Introduction
Polo-like kinase 1 (Plk1) is an evolutionarily
conserved serine/threonine protein kinase and a key regulator of
mitosis that is required for kinetochore-microtubule attachments,
centrosome maturation, chromosome dynamics, chromosome segregation,
spindle function and cytokinesis (1–3). Full
length Plk1 consists of the N-terminus kinase domain (KD) and the
C-terminus Polo-box domain (PBD) (4). Its functions are executed by binding
and phosphorylating proteins through its two domains. The different
subcellular location of Plk1 is based on the PBD and
phosphorylation of Thr210 (5,6).
Diverse functions of the Polo-like kinases (PLKs) have been
reported at different stages of the cell cycle. An association
between PLKs and the centrosome is implicated in regulating mitotic
entry. Sudden Plk1 activation during late G2 depends on cyclin
A/Cdk activity levels and induces phosphorylation of Cdc25C1 before
mitotic entry (7). Plk1
phosphorylates BubR1 in mitotic cells, and this phosphorylation is
associated not only with spindle checkpoint signaling but also with
stable microtubule-kinetochore attachment, probably by generating
the force that pulls each centrosome towards the two spindle poles
(8,9). Plk1 interacts with the key mitotic
kinase Aurora B. Aurora B-dependent phosphorylation of the Plk1
activation loop at the kinetochore regulates its function (10). Additionally, recruitment of Aurora B
to the centromeres depends partly on Plk1-dependent phosphorylation
of survivin and haspin. Therefore, Plk1 is a critical mitotic
kinase during the cell cycle.
Ring finger protein 2 (RNF2), also known as RING2 or
RING1B, was first identified as an interactor with Bmi1, a group II
polycomb group (PcG) protein. RNF2 acts as a ubiquitin E3 ligase to
mono-ubiquitinate histone H2A (11–13).
Moreover, RNF2 is involved in regulating different biological
processes by distinct molecular mechanisms. For example, RNF2
promotes MDM2-mediated p53 ubiquitination, and overexpression of
RNF2 increases the half-life of MDM2 and inhibits its
ubiquitination (14). Furthermore,
one study revealed that negative regulation of p53 by RNF2 promoted
tumor development in selective cancer cell types (15). RNF2 is ubiquitously expressed in
human tissues and is amplified or overexpressed in many human
tumors, such as ovarian, breast and pancreatic cancer (16–18).
RNF2 is considered to be a prognostic biomarker and potential
therapeutic target for these cancer types, as high expression of
RNF2 is positively correlated with tumor progression and shortened
survival. However, the extensive biochemical characterization of
RNF2 in mitotic regulation has not been elucidated.
In the present study, we identified for the first
time that Plk1 interacts with RNF2 using a yeast two-hybrid screen.
We sought to elucidate the relationship between Plk1 and RNF2. Our
results demonstrated that RNF2 co-localized with Plk1 at mitotic
chromosomes in the prometaphase and metaphase. In addition, Plk1
kinase activity was required for the ubiquitin-dependent
degradation of RNF2. These findings provide a new clue to our
understanding of the function of RNF2 in mitotic regulation and
tumorigenesis.
Materials and methods
Cell culture, reagents and
plasmids
HeLa and 293T cells (ATCC, Manassas, VA, USA)
were grown at 37°C in a 5% CO2 atmosphere in Dulbecco's
modified Eagle's medium (DMEM; HyClone Laboratories Inc.; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10%
fetal bovine serum (FBS; HyClone Laboratories Inc.; Thermo Fisher
Scientific, Inc.), 100 µg/ml streptomycin and 100
µg/ml penicillin. The cells were synchronized at the G1/S
phase with a 250 mM thymidine (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) block. The plasmid vectors 3× FLAG
(Sigma-Aldrich; Merck KGaA), pEGFP-C2 (BD Biosciences, San Jose,
CA, USA), pGBKT7 (Clontech Laboratories, Inc., Palo Alto, CA, USA)
and pGADT7 (Clontech Laboratories, Inc.) were used to generate
mammalian and yeast expression constructs carrying Plk1 or RNF2.
Plk1 or RNF2 was cloned into the bacterial expression plasmids
pET-28a (Novagen, Wilmington, DE, USA) and pGEX-6P-1 (GE
Healthcare, Parsippany, NJ, USA). MG132 (EMD Millipore, Bedford,
MA, USA), anti-RNF2 antibody (cat. no. ab101273) and anti-Plk1
antibody (cat. no. ab17057) were both purchased from Abcam
(Cambridge, MA, USA). GFP (cat. no. G1546) and FLAG (cat. no.
F1804) antibodies were purchased from Sigma-Aldrich (Merck
KGaA).
Yeast two-hybrid analysis
The specificity of the interaction between Plk1 and
RNF2 was verified by yeast two-hybrid. RNF2 was fused to pGBKT7 to
generate the BD-RNF2 vector. Plk1 was fused to pGADT7 to generate
the AD-Plk1 vector. Yeast two hybrid screening and yeast two hybrid
co-transformation assays were performed following the
manufacturer's instructions for the Yeast transformation system 2
(Clontech Laboratories, Inc.). The yeast was detected on yeast
dropout medium lacking tryptophan, leucine and histidine.
