Introduction
Human esophageal cancer, as one of the most common
malignancies, it ranks eighth in incidence and sixth in
cancer-related death worldwide (1).
It contains two principle histopathologic subtypes, including
esophageal adenocarcinoma and esophageal squamous cell carcinoma
(ESCC) (2), the latter
predominantly occurs in eastern and southern Africa, and eastern
Asia (3). Despite great advances in
early diagnosis and treatment strategies of human ESCC during the
past twenty years, the clinical outcome of patients with this
malignancy remains very poor, with estimated 5-year survival rate
at <25% (4). Emerging evidence
indicates that esophageal-carcinogenesis is governed by various
molecular processes such as dysregulation of oncogenes or tumor
suppressor genes which were observed frequently in ESCC tissues
leading to their aberrant expression and increased cellular
proliferation, cell cycle progression and cell motility (5,6).
Therefore, it is crucial to identify novel markers for early
detection and efficient treatment, and to understand the underlying
mechanisms of carcinogenesis and cancer progression of human
ESCC.
MicroRNAs (miRNAs), a group of newly discovered
endogenous, non-coding, single stranded and short RNAs with 18–29
nucleotides in length, has been proved to play roles in various
cellular processes, including cell development, differentiation,
proliferation, metabolism and malignant transformation (7). miRNAs can negatively regulate
expression levels of the corresponding target genes at a
post-transcriptional level by promoting mRNA degradation or
suppressing translation (8).
Accumulating studies have identified a number of miRNAs which may
be involved into the initiation and progression of human cancers
(9). Especially, Liu et al
(10) proved that miR-373 could
promote migration and invasion in human ESCC by inhibiting TIMP3
expression. Results of a previous study reported by Nie et
al (11) showed that miR-34a
was associated with ESCC migration and inhibited the migration and
invasion of ESCC cells by targeting Yin Yang-1; Mao et al
(12) indicated that miR-1290 might
function as an oncogene in the progression of ESCC by targeting
nuclear factor I/X. These findings imply that the dysregulation of
miRNAs may contribute to several malignant processes including
cancer cell cycle, apoptosis, invasion, migration and metastasis by
affecting the corresponding transcripts (13). miR-615-5p, located in CpG islands of
the HOX gene cluster on chromosome 12q13.13, has been determined to
be frequently downregulated and functions as a tumor suppressor in
various human cancers via specially binding to the complimentary
recognition sequences in the 3′-untranslated region (3′UTR) of the
corresponding target mRNAs (14–16).
However, little is known regarding its involvement in human
ESCC.
To address this problem, we performed quantitative
real-time PCR to detect expression levels of miR-615-5p in both
ESCC tissues and cell lines. Then, associations between miR-615-5p
expression and various clinicopathological features of ESCC
patients were statistically evaluated. Then the candidate targets
of miR-615-5p were identified by integrating bioinformatics miRNA
target prediction, western blot analysis and luciferase reporter
assay. In addition, the functions of miR-615-5p in ESCC cell
migration and invasion were determined using the transfection of
miRNA mimics, or co-transfected with miRNA mimics and the
expression vector of its target gene.
Materials and methods
Ethics approval
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Institutional and/or National Research Committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards.
Ethic statement
The present study was approved by the Ethic
Committee of Huai'an First People's Hospital. Prior informed
consent was signed by all the patients enrolled in this study based
on the guidelines of Huai'an First People's Hospital. All clinical
tissue specimens were handled and made anonymous according to the
ethical and legal standards.
Patients and tissue samples
Sixty primary ESCC and matched adjacent
non-cancerous esophageal mucosa tissues were obtained from 60 ESCC
patients in the Department of Gastroenterology, Huai'an First
People's Hospital between January 2012 and December 2015. All ESCC
patients underwent esophagectomy, and none of them received
pre-operative radiotherapy or chemotherapy. The diagnosis of ESCC
patients was confirmed by clinical examination and
histopathological analysis of the tissue specimens. The
clinicopathological characteristics of all 60 ESCC patients,
including age, sex, tumor location, lymph node metastasis,
Tumor-node-metastasis (TNM) stage and pathological grade are
summarized in Table I. All the
samples were collected and immediately snap-frozen in liquid
nitrogen and stored at −80°C until further use.
 | Table I.Associations between miR-615-5p
expression and various clinicopathological characteristics of 60
patients with ESCCs. |
Table I.
Associations between miR-615-5p
expression and various clinicopathological characteristics of 60
patients with ESCCs.
| Clinical
variables | No. of patients
(%) | Low miR-615-5p
expression | P-value |
|---|
| Age (year) |
|
|
|
| ≤60 | 25 (41.67) | 13 (52.00) | NS |
|
>60 | 35 (58.33) | 18 (51.43) |
|
| Sex |
|
|
|
| Male | 40 (66.67) | 21 (52.50) | NS |
|
Female | 20 (33.33) | 10 (50.00) |
|
| Tumor location |
|
|
|
| Upper
1/3-middle 1/3 | 41 (68.33) | 22 (53.66) | NS |
| Lower
1/3 | 19 (31.67) | 9
(47.37) |
|
| Lymph node
metastasis |
|
|
|
|
Negative | 24 (40.00) | 6
(25.00) | 0.01 |
|
Positive | 36 (60.00) | 25 (69.44) |
|
| TNM stage |
|
|
|
|
Absent | 20 (33.33) | 2
(10.00) | <0.001 |
|
Present | 40 (66.67) | 29 (72.50) |
|
| Histological
differentiation |
|
|
|
| Well | 18 (33.33) | 6
(33.33) | 0.03 |
|
Moderate-poor | 42 (66.67) | 25 (59.52) |
|
Cell lines and culture
Human normal esophageal cell line (HEEC) and two
human ESCC cell lines (ECA109 and KYSE410) were purchased from the
Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). All cell lines were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum.
Cells were cultured at 37°C with 5% CO2 for further
use.
