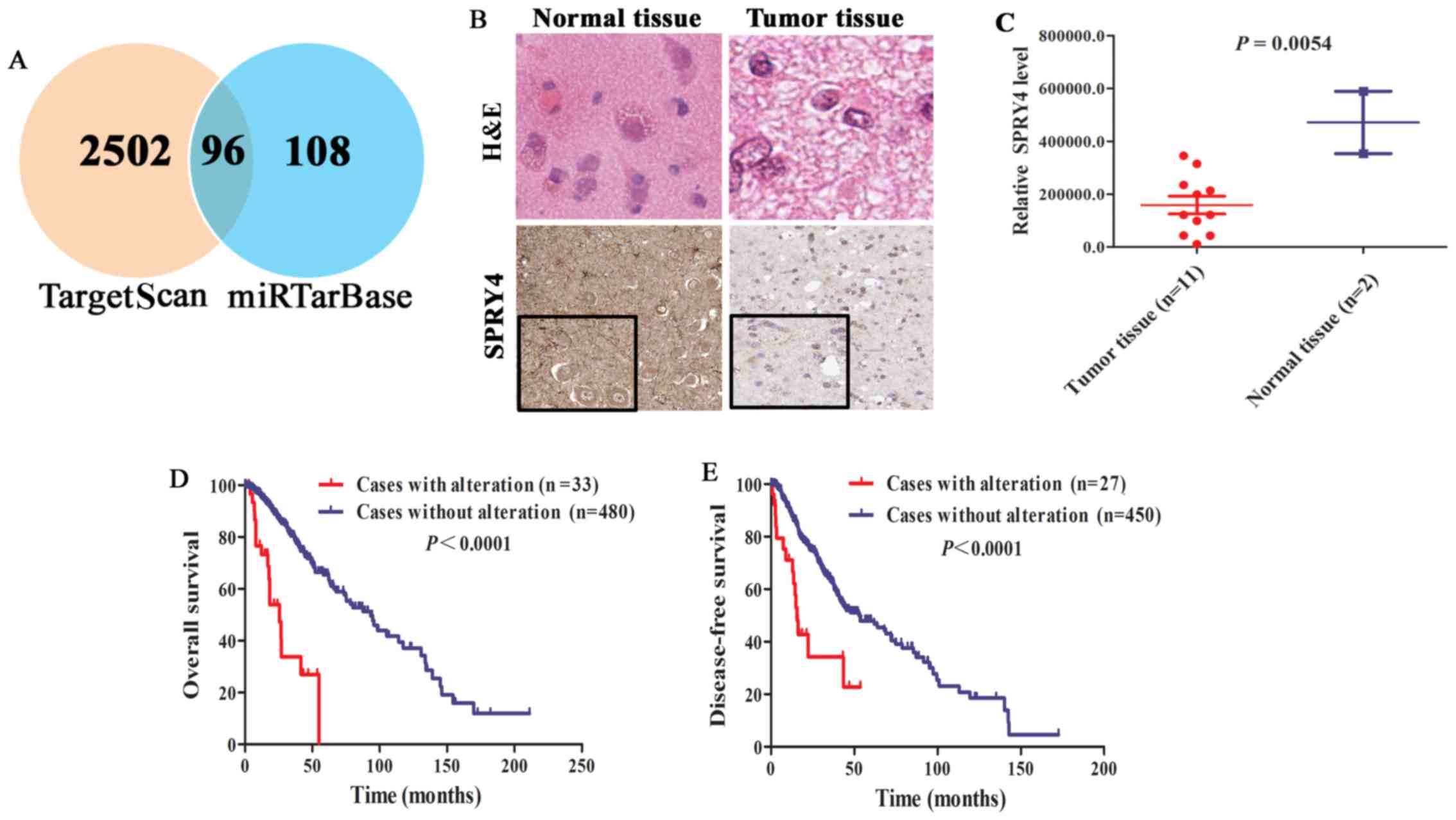
our interest (Fig. 1A and Table I).
 | Table I.The main dysregulated miRNAs in glioma
tissue. |
Table I.
The main dysregulated miRNAs in glioma
tissue.
| No. | Name | log2 fold
change | Average
expression | P-value |
|---|
| 1 | hsa-miR-129-1-3p | –1.182734902 | 1.36366824 | 0.026579 |
| 2 | hsa-miR-6749-5p | 1.063017222 | 6.664388833 | 0.027495 |
| 3 |
hsa-miR-6789-5p | 1.538438528 | 6.707455986 | 0.027804 |
| 4 | hsa-mir-6800 | 1.041350944 | 8.824345889 | 0.027867 |
| 5 | hsa-miR-4310 | 1.023917706 | 2.157122464 | 0.028345 |
| 6 | hsa-mir-628 | –1.084536185 | 1.309829702 | 0.029722 |
| 7 |
hsa-miR-1908-5p | 1.031185667 | 8.369524444 | 0.030411 |
| 8 | hsa-miR-7977 | –1.52912775 | 7.489592208 | 0.033013 |
| 9 |
hsa-miR-1238-3p | 1.018972278 | 1.632209472 | 0.033941 |
| 10 |
hsa-miR-3162-5p | 1.320415407 | 5.361444565 | 0.033947 |
| 11 | hsa-mir-5095 | 1.224927278 | 4.761240056 | 0.036548 |
| 12 |
hsa-miR-6503-3p | 1.830256801 | 2.51717765 | 0.03752 |
| 13 | hsa-miR-4767 | 1.9436247 | 2.648728206 | 0.037793 |
| 14 | hsa-miR-8089 | 1.274433444 | 4.688330611 | 0.039185 |
| 15 |
hsa-miR-6724-5p | 1.074298778 | 9.620908778 | 0.043217 |
To further validate the abnormal regulated
expression level of miR-1908, we compared the expression level of
miR-1908 in glioma tissue and normal tissue by GEO database.
