Introduction
Renal cell carcinoma (RCC) is a disease in which
cells in the tubules of the kidney undergo oncogenic
transformation. The most common subtype (approximately 80% of
cases) of RCC is clear cell RCC (ccRCC) (1). The 5-year survival rate of
advanced-stage RCC patients is extremely poor (5–10%) due to
recurrence or distant metastasis (2). At diagnosis, nearly 30% of RCC
patients present with metastasis (3). Current treatments for RCC include
molecular-targeted therapeutics such as anti-angiogenic
multi-tyrosine kinase inhibitors or mTOR inhibitors, which are
being widely used for patients with metastatic or recurrent RCC.
However, these types of therapies are not expected to have curative
effects as they only slightly extend progression-free survival
(4). Therefore, to improve the
outcome of patients with RCC, it is necessary to fully elucidate
the molecular mechanisms underlying the development and progression
of RCC and the associated oncogenic pathways.
The discovery of non-coding RNAs (ncRNAs) in the
human genome was an important conceptual breakthrough, and the
understanding of ncRNAs is important for progress in cancer
research. MicroRNAs (miRNAs) are endogenous small ncRNA molecules
(19–22 bases in length) that regulate protein-coding gene
expression by repressing translation or cleaving RNA transcripts in
a sequence-specific manner (5).
miRNAs are predicted to regulate more than 60% of the
protein-coding genes in the human genome (6). Accumulating evidence suggests that
miRNAs are aberrantly expressed in many human cancers and play
significant roles in human oncogenesis and metastasis (7). Therefore, identifying aberrantly
expressed miRNAs is an important first step to elucidate the
details of miRNA-mediated oncogenic pathways in cancer cells.
Previously, our miRNA expression signature of ccRCC
revealed that microRNA-1274a (miR-1274a) was
significantly upregulated in cancer tissues, suggesting that this
miRNA functions as an oncogenic miRNA (8). Therefore, the aim of the present study
was to investigate the functional significance of miR-1274a
and to identify the molecular targets and pathways mediated by
miR-1274a in ccRCC cells. Our data demonstrated that
downregulation of miR-1274a inhibited cancer cell
proliferation and induced apoptosis. Moreover, gene expression data
and in silico database analysis showed that bone
morphogenetic protein receptor type 1B (BMPR1B) gene was identified
as a candidate target of miR-1274a. The discovery of genes
and pathways mediated by oncogenic miR-1274a provides
important insights into the potential mechanisms of ccRCC
oncogenesis and suggests novel therapeutic strategies for the
treatment of ccRCC.
Materials and methods
Clinical specimens and cell
culture
The ccRCC and adjacent normal tissues were collected
from 40 ccRCC patients immediately after undergoing radical
nephrectomies at Kagoshima University Hospital between 2003 and
2013. The clinicopathological information of the patients is shown
in Table I. The specimens were
staged according to the American Joint Committee on Cancer-Union
Internationale Contre le Cancer (UICC) tumor-node-metastasis (TNM)
classification and histologically graded (9). This study was approved by the
Bioethics Committee of Kagoshima University; prior written informed
consent and approval were obtained from all patients. We used human
ccRCC cell lines, 786O, A498, ACHN and Caki1, obtained from the
American Type Culture Collection (ATCC; Manassas, VA, USA). These
cell lines were incubated in RPMI-1640 medium (Thermo Fisher
Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine
serum (FBS) and maintained in humidified incubators (5%
CO2) at 37°C.
 | Table I.Clinicopathological characteristics
of the ccRCC patients. |
Table I.
Clinicopathological characteristics
of the ccRCC patients.
|
Characteristics | Data |
|---|
| Total number of
cases | 40 |
| Mean age ± SD
(years) | 64.4±13.5 |
| Sex, n (%) |
|
|
Male | 14 (35.0) |
|
Female | 26 (65.0) |
| Stage, n (%) |
|
|
pT1a | 18 (45.0) |
|
pT1b | 13 (32.5) |
|
pT2 | 2 (5.0) |
|
pT3a | 4 (10.0) |
|
pT3b | 3 (7.5) |
|
pT4 | 0 (0.0) |
| Grade, n (%) |
|
| G1 | 8 (20.0) |
| G2 | 30 (75.0) |
| G3 | 0 (0.0) |
|
Unknown | 2 (5.0) |
| Infiltration, n
(%) |
|
| α | 27 (67.5) |
| β | 13 (32.5) |
| γ | 0 (0.0) |
| Venous invasion, n
(%) |
|
| v
(−) | 29 (72.5) |
| v
(+) | 11 (27.5) |
Tissue collection and RNA
extraction
Tissues were immersed in RNAlater (Thermo Fisher
Scientific) and stored at −20°C until RNA extraction was conducted.
Total RNA, including miRNA, was extracted using the mirVana™ miRNA
isolation kit (Thermo Fisher Scientific) following the
manufacturers protocol.
Quantitative real-time reverse
transcriptase-polymerase chain reaction (qRT-PCR)
Stem-loop RT-PCR (TaqMan MicroRNA Assays; product
ID: 2883 for miR-1274a; Applied Biosystems, Foster City, CA, USA)
was used to quantify miRNAs according to the manufacturers protocol
for PCR conditions. We used human RNU48 (product ID: 001006;
Applied Biosystems) as internal control. For investigating the
genes, we applied SYBR-Green quantitative PCR-based array approach,
and the following primers were used: BMRP1B forward primer,
5′-CTTTTGCGAAGTGCAGGAAAAT-3′ and reverse primer,
5′-TGTTGACTGAGTCTTCTGGACAA-3′; GUSB forward primer,
5′-CGTCCCACCTAGAATCTGCT-3′ and reverse primer,
5′-TTGCTCACAAAGGTCACAGG-3′. qRT-PCR was performed with 500 ng of
total RNA using the Power SYBR-Green Master Mix (cat. no. 4367659;
Applied Biosystems) on the 7300 Real-Time PCR System (Applied
Biosystems). The experimental procedures were conducted according
to the protocol recommended by the manufacturer. The specificity of
amplification was monitored using the dissociation curve of the
amplified product. BMPR1B data values were normalized to
GUSB. The ∆∆Ct method was employed to calculate the
fold-change of expression level relative to the internal
controls.
miRNA inhibitor transfection
ccRCC cell lines were transfected with Lipofectamine
RNAiMAX transfection reagent and Opti-MEM (Thermo Fisher
Scientific) with 10–60 nM miRNA inhibitor (hsa-miR-1274a;
product ID: AM13432, cat. no. AM17000) and negative control #1
anti-miRNA (cat. no. AM17010; Thermo Fisher Scientific).
