Introduction
Bladder cancer (BCa) is the second most common
malignant tumor of the urogenital system and has the fourth highest
incidence among all tumors diagnosed in males in Western countries
(1). Local perfusion chemotherapy
can significantly reduce the recurrence rate in patients with
non-muscle-invasive BCa. However, nearly half of patients still
experience tumor recurrence and metastasis after the second round
of chemotherapy (2).
Muscle-invasive BCa is also resistant to chemotherapeutic drugs and
often progresses with poor prognosis. Although different patients
exhibit varying sensitivities to chemotherapy drugs, BCa is
resistant to multiple drugs in most cases. These resistance
characteristics significantly influence the effectiveness of
clinical chemotherapy (3).
Therefore, elucidating the mechanisms involved in the
chemoresistance of BCa has become increasingly urgent.
MicroRNAs (miRNAs) are endogenous single-stranded,
small-molecule non-coding RNAs with lengths of 19–25 nucleotides
(4). These molecules participate in
the regulation of cell formation, differentiation, proliferation,
apoptosis and other basic biological processes. miRNAs bind to the
3′ untranslated region (3′UTR) of target mRNAs as completely or
incompletely complementary pairs and cause mRNA degradation or
protein expression inhibition. Ultimately, target gene processes
are negatively regulated (5). By
regulating different downstream target genes, miRNAs participate in
a wide range of key signaling pathways, such as the DNA damage,
Notch and nuclear factor (NF)-κB pathways, and consequently affect
mechanisms of tumor cell chemoresistance. As such, miRNAs serve as
promising candidate molecular markers (6). For example, in BCa, various miRNAs,
including miR-34a (7), miR-21
(8), miR-30d, miR-181, miR-199a-5p
(9) and miR-193a-3p, are reportedly
associated with chemoresistance (10–13).
miR-22-3p is located on chromosome 17 (17p13.3) and
is adjacent to the p53 gene (14).
Recent studies have shown that miR-22-3p plays a vital role in
different types of cancer, including breast, rectal, pancreatic,
hepatocellular (15) and gastric
cancers (16). As a potent
proto-oncogenic miRNA, miR-22 is noted for its role in chromatin
remodeling. miRNAs were found to trigger epithelial-mesenchymal
transition, enhance stemness and promote breast cancer development
and metastasis in mouse models by increasing methylation of the
miR-200 promoter. This process suppressed expression of the
anti-metastatic miR-200 family (17). In hematological malignancies, miR-22
was also reported to reduce global levels of
5-hydroxymethylcytosine (5-hmC) in the genome. 5-hmC was found to
trigger an increase in cell self-renewal capability and defective
differentiation and tumorigenesis by targeting tet methylcytosine
dioxygenase 2 (18). miR-22 has
also been noted to promote tumorigenesis by suppressing expression
of phosphatase and tensin homolog (PTEN) in prostate cancer
(19). However, some in
vitro studies have revealed the tumor-suppressor functions of
miR-22; these functions included restraining invasion and
metastasis of hepatocellular carcinoma by inhibiting expression of
cluster of differentiation (CD)147 (20), CD151 (gastric cancer) (16) and lysine-specific demethylase 3A
(KDM3A) (Ewing's sarcoma) (21). In
osteosarcoma, expression of high mobility group box protein 1
(HMGB1), a direct target of miR-22, and subsequent HMGB1-mediated
autophagy were found to be upregulated and to contribute to
chemotherapy resistance in vitro. However, as a compensatory
effect, miR-22 is presumably upregulated during chemotherapy
(22).
In the present study, we performed miR-omic and
RNA-seq assays and discovered differentially expressed genes in
multi-drug chemosensitive (5637) and multi-drug chemoresistant
(H-bc) BCa cell lines. Additional in-depth studies demonstrated
that miR-22-3p promoted chemoresistance in BCa by inhibiting the
neuroepithelial cell transforming 1 (NET1) gene, a target gene of
miR-22-3p that is a RhoA guanine exchange factor, via cysteine
residues in its membrane domain, which mediates extracellular
signal transduction and is also associated with tumor invasion,
metastasis and chemoresistance (23,24).
We also further revealed the role of NET1 and miR-22a-3p in
multi-drug chemotherapy of BCa by plate colony formation assay and
apoptotic experiments.