Consistent with previous reports, miR-1908 was significantly
upregulated in tumor tissue specimens (n=89) compared with matched
normal controls (n=32) (Fig. 1B;
P=0.0343). To further determine the predictive value of miR-1908
expression level as prognostic marker, 206 glioma patients from
TCGA database were divided into miR-1908 high expression and
miR-1908 low expression groups according to median miR-1908
expression level. K-M curve showed that miR-1908 high expression
group had shorter OS and DFS time than miR-1908 low expression
group in terms of survival duration (Fig. 1C and D; P<0.0001 and P=0.0028,
respectively), particularly in overall survival. The result
suggests that upregulated miR-1908 may be a novel prognostic marker
for glioma patients.
miR-1908 is involved in multiple types
of cancer-related pathways
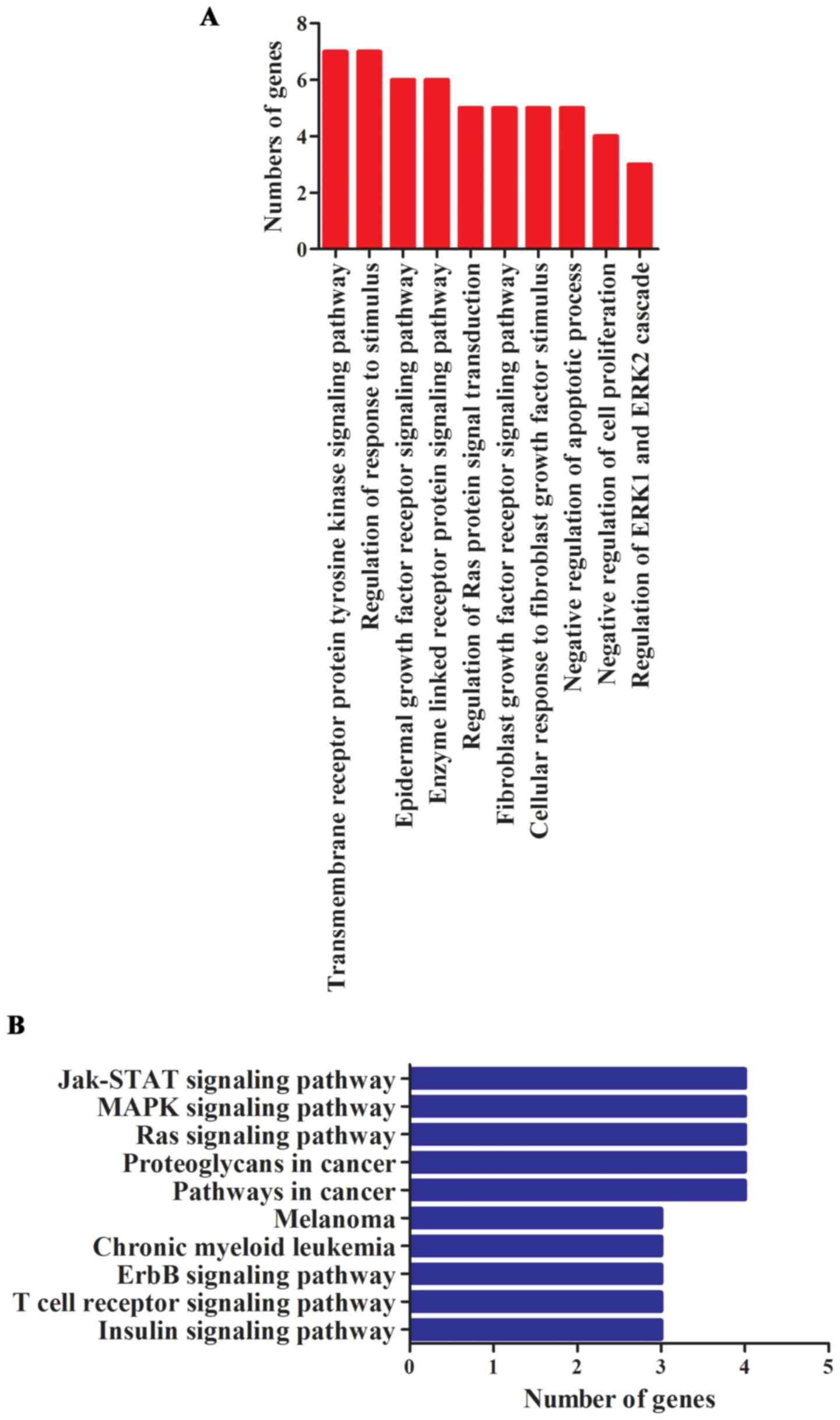
In order to understand the function and role of
miR-1908 in tumor progression, GO and KEGG pathway analysis were
performed. We noted that these genes were especially enriched in
functions of regulating cell proliferation, invasion and apoptosis
(Fig. 2A). miRNAs were reported to
promote cancer occurrence and progression through activating
related signaling pathways. To better understand underlying
molecular mechanisms in which miR-1908 plays roles, we investigated
miR-1908 relevant KEGG pathways. Results revealed that miR-1908 was
involved in 54 significant KEGG pathways, the main signaling
pathways are shown in Fig. 2B.