Cell proliferation and apoptosis
assays following anti-miR-1274a transfection
Seventy-two hours after transfection, cell
proliferation was determined with an XTT assay using the Cell
Proliferation Kit II (Roche Molecular Biochemicals, Mannheim,
Germany) according to the manufacturers instructions. Cell
apoptosis assays were carried out using ccRCC cell lines
transfected with the transfection reagent, anti-miR-control and
anti-miR-1274a, in 6-well tissue culture plates. Cells were
harvested 72 h after transfection by trypsinization and washed in
cold phosphate-buffered saline (PBS). Double staining with
FITC-Annexin V and propidium iodine (PI) was carried out using the
FITC Annexin V apoptosis detection kit (BD Biosciences, Bedford,
MA, USA) according to the manufacturer's recommendations and
analyzed within 1 h by flow cytometry (FACScan; BD Biosciences).
Cells were discriminated into viable cells, dead cells, early and
late apoptotic cells by CellQuest software (BD Biosciences), and
then the percentages of total apoptotic cells from each samples
were compared. Experiments were conducted in triplicate.
Putative target gene analysis of
miR-1274a
To investigate the expression status of candidate
miR-1274a target genes in ccRCC clinical specimens, we
examined Oligo Microarray human 44K (Agilent Technologies, Santa
Clara, CA, USA) expression profiling of ACHN with anti-miR-control
and anti-miR-1274a transfectants. Then, gene expression data
were adapted to Kyoto Encyclopaedia of Genes and Genomes (KEGG)
pathway categories by the GENECODIS program (http://genecodis.cnb.csic.es/). Publicly available
expression data from ccRCC specimens in the GEO database (accession
number: GSE36895 + GSE22541) were used. We merged these data sets
and selected putative genes targeted by miR-1274a according
to the TargetScan algorithm (June, 2011 release, http://www.targetscan.org/vert_50/).
Plasmid construction and
Dual-Luciferase reporter assays
A partial wild-type sequence of the BMPR1B
3-untranslated region (UTR) or the same region lacking its
miR-1274a target site (positions 3237–3243 of the BMPR1B
3-UTR, according to the TargetScan program) was inserted between
the XhoI and PmeI restriction sites in the 3-UTR of
the hRluc gene in the psi-CHECK-2 vector (C8021; Promega, Madison,
WI, USA). ACHN cells were transfected with 50 ng of the vector and
10 nM miR-1274a (product ID: AM13432, cat. no. AM17100;
Thermo Fisher Scientific) or control miRNAs (product ID: AM17111;
Thermo Fisher Scientific) using Lipofectamine 2000 (Invitrogen).
The activities of firefly and Renilla luciferases in cell
lysates were determined with a Dual-Luciferase assay system (E1960;
Promega). Normalized data were calculated as the ratio of
Renilla/firefly luciferase activities.
Overall survival analysis of a ccRCC
cohort (KIRC) from The Cancer Genome Atlas (TCGA)
The TCGA cohort database for 72 normal kidneys and
534 ccRCC patients (KIRC) was used to determine the relationship
between BMPR1B expression in normal and in ccRCCs, and
between BMPR1B expression of ccRCC patients and overall
survival. Normalized RNA-seq by expectation-maximization (RSEM)
value from RNA-seq expression data were used for gene expression
quantification (10). Full
sequencing information and clinical information were acquired from
the cBioPortal (http://www.cbioportal.org/public-portal/) and the TCGA
(https://tcga-data.nci.nih.gov/tcga/)
(11–13).
Immunohistochemistry
A tissue microarray of 67 ccRCC samples and 10
normal renal tissues was obtained from US Biomax, Inc. (KD806;
Rockville, MD, USA). Immunostaining was carried out on the tissue
microarray according to the manufacturers protocol using the
UltraVision Detection System (Thermo Fisher Scientific). Primary
rabbit polyclonal antibodies against BMPR1B (GTX102453; GeneTex,
Inc., Irvine, CA, USA) were diluted 1:500. Slides were treated with
biotinylated goat anti-rabbit. Diaminobenzidine hydrogen peroxidase
was the chromogen and counterstaining was conducted with 0.5%
hematoxylin. Immunostaining was evaluated according to a previously
described scoring method (14).
Statistical analysis
Relationships between two or three variables and
numerical values were analyzed using the Mann-Whitney U test or
Bonferroni-adjusted Mann-Whitney U test. Overall survival of ccRCC
patients from the TCGA cohort was evaluated by the Kaplan-Meier
method. Patients were numerically divided equally into two groups
according to BMPR1B expression, and the differences between
the two groups were evaluated by log-rank tests. We used Expert
StatView software, version 5.0 (SAS Institute, Inc., Cary, NC, USA)
for these analyses.
Results
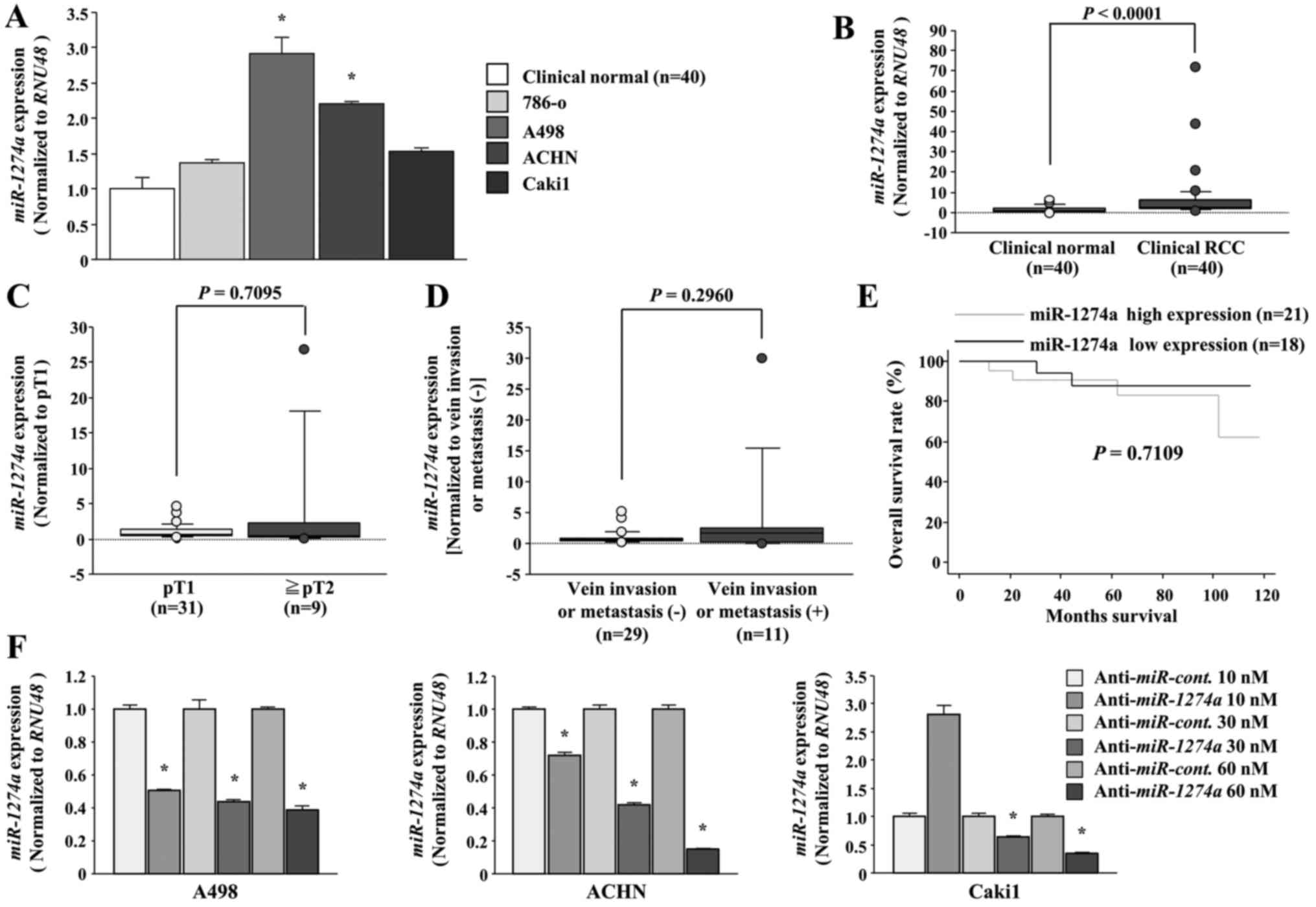
Expression levels of miR-1274a in
ccRCC specimens and ccRCC cell lines
We evaluated the expression level of
miR-1274a in ccRCC tissues (n=40), adjacent normal kidney
tissue (n=40) and ccRCC cell lines (786-O, A498, ACHN and Caki1).