Materials and methods
Cell culture and reagents
Five BCa cell lines were purchased from the Chinese
Academy of Cell Resource Centre (Shanghai, China). These cell lines
included 5637 (ATCC No. HTB-9), T24 (ATCC No. HTB-4), Um-Uc-3 (ATCC
No. CRL-1749), J82 (ATCC No. HTB-1) and H-bc (established by the
Cancer Research Institute of Kunming Medical College in 1986). The
Um-Uc-3 cell line was cultured in Minimum Essential Medium (MEM;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) + 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.) and 1% glutamine at 37°C in 5% CO2. The remaining
cell lines were cultured in Roswell Park Memorial Institute
(RPMI)-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.) +
10% FBS and 1% glutamine at 37°C in 5% CO2. miR-22-3p
mimic and antagomiR, NET1 siRNA, scrambled sequences (including
mimic-negative control (NC), antagomiR-NC and siRNA-NC) and the
riboFECT CP Transfection kit were supplied by Guangzhou Ribobio
Co., Ltd. (Guangzhou, China). Control check (CK) represents the use
of cell culture media only. pINDUCER21 was provided by Professor
Yang Wang (Dalian Medical University, Dalian, China).
pINDUCER21-enhanced green fluorescent
protein (EGFP)-NET1 construction
pINDUCER21-EGFP and OmicsLink full-length open
reading frame (ORF) (GeneCopoeia, Rockville, MD, USA) expression
clone plasmid (NET1) were transformed into DH5α Escherichia coli
cells for DNA amplification and extraction. OmicsLink full-length
ORF of NET1 was digested with restriction endonucleases HindIII and
XhoI. Then, small fragments (insert) were recovered after digestion
(1.7 kb), and ends were reinforced. pINDUCER21-EGFP plasmid was
digested as a vector via restriction endonucleases MluI and Klenow.
The insert was then ligated into a new expression vector by T4 DNA
ligase, and ligation products were used to transform DH5α E. coli
bacteria. Positive clones pINDUCER21-EGFP-NET1 were screened for
resistance to ampicillin. Then, plasmids were extracted by Gel
Recovery kit (Omega Bio-tek, Inc., Norcross, GA, USA) and verified
by restriction enzyme digestion. Finally, the correct clone was
selected for sequencing.
Luciferase reporter assay
The 3′UTR fragment of NET1
(range=chr10:5498958-5500183) was amplified with primers NET1 UTR
sense 5′-GCGCTAGCAGAAGGCTCTGTGTGTTAACTGAT-3′ and NET1 UTR antisense
5′-GCGGATATCTTCCATCTTATGACTAAATCCACC-3′. Polymerase chain reaction
(PCR) products were cloned into a firefly luciferase reporter
vector pGL3-control (Invitrogen; Thermo Fisher Scientific, Inc.)
termed as pGL3-Luc-NET1-3′UTR-wt. The plasmid carrying the mutated
sequence for the seed region of miR-22-3p in complementary sites
(range=chr10:5499107-5499114) was generated based on the
pGL3-NET1-3′UTR plasmid by primer design for point-directed
mutagenesis experiments. This plasmid was designated as
pGL3-Luc-NET1-3′UTR-mut. Plasmids were confirmed by sequencing
analysis. Cells were seeded into 96-well plates at 1×104
cells/well and transfected with a mixture of 50 ng
pGL3-luc-NET1-wt/mut, 5 ng Renilla and 15 pmol antagomiR or
unrelated control (NC) oligo nucleotides by using the riboFECT CP
transfection kit in accordance with the manufacturer's
instructions. Firefly and Renilla luciferase activities were
measured 18 h after transfection by the Dual-Luciferase Reporter
Assay system (Promega, Madison, WI, USA) through a Promega GloMax
20/20 luminometer (Promega).
Chemoresistance studies
Clinical-grade pirarubicin (Pi; Shenzhen Wanle
Pharmaceutical Co., Ltd., Shenzhen, China), paclitaxel (Pa; Taiji
Pharmaceutical Co., Ltd., Sichuan, China), adriamycin (Ad; Pfizer,
Suzhou, China), epirubicin hydrochloride (EH; Zhejiang Haizheng
Co., Ltd., Zhejiang, China), hydroxycamptothecin (Hy; Lishizhen
Pharmaceutical Co., Ltd., Wuhan, China), cisplatin (Ci; Haosen
Pharmaceutical Co., Ltd., Jiangsu, China) and gemcitabine (Ge;
Dihao Pharmaceutical Co., Ltd., Jiangsu, China) (NCI Dictionary of
Cancer Terms, http://www.cancer.gov/dictionary) were used in the
present study. Cells in the logarithmic growth phase were seeded in
triplicate in 96-well plates at a density of 0.4×104
cells/well and treated with the dose of the agents required for
inhibition of 50% of the cells (IC50) at 72 h. Cell
survival was then measured by Vita-Blue (Biotool LLC, Houston, TX,
USA; B34102; Ex 530–570 nm/Em 590–620 nm reading)-based cell
proliferation assay. Relative survival was presented as the
percentage relative to the results in the control group (no drugs
added).
Cell proliferation assay
Cells in the logarithmic growth phase were seeded in
96-well plates at a cell density of 0.2×104 cells/well
(in triplicate) to allow adhesion. At 0, 24, 48, 72 and 96 h,
Vita-Blue-based cell density assays were performed. Cells were
incubated with 5 µl Vita-Blue (Biotool, LLC; B34102) at 37°C for 4
h, and fluorescent intensity was measured at Ex 535 nm/Em 595 nm.
