Introduction
Ovarian cancer is one of the most common malignant
tumors of the gynecologic system. In particular, epithelial ovarian
cancer (EOC) is the most common type of ovarian cancer and is a
serious gynecological disease due to its asymptomatic nature,
accounting for 50–70% of ovarian cancers (1). Platinum and paclitaxel are the most
frequently used chemotherapeutic agents for treatment of EOC.
However, frequent recurrence and drug resistance often affect
therapeutic outcomes.
Methyl methanesulfonate and ultraviolet sensitive
gene clone 81 (MUS81), which is critical for the repair of DNA
double-strand breaks (DSBs), is a highly conserved gene across
species and encodes a structure-specific DNA endonuclease (2,3). DNA
DSBs resolve Holliday junctions (HJs) by constituting a heterodimer
with Eme1/Mms4 and maintaining genetic stability (4). HJs are co-regulated by the BTR
(BLM-topoisomerase IIIa-RMI1-RMI2) complex, the SLX-MUS
(SLX1-SLX4-MUS81-EME1) complex and GEN1 (5). Changes in HJs affect both the activity
and DNA stability of homologous recombination (HR) in the repair of
DSBs. Our previous studies demonstrated that downregulation of
MUS81 inhibits ovarian tumor growth and enhances platinum drug
sensitivity both in vivo and in vitro (6). Moreover, MUS81-targeting represents a
novel approach for promoting sensitivity to chemotherapy and has
recently attracted significant research attention. Studies have
reported that embryonic stem cells and mice that are deficient in
the MUS81 gene are significantly more sensitive to mitomycin C
(MMC). The survival of mice with MUS81+/− and
MUS81−/− genotypes was significantly lower than that of
wild-type mice treated with the same dose of MMC (7). Disruption of the MUS81 gene increases
sensitivity of MMC and cisplatin, returning to normal after MUS81
is re-expressed. These results indicate that MUS81 plays an
essential role in the chemosensitivity of human malignancies
(8). Although previous studies have
found that MUS81 is associated with chemotherapeutic drug
sensitivity in ovarian cancer and potentially inhibits HR activity
via MUS81 depletion, the role of MUS81 in regulating
chemosensitivity in ovarian cancer remains unclear.
To overcome the challenge of resistance to
chemotherapy agents, poly(ADP-ribose) polymerase PARP inhibitors
have been applied for the treatment of BRCA-mutated ovarian cancer
and have resulted in satisfactory treatment effects (9,10).
Recent studies found that, olaparib, a potent oral PARP inhibitor,
can be used as a targeted therapeutic drug for BRCA-mutated ovarian
cancer treatment (11,12). The antitumor effect of olaparib was
demonstrated in ovarian cancer and was associated with HR
deficiency (13). However, the role
of olaparib in the treatment of BRCA wild-type ovarian cancer,
potentially via the modulation of pathways to induce HR deficiency,
has not yet been fully elucidated. In this study, the role of MUS81
on the chemosensitivity of olaparib in EOC, its underlying
molecular mechanisms and its association with HR were
investigated.
Materials and methods
Cell lines
Human ovarian cancer A2780 and SKOV3 cells were
purchased from the Chinese Academy of Sciences Committee (Shanghai,
China). All ovarian cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) and 1%
penicillin-streptomycin. Cells were incubated at 37°C in a
humidified atmosphere with 5% CO2.
Immunofluorescence
For immunofluorescence studies, 5×105
cells were seeded in 24-well plates on glass coverslips. After the
cells adhered, they were washed with PBS and fixed with 4%
formaldehyde at room temperature. The fixed cell slides were washed
with PBS, and then methanol and acetone were added at a 1:1 ratio,
followed by blocking with 1% BSA. The cells were then incubated
with antibodies against MUS81 (Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and MCM2 [Cell Signaling Technology, (CST) Inc.,
Beverly, MA, USA)] overnight at 4°C. After washing, the cells were
labeled with Alexa Fluor 488-conjugated secondary antibody
(Invitrogen, Carlsbad, CA, USA) and examined under a fluorescence
microscope (SP5; Leica Microsystems, Wetzlar, Germany).
Co-immunoprecipitation
Cultured cells were carefully washed twice with 4°C
PBS, and IP lysis buffer (Beyotime Biotechnology, Shanghai, China)
was added. To extract protein, cells were scraped off, and 50%
protein A/G agarose (Santa Cruz Biotechnology, Inc.) was added to
the sample solution at a ratio of 1:10. The cells were placed on a
horizontal shaker for 1 h at 4°C. Then, the protein A/G-agarose
beads were discarded and the appropriate amount of primary MUS81
antibody was added according to the protein content. The
antigen-antibody complex was slowly shaken in a rotating shaker at
4°C overnight. The antigen-antibody complexes were precipitated
with protein A/G-agarose beads, and the supernatant was collected
for western blotting.
Cells treated with UV and CPT
Cells were seeded in 6-well plates at a density of
4×105 cells/well. CPT (20/40/80 nmol/l) was added to
A2780 and SKOV3 cells, and a well without CPT was taken as a
control, and the cells were cultured for 24 h. Cells were collected
and lysed for total protein extraction using IP lysis buffer
(Beyotime Biotechnology). The expression of MUS81 and MCM2 were
verified by western blotting, and pH2AX was used as a sign of the
repair of double-strand breaks. The cells were irradiated with
different degrees of ultraviolet radiation using a UV cross-linking
instrument. The irradiation degree was calculated as number × area
× 1×106. Protein was extracted using the same method and
verified with western blotting.
Establishment of cell lines with
downregulation of MUS81 and MCM2
A MUS81 lentivirus expressing a specific RNAi
interference sequence was first designed. The nucleotide sequences
were cloned into the AgeІ and EcoRІ sites of the
pLKO.1-puro vector (Addgene, Cambridge, MA, USA) to generate the
pLKO.1-puro-MUS81 (shMUS81-1 and shMUS81-2) and pLKO.1-puro-control
(shCtrl) (Table I) recombinant
vectors. Lentiviruses were packaged by transfecting 293T cells with
the above lentivirus recombinant vectors, the packing plasmid
psPAX2, and the envelop vector pMD2.G (Addgene) using Lipofectamine
2000 transfection reagent (Invitrogen). Culture medium containing
lentivirus particles was collected after 48 h. Ovarian cancer cells
were transfected with 1×106 IFU/ml lentivirus in 8 mg/ml
Polybrene (Sigma-Aldrich, St. Louis, MO, USA) for 24 h. Stably
transduced cells were selected using 1 mg/ml puromycin
(Sigma-Aldrich) for 7 days, and the RNAi knockdown efficiency was
detected by western blotting. A2780 cells with MCM2 downregulation
were established via the same method.
 | Table I.Sequences of primers and targets. |
Table I.