The expression levels of miR-1274a were significantly higher
in tumor tissues and two ccRCC cell lines (A498 and ACHN) than in
normal kidney tissue (P<0.05, Fig.
1A, and P<0.0001, Fig. 1B).
miR-1274a expression in Caki1 was higher than that noted in
the normal kidneys even though there was no significant difference.
Therefore, we used A498, ACHN and Caki1 cells for the next
experiments. There were no significant correlations between any of
the clinicopathological parameters or overall survival rate and the
expression of miR-1274a (Fig.
1C-E). Next, we evaluated the miR-1274a expression level
following anti-miR-1274a transfection. The expression of
miR-1274a in the A498, ACHN and Caki1 cells was sufficiently
downregulated following transfection of 30 and 60 nM
anti-miR-1274a (P<0.05; Fig.
1F).
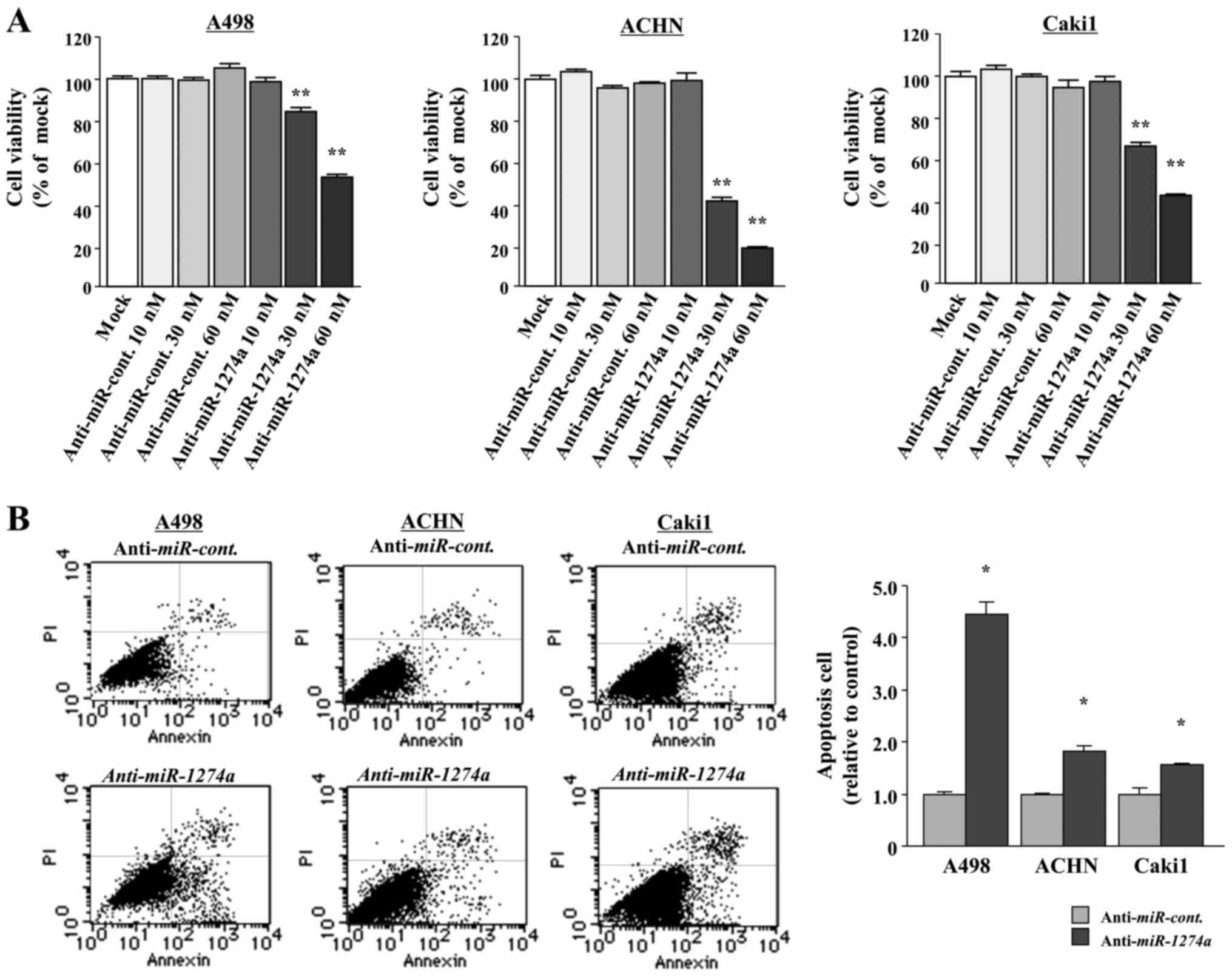
Effects of the inhibition of miR-1274a
on cell proliferation and apoptosis in ccRCC cell lines
To examine the functional roles of miR-1274a,
we performed loss-of-function studies by using
anti-miRNA-transfected A498, ACHN and Caki1 cells. XTT assays
revealed that there was a significant inhibition of cell
proliferation in the 10, 30 and 60 nM
anti-miR-1274a-transfectants (P<0.0001; Fig. 2A). We also analyzed the numbers of
apoptotic cells and found that miR-1274a inhibition
significantly induced apoptosis of the A498, ACHN and Caki1 cells
(P<0.05; Fig. 2B).