Cell transfection
miR-615-5p and negative control mimics
(miR-615-5p/NC_mimics) were purchased from GeneCopoeia (Guangzhou,
China). The IGF2 expression plasmid was constructed using a
PCR-generated fragment from the mRNA and the Lv-CMV-GFP vector
(en-IGF-2), and the negative control (en-NC) had no insert. Two
human ESCC cell lines were transfected with miR-615-5p/NC_mimics or
en-IFG-2/en-NC by Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer's instructions. Forty-eight
hours after the transfection, ESCC cells were harvested for western
blot or real-time quantitative PCR analyses.
Western blot analysis
Proteins were extracted from fresh clinical tissue
specimens and cells using cell lysis buffer (50 mM Tris-HCl, pH
8.0, 2 mM EDTA, 1 mM DTT, 10 mM NaCl, 5 mg/ml leupeptin, 1% NP-40,
2 mg/ml pepstatin, 2 mg/ml aprotinin, 0.1% SDS and 1 mM
phenylmethylsulfonyl fluoride). Equal amounts of protein (50 µg)
were separated by 10% SDS PAGE and then transferred onto
polyvinylidene difluoride membranes (Qiagen China Co., Ltd.). Then,
the membranes were incubated with the primary antibodies: anti-IGF2
(dilution 1:500, Abcam Inc., MA, USA) and anti-GAPDH (dilution
1:500, Abcam Inc.), after blocking with 8% milk in
phosphate-buffered saline (PBS; pH 7.5). After that, the membranes
were incubated with the appropriate horseradish
peroxidase-conjugated secondary antibodies (dilution 1:1,000, Abcam
Inc.) after the incubation at 4°C overnight. Finally, protein was
visualized using enhanced chemiluminescence reagent (Santa Cruz
Biotechnology, Santa Cruz, CA, USA). The expression level of IGF2
protein was normalized to that of GAPDH protein. Each sample was
examined in triplicate.
RNA extraction and real-time
quantitative RT-PCR
Total RNA in fresh clinical tissue specimens and
cells were extracted using the RNeasy RNA Mini kit (Qiagen GmbH,
Hilden, Germany) according to the manufacturer's instructions.
First strand cDNA was synthesized using SuperScript reverse
transcriptase (Clontech Laboratories, Inc., Mountainview, CA, USA)
according to the manufacturer's instructions. Following the cDNA
synthesis, the real-time PCR was performed using a Fast Start
Master SYBR Green kit (Roche Molecular Systems, Indianapolis, IN,
USA) on a LightCycler (Roche Molecular Systems) according to the
manufacturer's instructions. The sequence-specific primer pairs
were used for quantitative PCR and are as follows: miR-615-5p
forward, 5′-TCC GAT TCT CCC TCT GGG TC-3′; reverse, 5′-GTG CAG GGT
CCG AGG T-3′. U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′; reverse,
5′-AAC GCT TCA CGA ATT TGC GT-3′. IGF2 forward, 5′-CCT TGG ACT TTG
AGT CAA ATT-3′; reverse, 5′-GGT CGT GCC AAT TAC ATT TCA-3′. GAPDH
forward, 5′-GCT GAG TAT GTC GTG GAG TC-3′; reverse, 5′-AGT TGG TGG
TGC AGG ATG C-3′. Relative expression levels of miRNA and mRNA
expression were calculated with the 2−ΔΔCt method in
fresh tissues and cells. Each sample was examined in
triplicate.
Cell invasion and migration
assays
Cell migration and invasion abilities of human ESCC
cell lines with the transfection of miR-615-5p/NC-mimics or with
the co-transfection of miR-615-5p/en-IGF2 were evaluated using a
Millicell Transwell chamber (Millipore, Billerica, MA, USA), with
or without Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). For
the invasion and migration assays, 48 h following the transfection,
Transwell chambers were placed into 24-well plates which were
respectively precoated with or without a 5-ml mixture of BD
Matrigel and DMEM (1:1, v/v). Following incubation at 37°C in a
humidified incubator with 5% CO2 for 40 min, for cell
migration and invasion assays, 1×105 tumor cells in 0.1
µl of media without FBS were plated in the upper chamber. In the
lower chamber, 0.6 µl of the medium with 10% FBS was added.
Forty-eight hours after the incubation, cells on the upper surface
of the Millicell chambers, non-invasive or migrated cells, were
removed with a cotton swab. Tumor cells on the bottom surface of
the membrane were fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 0.1% crystal violet for 15
min. The number of migrated or invasive cells was counted in five
randomly selected fields under an inverted microscope (Olympus
CKX41; Olympus Corporation, Tokyo, Japan). Each sample was examined
in triplicate.
miRNA target prediction
miRTarBase (Release 6.1; http://mirtarbase.mbc.nctu.edu.tw/) was used to
collect the validated target genes of miR-615-5p. miRTarBase has
accumulated more than 360,000 MTIs validated experimentally by
reporter assay, western blot analysis, qPCR, microarray and
next-generation sequencing experiments (17). This study only collected the MTIs
which are validated experimentally by reporter assay, western blot
and qPCR analyses.
Luciferase reporter assay
Two human ESCC cell lines ECA109 and KYSE410
(4.0×104) were cultured in 24-well plates. The fragments
of ligating oligonucleotides with the wild-type (WT) or mutant-type
(MUT) putative binding site of the human IGF2 3′-UTR were cloned
between the Fse1 and Xba1 sites of pGL3-control
(Promega, Madison, WI, USA). Then, the cells were co-transfected
with miR-615-5p/NC-mimics and pGL3-IGF2-3′-UTR-WT/MUT using
Lipofectamine 2000 (Invitrogen). Forty-eight hours following the
transfection, ECA109 and KYSE410 cells were collected and detected
with the Dual-Luciferase Reporter assay system (Promega).