These pathways mainly included multiple types of cancer-related
signaling pathways, especially the Jak-STAT, MAPK and Ras signaling
pathways.
miR-1908 promotes cell proliferation
and invasion of glioma
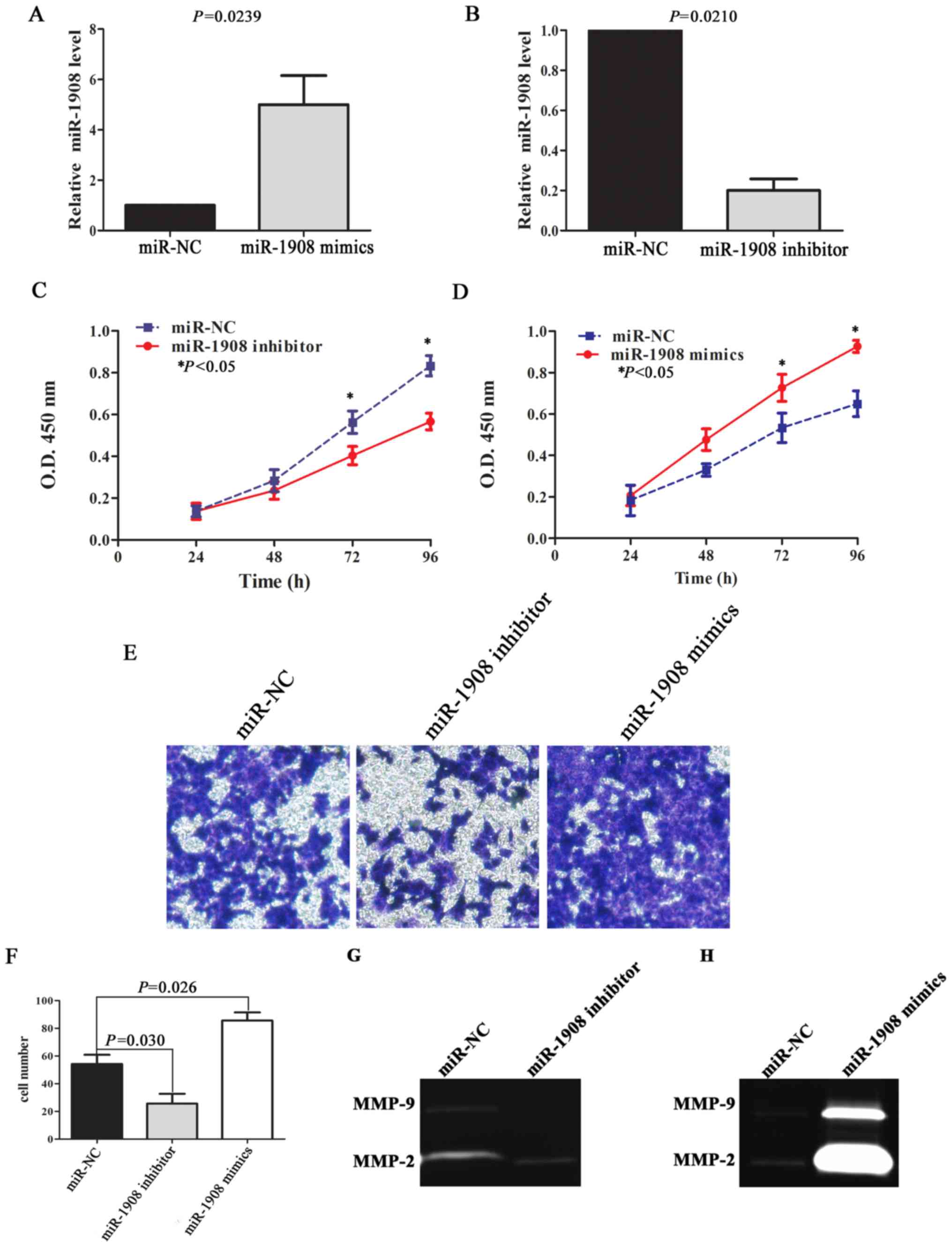
In order to demonstrate the effect of miR-1908 on
glioma malignant phenotype, we performed cell proliferation and
invasion assays. qRT-PCR results reveal that relative miR-1908
level was upregulated in U251 cells transfected with miR-1908
mimics (Fig. 3A; P=0.0239), and
miR-1908 level was downregulated after miR-1908 inhibitor
transfection (Fig. 3B; P=0.0210).
The ability of miR-1908 to modulate the proliferation of U251 cells
was analyzed using a CCK-8 assay. The results indicated that
downregulated miR-1908 could inhibit cell proliferation in 72 and
96 h (Fig. 3C; P=0.0285 and
P=0.0152, respectively), while upregulated miR-1908 significantly
enhanced the ability of cell proliferation (Fig. 3D; P=0.0254 and P=0.0196,
respectively). Similarly, upregulated miR-1908 also significantly
promoted cell invasion compared with control group through
Transwell assay, on the contrary, downregulated miR-1908 in U251
cells inhibited invasion (Fig. 3E and
F; P=0.026 and P=0.030, respectively).