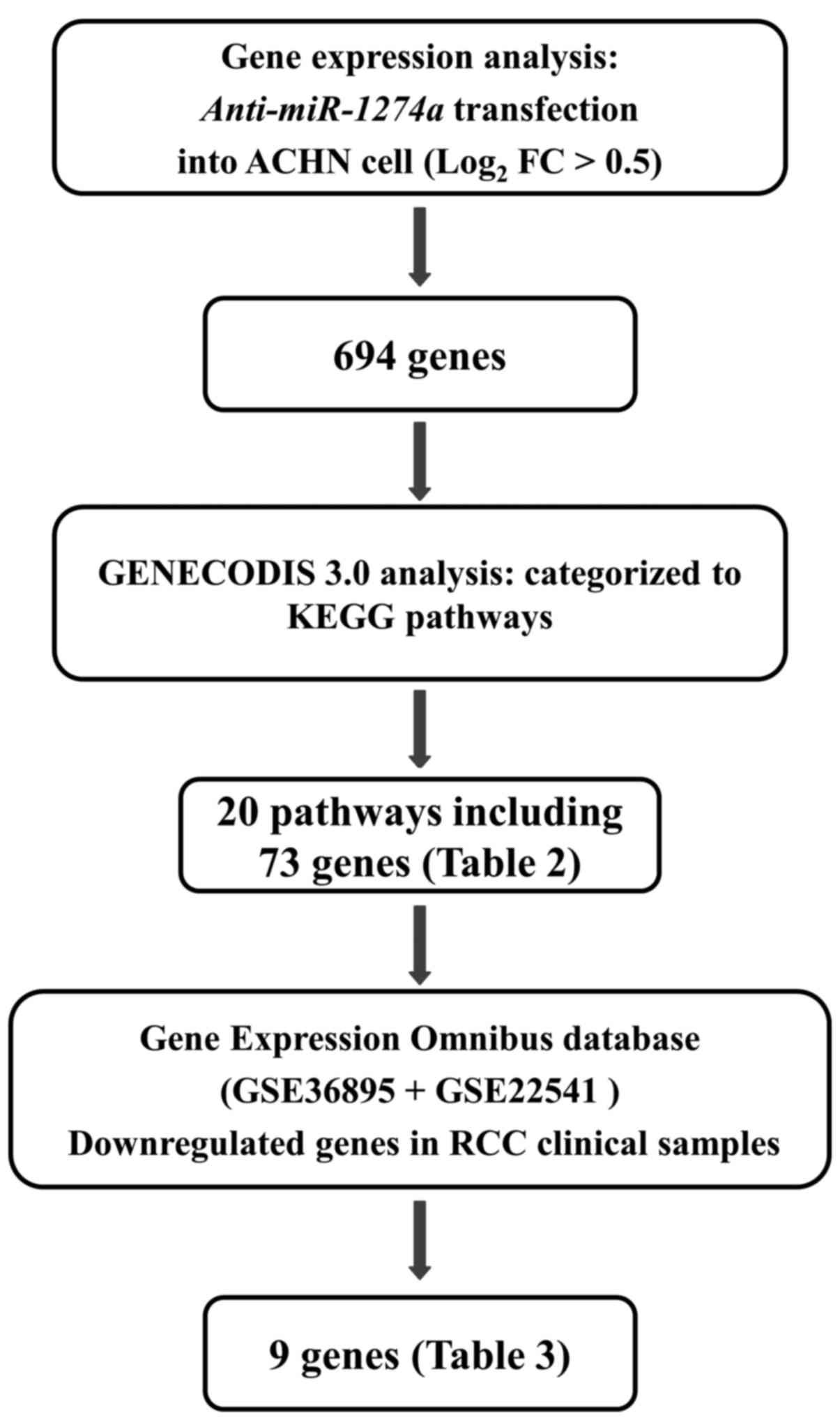
Screening of putative target genes of
miR-1274a in ccRCC
To gain further insight into the molecular
mechanisms regulated by oncogenic miR-1274a in ccRCC cells,
we screened miR-1274a-regulated genes by using in
silico and genome-wide gene expression analysis (Fig. 3). As for the target search by oligo
microarray, we used ACHN as the knockdown efficiency and cell
proliferative inhibition rate by anti-miRNA was the
strongest in the ACHN cells. First, we found that 694 genes were
upregulated in the oligo microarray by
anti-miR-1274a-transfectants of ACHN. These 694 genes were
assigned KEGG annotations using singular enrichment analysis of
GeneCodis to identify the molecular pathways regulated by
miR-1274a. Twenty signaling pathways and 73 genes were
identified in this analysis (Table
II). Furthermore, we examined gene expression signatures that
were downregulated in clinical ccRCC specimens from the GEO
database and found that 9 genes were significantly downregulated in
53 ccRCCs compared to 23 normal kidney tissues. Among these, we
focused on BMPR1B, as the BMPR1B expression level in ccRCC
tissues was most significantly downregulated compared with the
level noted in normal tissues (Table
III) and TargetScan 5.2 database predicted that there was a
binding site for miR-1274a in BMPR1B 3UTR.
 | Table II.Significantly enriched annotations
regulated by miR-1274a (top 20 pathways). |
Table II.
Significantly enriched annotations
regulated by miR-1274a (top 20 pathways).