Luciferase activity was measured by a Lumat LB 9507 apparatus
(Berthold Technologies GmbH & Co. KG, Bad Wildbad, Germany).
Each sample was examined in triplicate.
Statistical analysis
All data were described as mean ± standard deviation
(SD) and statistically analyzed using SPSS software (version 11.0,
SPSS, Inc., Chicago, IL, USA). The differences between groups were
analyzed using Student's t-test. Spearman's correlation analysis
was performed to assess the correlation between miR-615-5p
expression and IGF2 mRNA expression in ESCC tissues. The
associations between miR-615-5p expression and various
clinicopathological characteristics of ESCC patients were performed
using the χ2 test for categorical variables. Difference
between groups was considered to be statistically significant at
P<0.05.
Results
Reduced expression of miR-615-5p in
both ESCC tissues and cells
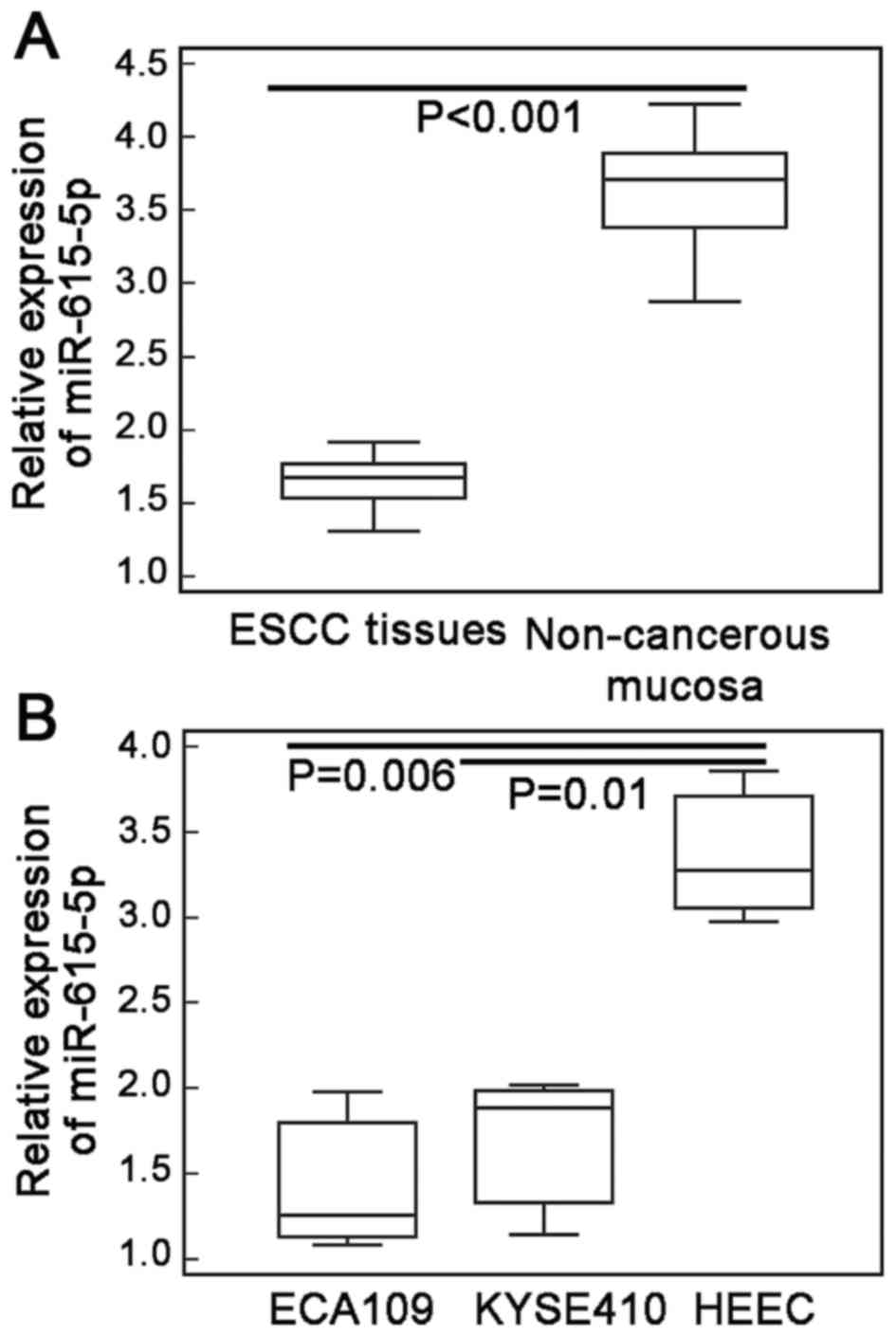
miR-615-5p expression in ESCC tissues, and two human
ESCC cell lines ECA109 and KYSE410 were markedly lower than those
in adjacent non-cancerous esophageal mucosa and human normal
esophageal cell line (HEEC) (ESCC vs. non-cancerous tissues:
1.70±0.30 vs. 3.62±0.39, P<0.001, Fig. 1A; ECA109 vs. HEEC cells: 1.46±0.42
vs. 3.38±0.45, P=0.006; KYSE410 vs. HEEC cells: 1.69±0.46 vs.
3.38±0.45, P=0.01; Fig. 1B).
Reduced expression of miR-615-5p
associates with aggressive tumor progression of ESCC patients
To evaluate the associations between miR-615-5p
expression and various clinicopathological features of ESCC
patients, we divided the 60 patients into low miR-615-5p expression
(n=31) and high miR-615-5p expression (n=29) groups using the
median value of miR-615-5p (1.68) expression levels in ESCC tissues
as a cutoff point. As shown in Table
I, ESCC patients with positive lymph node metastasis (P=0.01,
Table I), moderate-poor
differentiation (P=0.03, Table I)
and advanced TNM stage (P<0.001, Table I) had lower expression of miR-615-5p
than those with negative lymph node metastasis, well
differentiation and early TNM stage. However, no significant
association between miR-615-5p expression and the patient age or
sex, and tumor location was found (all P>0.05, Table I).