Glutathione S-transferase (GST)
pull-down assay
Purified RNF2-His was incubated with Glutathione
Sepharose 4B beads carrying the fusion proteins (PLK1-GST or GST)
at 4°C for 4 h. After washing with pre-cooled phosphate-buffered
saline (PBS) containing 1% Triton X-100, the beads were fixed in a
sample loading buffer at 100°C for 10 min and then subjected to
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) followed by immunoblot assay.
Immunoblot assay and
immunoprecipitation
293T or HeLa cells transformed with the
corresponding plasmids were harvested and resuspended in lysis
buffer (pH 7.4, 50 mM HEPES, 150 mM NaCl, 2 mM EGTA, 0.1% Triton
X-100, 1 mM phenylmethylsulfonyl fluoride, 10 g/ml leupeptin and 10
g/ml pepstatin A) containing a protease inhibitor mixture
(Sigma-Aldrich; Merck KGaA). After separation by SDS-PAGE, the
proteins were transferred to nitrocellulose membranes. After
blocking in TBST buffer with 5% non-fat milk, the membranes were
probed with the corresponding antibodies, and developed using an
enhanced chemiluminescence reagent. The cell lysates were purified
using anti-FLAG M2 affinity gel at 4°C for 4 h with rotation for
immunoprecipitation. After washing with lysis buffer and PBS, the
immunoprecipitants were eluted by boiling with a sample loading
buffer at 100°C for 10 min and then subjected to SDS-PAGE followed
by immunoblot assay.
Immunofluorescence
HeLa cells were harvested and plated on
poly-L-lysine-coated sterile glass slides. Then, the cells were
fixed in 4% paraformaldehyde for 15 min and permeabilized with PBS
containing 0.1% Triton X-100 for 15 min. After washing in PBS, the
coverslips were blocked with PBS containing 5% bovine serum albumin
(BSA). Subsequently, the coverslips were incubated with anti-Plk1
(1:100; cat. no. ab17057; Abcam) and anti-RNF2 (1:100; cat. no.
ab101273; Abcam) overnight at 4°C and incubated with the secondary
antibodies FITC-conjugated goat anti-mouse IgG (1:500; cat. no.
F-2761; Thermo Fisher Scientific, Inc.) and rhodamine-conjugated
goat anti-rabbit IgG (1:500; cat. no. R-6394; Thermo Fisher
Scientific, Inc.) and DAPI dye. Images were captured using an
Olympus BX60 upright fluorescence microscope (Olympus Corp., Tokyo,
Japan) with identical acquisition parameters for each
experiment.
Results
Identification of Plk1 as a
RNF2-binding protein
To identify host proteins targeted by RNF2, the
yeast two-hybrid screen was undertaken to screen the human
HeLa cDNA library. Surprisingly, Plk1, an important mitotic
regulator was found among the positive clones. To confirm the
interaction between Plk1 and RNF2, we co-transformed AH109 yeast
competent cells with Plk1-pGADT7 and RNF2-pGBKT7. The results
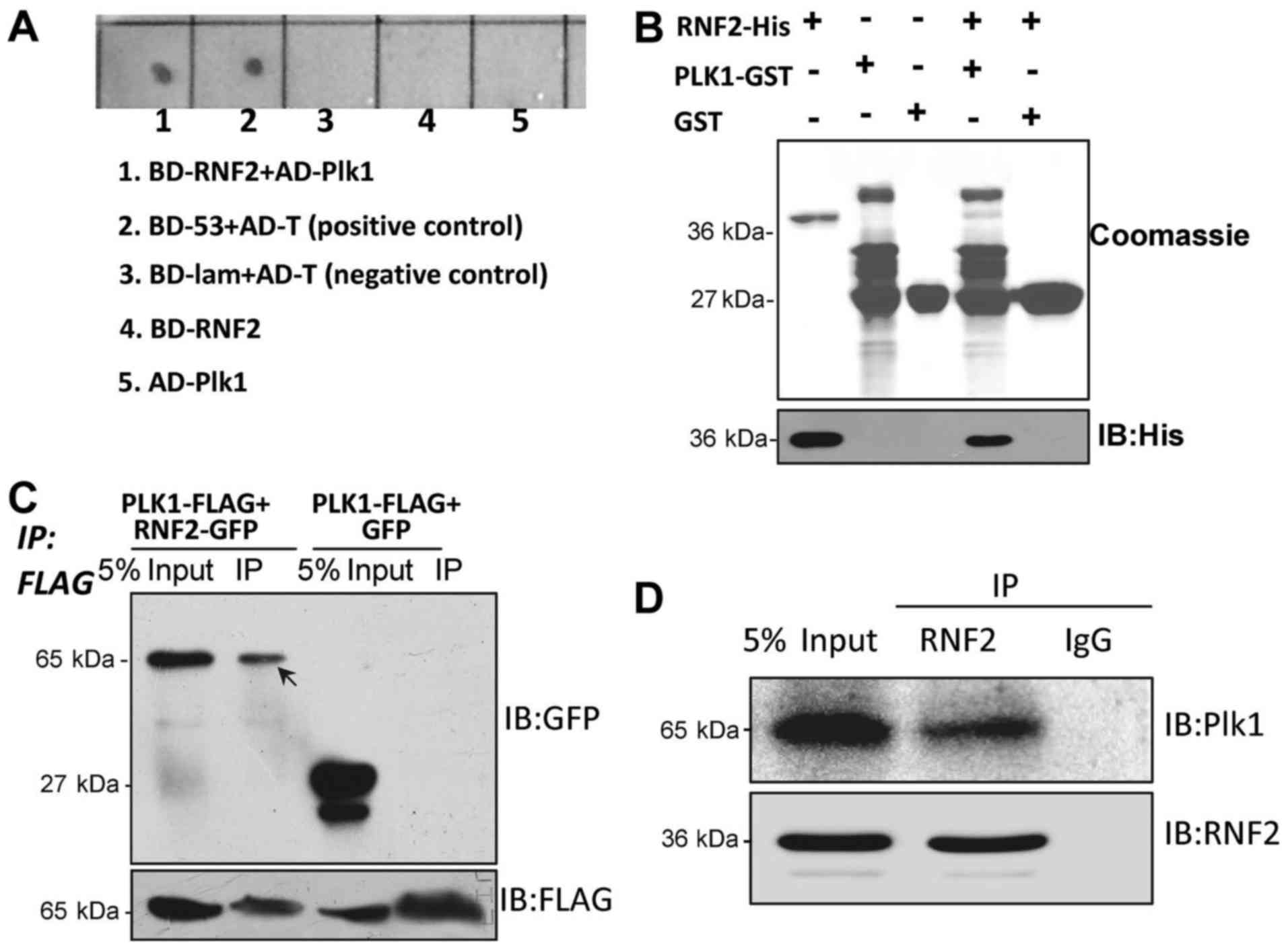
demonstrated that Plk1 interacted with RNF2 (Fig. 1A). To further confirm this
interaction, we performed a GST pull-down assay in vitro
using recombinant RNF2-His and Plk1-GST proteins expressed in E.
