Introduction
Esophageal cancer, located in the area from the
esophagus between the throat and stomach, has become the eighth
most commonly diagnosed cancer in the past decade. The 5-year
survival rate of esophageal cancer patients is only approximately
17.5% as reported by data of SEER 18 2004–2010 (1). Based on the location and ancestral
precancerous cells, esophageal cancer is mainly divided into two
subtypes: adenocarcinoma and squamous cell carcinoma (2,3).
Alcohol consumption and smoking are related to both subtypes.
Gastroesophageal reflux disease (GERD) and obesity are associated
with adenocarcinoma, while nitrosamines and nutritional
deficiencies could lead to squamous cell carcinoma (4). In the clinic, surgery, chemotherapy
and radiotherapy are commonly used to treat esophageal cancer
(5).
It has been reported that mutations of several genes
are associated with esophageal cancer, such as p53, FasL and EGFR
(6–9). Recent research has demonstrated that
miRNAs play essential roles in esophageal cancer (10). MicroRNAs are a class of non-coding
RNAs of 17–24 nucleotides, which regulate gene expression by
targeting specific mRNAs (11–13).
Primary transcripts are firstly cleaved by Drosha and clipped by
Dicer (14). MicroRNAs act as
oncogenes or tumor-suppressor genes in cancers to participate in
processes of tumorigenesis, differentiation and apoptosis (15,16).
There are various reports concerning the aberrant expression of
microRNAs such as miR-21, miR-127 and miR-377 in esophageal
squamous cell carcinoma (17–20).
miR-486, located in the 40th intron of the ankyrin-1 gene, was
firstly identified in human fetal liver (21,22).
It has been reported that aberrant expression of miR-486 is present
in several types of cancer including gastric cancer, cutaneous T
cell lymphomas and kidney cancer (23,24).
It has been found that miR-486 functions as a tumor suppressor in
several types of cancer (25–27).
Yet, reports of miR-486 in esophageal cancer are rare. In
esophageal cancer tissues, miR-486-5p was found to be suppressed
and it affected cell proliferation, migration and apoptosis to
suppress cancer (28). In
esophageal cancer tissues, we found aberrantly downregulated
expression of miR-486.
In the present study, we investigated the miR-486
expression alteration between esophageal squamous carcinoma and
normal tissues and assayed the effects of miR-486 overexpression on
esophageal cancer cell proliferation, invasion and apoptosis. Based
on the findings of the function of miR-486 in the present study, we
conclude that miR-486 plays an essential role in the progression of
esophageal cancer and may be a potential therapeutic target for
esophageal squamous carcinoma.
Materials and methods
Sample collection
A total of 20 histopathologically confirmed
esophageal squamous carcinoma and corresponding normal samples were
collected from patients. All the procedures were approved by the
institutional review boards of the participating hospitals
affiliated with Zhengzhou Univeristy. The fresh specimens were
obtained and stored at −80°C immediately. All specimens were
obtained by surgical resection, fixed in 10% neutral formalin and
embedded in paraffin. After hematoxylin and eosin (H&E)
staining, the samples were pathologically diagnosed under a
microscope according to the criteria for diagnosis and treatment of
esophageal cancer (2011) (29).
Relevant clinical pathological information was obtained from the
hospital.
Cell culture
Three esophageal cell lines (KYSE150, EC9706 and
TE-9) and human normal esophageal epithelial cell line Het-1A were
purchased from the Cell Bank of Shanghai Institute of Cell Biology
(Chinese Academy of Medical Sciences, Shanghai, China). KYSE150,
EC9706 and TE-9 cells were cultured in RPMI-1640 medium (Gibco,
Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco) and a 1% penicillin (Invitrogen, Shanghai, China). Het-1A
cells were cultured in Dulbeccos modified Eagles medium (DMEM;
Gibco) supplemented with 10% FBS and 1% penicillin. All the cell
lines were grown at 37°C in an incubator.
H&E staining of tissue
sections
Before staining, the tissues were sliced into 5-µm
thick sections and dewaxed in xylene, and rehydrated through a
serial of decreasing concentrations of ethanol. After washing in
phosphate-buffered saline (PBS), the sections were stained with
H&E. Finally, the sections were dehydrated in increasing
concentrations of alcohol and xylene.
RT-PCR
Total RNA was extracted from the samples and cell
lines using TRIzol reagent (Takara Bio, Shiga, Japan). cDNA of
miRNA was synthesized by miRNA cDNA Synthesis kit (Abm Canada Inc.,
Milton, ON, Canada). cDNA from mRNA was synthesized by RT Master
Mix (Abm Canada). The RT-PCR was performed using SYBR®
Green Master Mixes (SR1110; Thermo Fisher Scientific, Darmstadt,
Germany) according to the manufacturers instructions. The primers
were: GAPDH forward, 5-TGTTCGTCATGGGTGTGA AC-3 and reverse,
5-ATGGCATGGACTGTGGTCAT-3; CDK4 forward, 5-TGACATTCCCCTCCCACCTCTCC-3
and reverse, 5-ATCCTCCTGCCTCAGTCTCCCAAGTA-3; BCAS2 forward,
5-AAGGACAACAGCATCTTCCCAAA AC-3 and reverse,
5-TCACATACATCTAGTTCATCTACC TAAAGTGTTC-3; miR-486-5p forward,
5-ACACTCCAG CTGGGTCCTGTACTGAGCTGCCC-3 and reverse, 5-CT
CAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGC CCCGAG-3; U6 forward:
5-CTCGCTTCGGCAGCACA-3 and reverse, 5-AACGCTTCACGAATTTGCGT-3.
Relative gene expression was calculated by the 2−ΔΔCt
method.
Western blotting
The tissues and cells were lysed with RIPA buffer
supplemented with phenlymethanesulfonyl fluoride (PMSF). After the
concentration was quantified, the proteins were subjected to
SDS-PAGE and transferred to polyvinylidene difluoride membranes.
The membranes were immunoblotted with the primary antibodies: CDK4
(1:500, ab108357; Abcam, Cambridge, UK), BCAS2 (1:500, ab151293;
Abcam), GAPDH (1:500, ab8245; Abcam), p21 (1:500, ab109520; Abcam)
and caspase-3 (1:300, ab2171; Abcam) overnight at 4°C. After being
rinsed with TBST, the membranes were incubated with secondary
antibodies (at a dilution of 1:5,000) conjugated to horseradish
peroxidase. The protein bands were visualized and exposed to X-ray
film. GAPDH was used as the internal control.
Immunohistochemistry
The sections were rehydrated in xylene and
decreasing concentrations of alcohol. Then, the slices were
incubated with 3% H2O2 for blocking and
incubated with 10 mM citric acid for antigen retrieval. After a
second blocking in TBS with 5% BSA, the sections were incubated
with the primary antibody CDK4 (1:500) or BCAS2 (1:500), at 4°C
overnight. Subsequently, the sections were treated with the
secondary antibody for 60 min at room temperature (RT). Finally, a
colorizing reagent was used to stain and hematoxylin was used to
counterstain cell nuclei.
miRNA/siRNA synthesis and
transfection
miR-486 mimics/NC, target siRNA and control siRNA
were synthesized in Shanghai GenePharma Co., Ltd. (Shanghai,
China). Cells were transfected with siRNA diluted by Opti-MEM I
Reduced Serum Medium using Lipofectamine 2000 (Abm Canada)
according to the manufacturers protocol.