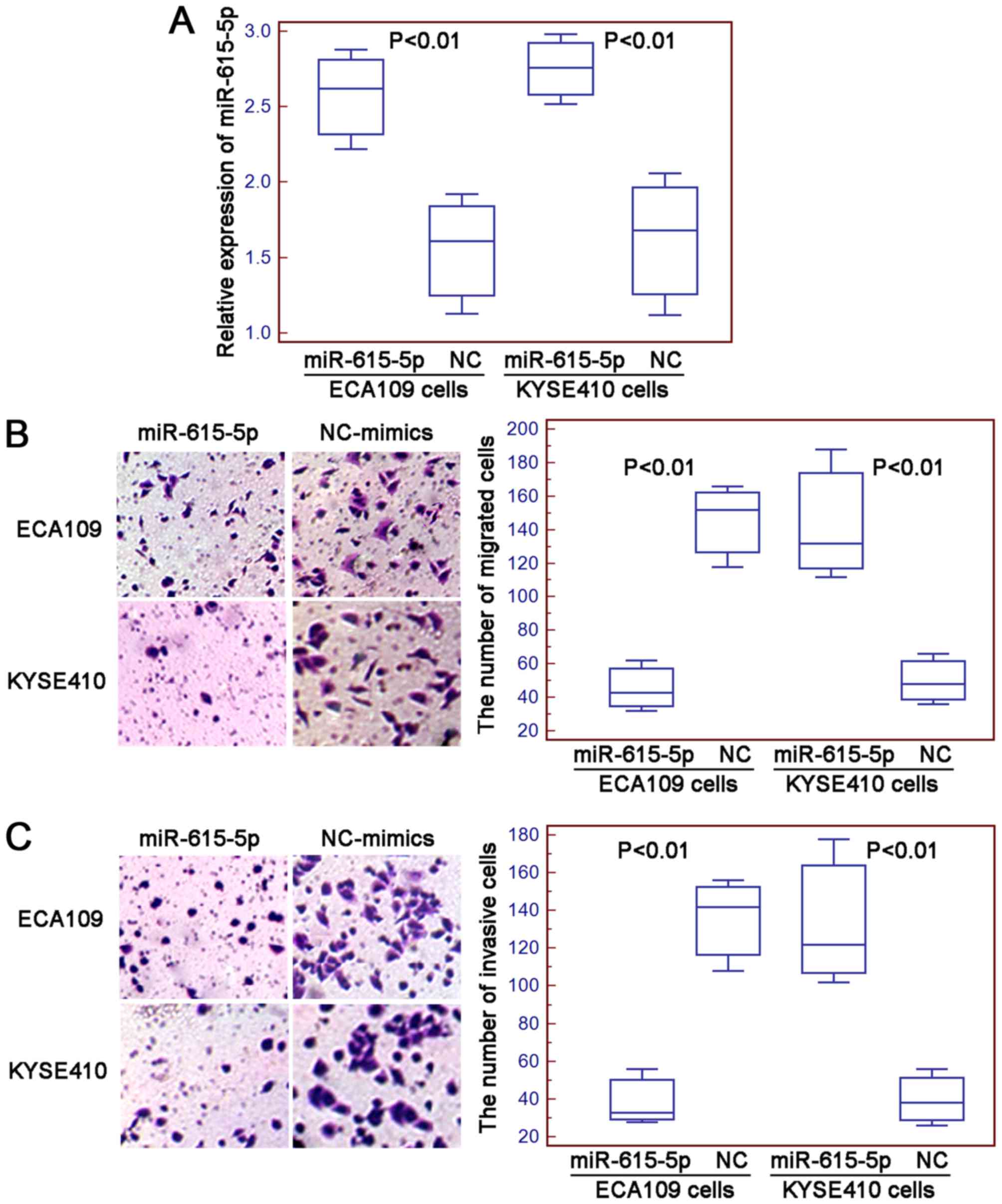
miR-615-5p suppresses cell motility of
human ESCC cells in vitro
The migration and invasion of ECA109 and KYSE410
cells after the transfection with miR-615-5p mimics or negative
control mimics were determined using the Transwell assays. The
transfection efficiency was evaluated using quantitative real-time
PCR and the results indicated that the expression levels of
miR-615-5p in ECA109 and KYSE410 cells were both significantly
increased by the transfection with miR-615-5p mimics compared with
negative control mimics (both P<0.01, Fig. 2A). Then, the numbers of migrated and
invasive ESCC cells were markedly decreased in cells transfected
with miR-615-5p mimics compared to the cells transfected with
negative control mimics (for ECA109 and KYSE410 cells: both
P<0.01, Fig. 2B and C).