coli. Plk1-GST, but not GST, was able to pull down RNF2-His,
demonstrating that Plk1 directly bound to RNF2 (Fig. 1B). To further confirm this result,
we co-transfected Plk1-FLAG and RNF2-GFP into 293T cells for an
immunoprecipitation assay using an anti-FLAG antibody. The results
revealed that RNF2-GFP was pulled down by the FLAG antibody via
Plk1-FLAG (Fig. 1C). To test if
endogenous RNF2 formed a complex with Plk1 in 293T cells, we
carried out an immunoprecipitation with RNF2 antibody and control
IgG. The result indicated that endogenous RNF2 interacted with
Plk1, suggesting that RNF2 interacts with Plk1 in vivo
(Fig. 1D).
RNF2 and Plk1 are co-localized in HeLa
cells during mitosis
Dynamic localization during mitosis is one of the
most striking aspects of Plk1. To evaluate if there is specific
co-localization and the role of the interaction between RNF2 and
Plk1 during mitosis, we conducted immunofluorescence studies to
assess the localization of the two proteins at different mitotic
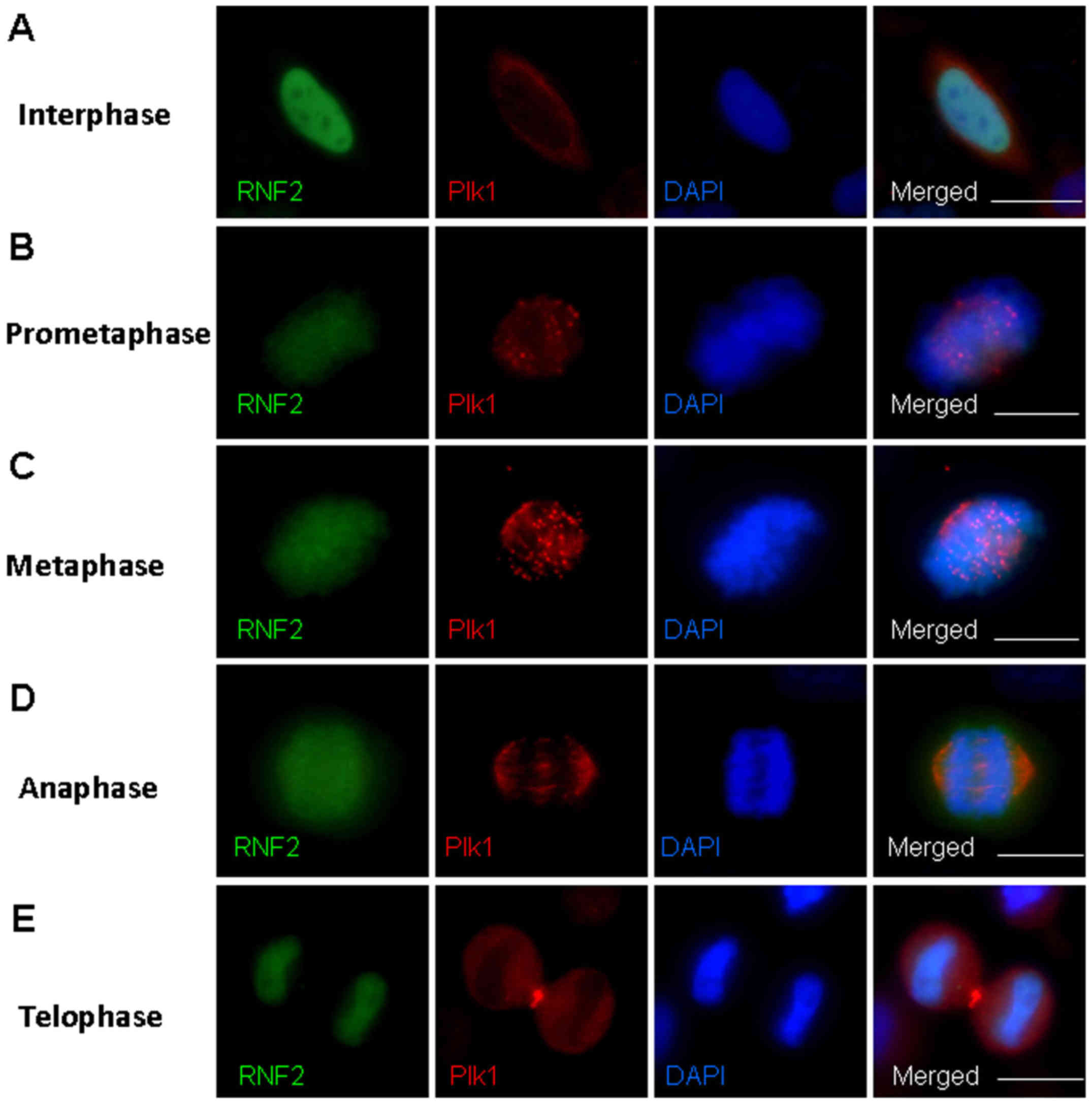
stages in HeLa cells. Notably, we found a defined
co-localization of RNF2 and Plk1 at the mitotic chromosomes in the
prometaphase and metaphase during mitosis (Fig. 2). RNF2 was diffusely localized at
the nucleus during interphase, which is consistent with its known
role in transcriptional regulation. Plk1, as expected, was found
scattered in the cytoplasm (Fig.