Previous studies have shown that proteolysis is a
necessary part of the invasion process (14), and that increased expression of
several members of the MMP family are correlated with high-grade
gliomas, especially MMP-2/9 (15).
We therefore detected the activity of MMP-2/9 by using gelatin
zymography assay. The results revealed that upregulated miR-1908
expression significantly enhanced the activity of MMP-2, but there
was no significant difference in MMP-9 activity between miR-1908
mimics group and control group (Fig. 3G
and H).
miR-1908 inhibits cell apoptosis via
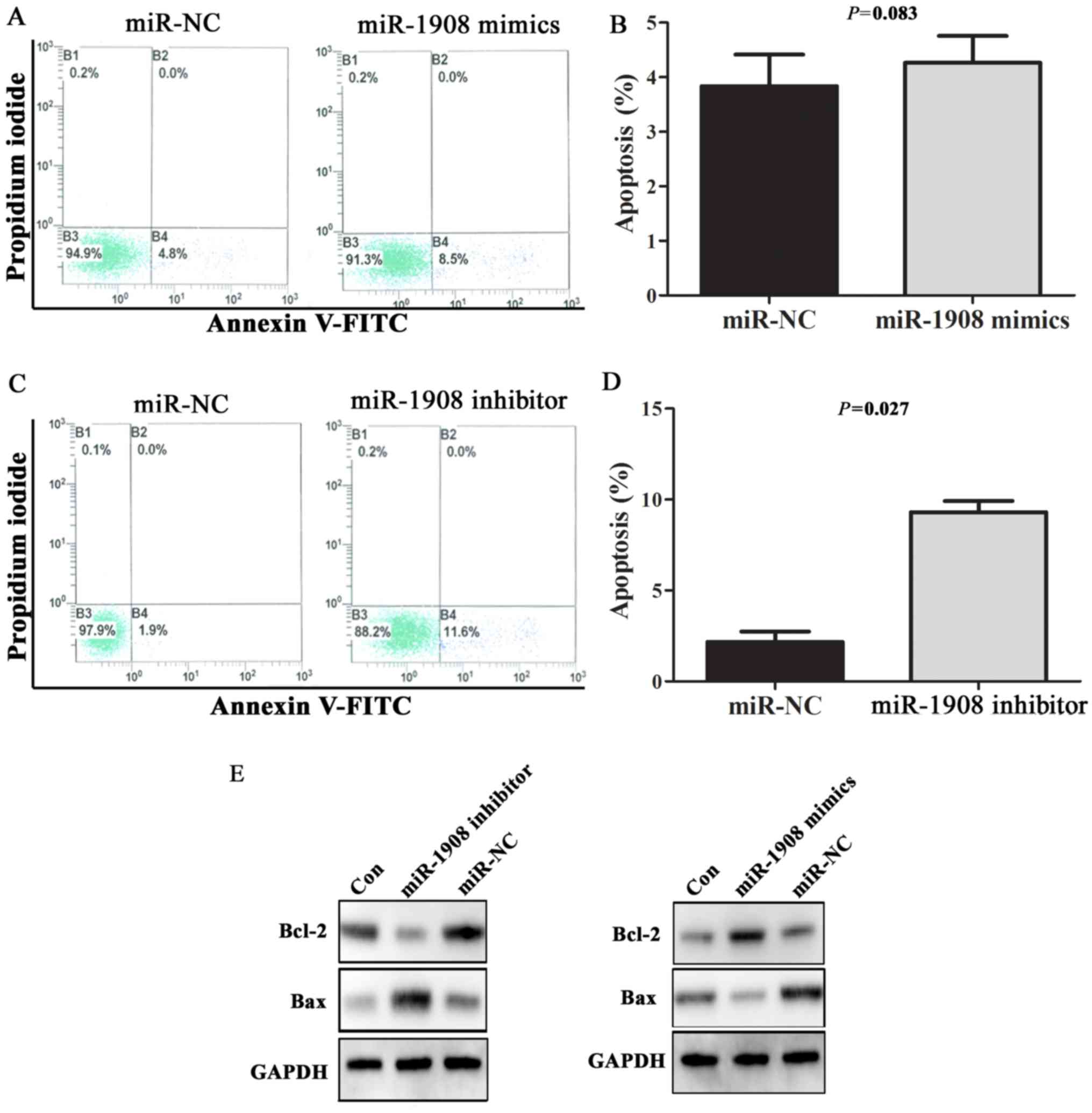
regulating Bax/Bcl-2 expression
The processes of apoptosis, which induce degradation
of proteins and organelles or cell death upon cellular stress, are
crucial in the pathophysiology of various tumors. In order to
observe the effect of miR-1908 overexpression on cell apoptosis,
miR-1908 mimics or inhibitor were transfected into U251 cells, and
apoptosis rate and the expression of apoptosis-associated protein
Bax/Bcl-2 were detected by flow cytometry and western blot
analysis, respectively. Representative results of flow cytometric
analysis are shown in Fig. 4A and
C. In general, the rates of cell death were low (<5%).
Apoptosis rate was not statistically different in cells transfected
with miR-1908 mimics and negative control sequences (Fig. 4B; P=0.083). However, U251 cells
transfected with miR-1908 inhibitor had significantly higher rate
of apoptosis when compared with cells transfected with negative
control sequences (Fig. 4D;
P=0.027). Furthermore, transfection of miR-1908 inhibitor caused an
increase in the expression of Bax, a pro-apoptotic protein, but a
marked decrease in the expression of Bcl-2, an anti-apoptotic
protein. In contrast, transfection of miR-1908 mimics downregulated
the expression level of Bax. These data supported that miR-1908
enhances the ability of cell anti-apoptosis via regulating
Bax/Bcl-2 expression.