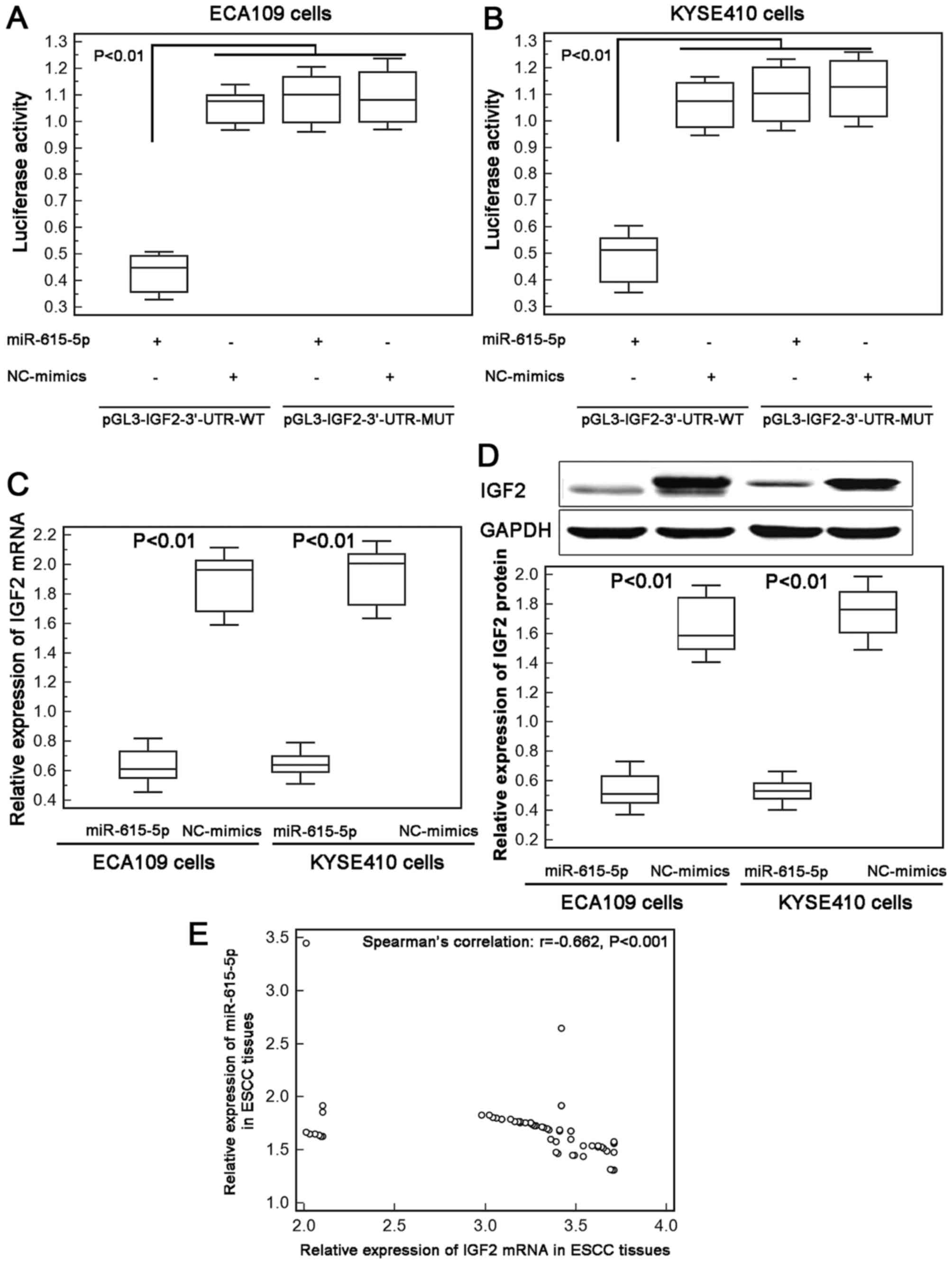
IGF2 is a direct target gene of
miR-615-5p
To investigate the underlying mechanism of the
inhibitory effect of miR-615-5p in ESCC cell motility, we collected
its candidate targets from miRTarBase and found that the
interaction between miR-615-5p and IGF2 were validated
experimentally by reporter assay in hepatocellular carcinoma cells
(18).
Next, we performed luciferase report assay based on
two human ESCC cell lines to confirm the binding efficiency between
miR-615-5p and IGF2 in ECA109 and KYSE410 cells. As a result, ESCC
cells co-transfected with miR-615-5p-mimics and pGL3-IGF2-3′-UTR-WT
both showed significantly lower luciferase reporter activity than
those co-transfected with NC-mimics and pGL3-IGF2-3′-UTR-WT
plasmid. However, there was no difference with statistical
significance in the reporter activity between ECA109 and KYSE410
cells co-transfected with miR-615-5p-mimics and
pGL3-IGF2-3′-UTR-MUT plasmid and cells co-transfected with
NC-mimics and pGL3-IGF2-3′-UTR-MUT plasmid (Fig. 3A and B).
Moreover, the re-expression of miR-615-5p
efficiently suppressed the endogenous expression of IGF2 at both
mRNA and protein levels in ECA109 and KYSE410 cells (Fig. 3C and D). Further Spearman's
correlation analysis displayed a negative correlation between
miR-615-5p and IGF2 mRNA expression in ESCC tissues (Spearman's
correlation: r=−0.662, P<0.001, Fig.
3E). These data offer evidence that IGF2 may be a direct target
of miR-615-5p in human ESCC cells.