| No. of genes | P-value | KEGG pathway | Genes |
|---|
| 12 | 1.35E-08 | NOD-like receptor
signaling pathway | IL6, BIRC2, CXCL2,
TNFAIP3, NFKBIA, CXCL1, BIRC3, IL8, MAPK9, CARD9, CCL2, NLRP3 |
| 19 | 3.52E-06 | Cytokine-cytokine
receptor interaction | IL12RB1, IL6,
TNFRSF10D, CXCL2, BMPR1B, CXCL3, FAS, CXCL1, CCL20, CNTF, CXCL16,
IL8, EDA2R, CCL2, LTB, IL18RAP, TNFRSF9, VEGFA, CSF2 |
| 11 | 5.04E-06 | Rheumatoid
arthritis | IL6, FOS, CXCL1,
CCL20, IL8, ICAM1, CCL2, LTB, VEGFA, CSF2, MMP1 |
| 19 | 5.47E-05 | Pathways in
cancer | IL6, MDM2, FOS,
BIRC2, NFKBIA, ITGA2, FAS, ETS1, BIRC3, RUNX1, IL8, MAPK9, LAMB3,
MECOM, TRAF1, VEGFA, WNT1, STAT5A, MMP1 |
| 16 | 1.74E-04 | MAPK signaling
pathway (American trypanosomiasis) | GADD45A, NLK, FOS,
DUSP1, DUSP16, RELB, DDIT3, FAS, PPM1A, HSPA6, CACNA1B, DUSP5,
MAPK9, MECOM, DUSP10, MAP3K8 |
| 9 | 8.25E-04 | Chagas disease
(American trypanosomiasis) | IL6, FOS, NFKBIA,
FAS, SERPINE1, IL8, MAPK9, CCL2, GNA15 |
| 10 | 9.08E-04 | Osteoclast
differentiation | FOS, SIRPG, NFKBIA,
RELB, SQSTM1, LILRB3, SOCS3, FOSB, MAPK9, JUNB |
| 8 | 6.08E-03 | T cell receptor
signaling pathway | FOS, NFKBIA, PAK6,
MAPK9, NFKBIE, PTPRC, CSF2, MAP3K8 |
| 7 | 8.12E-03 | ErbB signaling
pathway | NRG1, EREG, PAK6,
ABL2, HBEGF, MAPK9, STAT5A |
| 10 | 1.29E-02 | Chemokine signaling
pathway | CXCL2, NFKBIA,
CXCL3, CXCL1, CCL20, CXCL16, IL8, GRK5, CCL2, GNGT1 |
| 7 | 1.62E-02 | Toll-like receptor
signaling pathway | IL6, FOS, NFKBIA,
IL8, IRF5, MAPK9, MAP3K8 |
| 6 | 2.49E-02 | TGF-β signaling
pathway | CHRD, BMPR1B, BMP6,
ID3, SMURF1, ACVR1C |
| 6 | 2.60E-02 | Small cell lung
cancer | BIRC2, NFKBIA,
ITGA2, BIRC3, LAMB3, TRAF1 |
| 6 | 2.72E-02 | Apoptosis | TNFRSF10D, BIRC2,
NFKBIA, FAS, BIRC3, IRAK2 |
| 4 | 3.84E-02 | Bladder cancer | MDM2, IL8, VEGFA,
MMP1 |
| 5 | 4.01E-02 | p53 signaling
pathway | MDM2, GADD45A, FAS,
SERPINE1, ZMAT3 |
| 5 | 4.01E-02 | Epithelial cell
signaling in Helicobacter pylori infection | NFKBIA, CXCL1,
HBEGF, IL8, MAPK9 |
| 7 | 4.09E-02 | Measles | IL6, TNFRSF10D,
TNFAIP3, NFKBIA, FAS, HSPA6, STAT5A |
| 5 | 4.62E-02 | Chronic myeloid
leukemia | MDM2, NFKBIA,
RUNX1, MECOM, STAT5A |
| 6 | 4.63E-02 | Amoebiasis | IL6, CXCL1, IL8,
LAMB3, GNA15, CSF2 |
 | Table III.Expression of miR-1274a target
genes involved in significantly enriched annotations according to
gene expression data in ACHN cells. |
Table III.
Expression of miR-1274a target
genes involved in significantly enriched annotations according to
gene expression data in ACHN cells.
|
|
|
|
| GEO |
|
|---|
|
|
|
|
|
|
|
|---|
| Gene symbol | Fold change (ACHN:
anti-miR-1274/control) | Entrez Gene ID | Gene name | P-value | Fold change | TargetScan 5.2 |
|---|
| BMPR1B | 1.58 | 658 | Bone morphogenetic
protein receptor type 1B | 2.31E-09 | −9.75 | ○ |
| PAK6 | 1.43 | 56924 | p21 protein
(Cdc42/Rac)-activated kinase 6 | 2.00E-09 | −7.08 | ○ |
| MECOM | 1.47 | 2122 | MDS1 and EVI1
complex locus | 4.61E-09 | −4.85 |
|
| ACVR1C | 3.36 | 130399 | Activin A receptor
type 1C | 4.94E-06 | −2.18 |
|
| GADD45A | 2.62 | 1647 | Growth arrest and
DNA damage inducible alpha | 7.83E-05 | −1.99 |
|
| PPM1A | 1.44 | 5494 | Protein
phosphatase, Mg2+/Mn2+ dependent 1A | 6.08E-06 | −1.73 |
|
| NRG1 | 1.41 | 3084 | Neuregulin 1 | 1.46E-03 | −1.62 |
|
| ITGA2 | 1.49 | 3673 | Integrin subunit α
2 | 7.67E-03 | −1.59 |
|
| NLK | 1.56 | 51701 | Nemo-like
kinase | 2.37E-02 | −1.15 |
|
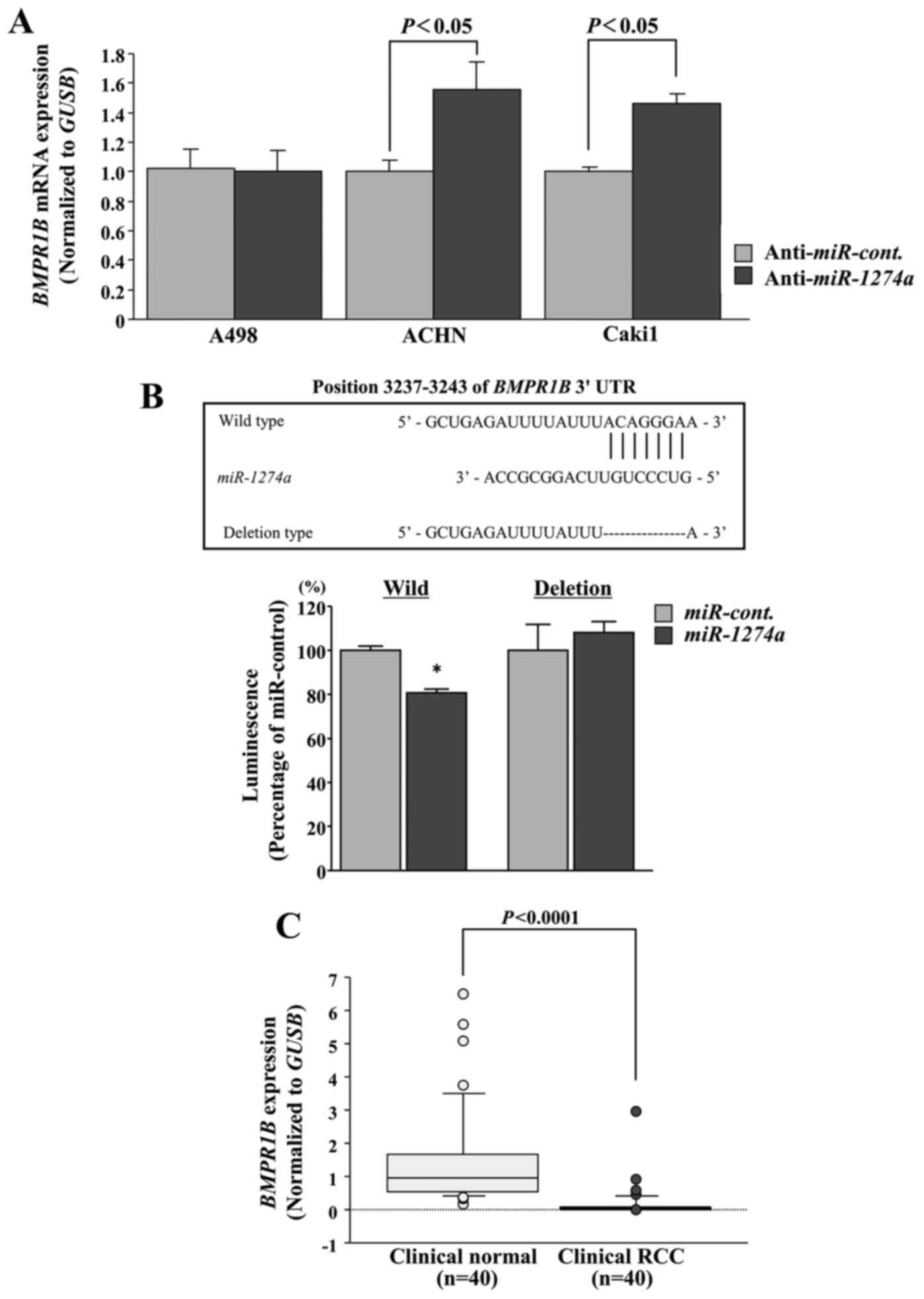
BMPR1B is regulated by miR-1274a
We performed qRT-PCR to determine whether inhibition
of miR-1274a resulted in upregulation of BMPR1B mRNA
expression in ccRCC cells. The mRNA levels of BMPR1B were
significantly upregulated in the anti-miR-1274a
transfectants in comparison with the anti-miR-control transfectants
in ACHN and Caki1 cells (P<0.05; Fig. 4A). Next, we performed luciferase
reporter assays to determine whether the 3′-UTR of BMPR1B
contained an actual binding site of miR-1274a. The
TargetScan database predicted that one putative miR-1274a
binding site existed in the 3′-UTR of BMPR1B (positions
3237–3243; Fig. 4B). We found that
the luminescence intensity in ACHN cells was significantly reduced
by transfection of miR-1274a with the wild-type vector
carrying the 3′-UTR of BMPR1B, whereas transfection with a
deletion vector showed no decrease in luminescence (P<0.05;
Fig. 4B). The expression levels of
BMPR1B were significantly lower in ccRCCs than in normal kidneys
using our clinical samples (P<0.0001; Fig. 4C). These data were consistent with
the in silico database analysis (Fig. 3) indicating that downregulated
BMPR1B as reported in the GEO data analysis was upregulated when
the cells were treated with anti-miR-1274a in the microarray.