Sequences of primers and targets.
|
Primers/targets | Sequences |
|---|
| MUS81 qRT-PCR
forward |
5′-CTGAAGCGCTGTGGTCTG-3′ |
| MUS81 qRT-PCR
reverse |
5′-AGTGTTGGTGACAGCCTG-3′ |
| β-actin qRT-PCR
forward |
5′-AAGGTGACAGCAGTCGGTT-3′ |
| β-actin qRT-PCR
reverse |
5′-TGTGTGGACTTGGGAGAGG-3′ |
| MCM2 qRT-PCR
forward |
5′-AGAGGATCGTGGTACTGCTATGGC-3′ |
| MCM2 qRT-PCR
reverse |
5′-TTATGGATGGCATAGGGCCTCAGA-3′ |
| shMUS81-1 |
5′-ACACTGCTGAGCACCATTAAG-3′ |
| shMUS81-2 |
5′-GCAGCCCTGGTGGATCGATAC-3′ |
| shCtrl/SRC
(scrambled sequence) |
5′-CCTAAGGTTAAGTCGCCCTCG-3′ |
|
|
5′-GCTCTTCATACTGAAGCAGTTCT |
| MCM2-1 |
CGAGAACTGCTTCAGTATGAAGAGC-3′ |
|
|
5′-CTATCAGAACTACCAGAGTATCTCG |
| MCM2-3 |
AGATACGCTGGTAGTTCTGATAG-3′ |
Western blotting
Total protein was collected as described above. Cell
lysates were resolved by SDS-PAGE, and proteins were
electro-transferred to polyvinylidene fluoride (PVDF) membranes
(Millipore, USA). The PVDF membranes were blotted with 10% non-fat
milk (Solarbio, Beijing, China). The primary antibodies included
MUS81 (1:200 dilution; Santa Cruz Biotechnology, Inc.), β-actin
(1:3,000 dilution; Abcam, Cambridge, MA, USA), pH2AX (1:400
dilution; CST), cyclin B (1:3,000 dilution; Abcam), POLE (1:200
dilution; Santa Cruz Biotechnology, Inc.), NBS1 (1:3,000 dilution;
Abcam) and MCM2 (1:1,000 dilution; CST).
Quantitative RT-PCR
Preparation of MUS81 and MCM2 gene downregulated
cell lines. RNA was isolated from cells using TRIzol®
reagent (Thermo Fisher Scientific, Shanghai, China) and reverse
transcribed using a PrimeScript® RT reagent kit (Takara,
Shiga, Japan) according to the manufacturer's protocol. RT-PCR was
performed in a 10 µl reaction solution containing 10 ng cDNA, 0.1
mmol/l primer (Table I), and 5 ml
2X SYBR Premix Ex Taq (Takara). PCR amplification was performed at
95°C for 5 min, followed by 40 cycles at 95°C for 15 sec, and 65°C
for 40 sec, using a Mastercycler® ep realplex
(Quantstudio dx, Thermo Fisher Scientific). The abundance of MUS81
and MCM2 transcripts was expressed relative to β-actin as a
control.
Measurement of HR efficiency
HR efficiency was detected using a qPCR-based HR
assay kit (Norgen BiotekCorp., Thorold, ON, Canada). The dl-1 and
dl-2 plasmids were co-transfected into shCtrl, shMUS81-1 and
shMUS81-2 cells, and the total cellular DNA was isolated using a
Blood & Cell Culture DNA Mini kit (Qiagen, Hilden, Germany)
after 48 h. qPCR was performed using a set of universal primers and
the assay primer. The difference in the number of cycles to reach
the amplification curve inflection point generated using
HR-specific primers and the number of cycles to reach the universal
primer amplification curve inflection point was converted to the
difference in DNA content based on a standard DNA content curve.
This value was set to 1 for the control cell populations, and the
amount of recombinant HR DNA produced in the experimental groups
was normalized to this value.
Olaparib sensitivity assay
Single-cell suspensions were prepared and seeded in
96-well plates (1×104 cells/well). After treatment with
serial dilutions of olaparib (MCE, Shanghai, China) ranging from 2
to 10 µmol/l for 48 h, cell viability was assessed using a CCK-8
kit (Dojindo, Kumamoto, Japan). Each well was read at a wavelength
of 450 nm (Synergy H4; Bio-Tek). Cell viability was calculated as
follows: Viability of cells (%) = (drug group-blank) OD450/(no drug
group-blank) OD450 × 100%.
Colony formation assay
For the colony formation assay, 1,000 cells/well
were seeded into 6-well plates and incubated for 10–14 days until
visible clones appeared. The control group was cultured in
RPMI-1640 medium supplemented with 10% FBS and 1%
penicillin-streptomycin, and the experimental group was treated
with 5 µmol/l olaparib. Colonies were stained with 0.5% crystal
violet. The number of colonies was determined using a
microscope.
Cell cycle and apoptosis assay
Cells were seeded onto 6-well plates at a density of
4×105 cells/well, and then treated with olaparib (5
µmol/ml) for 48 h. To examine apoptosis, 1×105 cells
were collected and washed twice with 4°C PBS. Then, the cells were
resuspended in 1X Annexin V binding buffer and stained with
propidium iodide (PI) and Annexin V according to the instructions
of the Annexin V-fluorescence apoptosis detection kit I (BD
Biosciences; Pharmingen, San Diego, CA, USA). For cell cycle
analysis, transduced cells were harvested, fixed in 70% alcohol
overnight at 4°C, and then incubated with 500 ml of PI (BD
Pharmingen™) for 15 min in the dark. Finally, apoptosis and the
cell cycle were analyzed by flow cytometry (BD, Caliburn), and the
assays were repeated three times.
Statistical analysis
Data are presented as the mean ± SD of three
independent experiments, and SPSS 17.0 software was used for
statistical analyses. The differences between groups were examined
by analysis of variance (ANOVA) and Student's t-tests. P<0.05
was considered to indicate statistical significance.