2A). As the cells entered prometaphase, Plk1 began to localize
to the spindle poles and the kinetochores. RNF2 demonstrated
increasing localization at the mitotic chromosome that overlapped
Plk1 at the kinetochores (Fig. 2B).
Plk1 was concentrated at the kinetochores in the metaphase. The
examination of RNF2 labeling in the same cells revealed typical
mitotic chromosome localization. Thus, RNF2 and Plk1 co-localized
at the mitotic chromosomes during the prometaphase and metaphase
(Fig. 2C). Furthermore, as
chromosome segregation occurred in the anaphase and telophase, Plk1
was maintained at the mid-body and RNF2 extended into the nucleus
of each sister cell (Fig. 2D and
E). These results indicate that RNF2 and Plk1 are co-localized
at mitotic chromosomes during mitotic progression.
Plk1 kinase activity is responsible
for degradation of RNF2
We demonstrated that Plk1 is a bona fide
RNF2-interacting protein, thus we aimed to address the functional
relevance of this interaction. We overexpressed GFP-tagged
wild-type Plk1 in HeLa cells and subsequently monitored the
relative levels of endogenous RNF2 in these cells via western
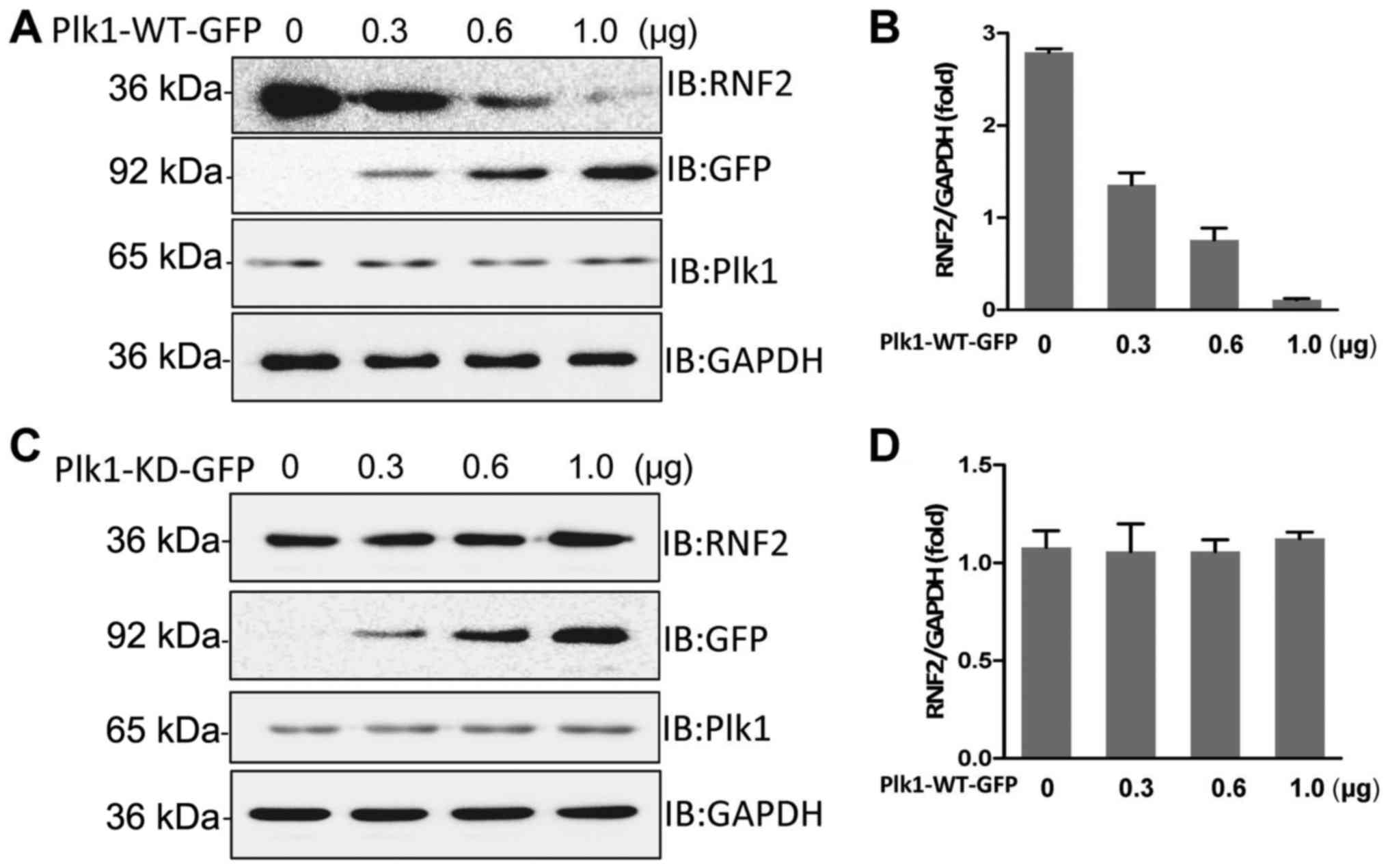
blotting. The result revealed that overexpression of wild-type Plk1