SPRY4 is a key target gene of miR-1908
in glioma
miRNAs exert important functions in various
biological processes via directly suppressing the expression of
their target genes. According to integration analysis from
TargetScan and miRTarBase database, 96 verified target genes
(5) of miR-1908 were identified
(Fig. 5A and Table II). Of which, SPRY4 was
significantly downregulated in glioma tissue (n=11) compared with
normal tissue (n=2) (Fig. 5B and C;
P=0.0054). Moreover, patients with SPRY4 alteration had lower OS
and DFS compared with patients without alteration (Fig. 5D and E; P<0.0001). Hence, SPRY4
may be a key and specific target gene of miR-1908 in glioma.
 | Table II.The main target genes of
miR-1908. |
Table II.
The main target genes of
miR-1908.
| No. | Gene name | Cumulative weighted
context score | Total context
score | Aggregate PCT |
|---|
| 1 | PRSS22 | –1.51 | –1.51 | N/A |
| 2 | TOR4A | –1.02 | –1.02 | N/A |
| 3 | ZNF385A | –0.9 | –0.9 | N/A |
| 4 | DPF1 | –0.9 | –0.9 | N/A |
| 5 | SRCIN1 | –0.8 | –0.8 | N/A |
| 6 | SCAMP4 | –0.79 | –0.79 | N/A |
| 7 | RAB11B | –0.75 | –0.97 | N/A |
| 8 | GNAI2 | –0.74 | –0.74 | N/A |
| 9 | NRGN | –0.7 | –0.7 | N/A |
| 10 | NKX2–5 | –0.7 | –0.78 | N/A |
| 11 | H2AFX | –0.62 | –0.62 | N/A |
| 12 | POU3F1 | –0.54 | –0.54 | N/A |
| 13 | BARHL1 | –0.53 | –0.53 | N/A |
| 14 | FAM83H | –0.49 | –0.49 | N/A |
| 15 | SLC11A1 | –0.49 | –0.49 | N/A |
| 16 | GRB2 | –0.45 | –0.45 | N/A |
| 17 | CALR | –0.43 | –0.43 | N/A |
| 18 | PRX | –0.42 | –0.42 | N/A |
| 19 | POFUT2 | –0.41 | –0.41 | N/A |
| 20 | TACC3 | –0.41 | –0.41 | N/A |
| 21 | CASZ1 | –0.41 | –0.41 | N/A |
| 22 | GPR20 | –0.4 | –0.48 | N/A |
| 23 | SIGLEC12 | –0.4 | –0.4 | N/A |
| 24 | MYADM | –0.38 | –0.45 | N/A |
| 25 | SPRY4 | –0.38 | –0.47 | N/A |
| 26 | SYNGR1 | –0.37 | –0.37 | N/A |
| 27 | MARVELD1 | –0.36 | –0.36 | N/A |
| 28 | PHF15 | –0.36 | –0.53 | N/A |
| 29 | APOE | –0.33 | –0.33 | N/A |
| 30 | NACC1 | –0.33 | –0.36 | N/A |
SPRY4/RAF1 axis is regulated by
miR-1908 in glioma
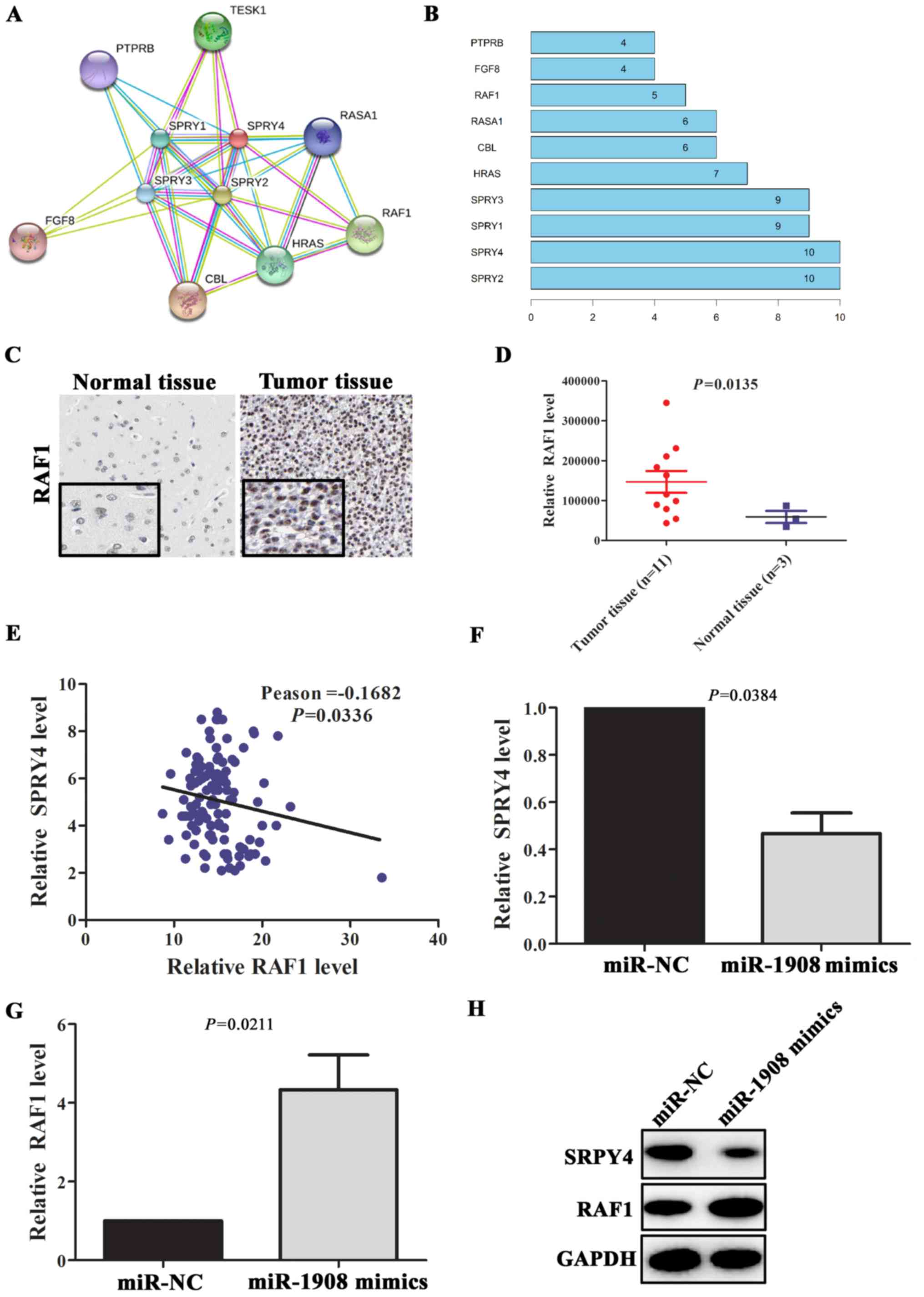
PPI analysis revealed that SPRY4 can interact with
multiple proteins, included SPRY2, SPRY1, SPRY3, HRAS, CBL, RASA1,
RAF1, FGF8 and PTPRB (Fig. 6A and
B). Above all, RAF1 is a well-known pro-oncogene in multiple
types of tumors (16). The abnormal
upregulation of RAF1 protein plays an important role in the
malignant transformation of tumor. In glioma tissues (n=11), RAF1
protein expression levels was significantly upregulated (Fig. 6C and D; P=0.0135) compared with
normal tissues (n=3). To further verify the relationship between