Results
MUS81 interacts with MCM2 to
participate in the DNA damage repair process
MUS81 induces an important endonuclease effect on
the repair of damaged replication forks of HJ. MCM2, a pivotal
component of MCM2-7 helicase, induces helicase activity at the
replication fork and regulates the formation of DNA double strands
with downstream molecules. In this study, we found that both MUS81
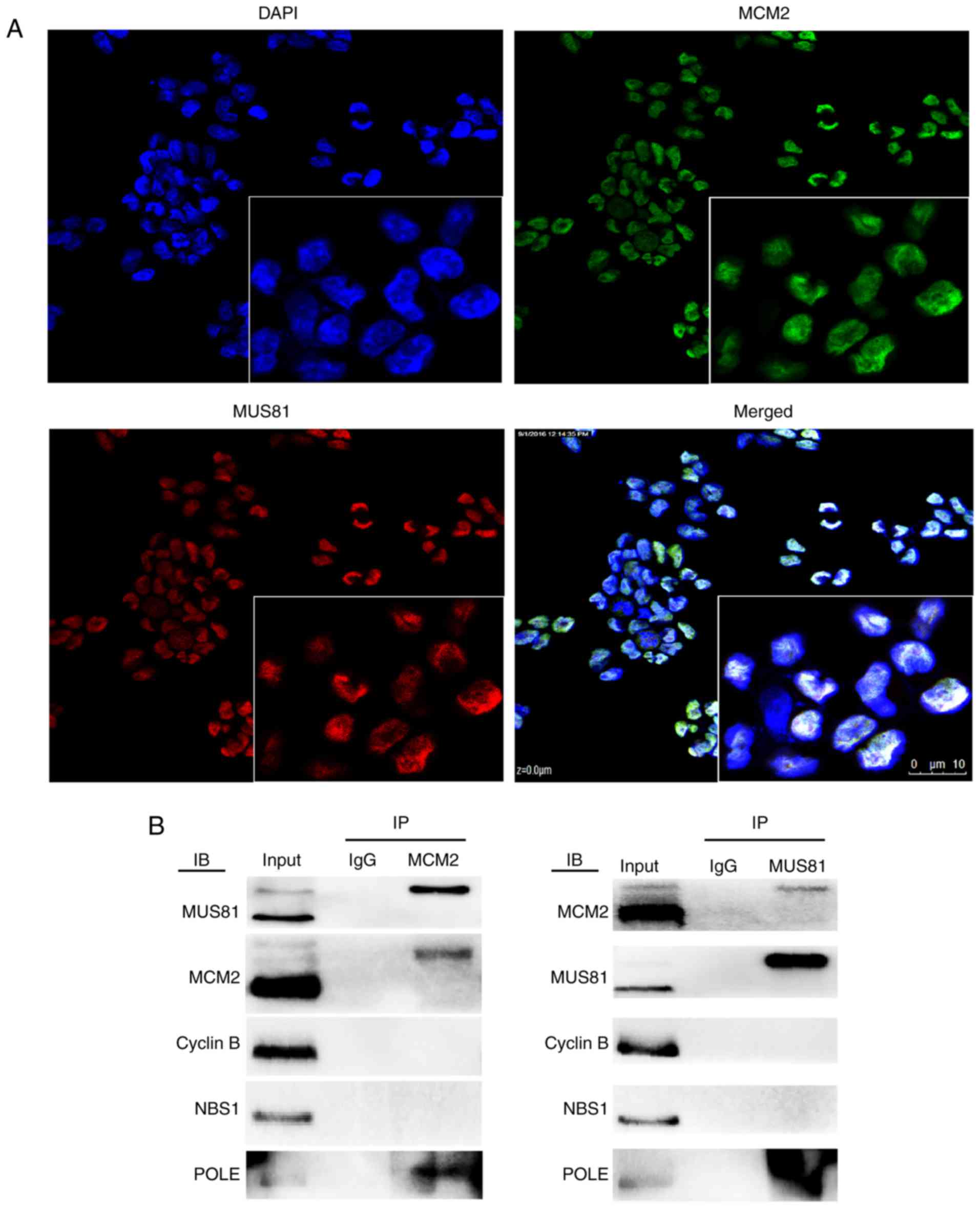
and MCM2 were localized in the nucleus, as shown by
immunofluorescence assays (Fig.
1A). Our previous data generated using protein interaction
chips revealed five proteins that bind to MUS81, of which MCM2 had
the highest correlation. In A2780 cells, MUS81 co-existed with MCM2
and POLE in co-immunoprecipitation experiments, and therefore, we
speculated that the interaction between MUS81 and MCM2 is regulated
by POLE (Fig. 1B). To study the
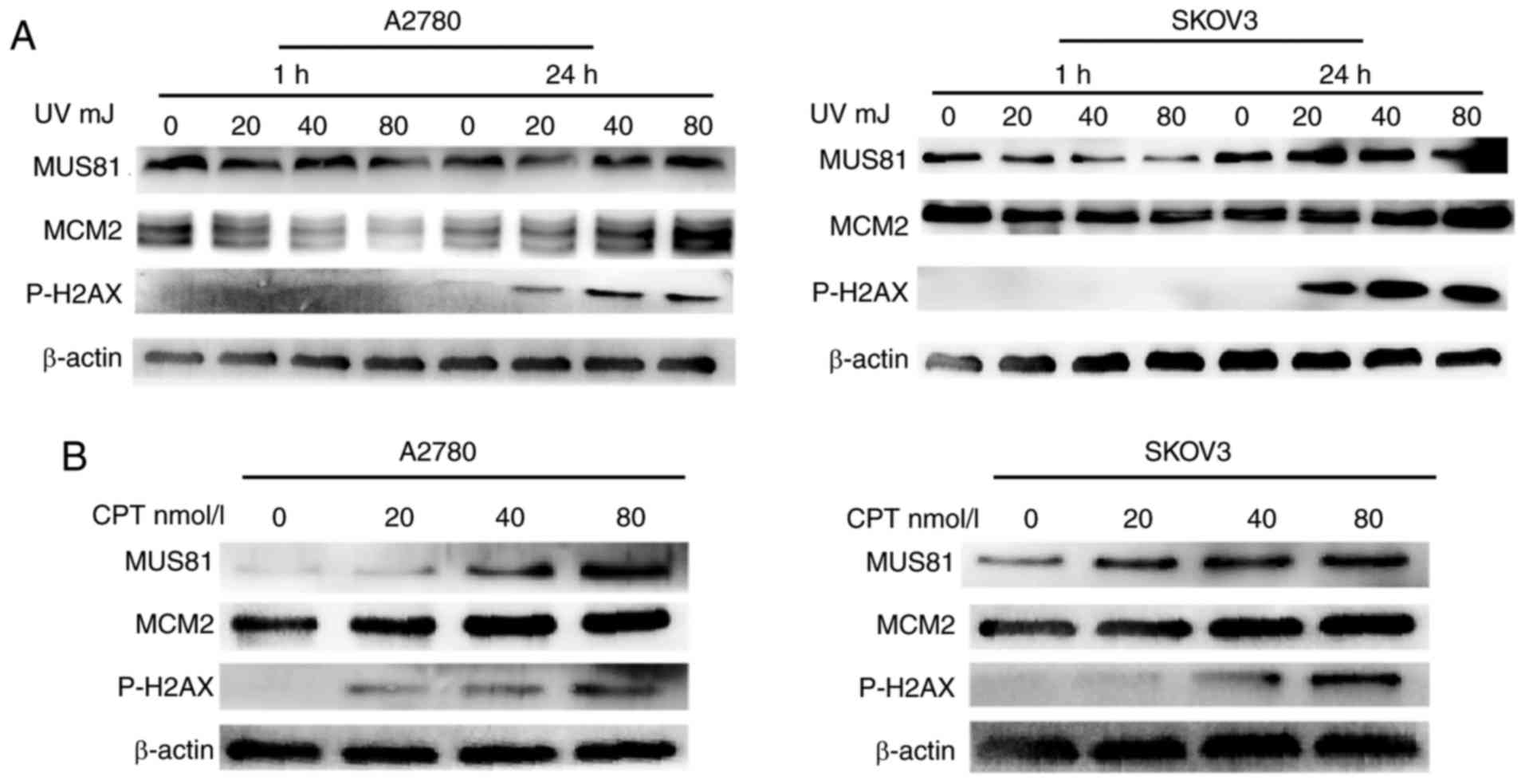
roles of MUS81 and MCM2 in DSB repair in vitro, a DNA DSB
model was established via physical treatment with UV irradiation,
and chemical treatment with camptothecin (CPT). The results
indicated that DNA DSBs occurred after 24 h of UV irradiation, at
which time MUS81 and MCM2 were upregulated with increasing degrees
of UV irradiation-induced damage (Fig.