Colony formation assay
After transfection with the miR-486 mimics/NC, the
cells were cultured for 48 h and collected. The cell density was
adjusted to 2000 cells/ml and then the cells were seeded in 6-well
plates. The cells were observed under a microscope each day and
medium was changed every two days. After fixation with 4%
paraformaldehyde, the cells were strained using crystal violet.
After washing with PBS, the number of cell colonies was counted for
data analysis.
Scratch wound healing assay
After transfection with the miR-486 mimics/NC, the
cells were cultured overnight. Cell scratches were made with 1-ml
tips and the width of the scratch wounds were kept the same. After
washing with PBS, the cells were cultured in an incubator and
images of the scratch wounds were captured after 48 h.
Flow cytometry
The cells were cultured for 48 h and fixed in
ice-cold 70% ethanol for 12 h. After washing, the cells were
incubated with 0.25 mg/ml RNase at 37°C for 30 min. The cells were
resuspended in PI solution (50 µg/ml) and subjected to cell cycle
analysis by flow cytometric analysis.
The cells were incubated for 48 h, cells were washed
and resuspended in binding buffer and incubated with FITC-Annexin V
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). After 15 min, 5
µl PI and 300 µl were added to each sample. The cells were
incubated for 30 min at RT in the dark and subjected to flow
cytometric analysis within 1 h.
Transwell invasion assay
Transwell assay was performed to detect the invasive
ability of the cells. The upper chambers of Transwell plates (BD
Biosciences, San Jose, CA, USA) were coated with Matrigel and
~5×104 transfected cells were seeded into the chambers.
After a 48-h incubation, the cells on the upper chambers were wiped
off and the cells on the lower surface were washed. After being
fixed, the cells on the lower surface of the membrane were stained
with crystal violet to test the extent of invasion.
Dual-Luciferase experiment
The cells were cultured at 60–80% confluence in
24-well plates for 12 h. The reporter plasmids and miR-486-5p were
transiently transfected into the cells. After 48 h, the
Dual-Luciferase reporter assay (Promega, Madison, WI, USA) was used
to measure the Renilla luciferase activity and the data were
normalized with firefly luciferase.
Statistical analysis
The Spearmans rank correlation coefficient was used
to analyze the ranked data and the Students t-test was performed to
analyze the RT-PCR data. One-way ANOVA was used to analyze the
multiple group comparisons. p<0.05 was considered indicative of
statistical significance. Each sample was assayed at least three
times.
Results
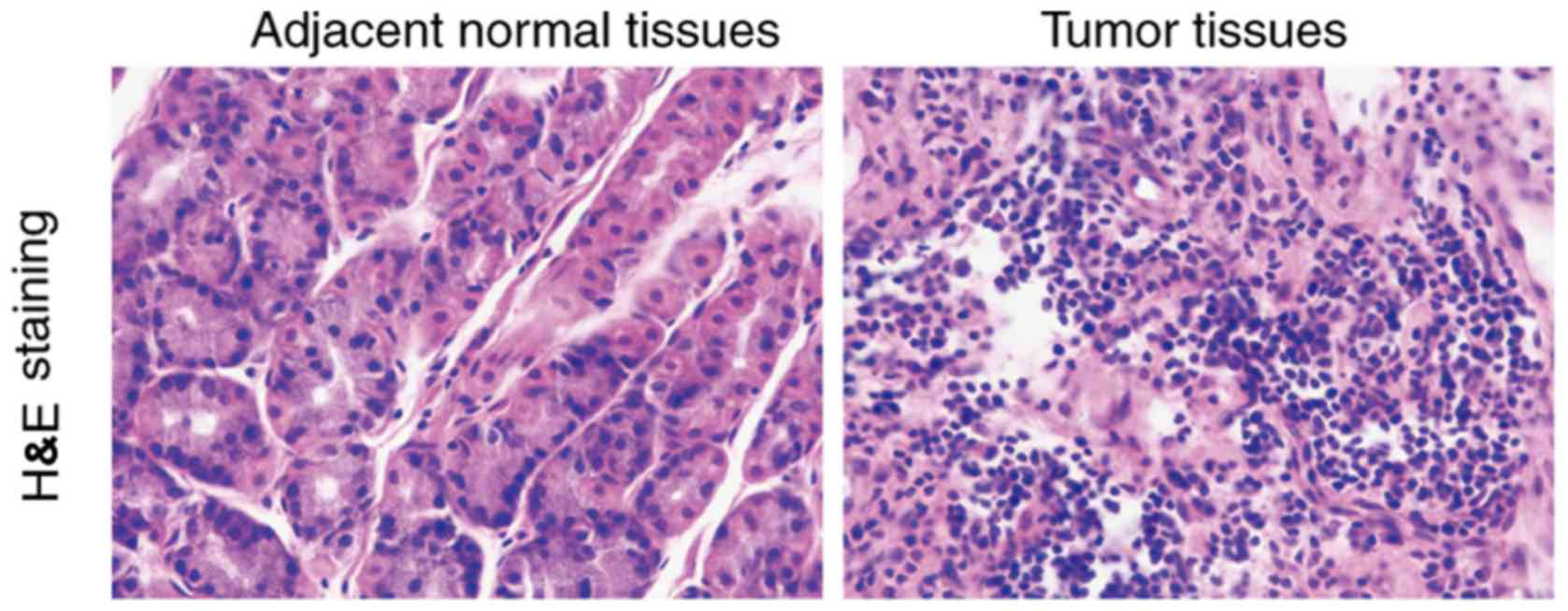
H&E staining of esophageal
squamous carcinoma tissues and adjacent tissues
According to histological type, esophageal cancer
can be divided into esophageal squamous cell carcinoma and
adenocarcinoma. Compared to the normal tissues, cancer cells are
larger in size than normal cells and the morphology is not
consistent. It was observed that giant nuclei, dual nuclei, multi
nuclei or heteromorphic nuclei were present in the cancer cells.
The ratio of the nucleus and cytoplasm of the cancer cells was
different from that of the normal cells (Fig. 1).