BMPR1B expression as a prognostic
marker of ccRCC
The expression levels of BMPR1B were
significantly lower in ccRCC tissue samples than that noted in the
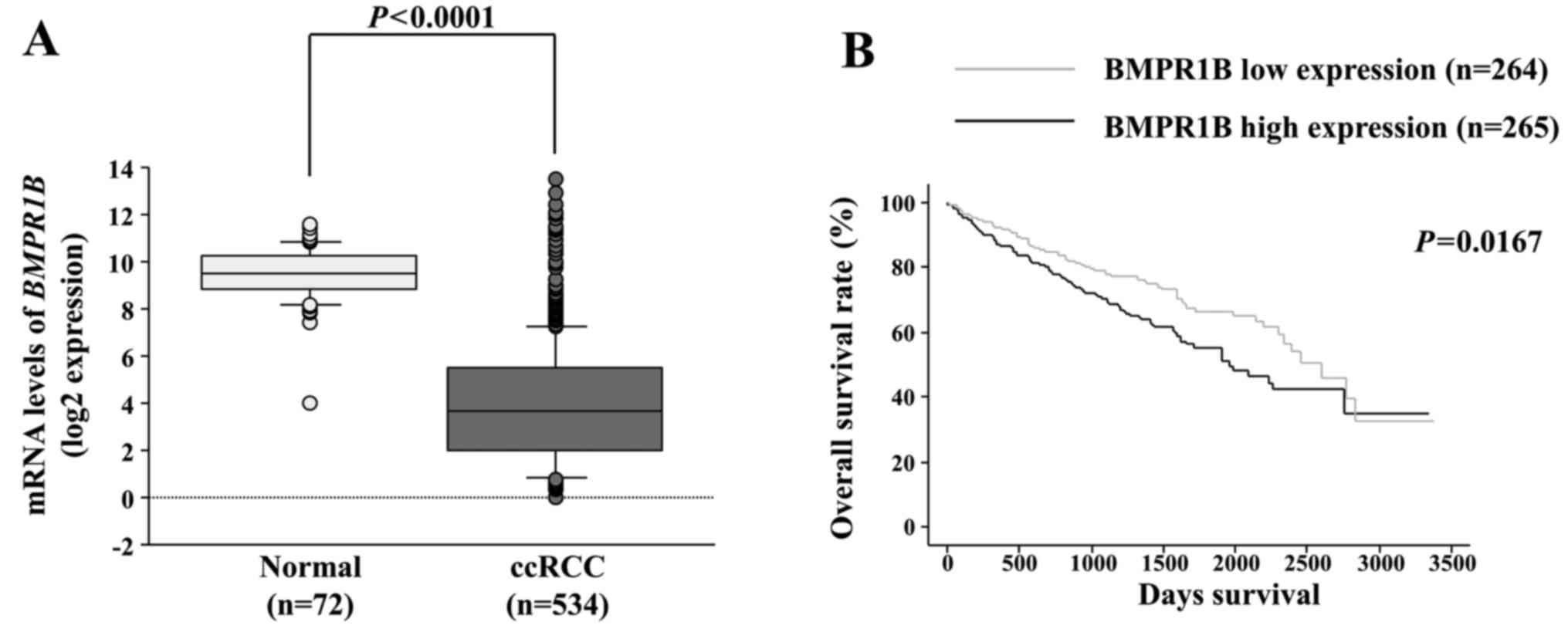
normal kidneys using the TCGA database (P<0.0001; Fig. 5A). We then evaluated the correlation
of BMPR1B expression levels with overall survival of the
ccRCC patients. The cohort was numerically divided equally into two
groups based on BMPR1B expression. Unexpectedly, we found
that the low BMPR1B expression group (n=264) had better
median overall survival in comparison with the high BMPR1B
expression group (n=265) (P=0.0167; Fig. 5B). This findings were supported by
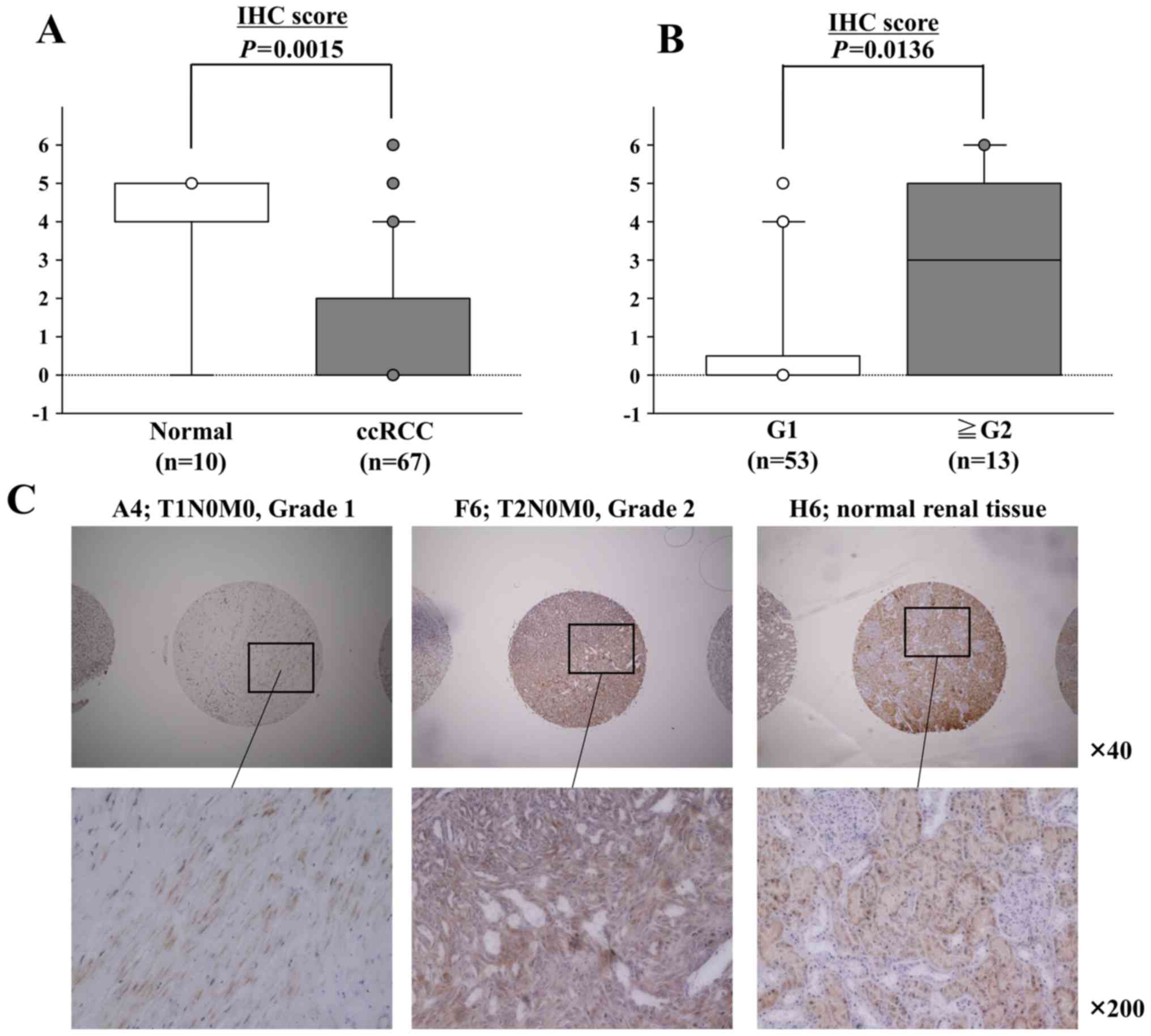
the results of the immunohistochemical staining of BMPR1B in tissue
specimens (Fig. 6). The expression
scores of BMPR1B were significantly lower in 67 ccRCCs in
comparison with 10 normal renal tissues (P=0.0015; Fig. 6A). On the other hand, the scores
were significantly higher in ccRCCs with high-grade tumors (more
than G2, n=13) in comparison with scores in ccRCCs with low-grade
tumors (G1, n=53) (P=0.0136; Fig.
6B).
Discussion
There is considerable evidence that normal miRNA
regulatory mechanisms are disrupted in cancer cells (7). However, the regulatory mechanisms by
which upregulated miRNAs exert their effects has not been fully
understood compared to downregulated miRNAs. Upregulated miRNAs
have been mainly viewed as markers (15), and functional studies have not been
performed in detail, even though many studies have successfully
inhibited miRNA expression using miRNA inhibitors, morpholinos,
sponges, or CRISPR/Cas9 (8,16–18).
One of the possible reasons is that miRNA gain-of-function studies
in downregulated miRNAs can be easily performed by transfection of
precursor miRNAs with sufficient efficiency. In the present study,
we suppressed upregulated miR-1274a with miRNA antisense
inhibitor. Even though we obtained nearly 85% knockdown efficiency
at most, high concentration of the miRNA antisense inhibitor was
needed. Therefore, other approaches such as CRISPR/Cas9 to gain
adequate knockdown efficiency without off-target effects will be
promising in subsequent research.
Our previous studies of miRNA expression signatures
in ccRCC identified several upregulated miRNAs (miR-885-5p,
miR-1274a, miR-210-3p, miR-224 and
miR-1290) (8). In this
study, we focused on miR-1274a, as increased expression of