2A). After 1 h of irradiation, nuclear MUS81 and MCM2 were
involved in the process of repairing gene damage, resulting in no
rupture of the DNA double strand. CPT treatment can lead to DNA
DBSs in cells and induce the formation of replication forks, which
can be assessed by measuring P-H2AX as an indicator of represent
damage repair. MCM2 and MUS81 expression in A2780 and SKOV3 cells
dose-dependently increased with CPT treatment after 24 h (Fig. 2B), suggesting that both MUS81 and
MCM2 are involved in the formation of replication forks and HR.
HR activity decreases after MUS81
downregulation, and there is negative feedback regulation between
MUS81 and MCM2
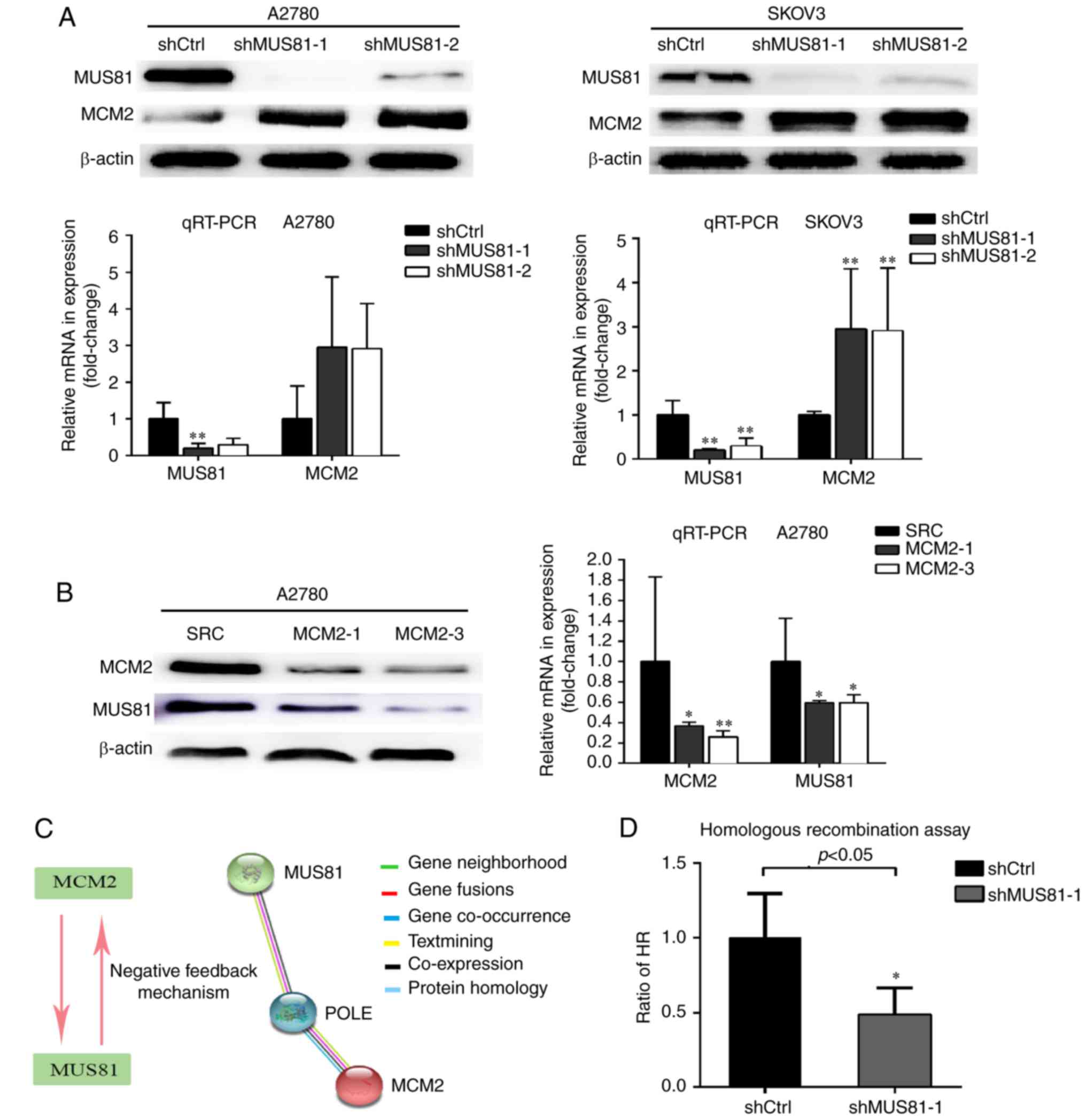
A2780 and SKOV3 cells with downregulation of MUS81
expression were constructed using a lentivirus-mediated method, and
the regulatory relationship between MUS81 and MCM2 at the protein
level in ovarian cancer was further investigated in vitro.
Increased expression of MCM2 was found in shMUS81-1 and shMUS81-2
ovarian cells compared with shCtrl cells according to western
blotting, and MUS81 played a negative role in the regulation of
MCM2. Consistent results were observed at the RNA level by RT-PCR
(Fig. 3A). To further define the
regulatory relationship between MUS81 and MCM2, we constructed
MCM2-downregulated A2780 cell lines and found that MUS81 levels
decreased following the downregulation of MCM2 (Fig. 3B). In accordance with these results,
we hypothesized that MCM2, as a molecule upstream of MUS81, is
involved in regulating MUS81 expression levels and that MUS81
expression exerts negative feedback regulation on MCM2. STRING
database results revealed that POLE may regulate the interaction
between MUS81 and MCM2 (Fig. 3C).
As the main factor of HJ decomposition and involved in HR, the
association of MUS81 with HR activity was further investigated. A
significant reduction in the HR efficiency of shMUS81 cells
compared with shCtrl cells was observed via qPCR using an HR assay
kit (P<0.05) (Fig. 3D). These
results suggested that MUS81 and MCM2 are involved in HR of DSBs.
The suspension of HJ formation and dissociation of the replication
fork may be inhibited by the activation of MCM2 helicase function
through downregulation of MUS81 and subsequent inhibition of HR
activity.