reduced the level of RNF2 in a dose-dependent manner (Fig. 3A and B). Next, we aimed to ascertain
whether Plk1 kinase activity is essential for degradation of RNF2;
thus, we overexpressed GFP-tagged kinase-deficient Plk1 in
HeLa cells and detected the relative levels of endogenous
RNF2. Notably, overexpression of kinase-deficient Plk1 did not
affect the level of endogenous RNF2 (Fig. 3C and D). Thus, we concluded that
Plk1 kinase activity is responsible for RNF2 degradation.
Plk1 interacts with RNF2 and promotes
its proteasome-ubiquitin-dependent degradation
Western blot analysis indicated that wild-type Plk1
reduced the level of RNF2, but kinase-deficient Plk1 failed to do
so. To explore whether Plk1 reduces RNF2 mRNA, we performed a
quantitative reverse transcription-polymerase chain reaction
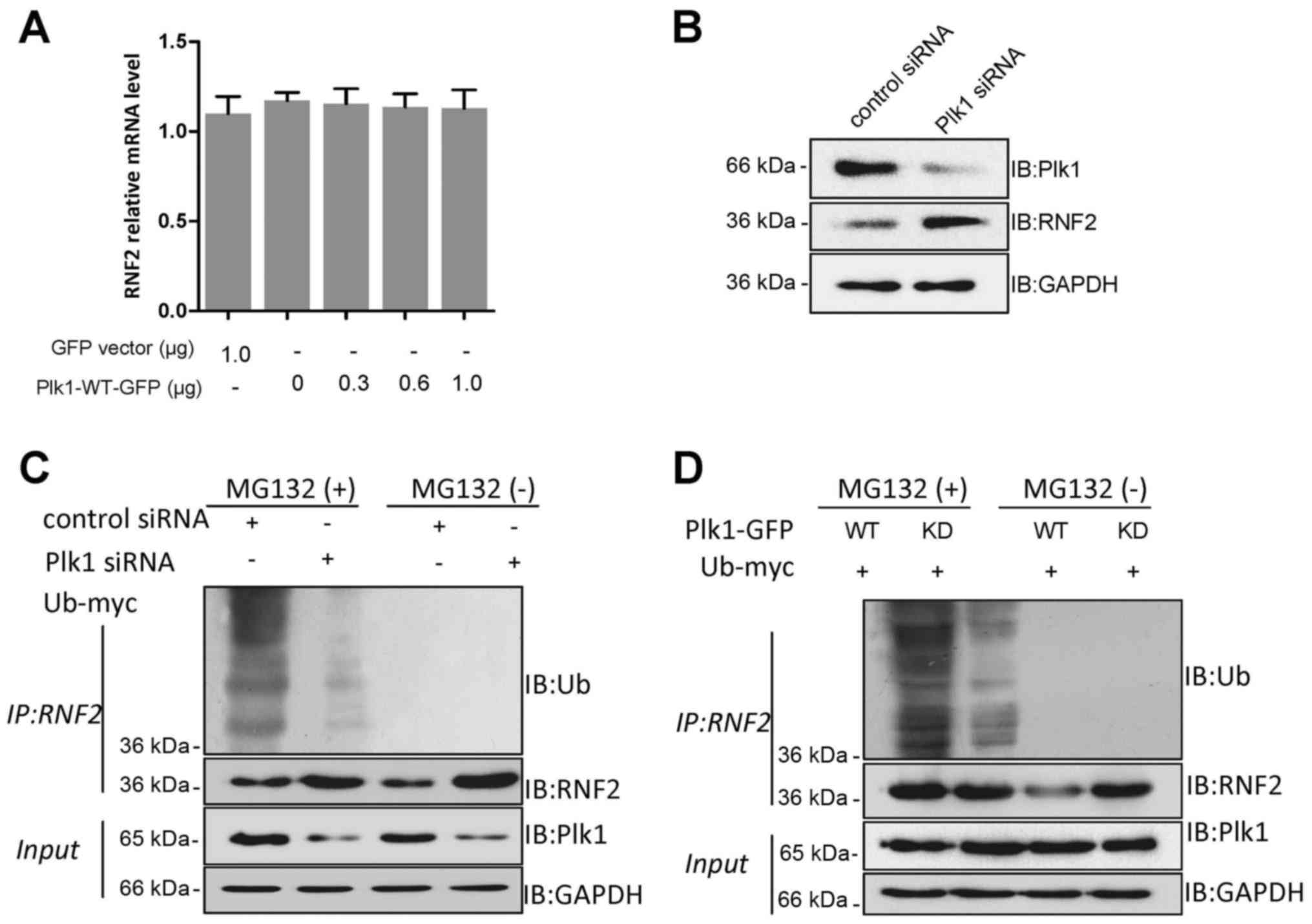
(qRT-PCR) analysis. The results revealed that the RNF2 mRNA level
did not change significantly after overexpressing Plk1, suggesting
a Plk1-dependent reduction in the RNF2 protein level (Fig. 4A). Moreover, since overexpression of
wild-type Plk1 resulted in lowering RNF2 protein levels, we
anticipated that Plk1 knockdown may create an opposite effect. The
results showed that Plk1 knockdown resulted in a significant
increase in the RNF2 level (Fig.