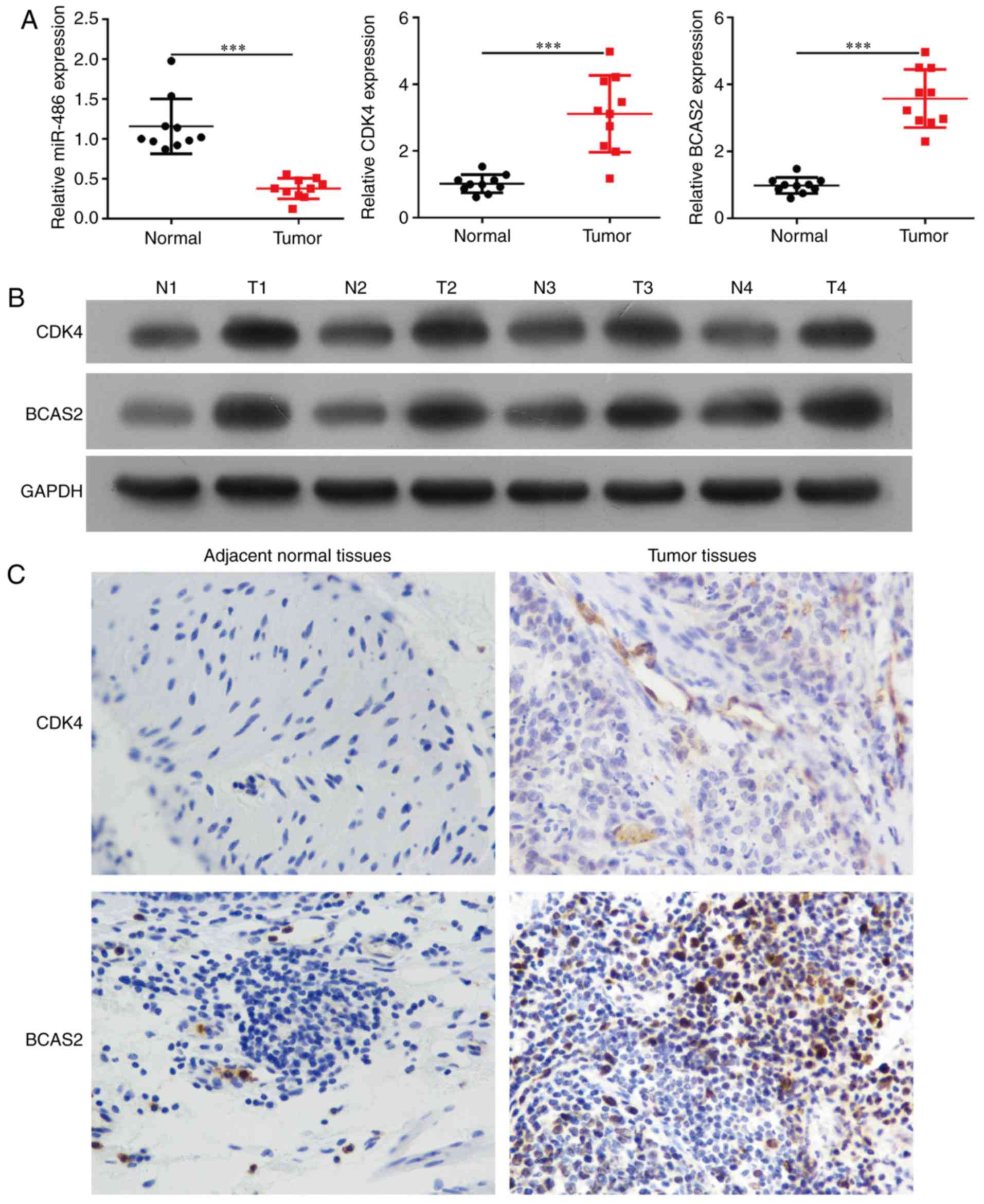
Expression of miR-486, CDK4 and BCAS2
in esophageal squamous cell carcinoma tissues and cells
miR-486 expression was significantly downregulated
in esophageal squamous carcinoma tissues compared to the
corresponding esophageal normal tissues of 20 esophageal cancer
patients (Fig. 2A, p<0.0001).
The mRNA and protein levels of CDK4 and BCAS2 were upregulated in
the esophageal cancer tissues (Fig. 2A
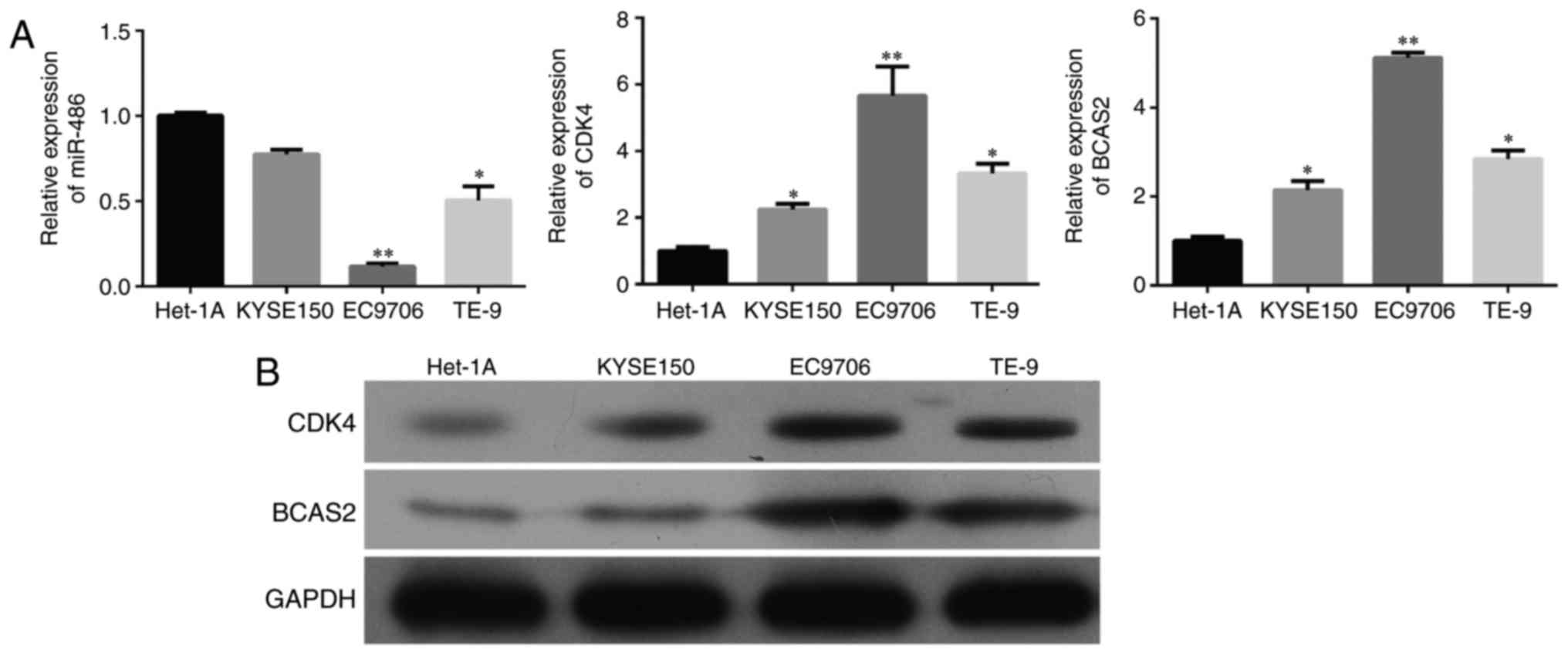
and B, p<0.001). We selected three esophageal cancer cell
lines, KYSE150, EC9706 and TE-9, and a normal esophageal epithelial
cell line Het-1A. miR-486 exhibited decreased expression while CDK4
and BCAS2 had increased mRNA and protein levels in the three
esophageal cancer cell lines than that in the normal cells
(Fig. 3 and B).
Through immunohistochemical staining, it was
observed that CDK4 and BCAS2 exhibited stronger staining in
esophageal squamous carcinoma tissues compared to that noted in the
normal tissues (Fig. 2C). The
expression of CDK4/BCAS2 was increased in the cell nucleus and
cytoplasm.