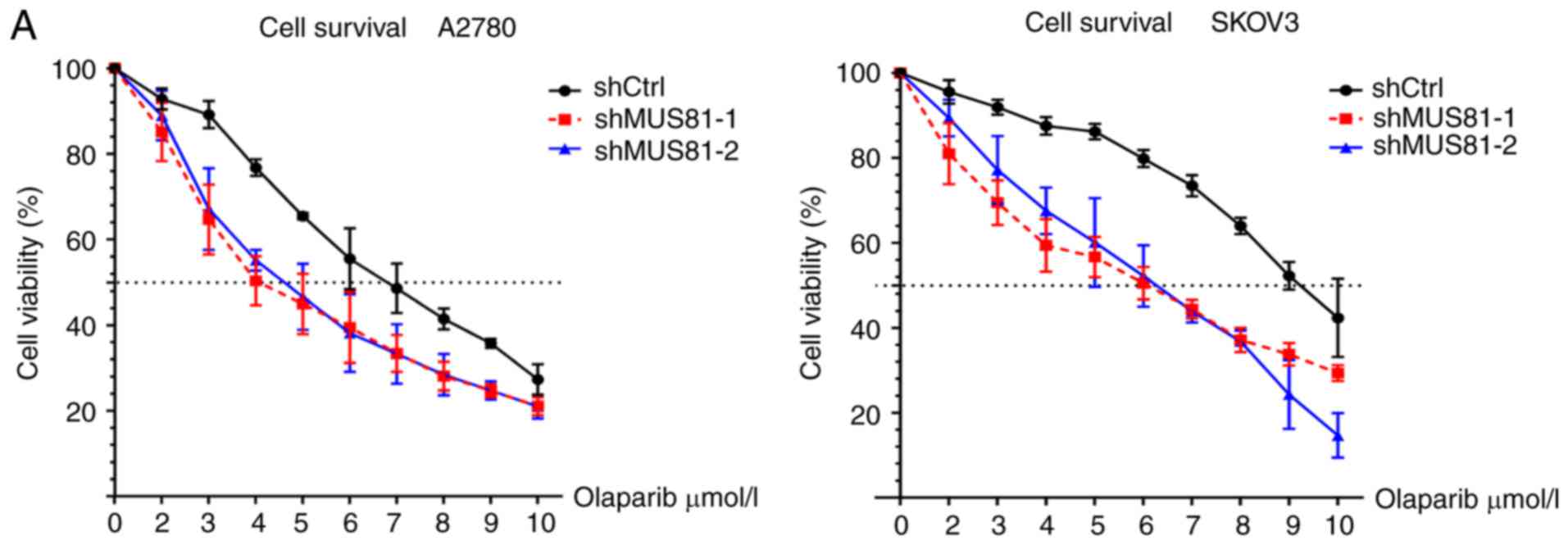
Downregulation of MUS81 induces HR deficiency that
promotes olaplarib-mediated inhibition of ovarian cancer cell
growth. We found that olaparib acts on BRCA wild-type ovarian tumor
cells with HR pathway defects and increases sensitivity to PARP
inhibitors. No BRCA gene mutation was found in A2780 and SKOV3 cell
lines in the COSMIC database. In MUS81-downregulated ovarian cancer
cells, olaparib dose-dependently inhibited cell growth and
significantly increased drug sensitivity in shMUS81-1 and shMUS81-2
cells compared to shCtrl cells (Fig.
4A). Furthermore, we performed colony formation assays and
found significantly fewer stably transduced shMUS81-1/shMUS81-2
colonies than shCtrl cell colonies after treatment with olaparib
(P<0.05) (Fig. 4B). These
results indicate an impairment of the repair process in ovarian
cancer cells resulting from HR deficiency mediated DSBs produced by
the inhibition of MUS81 in BRCA wild-type cells.
Increased MCM2 expression resulting
from MUS81 downregulation enhances drug sensitivity to
olaparib
To investigate the mechanism of MUS81 activity on
enhancing drug sensitivity to olaparib through the negative
regulation of MCM2, flow cytometry was implied for cell cycle and
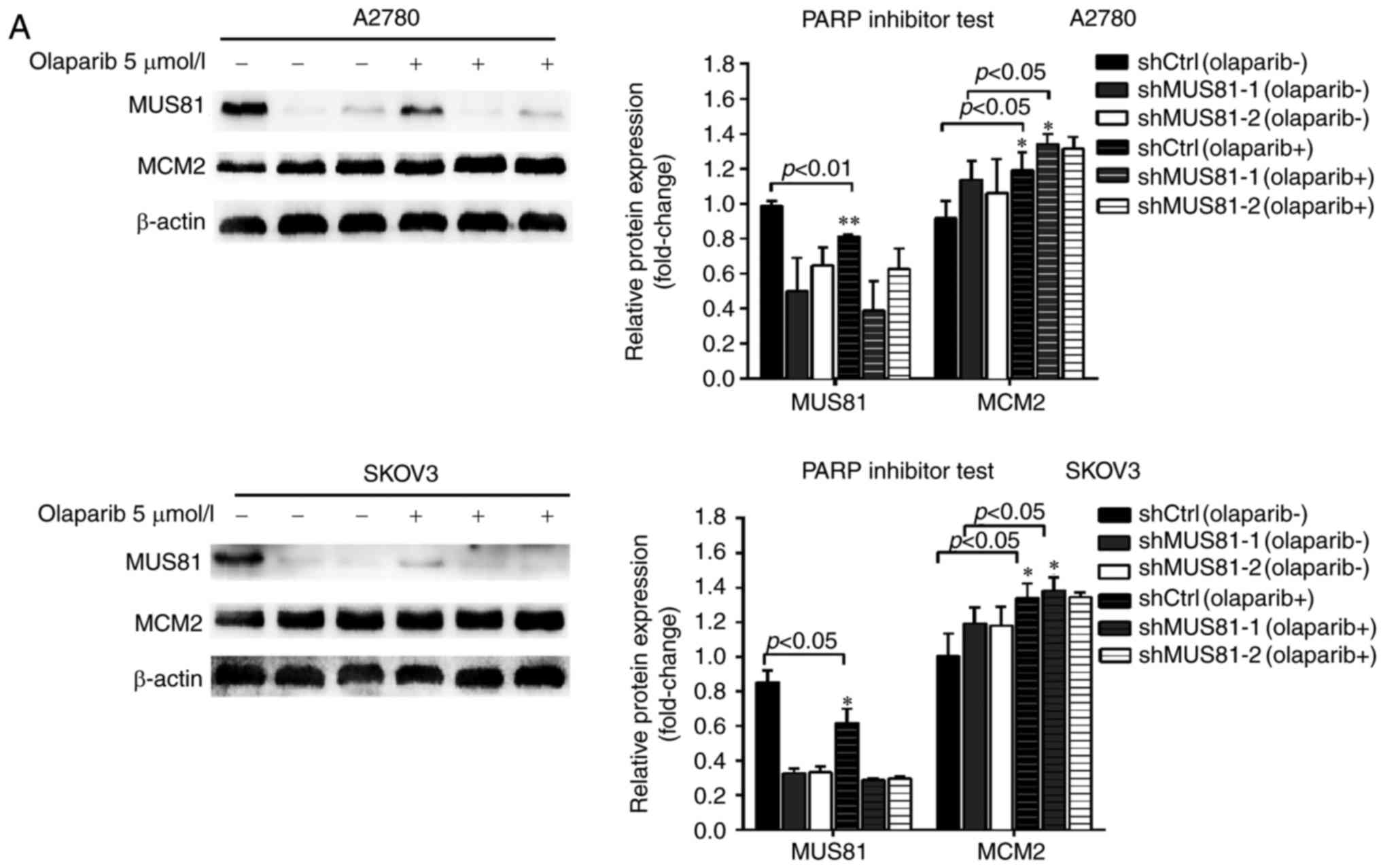
apoptosis analysis. According to western blotting, 5 µmol/l