4B). To determine whether Plk1-induced RNF2 degradation was
proteasome-ubiquitin-dependent, 293T cells were cotransfected with
control or Plk1 siRNA and myc-tagged ubiquitin. The proteasome
inhibitor MG132 was used to block degradation of possibly
ubiquitinated RNF2. The results demonstrated that ubiquitination of
RNF2 decreased significantly in response to depleted Plk1 in the
presence of MG132, suggesting that Plk1 is required for
ubiquitin-dependent degradation of RNF2 (Fig. 4C). In addition, to further test the
possible effect of Plk1 kinase on the ubiquitin-dependent
degradation of RNF2, we repeated the above experiment using
wild-type or kinase-deficient Plk1 plasmids. As expected,
ubiquitination of RNF2 was increased significantly by Plk1-WT-GFP,
compared with that of Plk1-KD-GFP in the presence of MG132
(Fig. 4D). Therefore, we concluded
that Plk1-dependent decrease in RNF2 protein level is mediated
through proteasome-ubiquitin-dependent degradation.
Discussion
The polycomb group (PcG) of proteins are epigenetic
transcriptional regulators that repress numerous developmental
regulators, including two repressive complexes, called PRC1 and
PRC2. RNF2, also known as RING2 or RING1B, is a member of the PRC1
complex. Previous studies have reported that RNF2 directly
associates with Bmi1, Cbx family proteins, Mel18, M33 and Phc1,
forming the core of the PRC1 complex (18–20).
The complex varies in its association with mitotic chromosomes.
Live-cell imaging and genome-wide sequencing demonstrates that most
PRC1 proteins dissociate from mitotic chromosomes, whereas a
quantitative subpopulation of PRC1 proteins remain associated with
mitotic chromosomes in Drosophila. Recent studies have shown
that canonical PRC1 exhibits various capacities of association with
mitotic chromosomes and that Cbx2 is required to recruit PRC1 to
mitotic chromosomes (20–22). Previous studies have revealed that
Plk1 phosphorylates numerous mitotic proteins, such as cdh1, HSF1
and Bora, and promotes their degradation by SCFβ−TRCP.
Additionally, Plk1 also phosphorylates Emi1 and promotes its
degradation during mitosis. All of these results suggest that Plk1
often collaborates with β-TRCP and the SCF complex to promote
protein degradation (23–25).
Abundant clinical and pathological evidence
indicates that RNF2 is expressed at high levels in several cancer
types compared with normal tissues (15,26).
Furthermore, RNF2 negatively regulates autophagy by promoting
AMBRA1 degradation (27). All of
these data imply that RNF2 could be considered a potential
biomarker and therapeutic target for these cancer types. However,
it remains elusive as to whether RNF2 is involved in mitotic
progression. During mitotic division, the multifunctionality of
Plk1 is relevant to its dynamic subcellular localization (3,28,29).
Plk1 is primarily located in the centrosome and kinetochore during
prophase, and then translocates to the spindle intermediate region
during meta-anaphase. Plk1 is concentrated at the equatorial plate
during cytokinesis (30,31). In the present study, we provide
evidence for the interaction between RNF2 and Plk1 through
co-immunoprecipitation, GST pull-down and yeast two-hybrid assays.
Immunofluorescence studies showed co-localization of RNF2 and Plk1
at mitotic chromosomes during the prometaphase and metaphase. Plk1
kinase activity is required for entry into mitosis during the
normal cell cycle. Some published results reveal that Plk1 is a key
mediator of mitotic checkpoint inactivation, as cells that cannot
activate Plk1 fail to properly dismantle the DNA damage checkpoint
during mitosis and instead show DNA damage-induced Chk2 kinase
activation (32). Plk1 is also
involved in stabilizing kinetochore-microtubule attachments,
whereas these attachments are more stable when kinetochore Plk1
levels decrease dramatically during metaphase (33). Furthermore, one study showed that
Plk1 interacts with and phosphorylates LSD1 at Ser126 and this
phosphorylation promotes release of LSD1 from chromatin during
mitosis (34). The Plk1 KD is
essential for creating the phosphorylated site that is bound by the
PBD (35). Additionally, the Plk1
KD contains a nuclear localization sequence that promotes access to
the nucleus by Plk1 before nuclear envelope breakdown (36). Inactivation or depletion of Plk1
after microinjection of the antibody, dominant-negative mutants,
small interfering RNAs, or antisense oligonucleotides leads to
defects in centrosomal maturation, mitotic failure and increased
apoptosis (3,37).