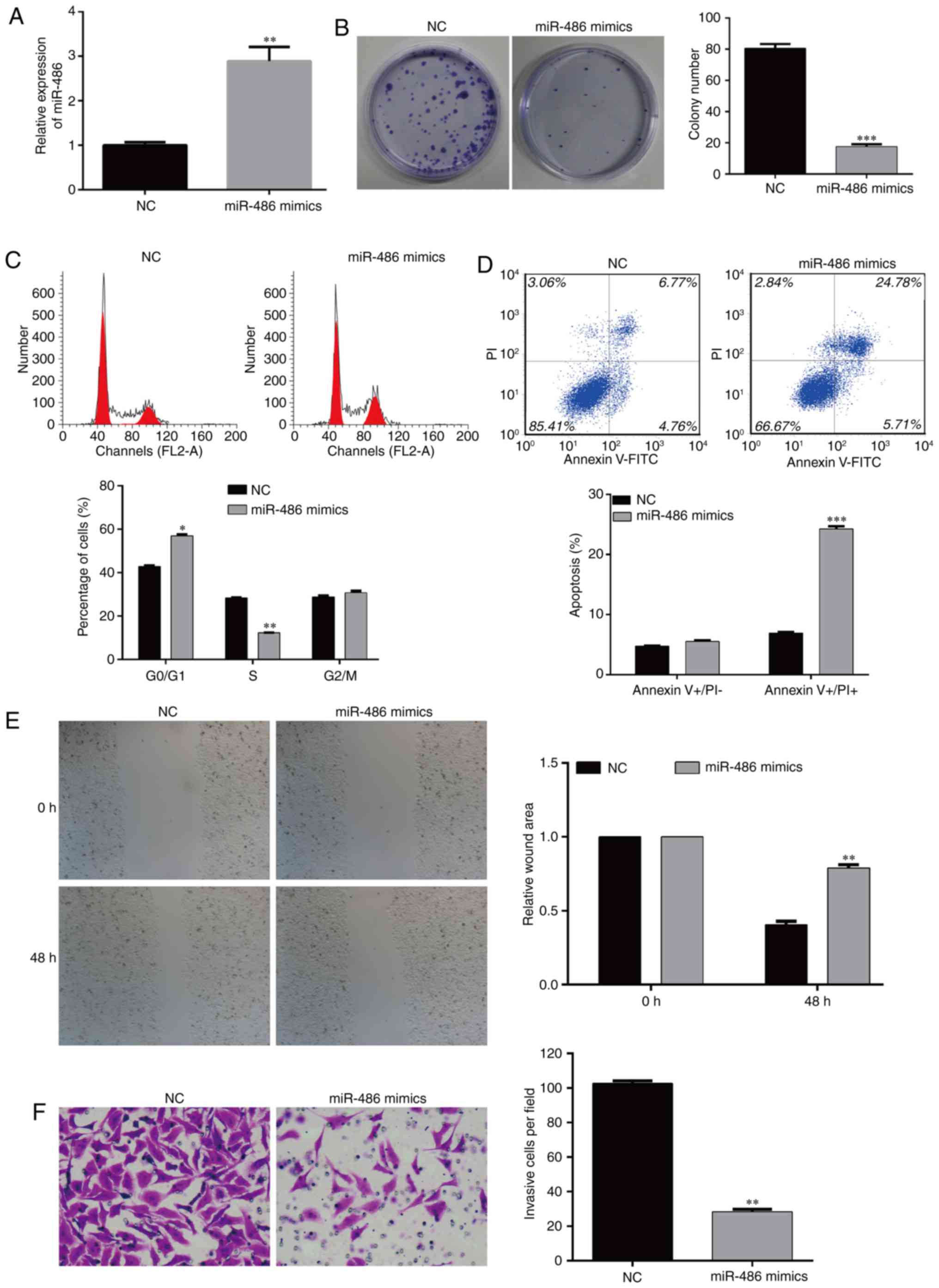
Overexpression of miR-486 inhibits the
colony formation of EC9706 cells
According to the above-mentioned results, the
expression of miR-486, CDK4 and BCAS2 in the three esophageal
cancer cell lines was the same, thus EC9706 cells were selected for
the following experiments. The cells were divided into two groups:
NC- and miR-486 mimic-transfected EC9706 cells. RT-PCR was
performed to detect the level of miR-486 (Fig. 4A). In order to observe whether
miR-486 affects the function of EC9706 cells, we detected the cell
viability of the two cell groups. The results showed that the
colony formation ability was significantly inhibited in the miR-486
mimic-transfected EC9706 cells; the number of colonies in the
experimental group was significantly lower than that in the
negative control (NC) group (Fig.
4B, p<0.001). These results suggest that miR-486 has the
ability to inhibit the colony formation of esophageal cancer
cells.
Overexpression of miR-486 induces cell
cycle arrest and apoptosis of EC9706 cells
Flow cytometry is a method by which to measure the
relative content of DNA, which confirms the cell proportion in each
stage of the cell cycle and the apoptotic rate. In order to observe
the effects of miR-486 on cell proliferation and apoptosis of the
EC9706 cells, we analyzed the percentage of cells in the different
stages of the cell cycle and apoptosis using flow cytometry. The
results showed that compared with the NC group, overexpression of
miR-486 significantly reduced EC9706 cell proliferation, which was
mainly reflected by the fact that a higher percentage of cells was
arrested in the G0/G1 phase (p<0.05). The corresponding
proportion of S phase cells was significantly decreased
(p<0.01), and cells in the G2/M phase were slightly changed
(Fig. 4C).
The change in anti-apoptosis ability is an important
characteristic of tumor cells. After overexpression of miR-486 in
EC9706 cells, flow cytometry was used to observe the proportion of
apoptosis of the two groups of cells. The results showed that the
miR-486 mimic-transfected EC9706 cells had a significantly higher
percentage of apoptosis compared with the NC group (Fig. 4D, p<0.001). These results suggest
that miR-486 inhibits the cell cycle progression and induces
apoptosis in esophageal cancer cells.
Overexpression of miR-486 suppresses
the migration and invasion of EC9706 cells
The migration and invasion ability of cancer cells
is an important factor related to tumor metastasis. Cell scratch
assay was used to observe the motile ability of the cells in the
miR-486 mimic-transfected and NC-transfected EC9706 cells. After 48
h, the migration ability of the EC9706 cells with overexpression of
miR-486 was significantly lower than that of the NC group, which
indicated that miR-486 could inhibit the motility of esophageal
cancer cells (Fig. 4E,
p<0.01).
Transwell assay was performed to test the effects of
miR-486 on the invasion ability of cancer cells. After
overexpression of miR-486 in EC9706 cells, the number of cells
invading the basement membrane was significantly lower than that in
the NC group (Fig. 4F, p<0.01).