SPRY4 and RAF1, correlation analyses between expression levels of
SPRY4 and RAF1 were performed in glioma tissues. The results showed
that the expression level of SPRY4 was negatively correlated with
the expression level of RAF1 (Fig.
6E; Pearson=-0.1682, P=0.0336). In addition, gene co-expression
analysis revealed that RAF1 was significantly co-expressed with
SPRY4 (Table III). To further
validate whether miR-1908 could promote the expression of RAF1 via
targeting SPRY4, miR-1908 mimics were transfected into U251 cells.
qRT-PCR assay showed that SPRY4 mRNA level was downregulated in
these cells, on the contrary, RAF1 mRNA level was significantly
upregulated (Fig. 6F and G;
P=0.0384 and P=0.0211, respectively). Similarly, compared with
negative group, the expression level of SPRY4 in U251 cells
transfected with miR-1908 mimics was significantly downregulated
and the expression level of RAF1 was increased (Fig. 6H). These data suggested that
miR-1908 contributed to tumor progression through modulation of
SPRY4/RAF1 axis in glioma.
 | Table III.The co-expession genes with SPRY4
ranked by cBioPortal database. |
Table III.
The co-expession genes with SPRY4
ranked by cBioPortal database.
| Gene | Mean protein
expression (Altered) | Mean protein
expression (Unaltered) | P-value | q-value |
|---|
| RAF1 | –0.11 | –0.44 | 9.61E-06 | 1.39E-03 |
| NRG1 | 0.13 | 0.33 | 1.21E-05 | 1.39E-03 |
| EIF4G1 | –0.31 | –0.61 | 2.93E-05 | 2.24E-03 |
| PRKAA1_PT172 | 0.25 | –0.05 | 1.06E-04 | 3.55E-03 |
| ARAF_PS299 | –0.13 | –0.03 | 1.15E-04 | 3.55E-03 |
| MYH11 | –1.75 | –0.8 | 1.26E-04 | 3.55E-03 |
| PGR | –0.1 | 0 | 1.30E-04 | 3.55E-03 |
| GATA3 | –0.52 | –0.28 | 1.35E-04 | 3.55E-03 |
| BAX | –0.1 | –0.51 | 1.40E-04 | 3.55E-03 |
| ADAR | –0.46 | –0.24 | 1.58E-04 | 3.61E-03 |
| RAB25 | –1.28 | –0.91 | 1.91E-04 | 3.97E-03 |
| IRS1 | –0.18 | 0.03 | 4.04E-04 | 6.74E-03 |
| ANXA7 | –0.02 | 0.17 | 4.88E-04 | 6.74E-03 |
| PARK7 | 0.12 | 0.37 | 5.00E-04 | 6.74E-03 |
Discussion
In the present study, integrative analysis of GEO
and TCGA data suggested that the expression level of miR-1908 was
significantly upregulated in glioma tissues and was associated with
shorter survival time of glioma patients. Furthermore, we verified
that miR-1908 could promote glioma cell proliferation, invasion,
anti-apoptosis and regulate SPRY4/RAF1 axis. These results
elucidated that miR-1908 is a novel prognosis marker via promoting
malignant phenotype and modulating SPRY4/RAF1 axis. To the best of
our knowledge, this is the first report showing prognostic value of
miR-1908 for glioma patients.