Some studies have revealed that RNF2 is degraded by
the ubiquitin system independent of its own ubiquitin ligase
activity in a process mediated by an exogenous, yet unidentified
E3; however, a clear mechanism is unknown (13). Our results confirmed that Plk1 is
associated with RNF2, and that Plk1 degraded RNF2 via a
ubiquitin-dependent degradation pathway. Kinase-deficient Plk1
formed a more stable interaction with RNF2 compared with wild-type
Plk1, suggesting that kinase-deficient Plk1 disabled the induction
of RNF2 degradation. Given that expression of RNF2 is upregulated
in many tumors, we speculate that the high expression of RNF2 in
tumors may be a consequence of deficient Plk1 with an inability to
degrade RNF2.
The present study has some limitations. First, the
clinical pathological sections should be used to clarify the
relationship between Plk1 and RNF2 in tumors. Second, the
possibility that β-TRCP and SCF complex may be involved in
Plk1-induced RNF2 degradation was not investigated. Finally, the
relationship between Plk1 and RNF2 could not be fully elucidated
based on the experiments that were performed.
Taken together, for the first time, our observations
demonstrated that Plk1 directly interacts with RNF2 and degrades
RNF2 via the ubiquitin-dependent degradation pathway. We conclude
that RNF2 may be used as a new target for mitotic regulation and
tumorigenesis.
Acknowledgements
Not applicable.
References
|
1
|
Zhang Z, Chen C, Cui P, Liao Y, Yao L,
Zhang Y, Rui R and Ju S: Plk1 inhibition leads to a failure of
mitotic division during the first mitotic division in pig embryos.
J Assist Reprod Genet. 34:399–407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu J and Zhang C: The equilibrium of
ubiquitination and deubiquitination at PLK1 regulates sister
chromatid separation. Cell Mol Life Sci. 74:2127–2134. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu D, Davydenko O and Lampson MA:
Polo-like kinase-1 regulates kinetochore-microtubule dynamics and
spindle checkpoint silencing. J Cell Biol. 198:491–499. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Archambault V, Lépine G and Kachaner D:
Understanding the polo kinase machine. Oncogene. 34:4799–4807.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen F, Zhuo X, Qin T, Guo X, Zhang C and
Lai L: Designed inhibitor for nuclear localization signal of
polo-like kinase 1 induces mitotic arrest. Chem Biol Drug Des.
89:732–740. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jang YJ, Ma S, Terada Y and Erikson RL:
Phosphorylation of threonine 210 and the role of serine 137 in the
regulation of mammalian polo-like kinase. J Biol Chem.
277:44115–44120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gheghiani L, Loew D, Lombard B, Mansfeld J
and Gavet O: PLK1 activation in late G2 sets up commitment to
mitosis. Cell Rep. 19:2060–2073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kishi K, van Vugt MA, Okamoto K, Hayashi Y
and Yaffe MB: Functional dynamics of Polo-like kinase 1 at the
centrosome. Mol Cell Biol. 29:3134–3150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sumara I, Giménez-Abián JF, Gerlich D,
Hirota T, Kraft C, de la Torre C, Ellenberg J and Peters JM: Roles
of polo-like kinase 1 in the assembly of functional mitotic
spindles. Curr Biol. 14:1712–1722. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carmena M, Pinson X, Platani M, Salloum Z,
Xu Z, Clark A, Macisaac F, Ogawa H, Eggert U, Glover DM, et al: The
chromosomal passenger complex activates Polo kinase at centromeres.
PLoS Biol. 10:e10012502012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bentley ML, Corn JE, Dong KC, Phung Q,
Cheung TK and Cochran AG: Recognition of UbcH5c and the nucleosome
by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 30:3285–3297.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qian T, Lee JY, Park JH, Kim HJ and Kong
G: Id1 enhances RING1b E3 ubiquitin ligase activity through the
Mel-18/Bmi-1 polycomb group complex. Oncogene. 29:5818–5827. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ben-Saadon R, Zaaroor D, Ziv T and
Ciechanover A: The polycomb protein Ring1B generates self atypical
mixed ubiquitin chains required for its in vitro histone H2A ligase
activity. Mol Cell. 24:701–711. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen W, Peng C, Kim MO, Jeong Ho C, Zhu F,
Yao K, Zykova T, Ma W, Carper A, Langfald A, et al: Knockdown of
RNF2 induces apoptosis by regulating MDM2 and p53 stability.