These results indicate that miR-486 has the function of reducing
the motility and invasion of esophageal cancer cells.
miR-486 regulates the expression of
CDK4 and BCAS2
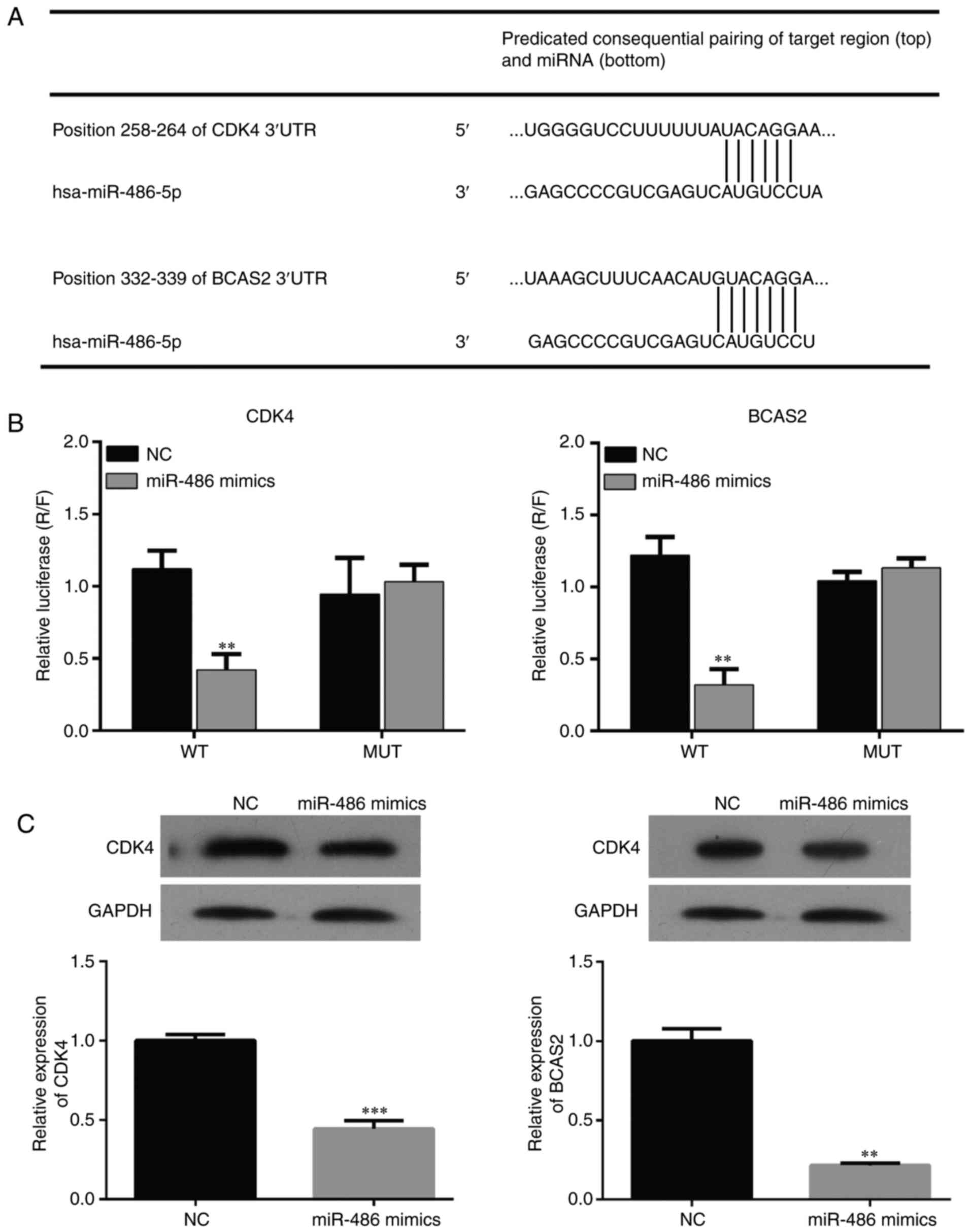
Bioinformatic method was used to predict the binding
sites of miR-486 and CDK4/BCAS2 (Fig.
5A). Dual-Luciferase assay was used to detect the interaction
between miR-486 and CDK4/BCAS2. The results showed that expression
of CDK4/BCAS2 was significantly suppressed in the miR-486
mimic-transfected EC9706 cells. When the binding site was mutated,
the interaction relationship disappeared and the expression of
CDK4/BCAS2 returned to normal, which indicated that miR-486 may be
a regulatory factor of the CDK4/BCAS2 sequence (Fig. 5B and C).
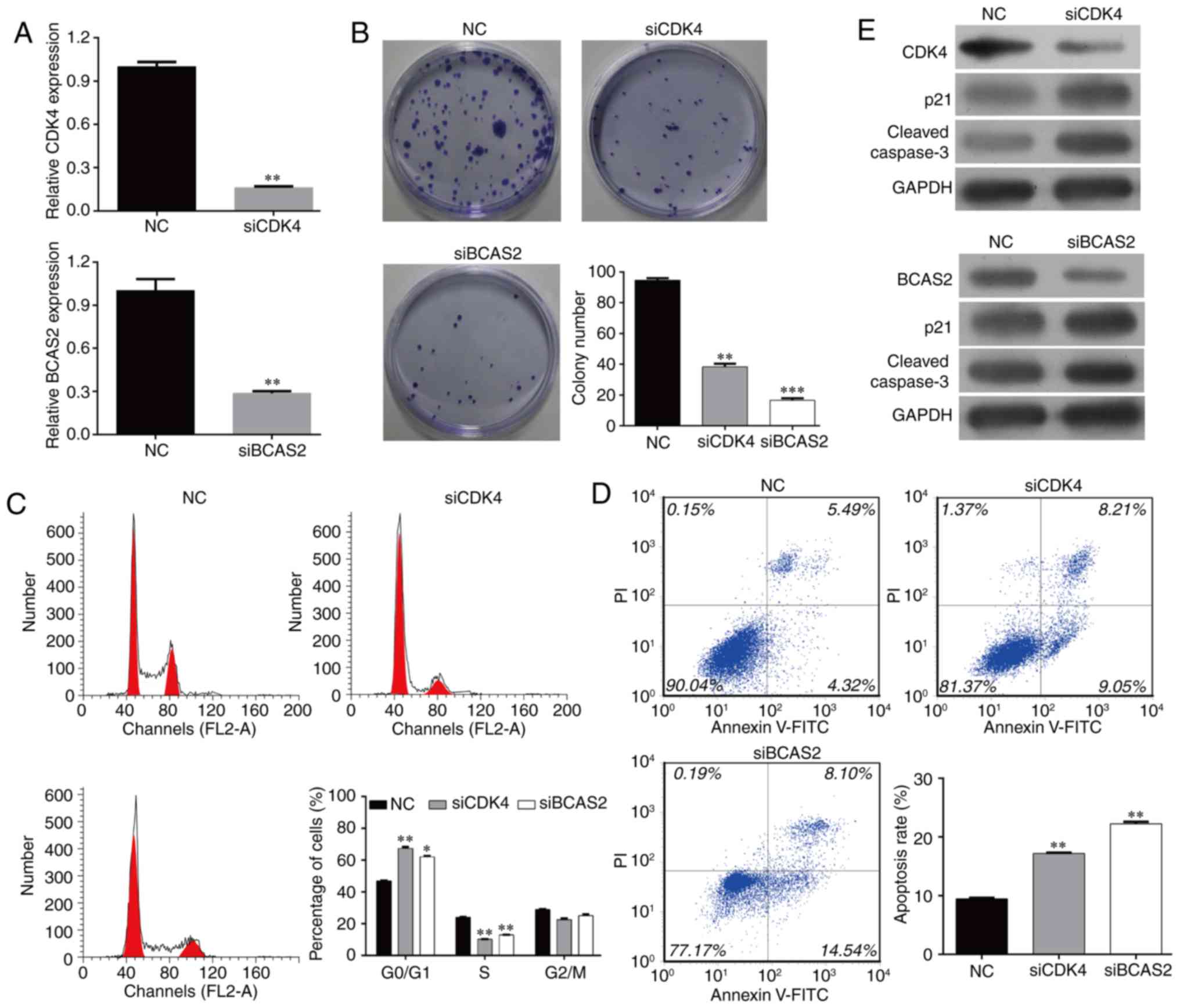
Knockdown of CDK4/BCAS2 downregulates
the colony formation of EC9706 cells
In order to study the effects of target genes
CDK4/BCAS2 on the behavior of esophageal cancer EC9706 cells, we
designed and synthesized siRNAs, siCDK4 and siBCAS2. The effects of
these two target genes on cell proliferation and colony formation
were detected following transfection. The cells were divided into
three groups: NC, siCDK4 and siBCAS2 EC9706 cells. SiCDK4 and
siBCAS2 effectively inhibited the expression of the target genes,
and the mRNA levels were significantly lower than levels noted in
the NC group (Fig. 6A, p<0.01).