miR-1274a has been reported in other types of human cancers
(19,20). Wang et al (19) reported that miR-1274a was
overexpressed in gastric cancer tissues or cancer cell lines, and
that miR-1274a promoted proliferation, migration and
epithelial-mesenchymal transition (EMT) by suppressing FOXO4, which
is a transcriptional factor and leads to cell cycle arrest or
apoptosis. In melanoma, high expression of miR-1274a was
also reported (20). On the other
hand, contrasting roles of miR-1274a have also been reported
in cancers to date. Zhou et al (21) reported that miR-1274a was
upregulated by sorafenib, which is a multi-kinase inhibitor
clinically used for cancer treatment. They also found that
miR-1274a could significantly suppress one of the protease
proteins ADAM9 and accelerate antitumor immunity in hepatocellular
carcinoma (HCC). Yan et al (22) also reported similar results in HCC
where miR-1274a was upregulated by paclitaxel. In fields
other than cancers, Ezzie et al (23) reported that miR-1274a was one
of the highly expressed miRNAs in lungs from subjects with chronic
obstructive pulmonary disease (COPD) compared with smokers without
COPD. Thus, diverse types of roles for miR-1274a have been
reported not only in cancers but also in chronic disease.
In the present study, we investigated the pathways
and targets that were regulated by oncogenic miR-1274a in
ccRCC cells. In the process of the identification of
miR-1274a-regulated molecular pathways and target genes, we
used a combination of expression data and in silico database
analysis (24). Using this
strategy, we have identified molecular targets and pathways in
several types of cancer that are regulated by tumor-suppressive
miRNAs. In this study, ‘Cytokine-cytokine receptor interaction’ and
‘TGF-β signaling pathway’ were significantly selected as candidate
pathways regulated by miR-1274a in ccRCC cells. In these
pathways, BMPR1B was selected as a candidate target of
miR-1274a. BMPR1B is a member of the BMP receptor family of
transmembrane serine/threonine kinases, and the ligands of this
receptor are BMPs (25,26). BMPs, members of the TGF-β
superfamily, bind their receptor including BMPR1B, which lead to
activation of the SMAD-dependent pathway subsequently through
phosphorylation of SMAD1/5/8 (25,26).
Then phosphorylated SMADs form a complex with the common signaling
transducer SMAD4, resulting in the induction of apoptosis (26–30).