olaparib inhibited MUS81 expression in shCtrl and shMUS81 ovarian
cancer cells. The expression of MCM2 in shMUS81 cells significantly
increased compared with shCtrl cells (Fig. 5A). Cell cycle analysis demonstrated
that PARP inhibitor treatment of the stably transfected shMUS81
SKOV3 and A2780 cells induced significant S phase arrest, and the
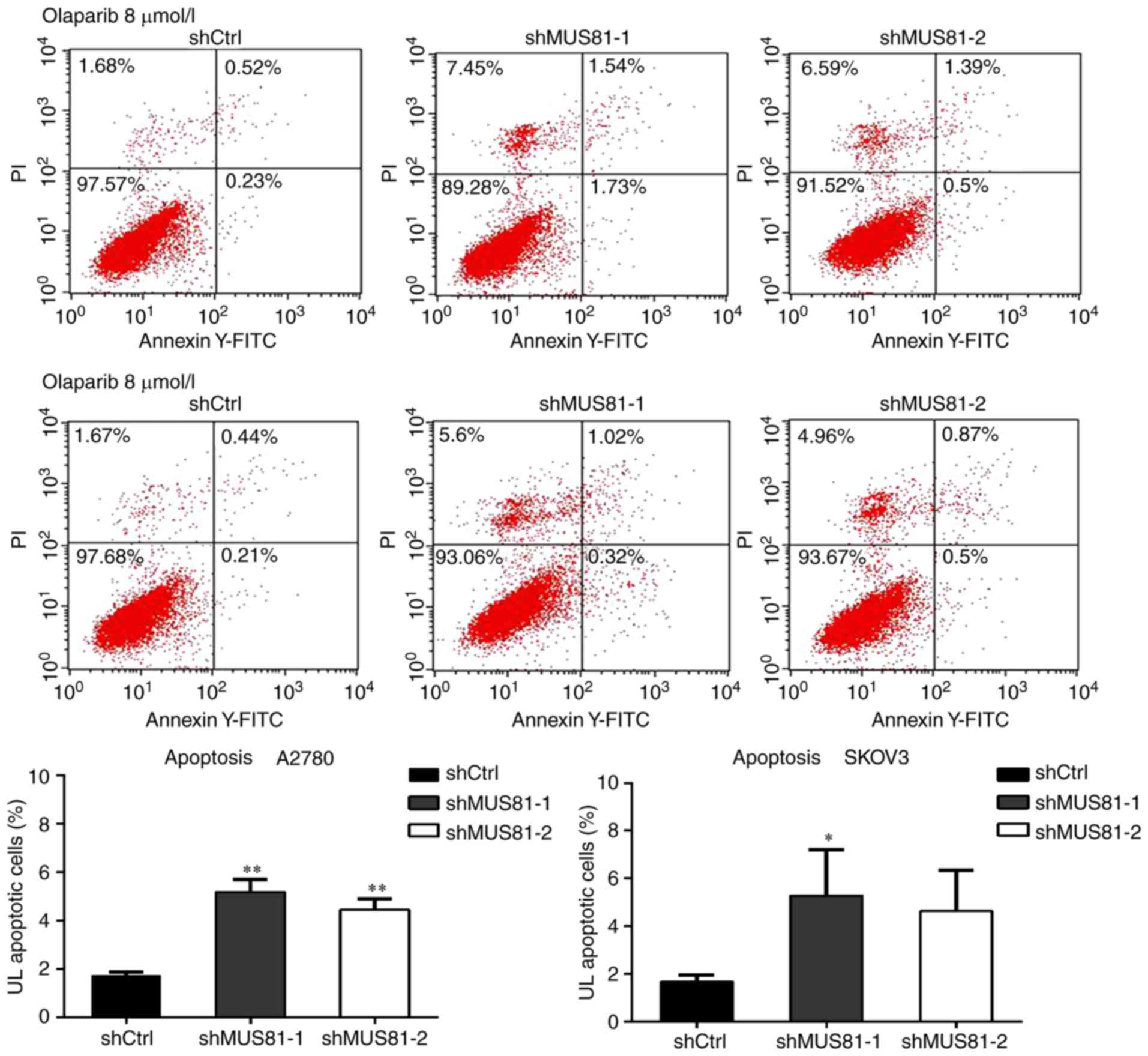
percentage of G0/G1 phase cells significantly decreased (Fig. 5B). Apoptosis assay also showed that
olaparib treatment significantly increased apoptosis of shMUS81
A2780 and SKOV3 cells (Fig. 6).
These results suggested that inhibition of MUS81 activates MCM2
expression and arrests cells in the S phase. Consistent with
previous studies, MCM2 is involved in promoting apoptosis. We
confirmed that downregulation of MUS81 can enhance the sensitivity
of BRCA wild-type ovarian cancer cells to olaparib.
Discussion
Eukaryotic cells rely on accurate DNA replication
and recombination to maintain genomic stability. HR and DNA
replication mechanisms are required to coordinate activity at DNA
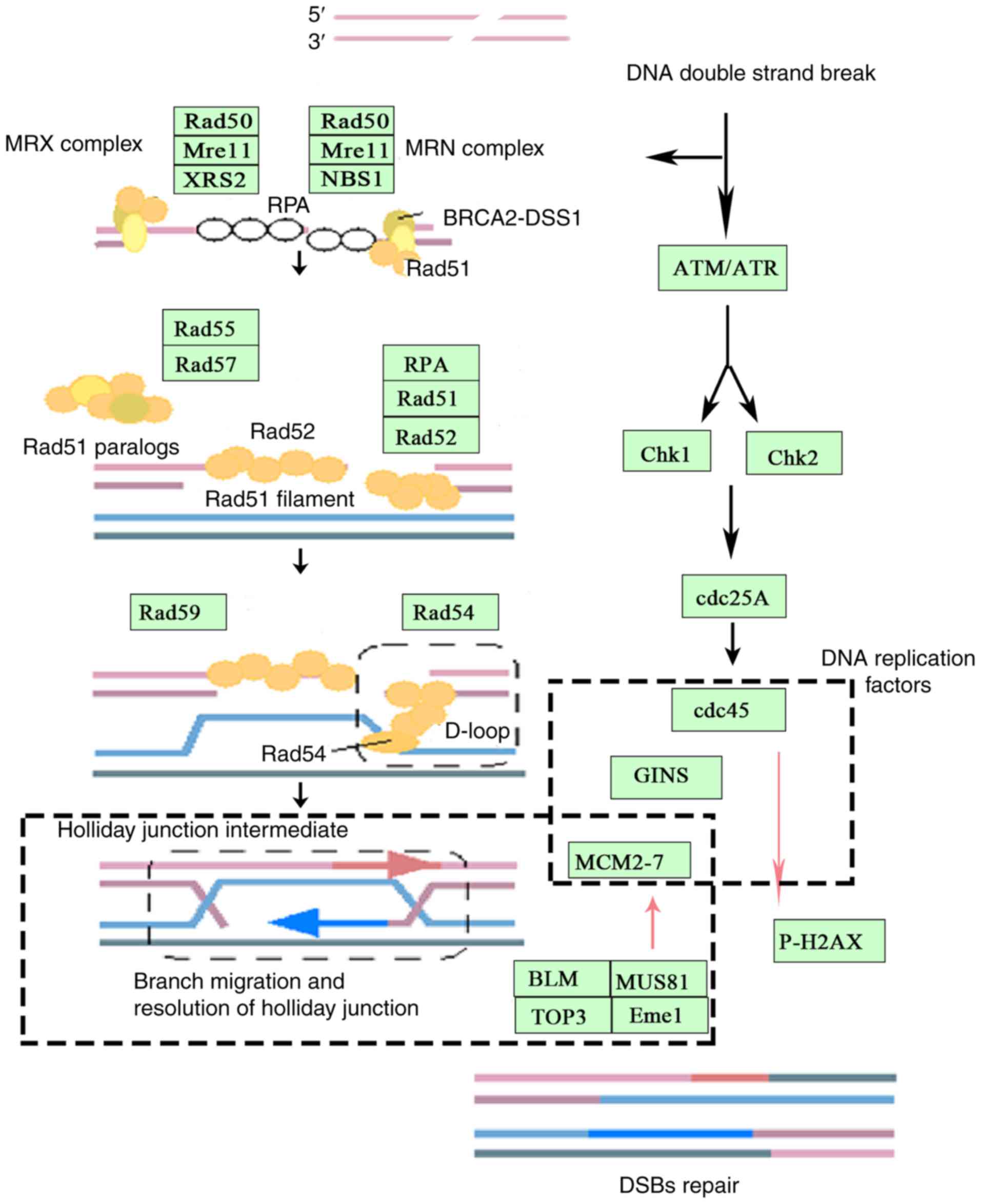
DSBs and stalled replication forks (Fig. 7). Genomic instability resulting from
either the loss or rearrangement of genetic material is closely
related to the development of various human diseases, such as
colorectal cancer and ovarian cancer (14,15).