After the three cell groups were cultured for 48 h, the number of
colonies was counted (Fig. 6B).
Compared with the NC group, the cell colony formation ability of
the siCDK4 and siBCAS2 EC9706 cells was significantly inhibited
(p<0.05; p<0.01).
Knockdown of CDK4/BCAS2 inhibits
EC9706 cell cycle progression and induces cell apoptosis
Compared with the NC group, the cell cycle
progression of the siCDK4 and siBCAS2 EC9706 groups was
significantly blocked. The percentages of cells in the G0/G1 phase
of the two target gene-knockout groups were increased
significantly, while the proportions of cells in the S phase and
G2/M phase were decreased significantly. The cell cycle was
arrested in the G0/G1 phase following the silencing of CDK4 or
BCAS2, and could not enter the cell cycle normally (Fig. 6C).
Compared with the NC group, the percentages of
apoptotic cells in the target gene knockout groups were increased
significantly (Fig. 6D, p<0.01).
These data suggest that knockdown of target genes CDK4/BCAS2 can
inhibit the cell cycle progression and induce the apoptosis of
esophageal cancer cells.
Knockout of CDK4/BCAS2 affects
expression of apoptosis-related proteins
The above results showed that the knockdown of
target genes CDK4/BCAS2 can induce the apoptosis of esophageal
cancer cells, but the apoptosis-related downstream proteins were
not clear. In order to study the downstream molecular changes in
the apoptotic signaling pathway, the protein levels of p21 and
caspase-3 were detected by western blotting. The results showed
that compared with the NC group, CDK4 and BCAS2 protein expression
was decreased, and expression levels of apoptotic signaling
molecules p21 and caspase-3 were also downregulated in the siCDK4
and siBCAS2 EC9706 cell groups (Fig.
6E).
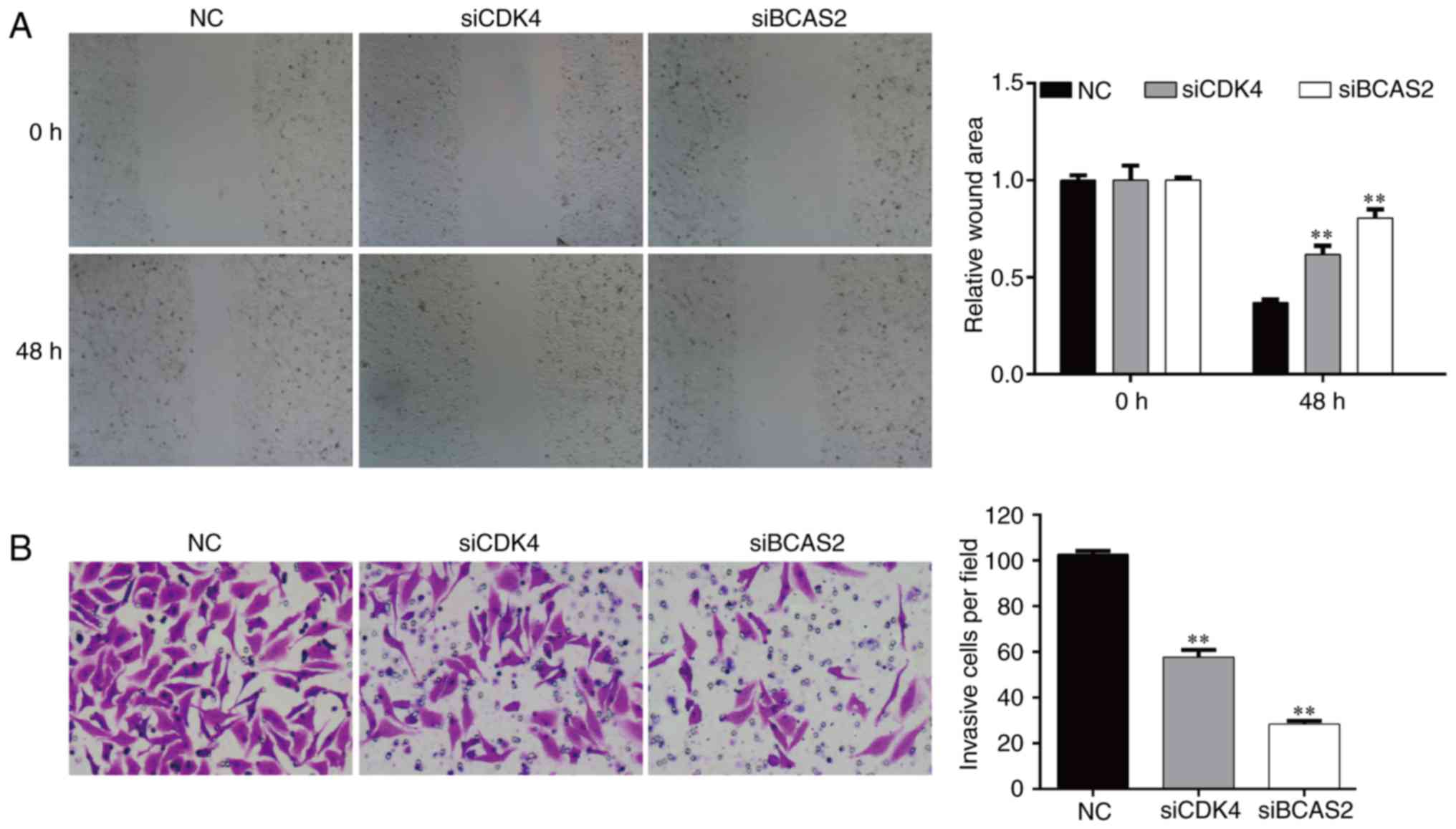
Knockdown of CDK4/BCAS2 inhibits the
invasion and migration abilities of EC9706 cells
After transfected for 48 h, the migration ability of
the EC9706 cells following silencing of CDK4/BCAS2 was
significantly lower than that noted in the NC group (both
p<0.01), which indicated that knockdown of target genes
CDK4/BCAS2 could inhibit the motility of esophageal cancer cells
(Fig. 7A).