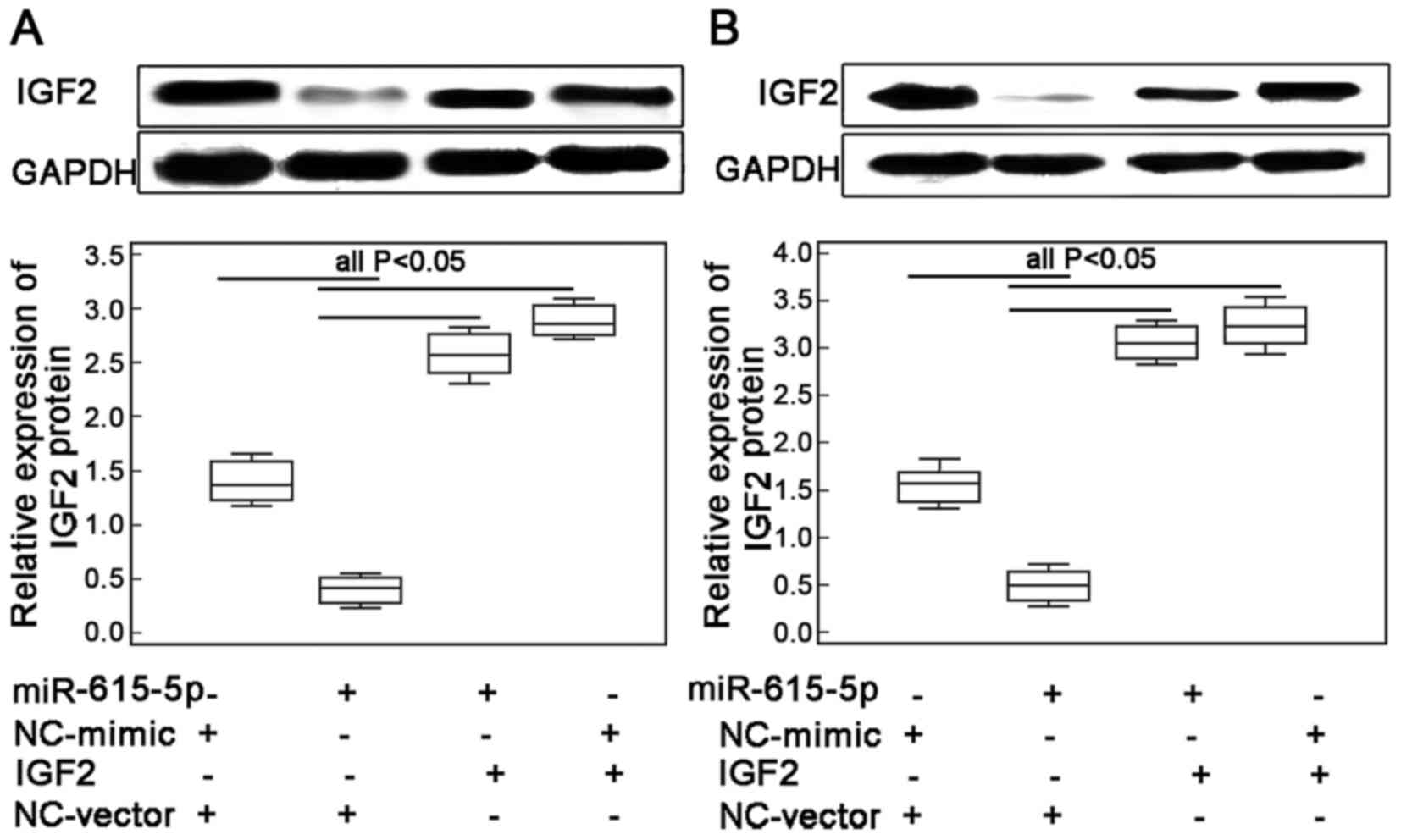
Restoration of IGF2 expression
reverses the inhibitory effects of miR-615-5p in ESCC cell
motility
To confirm whether miR-615-5p exerts its inhibitory
effects on ESCC cell motility through regulating its target gene
IGF2, the migrated and invasive capacities of ECA109 and KYSE410
cells with the co-transfection of miR-615-5p-mimics and
Lv-CMV-GFP-IGF2 expression vector were further evaluated. The
transfection efficiency was examined by western blot analysis. The
results indicated that the endogenous expression levels of IGF2
protein in ECA109 and KYSE410 cells were both significantly
increased following the transfection of the miR-615-5p mimic in the
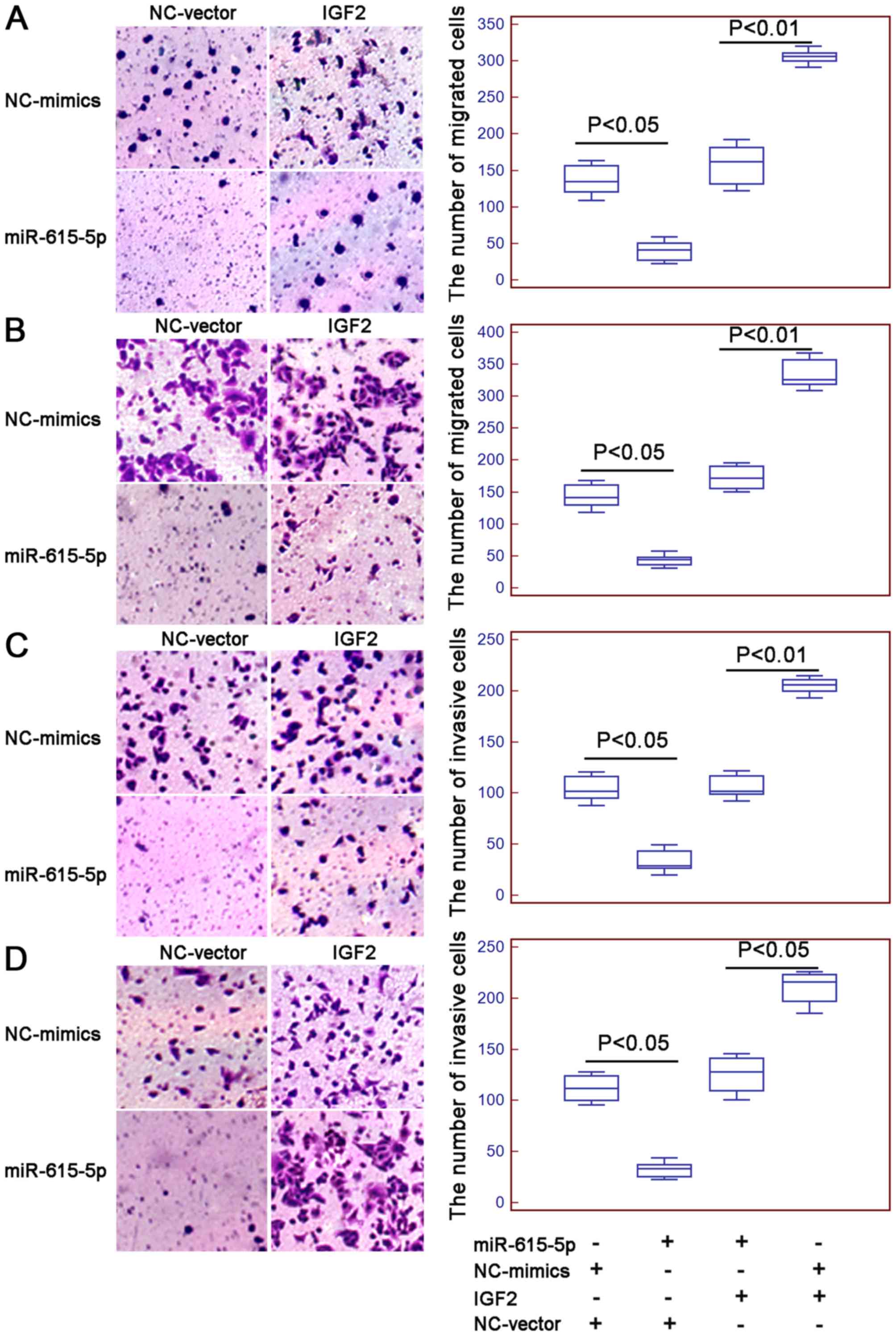
presence of IGF2 expression vector (both P<0.01, Fig. 4A and B, respectively).