Oncogene. 33:421–428. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su WJ, Fang JS, Cheng F, Liu C, Zhou F and
Zhang J: RNF2/Ring1b negatively regulates p53 expression in
selective cancer cell types to promote tumor development. Proc Natl
Acad Sci USA. 110:1720–1725. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei M, Jiao D, Han D, Wu J, Wei F, Zheng
G, Guo Z, Xi W, Yang F, Xie P, et al: Knockdown of RNF2 induces
cell cycle arrest and apoptosis in prostate cancer cells through
the upregulation of TXNIP. Oncotarget. 8:5323–5338. 2017.PubMed/NCBI
|
|
17
|
Qu C and Qu Y: Down-regulation of
salt-inducible kinase 1 (SIK1) is mediated by RNF2 in
hepatocarcinogenesis. Oncotarget. 8:3144–3155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Du J, An R, Chen L, Shen Y, Chen Y, Cheng
L, Jiang Z, Zhang A, Yu L, Chu D, et al: Toxoplasma gondii
virulence factor ROP18 inhibits the host NF-κB pathway by promoting
p65 degradation. J Biol Chem. 289:12578–12592. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhen CY, Duc HN, Kokotovic M, Phiel CJ and
Ren X: Cbx2 stably associates with mitotic chromosomes via a PRC2-
or PRC1-independent mechanism and is needed for recruiting PRC1
complex to mitotic chromosomes. Mol Biol Cell. 25:3726–3739. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Li X, Chen Z and Bepler G:
Ubiquitination and degradation of ribonucleotide reductase M1 by
the polycomb group proteins RNF2 and Bmi1 and cellular response to
gemcitabine. PLoS One. 9:e911862014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Steffen PA, Fonseca JP, Gänger C,
Dworschak E, Kockmann T, Beisel C and Ringrose L: Quantitative in
vivo analysis of chromatin binding of Polycomb and Trithorax group
proteins reveals retention of ASH1 on mitotic chromatin. Nucleic
Acids Res. 41:5235–5250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fonseca JP, Steffen PA, Müller S, Lu J,
Sawicka A, Seiser C and Ringrose L: In vivo Polycomb kinetics and
mitotic chromatin binding distinguish stem cells from
differentiated cells. Genes Dev. 26:857–871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fukushima H, Ogura K, Wan L, Lu Y, Li V,
Gao D, Liu P, Lau AW, Wu T, Kirschner MW, et al: SCF-mediated Cdh1
degradation defines a negative feedback system that coordinates
cell-cycle progression. Cell Rep. 4:803–816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YJ, Kim EH, Lee JS, Jeoung D, Bae S,
Kwon SH and Lee YS: HSF1 as a mitotic regulator: Phosphorylation of
HSF1 by Plk1 is essential for mitotic progression. Cancer Res.
68:7550–7560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seki A, Coppinger JA, Du H, Jang CY, Yates
JR III and Fang G: Plk1- and beta-TrCP-dependent degradation of
Bora controls mitotic progression. J Cell Biol. 181:65–78. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li XQ, He WP, Hou WH, Chen JW, Fan RR,
Yuan LJ, Yang GP, Cai MY, Chen L, Li J, et al: Overexpression of
RNF2 is positively associated with ovarian carcinoma aggressiveness
and indicative of poor patient survival. Oncotarget. 6:31181–31190.
2016.
|
|
27
|
Xia P, Wang S, Huang G, Du Y, Zhu P, Li M
and Fan Z: RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading
to downregulation of autophagy. Cell Res. 24:943–958. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lera RF, Potts GK, Suzuki A, Johnson JM,
Salmon ED, Coon JJ and Burkard ME: Decoding Polo-like kinase 1
signaling along the kinetochore-centromere axis. Nat Chem Biol.
12:411–418. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lu LY and Yu X: The balance of Polo-like
kinase 1 in tumorigenesis. Cell Div. 4:42009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wachowicz P, Fernández-Miranda G, Marugán
C, Escobar B and de Cárcer G: Genetic depletion of Polo-like kinase
1 leads to embryonic lethality due to mitotic aberrancies.
Bioessays. 38 Suppl 1:S96–S106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hasegawa H, Hyodo T, Asano E, Ito S, Maeda
M, Kuribayashi H, Natsume A, Wakabayashi T, Hamaguchi M and Senga
T: The role of PLK1-phosphorylated SVIL in myosin II activation and
cytokinetic furrowing. J Cell Sci. 126:3627–3637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
van Vugt MA, Gardino AK, Linding R,
Ostheimer GJ, Reinhardt HC, Ong SE, Tan CS, Miao H, Keezer SM, Li
J, et al: A mitotic phosphorylation feedback network connects Cdk1,
Plk1, 53BP1, and Chk2 to inactivate the G2/M DNA damage
checkpoint. PLoS Biol. 8:e10002872010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lénárt P, Petronczki M, Steegmaier M, Di
Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N and Peters JM: The
small-molecule inhibitor BI 2536 reveals novel insights into
mitotic roles of polo-like kinase 1. Curr Biol. 17:304–315. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Peng B, Shi R, Jiang W, Ding YH, Dong MQ,
Zhu WG and Xu X: Phosphorylation of LSD1 by PLK1 promotes its
chromatin release during mitosis. Cell Biosci. 7:152017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park JE, Soung NK, Johmura Y, Kang YH,
Liao C, Lee KH, Park CH, Nicklaus MC and Lee KS: Polo-box domain: A
versatile mediator of polo-like kinase function. Cell Mol Life Sci.
67:1957–1970. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taniguchi E, Toyoshima-Morimoto F and
Nishida E: Nuclear translocation of plk1 mediated by its bipartite
nuclear localization signal. J Biol Chem. 277:48884–48888. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tao YF, Li ZH, Du WW, Xu LX, Ren JL, Li
XL, Fang F, Xie Y, Li M, Qian GH, et al: Inhibiting PLK1 induces
autophagy of acute myeloid leukemia cells via mammalian target of
rapamycin pathway dephosphorylation. Oncol Rep. 37:1419–1429. 2017.
View Article : Google Scholar : PubMed/NCBI
|