After knockdown of target genes CDK4/BCAS2 in the
EC9706 cells, the number of cells invading the basement membrane
was significantly lower compared with that of the NC group
(Fig. 7B, p<0.01). These results
suggest that knockdown of target genes CDK4/BCAS2 can reduce the
migration and invasion abilities of the esophageal cancer
cells.
Discussion
Esophageal cancer is a common tumor, for which the
incidence differs according to race and geographical region. It has
been reported that the mortality and morbidity of esophageal cancer
is increasing yearly (30–32). Squamous cell carcinoma is one of the
main pathological types of advanced esophageal carcinoma.
Populations in some regions of northern China have a higher risk of
esophageal cancer and esophageal squamous cell carcinoma is the
main pathological type, accounting for more than 90% of all cases
(33). miRNAs are a type of
non-coding single-stranded RNAs, ranged from 20 to 25 nt, and
regulate the expression of target genes through interaction with
the mRNA 3′-UTR of genes (34). It
has been reported that miRNAs are critical diagnostic and
prognostic biomarkers in cancer (35,36).
miRNAs can function as oncogenes or as tumor-suppressor genes,
which play an important role in angiogenesis and
epithelial-mesenchymal transition and drug resistance in cancer
(37). In the present study, we
found that miR-486 exhibited decreased expression in esophageal
squamous carcinoma tissues and cell lines compared to that noted in
normal tissues and cells. In recent studies, it was found that
miR-486 functions as a tumor suppressor in breast, lung and
hepatocellular cancer (25–27). In esophageal cancer tissues,
miR-486-5p was found to be suppressed and exhibited an
anti-oncogene function by affecting proliferation, migration and
apoptosis (28). In non-small cell
lung cancer, miR-486-5p significantly inhibited cell growth and
cell cycle progression by targeting CDK4. The 3-UTR of CDK4 is the
direct interaction region between miR-486-5p and CDK4 (38). In non-small cell lung cancer,
miR-486-5p inhibited the progression and metastasis by targeting
ARHGAP5 (26). In breast cancer
cells, miR-486-5p exerted an anti-proliferative function by
targeting PIM-1, in addition to the fact that miR-486-5p inhibited
proliferation by interacting with PIK3R1 in hepatocellular
carcinoma (25,27). However, the function of miR-486 has
yet to be elucidated (27). The
expression of miR-486 was determined in three esophageal squamous
carcinoma cell lines and that of EC9706 had a significantly lower
level compared to Het-1A (human esophageal epithelial cell line).
Thus, we performed subsequent experiments using EC9706. The
differential levels of miR-486 may be due to the difference in cell
lines. In our results, overexpression of miR-486 inhibited the
ability of colony formation, arrested cell cycle progression and
induced apoptosis and its targets were CDK4 and BCAS2. CDK4
(cyclin-dependent kinase 4), a member of the CDK family, is a type
of serine/threonine kinase and is an essential signaling
transduction molecule which participates in the cell cycle and
apoptosis by binding with cyclin proteins. In the progression of
the cell cycle, the cyclin proteins are periodically expressed and
degraded, and the transient activation of CDK is used to catalyze
the phosphorylation of different substrates (39). The CDK family includes CDK1-13 and
the cyclin proteins are divided into cyclin A-L, in which cyclin D
includes cyclin D1, D2 and D3 (40,41).
Cyclin D is expressed in the G1 phase of the cell cycle, binds and
activates CDK4 and CDK6, which form the binding complex leading to
a series of substrate phosphorylation (42,43).
Downstream activated E2F induces a series of protein to transcript
and promotes cells enter into the S phase (44). In the meanwhile, the complex of
CDK-cyclin is negatively regulated by CKIs (cyclin kinase
inhibitors) (45).
BCAS2 (breast cancer amplified sequence 2), located
on chr1 p13.3–21, is a subunit of the prp19 complex, which is an
essential factor for the splicing of the cell nuclear precursor
mRNA (46,47). BCAS2 is mainly located in the
nucleus and has key roles in mitotic initiation (48). Knockdown of BCAS2 using RNAi was
found to lead to inhibition of the expression of the other three
subunits of PRP19, and the structure of the whole PRP19 complex is
shaken, which leads to abnormal mitosis (46–48).
In the present study, we found that the proliferation and clone
formation ability of esophageal cancer cells were inhibited after
knockdown of CDK4 and BCAS2 genes, and the ability of migration and
invasion was also reduced. This indicates that a low level of
miR-486 leads to the imbalance of CDK4 and BCAS2 in the process of
esophageal carcinogenesis, and abnormal increased CDK4 or BCAS2 can
promote the esophageal cancer cells to proliferate aberrantly. In
addition, p53 is also a target protein of BCAS2, while knockdown of
BCAS2 can enhance p53-induced apoptosis (49). These data are consistent with our
research. Knockdown of CDK4 and BCAS2 increased levels of the
apoptosis-related protein p21 and caspase-3, suggesting that the
apoptosis signaling pathway is activated, and the cells are induced
to enter apoptosis. After knockdown of CDK4, the level of p21 was
detected but the specific regulatory mechanism between CDK4 and p21
warrant further research. The expression of proteins associated
with cell cycle progression, invasion and apoptotic cell death in
response to miR-486 overexpression or downregulation need to be
detected in further research. These are the limitation of this
study.
In conclusion, the present study revealed that
miR-486 was downregulated in esophageal squamous carcinoma and
functions as a tumor suppressor by affecting cell proliferation,
colony formation and apoptosis by targeting CDK4 and BCAS2. This is
the first study concerning the biological function of miR-486 and
its target genes in esophageal squamous carcinoma. This novel
pathway has the potential for a greater understanding of the
pathogenesis of esophageal squamous carcinoma, and drug targets
could be developed for the diagnosis and therapeutic treatment of
esophageal squamous carcinoma in the future.
References
|
1
|
Beral V and Peto R: UK cancer survival
statistics. BMJ. 341(aug11 1): c41122010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Layke JC and Lopez PP: Esophageal cancer:
A review and update. Am Fam Physician. 73:2187–2194.
2006.PubMed/NCBI
|
|
3
|
Wu J, Wu X, Liang W, Chen C, Zheng L and
An H: Clinicopathological and prognostic significance of chemokine
receptor CXCR4 overexpression in patients with esophageal cancer: A
meta-analysis. Tumour Biol. 35:3709–3715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao YF, Yuan F, Liu J, Li LP, He YC, Gao
RJ, Cai YD and Jiang Y: Identification of new candidate Genes and
chemicals related to esophageal cancer using a hybrid interaction
network of chemicals and proteins. PLoS One. 10:e01294742015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al CROSS Group, :
Preoperative chemoradiotherapy for esophageal or junctional cancer.