Interestingly, the number of migrated and invaded ESCC cells with
the overexpression of miR-615-5p and the restoration of IGF2 were
both higher than those co-transfected with miR-615-5p mimics and
vector control (all P<0.05, Fig.
5).
Discussion
miRNAs have recently become a new ‘hot topic’ in the
research field of cancer-related molecular mechanisms. Increasing
evidence indicates that miRNAs play crucial roles in carcinogenesis
and cancer maintenance (8,19). Since it is still a major challenge
to identify molecular alterations in ESCC, we focused on this
malignancy in the present study. Our data showed that the
expression levels of miR-615-5p in ESCC tissues and cells were
markedly higher than those in non-cancerous esophageal mucosa and
human normal esophageal cells, respectively. Statistically, the
downregulation of miR-615-5p was closely correlated with various
aggressive clinicopathological features of patients with ESCC, such
as advanced TNM stage, positive lymph node metastasis and
moderate-poor differentiation. Functionally, we found that the
re-expression of miR-615-5p suppressed the invasion and migration
of ESCC cells. miRNAs exert the specific functions via inhibiting
translation or promoting degradation of the corresponding target
mRNAs using complementary sequences. Here, we identified IGF2 as a
direct target gene of miR-615-5p, and also confirmed that the
inhibitory effects of miR-615-5p in ESCC cell motility were
reversed by the restoration of IGF2 expression. To the best of our
knowledge, this is the first study to observe the expression
pattern of miR-615-5p, its clinical significance and functional
role in human ESCC.
miRNAs have been observed to be frequently expressed
abnormally and function as ‘onco(genic)-miRs’ or ‘tumor-suppressor
miRs’ in multiple malignant processes (20). These miRNAs may be good candidates
as biomarkers for cancer diagnosis and prognosis (21). In most recent studies, miR-615-5p
was identified as a tumor suppressor in various human cancers. For
example, Sun et al (22)
observed the downregulation of miR-615-5p in pancreatic ductal
adenocarcinoma, and confirmed that low miR-615-5p expression was an
independent prognostic marker for patients; Bai et al
(23) found the decreased
expression of miR-615 in breast cancer tissues and cell lines. In
contrast, Wu et al (24)
indicated that miR-615-5p was overexpressed in hepatocellular
carcinoma tissues and cell lines; however, its overexpression could
suppress both the cell proliferation and migration of
hepatocellular carcinoma cells in vitro, suggesting that it
is not necessarily the case that the overexpressed miRNAs act as
oncogenes or promote tumorigenesis, and the downregulated miRNAs
often play a tumor suppressive role. Consistent with these previous
studies, our results here confirmed the reduced expression of
miR-615-5p in both ESCC tissues and cell lines, and also indicated
its contributions to cancer progression. To investigate whether the
dysregulation of miR-615-5p is responsible for ESCC cell motility,
the transfection of miR-615-5p mimics was performed in ESCC cell
lines in vitro, and the Transwell assays showed that ESCC
cell migratory and invasive potentials were suppressed by
miR-615-5p, suggesting that the downregulation of miR-615-5p in
ESCC cells may be a prerequisite for metastasis.
Since the miRNA actions are exerted through
regulating the corresponding target genes, we also identified the
candidate target genes of miR-615-5p in order to investigate the
underlying mechanism of actions on how miR-615-5p abnormal
expression impacts ESCC. IGF2, as an imprinted gene in mammals, has
been observed to be overexpressed in multiple childhood and adult
malignancies (25). Its
upregulation is associated with resistance of chemotherapy and
unfavorable prognosis in patients (26–28).
Especially, Chava et al (29) performed immunohistochemistry to
observe the expression patterns of IGF2 in 39 archival tissue
samples of different esophageal pathologies, and found that IGF2
expression was enhanced in squamous cell carcinoma, adenocarcinoma,
and dysplasia of squamous epithelium samples when compared with
normal controls; Gao et al (30) discovered interaction between a long
non-coding RNA (lncRNA) 91H and IGF2, and revealed that 91H
contributed to the occurrence and progression of ESCC via
suppressing the expression of IGF2. In the present study, our data
demonstrated the IGF2 was directly regulated by miR-615-5p in ESCC.
Using a luciferase reporter assay, we demonstrated that miR-615-5p
directly targeted IGF2 by binding to its 3′UTR binding sites.
Moreover, the inverse correlation between miR-615-5p and IGF2 mRNA
expression in the ESCC tissues had statistical significance.
Concomitantly, the inhibitory effects of miR-615-5p in ESCC cell
migration and invasion were reversed by the enforced expression of
IGF2, implying that miR-615-5p downregulation may be an important
event related to IGF2 upregulation in human ESCC, and the reduced
expression of miR-615-5p may promote cancer cell motility by
influencing IGF2 expression.