A previous work demonstrated that miR-1908 was
aberrantly expressed in several types of tumors, including
osteosarcoma (17), lung (18) and liver cancer (19). miR-1908 is strongly associated with
cell proliferation and migration (17), and poor prognosis of osteosarcoma
patients (20). miR-1908 exerts
these effects via regulating many signaling pathways of tumor
formation. For example, overexpressed miR-1908 clusters
downregulate the MARK1 signaling pathway to alter cell
proliferation and differentiation in hepatoma cells, and serves as
a biomarker for poor prognosis in osteosarcoma (20). miRNA-1908 functions as an oncogene
in glioblastoma by repressing the PTEN signaling pathway (21). Notably, previous bioinformatics
analysis also found that miR-1908 may contribute to glioma
progression and be correlated with survival rate (22), but lacks sufficient experimental
bases. In this study, we not only showed that miR-1908 acts as a
novel prognostic marker for glioma, but also explored its possible
mechanisms. Clinical data showed that high expression of miR-1908
was correlated with shorter OS and DFS. Upregulation of miR-1908
in vitro promoted glioma cell proliferation and invasion,
and suppressed its apoptosis. These data indicated that miR-1908
contributes to poor prognosis of glioma patients via promoting
glioma cell malignant behavior.
miRNAs play their roles via suppressing the
expression of target mRNAs. Actually, the biological interactions
between miRNAs and their targets are very complex in vivo.
One miRNA may target multiple genes and target genes are tissue
specific (23,24). On the basis of known target gene
database, we further combined TargetScan and miRTarBase datasets
and used cBioPortal online analysis tool to find the tissue
specific and key target genes of miR-1908 in glioma. Among them,
SPRY4, a tumor suppressor, was downregulated in glioma patients and
patients with SPRY4 alteration had shorter OS and DFS. Hence, SPRY4
was identified as a key target gene of miR-1908.
Our further analysis revealed that mainly ten
proteins in glioma could interact and form the complex with SPRY4.
Among these ten interacting proteins, RAF1 is a well-known
pro-oncogene (25). In glioma
tissue, SPRY4 expression level was significantly downregulated, but
RAF1 expression level was significantly upregulated. The
relationship between SPRY4 and RAF1 were further verified by
correlation analysis of expression levels in tissues and in
vitro. miR-1908 could suppress the expression of SPRY4 and
upregulate RAF1 expression. Since proto-oncogene RAF1 serves as
part of the mitogen-activated protein kinases/extracellular
signal-regulated kinase signal transduction pathway and regulates
cell migration, apoptosis and differentiation (26), we speculate that miR-1908 may
promote glioma progression through regulating SPRY4/RAF1 axis.
Further experiments are necessary to verify our hypothesis.
In conclusion, miR-1908 is potentially a novel
prognostic biomarker for glioma via promoting glioma cell
proliferation, invasion, anti-apoptosis and regulating SPRY4/RAF1
axis. This study provides a novel prognostic marker and a new
treatment direction for glioma.
Acknowledgements
The present study was supported by funding from the
National Natural Science Foundation of China (no. 81102552), the
International Science and Technology Cooperation Project of Shanxi
Province (no. 2014081049-4), the Research Project Supported by
Shanxi Scholarship Council of China (no. 2017-129), the Returned
Chinese Scholars Technology Activities Preferred Project, Shanxi
Province of China (no. 2017-19) and the Great Science and
Technology Innovation Team Project of Traditional Chinese Medicine
of Shanxi University (no. 20150401).
Glossary
Abbreviations
Abbreviations:
|
SPRY4
|
sprouty RTK signaling antagonist 4
|
|
RAF1
|
Raf-1 proto-oncogene, serine/threonine
kinase
|
|
MMP-2/9
|
matrix metalloproteinase −2/9
|
|
BAX
|
BCL2 associated X protein
|
References
|
1
|
Lin L, Wang G, Ming J, Meng X, Han B, Sun
B, Cai J and Jiang C: Analysis of expression and prognostic
significance of vimentin and the response to temozolomide in glioma
patients. Tumour Biol. 37:15333–15339. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thompson EG and Sontheimer H: A role for
ion channels in perivascular glioma invasion. Eur Biophys J.