N Engl J Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ming Z, Jiang D, Hu Q, Li X, Huang J, Xu
Y, Liu Y, Xu C, Hua X and Hou Y: Diagnostic application of PIK3CA
mutation analysis in Chinese esophageal cancer patients. Diagn
Pathol. 9:1532014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Agarwal D, Pineda S, Michailidou K,
Herranz J, Pita G, Moreno LT, Alonso MR, Dennis J, Wang Q, Bolla
MK, et al kConFab Investigators; Australian Ovarian Cancer Study
Group, ; GENICA Network; TNBCC, : FGF receptor genes and breast
cancer susceptibility: Results from the Breast Cancer Association
Consortium. Br J Cancer. 110:1088–1100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hollstein MC, Metcalf RA, Welsh JA,
Montesano R and Harris CC: Frequent mutation of the p53 gene in
human esophageal cancer. Proc Natl Acad Sci USA. 87:pp. 9958–9961.
1990; View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao H, Zheng L, Li X and Wang L: FasL
gene −844T/C mutation of esophageal cancer in South China and its
clinical significance. Sci Rep. 4:38662014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng XR, Lu P, Mei JZ, Liu GJ and Fan QX:
Expression analysis of miRNA and target mRNAs in esophageal cancer.
Braz J Med Biol Res. 47:811–817. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zamore PD and Haley B: Ribo-gnome: The big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pillai RS: MicroRNA function: Multiple
mechanisms for a tiny RNA? RNA. 11:1753–1761. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M,
et al: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feber A, Xi L, Luketich JD, Pennathur A,
Landreneau RJ, Wu M, Swanson SJ, Godfrey TE and Litle VR: MicroRNA
expression profiles of esophageal cancer. J Thorac Cardiovasc Surg.
135:255–260, discussion 260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee
NPY, Law S, Xu LY, Li EM, Chan KW, et al: MicroRNA-377 suppresses
initiation and progression of esophageal cancer by inhibiting CD133
and VEGF. Oncogene. 36:3986–4000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao X, Wang X, Cai K, Wang W, Ju Q, Yang
X, Wang H and Wu H: MicroRNA-127 is a tumor suppressor in human
esophageal squamous cell carcinoma through the regulation of
oncogene FMNL3. Eur J Pharmacol. 791:603–610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y,
Jiang H, Sun Z and Zheng X: Identification of human fetal liver
miRNAs by a novel method. FEBS Lett. 579:3849–3854. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miya K, Shimojima K, Sugawara M, Shimada
S, Tsuri H, Harai-Tanaka T, Nakaoka S, Kanegane H, Miyawaki T and
Yamamoto T: A de novo interstitial deletion of 8p11.2 including
ANK1 identified in a patient with spherocytosis, psychomotor
developmental delay, and distinctive facial features. Gene.
506:146–149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oh HK, Tan AL, Das K, Ooi CH, Deng NT, Tan
IB, Beillard E, Lee J, Ramnarayanan K, Rha SY, et al: Genomic loss
of miR-486 regulates tumor progression and the OLFM4 antiapoptotic
factor in gastric cancer. Clin Cancer Res. 17:2657–2667. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goto K, Oue N, Shinmei S, Sentani K,
Sakamoto N, Naito Y, Hayashi T, Teishima J, Matsubara A and Yasui
W: Expression of miR-486 is a potential prognostic factor after
nephrectomy in advanced renal cell carcinoma. Mol Clin Oncol.
1:235–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang G, Liu Z, Cui G, Wang X and Yang Z:
MicroRNA-486-5p targeting PIM-1 suppresses cell proliferation in
breast cancer cells. Tumour Biol. 35:11137–11145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song
Z, Yu X and Lin B: Downregulated miR-486-5p acts as a tumor
suppressor in esophageal squamous cell carcinoma. Exp Ther Med.
12:3411–3416. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Allum W.H..et al: Guidelines for the
management of oesophageal and gastric cancer. Gut. 2011.60(11):
1449–72. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Trichopoulos D: The unequal burden of
cancer. BMJ. 320:3212000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37 Suppl
8:S4–S66. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mandard AM, Hainaut P and Hollstein M:
Genetic steps in the development of squamous cell carcinoma of the
esophagus. Mutat Res. 462:335–342. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zen K and Zhang CY: Circulating microRNAs:
A novel class of biomarkers to diagnose and monitor human cancers.
Med Res Rev. 32:326–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo J, Wang M and Liu X: MicroRNA-195
suppresses tumor cell proliferation and metastasis by directly
targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res.
34:912015.https://doi.org/10.1186/s13046-015-0209-7
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kawano M, Tanaka K, Itonaga I, Ikeda S,
Iwasaki T and Tsumura H: microRNA-93 promotes cell proliferation
via targeting of PTEN in osteosarcoma cells. J Exp Clin Cancer Res.
34:762015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX, et al: Direct repression of
the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small
cell lung cancer. Oncotarget. 7:34011–34021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weinberg RA: The retinoblastoma protein
and cell cycle control. Cell. 81:323–330. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meyerson M and Harlow E: Identification of
G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell
Biol. 14:2077–2086. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsushime H, Ewen ME, Strom DK, Kato JY,
Hanks SK, Roussel MF and Sherr CJ: Identification and properties of
an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type
G1 cyclins. Cell. 71:323–334. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kato J, Matsushime H, Hiebert SW, Ewen ME
and Sherr CJ: Direct binding of cyclin D to the retinoblastoma gene
product (pRb) and pRb phosphorylation by the cyclin D-dependent
kinase CDK4. Genes Dev. 7:331–342. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bates S, Bonetta L, MacAllan D, Parry D,
Holder A, Dickson C and Peters G: CDK6 (PLSTIRE) and CDK4 (PSK-J3)
are a distinct subset of the cyclin-dependent kinases that
associate with cyclin D1. Oncogene. 9:71–79. 1994.PubMed/NCBI
|
|
44
|
Botz J, Zerfass-Thome K, Spitkovsky D,
Delius H, Vogt B, Eilers M, Hatzigeorgiou A and Jansen-Dürr P: Cell
cycle regulation of the murine cyclin E gene depends on an E2F
binding site in the promoter. Mol Cell Biol. 16:3401–3409. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sherr CJ and Roberts JM: CDK inhibitors:
Positive and negative regulators of G1-phase progression. Genes
Dev. 13:1501–1512. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Neumann B, Walter T, Hériché JK,
Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel
U, et al: Phenotypic profiling of the human genome by time-lapse
microscopy reveals cell division genes. Nature. 464:721–727. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song EJ, Werner SL, Neubauer J, Stegmeier
F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW and Rape M:
The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control
reversible ubiquitination at the spliceosome. Genes Dev.
24:1434–1447. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kittler R, Surendranath V, Heninger AK,
Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H,
Kozak K, et al: Genome-wide resources of endoribonuclease-prepared
short interfering RNAs for specific loss-of-function studies. Nat
Methods. 4:337–344. 2007.PubMed/NCBI
|
|
49
|
Kuo PC, Tsao YP, Chang HW, Chen PH, Huang
CW, Lin ST, Weng YT, Tsai TC, Shieh SY and Chen SL: Breast cancer
amplified sequence 2, a novel negative regulator of the p53 tumor
suppressor. Cancer Res. 69:8877–8885. 2009. View Article : Google Scholar : PubMed/NCBI
|