Therefore, our finding that suppression of miR-1274a caused
apoptosis through BMPR1B upregulation is plausible. However, A498
ccRCC cells did not exhibit increased BMPR1B expression,
even though ACHN and Caki1 cells had rescued expression, and
miR-1274a suppression in all 3 cell lines induced apoptosis
in this study. Even though low expression of BMPR1B was observed in
ccRCC compared to normal kidneys, BMPR1B expression was
significantly higher in high-grade ccRCCs in comparison with
low-grade ccRCCs as detected by immunohistochemistry. In addition,
the low expression patient group had prolonged median overall
survival in comparison with the high expression patient group
according to the TCGA analyses. BMPR1B upregulation might be an
early event in ccRCC carcinogenesis, and its functional changes
might have occurred as the tumor acquires malignant potential at a
late stage. Further study is necessary to elucidate the further
functional role of BMPR1B. Concerning miR-1274a, we did not
find any significant correlation between expression of
miR-1274a and any of the clinicopathological parameters
including prognosis. This may be due to the small sample numbers
and short follow-up periods. A larger scale and longer follow-up
period of study should be considered in future research.
In conclusion, upregulation of miR-1274a was
observed in ccRCC clinical specimens and cell lines. Suppression of
miR-1274a significantly inhibited cancer cell proliferation
through apoptosis, suggesting that miR-1274a functions as an
oncogenic miRNA in ccRCC cells. The identification of
tumor-suppressive BMPR1B regulated by miR-1274a may
lead to a better understanding of ccRCC oncogenesis and the
development of new therapeutic strategies to treat this
disease.
Acknowledgements
We thank Mutsumi Miyazaki for the excellent
laboratory assistance and the critical editing of this manuscript.
The present study was supported by the KAKENHI (B) 16H05464, the
KAKENHI (C) 16K11015 and 17K11148, the KAKENHI (HOUGA) 16K15691,
and the KAKENHI (WAKATE)17K16799.
References
|
1
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hadoux J, Vignot S and De La Motte Rouge
T: Renal cell carcinoma: Focus on safety and efficacy of
temsirolimus. Clin Med Insights Oncol. 4:143–154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta K, Miller JD, Li JZ, Russell MW and
Charbonneau C: Epidemiologic and socioeconomic burden of metastatic
renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev.
34:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Margulis V, Master VA, Cost NG, Leibovich
BC, Joniau S, Kuczyk M, Mulders PF, Kirkali Z, Wirth MP, Hirao Y,
et al: International consultation on urologic diseases and the
European Association of Urology international consultation on
locally advanced renal cell carcinoma. Eur Urol. 60:673–683. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshino H, Yonemori M, Miyamoto K,
Tatarano S, Kofuji S, Nohata N, Nakagawa M and Enokida H:
microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis
through revival of TWIST1 in renal cell carcinoma. Oncotarget.
8:20881–20894. 2017.PubMed/NCBI
|
|
9
|
Sobin LH, Gospodarowicz MK and Wittenkind
C: TNM Classification of Malignant Tumour. 7th. Wiley-Liss Inc.;
New York: pp. 255–257. 2009
|
|
10
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Creighton CJ, Morgan M, Gunaratne PH,
Wheeler DA, Gibbs RA, Robertson A Gordon, Chu A, Beroukhim R,
Cibulskis K, Signoretti S, et al Cancer Genome Atlas Research
Network, : Comprehensive molecular characterization of clear cell
renal cell carcinoma. Nature. 499:43–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kloosterman WP, Lagendijk AK, Ketting RF,
Moulton JD and Plasterk RH: Targeted inhibition of miRNA maturation
with morpholinos reveals a role for miR-375 in pancreatic islet
development. PLoS Biol. 5:e2032007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stenvang J, Petri A, Lindow M, Obad S and
Kauppinen S: Inhibition of microRNA function by antimiR
oligonucleotides. Silence. 3:12012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang GJ, Liu GH, Ye YW, Fu Y and Zhang XF:
The role of microRNA-1274a in the tumorigenesis of gastric cancer:
Accelerating cancer cell proliferation and migration via directly
targeting FOXO4. Biochem Biophys Res Commun. 459:629–635. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sand M, Skrygan M, Sand D, Georgas D,
Gambichler T, Hahn SA, Altmeyer P and Bechara FG: Comparative
microarray analysis of microRNA expression profiles in primary
cutaneous malignant melanoma, cutaneous malignant melanoma
metastases, and benign melanocytic nevi. Cell Tissue Res.
351:85–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou C, Liu J, Li Y, Liu L, Zhang X, Ma
CY, Hua SC, Yang M and Yuan Q: microRNA-1274a, a modulator of
sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9)
down-regulation in hepatocellular carcinoma. FEBS Lett.
585:1828–1834. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan H, Wang S, Yu H, Zhu J and Chen C:
Molecular pathways and functional analysis of miRNA expression
associated with paclitaxel-induced apoptosis in hepatocellular
carcinoma cells. Pharmacology. 92:167–174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ezzie ME, Crawford M, Cho JH, Orellana R,
Zhang S, Gelinas R, Batte K, Yu L, Nuovo G, Galas D, et al: Gene
expression networks in COPD: microRNA and mRNA regulation. Thorax.
67:122–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshino H, Enokida H, Itesako T, Tatarano
S, Kinoshita T, Fuse M, Kojima S, Nakagawa M and Seki N:
Epithelial-mesenchymal transition-related microRNA-200s regulate
molecular targets and pathways in renal cell carcinoma. J Hum
Genet. 58:508–516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
García de Vinuesa A, Abdelilah-Seyfried S,
Knaus P, Zwijsen A and Bailly S: BMP signaling in vascular biology
and dysfunction. Cytokine Growth Factor Rev. 27:65–79. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heldin CH, Miyazono K and ten Dijke P:
TGF-beta signalling from cell membrane to nucleus through SMAD
proteins. Nature. 390:465–471. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jang CW, Chen CH, Chen CC, Chen JY, Su YH
and Chen RH: TGF-beta induces apoptosis through Smad-mediated
expression of DAP-kinase. Nat Cell Biol. 4:51–58. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Halvorsen AR, Kristensen G, Embleton A,
Adusei C, Barretina-Ginesta MP, Beale P and Helland Å: Evaluation
of prognostic and predictive significance of circulating MicroRNAs
in ovarian cancer patients. Dis Markers 2017.
30985422017.doi.org/10.1155/2017/3098542.
|
|
29
|
Slattery ML, Trivellas A, Pellatt AJ,
Mullany LE, Stevens JR, Wolff RK and Herrick JS: Genetic variants
in the TGFβ-signaling pathway influence expression of miRNAs in
colon and rectal normal mucosa and tumor tissue. Oncotarget.
8:16765–16783. 2017.PubMed/NCBI
|
|
30
|
Allison SE, Chen Y, Petrovic N, Zimmermann
S, Moosmann B, Jansch M, Cui PH, Dunstan CR, Mackenzie PI and
Murray M: Activation of the pro-migratory bone morphogenetic
protein receptor 1B gene in human MDA-MB-468 triple-negative breast
cancer cells that over-express CYP2J2. Int J Biochem. 80:173–178.
2017. View Article : Google Scholar
|