DNA DSBs, which primarily result from the collapse of replication
forks (16,17), are often repaired through HR. The
activity of HR machinery is closely related to the degree of injury
repair. HR may lead to the formation of a DNA structure called
Holliday junctions (HJs), which must be resolved for chromosome
segregation. The first nuclear enzyme identified in both fission
yeast and in human cells to possess a resolvase activity was MUS81,
which exists as the MUS81-Eme1 complex. MUS81 is the catalytic
subunit of a structural-specific endonuclease that has a preference
for cleaving branched DNA substrates, including structures that are
replication and recombination intermediates such as 3′flaps,
‘Y’-shaped structures, D-loops and ‘X’-shaped structures (HJs and
HJ incisions) (18,19). MUS81 also regulates the rate of DNA
replication during normal cell growth by facilitating replication
fork progression and reducing the frequency of replication
initiation events. In the absence of MUS81 endonuclease activity,
the rate of DNA synthesis declines, and the frequency of
replication initiation events increases (20). The key step of the selection of the
initiation replication site is the binding of the MCM helicase
complex, which triggers replication (21). MCM2, as the regulatory protein of
the replication initiation site, is an important component of
MCM2-7 and plays an important role in the formation and
decomposition of the replication fork (22,23).
Higher-order eukaryotes depend on high fidelity DNA replication and
recombination mechanisms to maintain genomic stability. The
dynamics of replisome components during fork collapse and restart
are poorly understood. The repair of severe lesions in DNA, such as
DSBs or stalled replication forks, requires coordinated activities
of HR and DNA replication machineries (24). Growing evidence has also indicated
that so-called ‘accessory proteins’ in two systems, such as helper
protein RAD52, BRCA, and DNA replication proteins MCM2-7 and RAD54,
are necessary for the effective coupling of recombination to
replication that is essential for restoring genomic integrity after
severe DNA damage (25). We
discovered that both MUS81 and MCM2 act on the replication fork
during HR, which in turn, activates MCM2 helicase when MUS81
endonuclease activity is blocked, leading to an unstable
replication fork that dissociates from the DNA strand and inhibits
the reformation of the DNA double strand.
HR is an important pathway contributing to DNA
repair and HR defects are found in many cancers, such as ovarian
cancer and hereditary breast cancer (26). Tumor cells with HR repair defects
are sensitive to chemotherapeutic agents (cisplatin, carboplatin,
etc.) (27,28). Recent studies have indicated that
HR-defective tumors are also exquisitely sensitive to novel agents,
such as PARP inhibitors (29,30).
PARP inhibitors have been identified as a new class of drugs for
the treatment of ovarian cancer, and they induce significant
effects in ovarian tumors with BRCA mutations. Olaparib is a
recently discovered targeting drug for the treatment of ovarian
cancer and exerts favorable effects against EOC with BRCA
mutations. PARP inhibitors have good sensitivity for HR-deficient
BRCA mutants in ovarian cancer and showed significant therapeutic
effects in the treatment of recurrent EOC in phase I clinical
trials (10). We found that the
inhibitory effect of olaparib was enhanced in BRCA wild-type
ovarian cancer cells and after downregulation of MUS81 DSBs. In
BRCA wild-type ovarian cancer cells, DSBs were caused by DNA
accumulation due to the inhibition of PARPs. The BRCA gene is
involved in the HR pathway, but downregulation of MUS81 can inhibit
the activity of endonucleases, reducing the efficiency of HR, and
thus, they cannot complete DSB repair or inhibit tumor cell growth.
Consequently, cell cycle and apoptosis assays revealed that
olaparib affected cell cycle arrest in the S phase and increased
cell apoptosis in MUS81+/BRCA-ovarian cancer cells.
We further found that downregulation of MUS81
improved the chemical sensitivity to olaparib. Treatment of cells
with olaparib increases MCM2 expression, and activation of MCM2 is
involved in regulation of the cell cycle and apoptosis. HR is
required to use the homologous sequence of the intact sister
chromatid as a template for its repair. Therefore, the repair
process of HR is generally limited from the S to the G2 phase of
the cell cycle. MCM2, as a cyclin-dependent protein, is present in
the G1 phase in the form of the MCM2-7 complex in a dormant state
(31). During S phase, many factors
are activated by binding to the MCM2-7 helicase or are chemically
modified to activate the replication site (32). Helicase activation depends on the
replisome component of Cdc45 (33),
in addition to MCM2-7 and concomitant GINS assembly (together with
the formation of CMG) (34).
Activation of MCM2-7 with Cdc45 and GINS (CMG) is involved in the
helicase, but inactivation of endonuclease activity results from
the downregulation of MUS81, which is unable to form a
3′-single-stranded end. The stalled replication fork is unable to
form HJs and separate from the replication fork, causing the cell
to arrest in the S phase. Furthermore, increasing MCM2 induces
marked increases in Cdc45. Previous studies have shown that
overexpression of Cdc45 in cells promotes cell apoptosis (35). Overexpression of Cdc45 significantly
reduced the rate of replication fork extension by two-fold, and the
replication fork was significantly asymmetric and arrested in the S
phase. In addition, increased Cdc45 leads to depletion of the
single-stranded binding protein RPA and accumulation of
single-stranded DNA. Single-stranded DNA accumulation is a marker
of replication disaster, which results in decreases in the ATR/Chk1
pathway response, followed by activation of ATM/Chk2 via a
replication fork break, which activates pH2AX expression and
promotes cell apoptosis (36).
We propose for the first time that MUS81 and MCM2
play important roles in the regulation of HR and clarify the
dynamic effect between stalled replication forks and DNA
replication during HR in DSB repair. Furthermore, the mechanisms of
MCM2 activation in the regulation of DNA damage repair and DNA
replication remain to be further explored.