In conclusion, our findings indicate miR-615-5p
downregulation as an underlying molecular mechanism of development
and progression, and as a potential therapeutic target of human
ESCC. Also, we showed that the miR-615-5p/IGF2 axis may bring
important contributions on cell motility of human ESCC.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan C, Qian X, Guan Z, Yang B, Ge Y, Wang
F and Cai J: Potential biomarkers for esophageal cancer.
Springerplus. 5:4672016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kang X, Chen K, Li Y, Li J, D'Amico TA and
Chen X: Personalized targeted therapy for esophageal squamous cell
carcinoma. World J Gastroenterol. 21:7648–7658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Makarova JA, Shkurnikov MU, Wicklein D,
Lange T, Samatov TR, Turchinovich AA and Tonevitsky AG:
Intracellular and extracellular microRNA: An update on localization
and biological role. Prog Histochem Cytochem. 51:33–49. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yan X, Xu H and Yan Z: Functional
perspective and implications of gene expression by noncoding RNAs.
Cancer Transl Med. 1:137–152. 2015. View Article : Google Scholar
|
|
8
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu W, Li M, Chen X, Zhang D, Wei L, Zhang
Z, Wang S, Meng L, Zhu S and Li B: MicroRNA-373 promotes migration
and invasion in human esophageal squamous cell carcinoma by
inhibiting TIMP3 expression. Am J Cancer Res. 6:1–14.
2015.PubMed/NCBI
|
|
11
|
Nie J, Ge X, Geng Y, Cao H, Zhu W, Jiao Y,
Wu J, Zhou J and Cao J: miR-34a inhibits the migration and invasion
of esophageal squamous cell carcinoma by targeting Yin Yang-1.
Oncol Rep. 34:311–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mao Y, Liu J, Zhang D and Li B: MiR-1290
promotes cancer progression by targeting nuclear factor I/X(NFIX)
in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother.
76:82–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang Y, Zhang Y, Li F, Du X and Zhang J:
CDX2 inhibits pancreatic adenocarcinoma cell proliferation via
promoting tumor suppressor miR-615-5p. Tumour Biol. 37:1041–1049.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M,
Kondo Y, Shinjo K, Zhu Y, Zhang J, et al: miR-615-5p is
epigenetically inactivated and functions as a tumor suppressor in
pancreatic ductal adenocarcinoma. Oncogene. 34:1629–1640. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song LJ, Zhang WJ, Chang ZW, Pan YF, Zong
H, Fan QX and Wang LXPU: PU.1 is identified as a novel metastasis
suppressor in hepatocellular carcinoma regulating the
miR-615-5p/IGF2 axis. Asian Pac J Cancer Prev. 16:3667–3671. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao ZG, Jin JY, Zhang AM, Zhang LP, Wang
XX, Sun JG and Chen ZT: MicroRNA profile of tumorigenic cells
during carcinogenesis of lung adenocarcinoma. J Cell Biochem.
116:458–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin
YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al: miRTarBase
2016: Updates to the experimentally validated miRNA-target
interactions database. Nucleic Acids Res. 44(D1): D239–D247. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
El Tayebi HM, Hosny KA, Esmat G, Breuhahn
K and Abdelaziz AI: miR-615-5p is restrictedly expressed in
cirrhotic and cancerous liver tissues and its overexpression
alleviates the tumorigenic effects in hepatocellular carcinoma.
FEBS Lett. 586:3309–3316. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gulyaeva LF and Kushlinskiy NE: Regulatory
mechanisms of microRNA expression. J Transl Med. 14:1432016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arora A, Singh S, Bhatt AN, Pandey S,
Sandhir R and Dwarakanath BS: Interplay between metabolism and
oncogenic process: Role of microRNAs. Transl Oncogenomics. 7:11–27.
2015.PubMed/NCBI
|
|
22
|
Sun Y, Zhang T, Wang C, Jin X, Jia C, Yu S
and Chen J: MiRNA-615-5p functions as a tumor suppressor in
pancreatic ductal adenocarcinoma by targeting AKT2. PLoS One.
10:e01197832015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bai Y, Li J, Li J, Liu Y and Zhang B:
MiR-615 inhibited cell proliferation and cell cycle of human breast
cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med.
8:3801–3808. 2015.PubMed/NCBI
|
|
24
|
Wu X, Deng L, Tang D, Ying G, Yao X, Liu F
and Liang G: miR-615-5p prevents proliferation and migration
through negatively regulating serine hydromethyltransferase 2
(SHMT2) in hepatocellular carcinoma. Tumour Biol. 37:6813–6821.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brouwer-Visser J and Huang GS: IGF2
signaling and regulation in cancer. Cytokine Growth Factor Rev.
26:371–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bruchim I, Sarfstein R and Werner H: The
IGF Hormonal Network in Endometrial Cancer: Functions, regulation,
and targeting approaches. Front Endocrinol (Lausanne).
5:762014.PubMed/NCBI
|
|
27
|
Chao W and D'Amore PA: IGF2: Epigenetic
regulation and role in development and disease. Cytokine Growth
Factor Rev. 19:111–120. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mu Q, Wang L, Yu F, Gao H, Lei T, Li P,
Liu P, Zheng X, Hu X, Chen Y, et al: Imp2 regulates GBM progression
by activating IGF2/PI3K/Akt pathway. Cancer Biol Ther. 16:623–633.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chava S, Mohan V, Shetty PJ, Manolla Ml,
Vaidya S, Khan IA, Waseem GL, Boddala P, Ahuja YR and Hasan Q:
Immunohistochemical evaluation of p53, FHIT, and IGF2 gene
expression in esophageal cancer. Dis Esophagus. 25:81–87. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao T, He B, Pan Y, Xu Y, Li R, Deng Q,
Sun H and Wang S: Long non-coding RNA 91H contributes to the
occurrence and progression of esophageal squamous cell carcinoma by
inhibiting IGF2 expression. Mol Carcinog. 54:359–367. 2015.
View Article : Google Scholar : PubMed/NCBI
|