45:635–648. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ho VK, Reijneveld JC, Enting RH, Bienfait
HP, Robe P, Baumert BG and Visser O: Dutch Society for
Neuro-Oncology (LWNO): Changing incidence and improved survival of
gliomas. Eur J Cancer. 50:2309–2318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liang AL, Zhang TT, Zhou N, Wu CY, Lin MH
and Liu YJ: MiRNA-10b sponge: An anti-breast cancer study in vitro.
Oncol Rep. 35:1950–1958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu J, Wang S, Zhang W, Qiu J, Shan Y,
Yang D and Shen B: Screening key microRNAs for castration-resistant
prostate cancer based on miRNA/mRNA functional synergistic network.
Oncotarget. 6:43819–43830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao R, Zhou M, Wang H, Xiong W, Li X and
Li G: MiRNA regulatory mechanism in tumor initiation and
progression. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 38:1282–1288.
2013.(In Chinese). PubMed/NCBI
|
|
8
|
Garzia L, Andolfo I, Cusanelli E, Marino
N, Petrosino G, De Martino D, Esposito V, Galeone A, Navas L,
Esposito S, et al: MicroRNA-199b-5p impairs cancer stem cells
through negative regulation of HES1 in medulloblastoma. PLoS One.
4:e49982009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Costa FF, Bischof JM, Vanin EF, Lulla RR,
Wang M, Sredni ST, Rajaram V, Bonaldo MF, Wang D, Goldman S, et al:
Identification of microRNAs as potential prognostic markers in
ependymoma. PLoS One. 6:e251142011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang L, Mao P, Song L, Wu J, Huang J, Lin
C, Yuan J, Qu L, Cheng SY and Li J: miR-182 as a prognostic marker
for glioma progression and patient survival. Am J Pathol.
177:29–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gal H, Pandi G, Kanner AA, Ram Z,
Lithwick-Yanai G, Amariglio N, Rechavi G and Givol D: MIR-451 and
Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells.
Biochem Biophys Res Commun. 376:86–90. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Malzkorn B, Wolter M, Liesenberg F,
Grzendowski M, Stühler K, Meyer HE and Reifenberger G:
Identification and functional characterization of microRNAs
involved in the malignant progression of gliomas. Brain Pathol.
20:539–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
You BR, Shin HR, Han BR and Park WH: PX-12
induces apoptosis in Calu-6 cells in an oxidative stress-dependent
manner. Tumour Biol. 36:2087–2095. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deakin NE and Chaplain MA: Mathematical
modeling of cancer invasion: The role of membrane-bound matrix
metalloproteinases. Front Oncol. 3:702013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fillmore HL, VanMeter TE and Broaddus WC:
Membrane-type matrix metalloproteinases (MT-MMPs): Expression and
function during glioma invasion. J Neurooncol. 53:187–202. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ram RR, Mendiratta S, Bodemann BO, Torres
MJ, Eskiocak U and White MA: RASSF1A inactivation unleashes a tumor
suppressor/oncogene cascade with context-dependent consequences on
cell cycle progression. Mol Cell Biol. 34:2350–2358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan H and Gao Y: MicroRNA-1908 is
upregulated in human osteosarcoma and regulates cell proliferation
and migration by repressing PTEN expression. Oncol Rep.
34:2706–2714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim HR, Shin CH, Lee H, Choi KH, Nam DH,
Ohn T and Kim HH: MicroRNA-1908-5p contributes to the oncogenic
function of the splicing factor SRSF3. Oncotarget. 8:8342–8355.
2017.PubMed/NCBI
|
|
19
|
Jin JC, Jin XL, Zhang X, Piao YS and Liu
SP: Effect of OSW-1 on microRNA expression profiles of hepatoma
cells and functions of novel microRNAs. Mol Med Rep. 7:1831–1837.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lian D, Wang ZZ and Liu NS: MicroRNA-1908
is a biomarker for poor prognosis in human osteosarcoma. Eur Rev
Med Pharmacol Sci. 20:1258–1262. 2016.PubMed/NCBI
|
|
21
|
Xia X, Li Y, Wang W, Tang F, Tan J, Sun L,
Li Q, Sun L, Tang B and He S: MicroRNA-1908 functions as a
glioblastoma oncogene by suppressing PTEN tumor suppressor pathway.
Mol Cancer. 14:1542015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shou J, Gu S and Gu W: Identification of
dysregulated miRNAs and their regulatory signature in glioma
patients using the partial least squares method. Exp Ther Med.
9:167–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aigner A: MicroRNAs (miRNAs) in cancer
invasion and metastasis: Therapeutic approaches based on
metastasis-related miRNAs. J Mol Med (Berl). 89:445–457. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mamoori A, Gopalan V, Smith RA and Lam AK:
Modulatory roles of microRNAs in the regulation of different
signalling pathways in large bowel cancer stem cells. Biol Cell.
108:51–64. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yde CW, Sehested A, Mateu-Regué À, Østrup
O, Scheie D, Nysom K, Nielsen FC and Rossing M: A new NFIA:RAF1
fusion activating the MAPK pathway in pilocytic astrocytoma. Cancer
Genet. 209:440–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang F, Jiang C, Sun Q, Yan F, Wang L, Fu
Z, Liu T and Hu F: miR-195 is a key regulator of Raf1 in thyroid
cancer. Onco Targets Ther. 8:3021–3028. 2015. View Article : Google Scholar : PubMed/NCBI
|