In this study, the roles of MUS81 in the regulation
of DNA damage repair in EOC and its potent effects on HR, the cell
cycle and apoptosis via MCM2 activation were observed in
vitro. These results suggest that MUS81 may be a new target of
olaparib for the treatment of EOC, as it potently improved the
sensitivity of BRCA wild-type EOC to PARP inhibitors, and that
MUS81 can be used in new therapeutic approaches for future ovarian
cancer treatment.
Acknowledgements
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
NSF-81572552).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Salani R and Bristow RE: Surgical
management of epithelial ovarian cancer. Clin Obstet Gynecol.
55:75–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haber JE and Heyer WD: The fuss about
Mus81. Cell. 107:551–554. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boddy MN, Lopez-Girona A, Shanahan P,
Interthal H, Heyer WD and Russell P: Damage tolerance protein Mus81
associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell
Bio. 20:8758–8766. 2000. View Article : Google Scholar
|
|
4
|
Hanada K, Budzowska M, Davies SL, van
Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID and
Kanaar R: The structure-specific endonuclease Mus81 contributes to
replication restart by generating double-strand DNA breaks. Nat
Struct Mol Biol. 14:1096–1104. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sarbajna S and West SC: Holliday junction
processing enzymes as guardians of genome stability. Trends Biochem
Sci. 39:409–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xie S, Zheng H, Wen X, Sun J, Wang Y, Gao
X, Guo L and Lu R: MUS81 is associated with cell proliferation and
cisplatin sensitivity in serous ovarian cancer. Biochem Bioph Res
Commun. 476:493–500. 2016. View Article : Google Scholar
|
|
7
|
McPherson JP, Lemmers B, Chahwan R, Pamidi
A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K,
Poonepalli A, et al: Involvement of mammalian Mus81 in genome
integrity and tumor suppression. Science. 304:1822–1826. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dendouga N, Gao H, Moechars D, Janicot M,
Vialard J and McGowan CH: Disruption of murine Mus81 increases
genomic instability and DNA damage sensitivity but does not promote
tumorigenesis. Mol Cell Biol. 25:7569–7579. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ledermann JA: PARP inhibitors in ovarian
cancer. Ann Oncol. 27 Suppl 1:i40–i44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gadducci A and Guerrieri ME: PARP
inhibitors in epithelial ovarian cancer: State of art and
perspectives of clinical research. Anticancer Res. 36:2055–2064.
2016.PubMed/NCBI
|
|
11
|
Liu FW and Tewari KS: New targeted agents
in gynecologic cancers: Synthetic lethality, homologous
recombination deficiency, and PARP inhibitors. Curr Treat Option
Oncol. 17:122016. View Article : Google Scholar
|
|
12
|
McLachlan J and Banerjee S: Olaparib for
the treatment of epithelial ovarian cancer. Expert Opin
Pharmacother. 17:995–1003. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gunderson CC and Moore KN: Olaparib: An
oral PARP-1 and PARP-2 inhibitor with promising activity in ovarian
cancer. Future Oncol. 11:747–757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Negrini S, Gorgoulis VG and Halazonetis
TD: Genomic instability - an evolving hallmark of cancer. Nat Rev
Mol Cell Biol. 11:220–228. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grady WM: Genomic instability and colon
cancer. Cancer Metastasis Rev. 23:11–27. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tercero JA, Longhese MP and Diffley JF: A
central role for DNA replication forks in checkpoint activation and
response. Mol Cell. 11:1323–1336. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sabatinos SA, Green MD and Forsburg SL:
Continued DNA synthesis in replication checkpoint mutants leads to
fork collapse. Mol Cell Biol. 32:4986–4997. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen XB, Melchionna R, Denis CM, Gaillard
PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J and
McGowan CH: Human Mus81-associated endonuclease cleaves holliday
junctions in vitro. Mol Cell. 8:1117–1127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boddy MN, Gaillard PH, McDonald WH,
Shanahan P, Yates JR III and Russell P: Mus81-Eme1 are essential
components of a holliday junction resolvase. Cell. 107:537–548.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu H, Martin MM, Regairaz M, Huang L, You
Y, Lin CM, Ryan M, Kim R, Shimura T, Pommier Y and Aladjem MI: The
DNA repair endonuclease Mus81 facilitates fast DNA replication in
the absence of exogenous damage. Nat Commun. 6:67462015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blow JJ and Dutta A: Preventing
re-replication of chromosomal DNA. Nat Rev Mol Cell Biol.
6:476–486. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sun J, Fernandez-Cid A, Riera A, Tognetti
S, Yuan Z, Stillman B, Speck C and Li H: Structural and mechanistic
insights into Mcm2-7 double-hexamer assembly and function. Genes
Dev. 28:2291–2303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cortez D, Glick G and Elledge SJ:
Minichromosome maintenance proteins are direct targets of the ATM
and ATR checkpoint kinases. Proc Natl Acad Sci USA. 101:pp.
10078–10083. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu M, Ba Z, Costa-Nunes P, Wei W, Li L,
Kong F, Li Y, Chai J, Pontes O and Qi Y: IDN2 interacts with RPA
and facilitates DNA double-strand break repair by homologous
recombination in arabidopsis. Plant Cell. 29:589–599. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maher RL, Branagan AM and Morrical SW:
Coordination of DNA replication and recombination activities in the
maintenance of genome stability. J Cell Biochem. 112:2672–2682.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mao P, Liu J, Zhang Z, Zhang H, Liu H, Gao
S, Rong YS and Zhao Y: Homologous recombination-dependent repair of
telomeric DSBs in proliferating human cells. Nat Commun.
7:121542016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan DS, Rothermundt C, Thomas K, Bancroft
E, Eeles R, Shanley S, Ardern-Jones A, Norman A, Kaye SB and Gore
ME: ‘BRCAness’ syndrome in ovarian cancer: A case-control study
describing the clinical features and outcome of patients with
epithelial ovarian cancer associated with BRCA1 and BRCA2
mutations. J Clin Oncol. 26:5530–5536. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krajewska M, Fehrmann RS, de Vries EG and
van Vugt MA: Regulators of homologous recombination repair as novel
targets for cancer treatment. Front Genet. 6:962015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Remus D, Beuron F, Tolun G, Griffith JD,
Morris EP and Diffley JF: Concerted loading of MCM2-7 double
hexamers around DNA during DNA replication origin licensing. Cell.
139:719–730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Labib K: How do Cdc7 and cyclin-dependent
kinases trigger the initiation of chromosome replication in
eukaryotic cells? Genes Dev. 24:1208–1219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tercero JA, Labib K and Diffley JF: DNA
synthesis at individual replication forks requires the essential
initiation factor Cdc45p. EMBO J. 19:2082–2093. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moyer SE, Lewis PW and Botchan MR:
Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for
the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci
USA. 103:pp. 10236–10241. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rodriguez R, Gagou ME and Meuth M:
Apoptosis induced by replication inhibitors in Chk1-depleted cells
is dependent upon the helicase cofactor Cdc45. Cell Death Differ.
15:889–898. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Köhler C, Koalick D, Fabricius A, Parplys
AC, Borgmann K, Pospiech H and Grosse F: Cdc45 is limiting for
replication initiation in humans. Cell Cycle. 15:974–985. 2016.
View Article : Google Scholar : PubMed/NCBI
|