Then, the cells were cultured with fresh medium until the next
round of measurement. Means and standard deviation (SDs) of
triplicate measurements were calculated and plotted.
RNA isolation and TaqMan-quantitative
reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated from cells at the logarithmic
phase by using TRIzol reagent (Tiangen Biotech Co., Ltd., Beijing,
China). Steady states of target and β-actin mRNA levels were
simultaneously quantified by dual-qRT-PCR assay with differentially
fluorescent-labeled TaqMan probes for the target (Cy5 or Rox) and
β-actin mRNA (HEX) (ShingGene, Shanghai, China). An FTC-3000P PCR
instrument (Funglyn Biotech Inc., Richmond Hill, ON, Canada) was
employed in the process. For miR analysis, cDNA was synthesized
with the specific stem-loop primer and quantified by TaqMan-qRT-PCR
assay. Through the 2−ΔΔCt method, U6 reads were
normalized for miR or with β-actin for mRNA. The primer and probe
sequences were: hNET1 F, 5′-AGCCAAGCAATAAAAGAGTTCG-3′; hNET1 R,
5′-GGTCACCACGAAGGGTAAATG-3′ and hNET1 probe,
5ROX-CACGTCCTTGGCAAATTTAATCTCTCCTG-3′; hACTB F,
5′-GCCCATCTACGAGGGGTATG-3′; hACTB R, 5′-GAGGTAGTCAGTCAGGTCCCG-3′
and hACTB probe, 5′HEX-CCCCCATGCCATCCTGCGTC-3′; miR-22-3p RT,
5′-GCGCGTGAGCAGGCTGGAGAAATTAACCACGCGCACAGTT-3′; miR-22-3p R,
5′-GAGCAGGCTGGAGAA-3′; miR-22-3p F, 5′-GGAAGCTGCCAGTTGA-3′ and
miR-22-3p probe, 5cy5′-CCACGCGCACAGTTC-3′; U6 RT,
5GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGATACGACAAAAATATG-3′; U6 F,
5′-GCTTCGGCAGCACATA-3′; U6 R, 5′-CTTCACGAATTTGCGTG-3′ and U6 probe,
5HEX-CCTTGCGCAGGGGCCATGC-MGB3′.
Western blot analysis
Cells at the logarithmic phase of culture were lysed
in a solution of 60 mM Tris-HCl (pH 6.8), 2% sodium dodecyl
sulphate, 20% glycerol, 0.25% bromophenol blue and 1.25%
2-mercaptoethanol and then heated at 100°C for 10 min before
electrophoresis/western blot analysis. Anti-NET1 antibody (1:1,000;
cat. no. 12740-1-AP), anti-glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) antibody (1:1,000; cat. no. 60004-1-Ig), anti-rabbit IgG
peroxidase-conjugated antibody (1:1,000; cat. no. SA00001-2) and
horseradish peroxidase-conjugated goat anti-mouse IgG antibody
(1:1,000; cat. no. SA00001-1) employed in the study were purchased
from Proteintech Group, Inc. (Chicago, IL, USA). Target bands were
revealed by enhanced chemiluminescence reaction (Pierce, Rockford,
IL, USA), and relative density of each protein over GAPDH was
quantified using Gel-Pro Analyser (Media Cybernetics, Inc.,
Rockville, MD, USA).
Colony formation assay
After 24 h of transfection, 2000 transfected cells
were placed in fresh 6-well plates in triplicate and maintained in
RPMI-1640 medium containing 10% FBS. The medium was replaced every
3 days, and samples were incubated for 2 weeks at 37°C and under 5%
CO2 in an incubator. Colony formation was observed with
the naked eye to determine the time for termination of experiments.
Clones were then fixed with methanol, stained with 0.1% crystal
violet for 30 min and then counted and recorded.
Cell apoptosis analysis
Apoptosis was analysed by Annexin V/propidium iodide
(PI) double staining. After transfection for 48 h, cells in the
logarithmic growth phase were harvested and rinsed twice with
ice-bathed PBS. Afterward, fluorescein isothiocyanate-labeled
enhanced Annexin V (3 µl) and PI (3 µl) were added to the cell
suspension at a final volume of 150 µl. After incubation for 30
min, flow cytometry was performed on a FACSCalibur instrument (BD
Biosciences, San Jose, CA, USA). The numbers of apoptotic and
necrotic cells were calculated by flow cytometry (Becton-Dickinson,
San Jose, CA, USA) and analysed by FlowJo v10 software (FlowJo LLC,
Ashland, OR, USA). Ratio of early apoptosis was adopted for the
test results. Experiments were independently performed thrice, and
a representative was subsequently obtained.
Statistical analysis
Data are presented as means, and error bars indicate
SD or standard error (SE) around the mean. All statistical analyses
were performed with GraphPad Prism v.6.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). Two-tailed Student's t-test, one-way ANOVA or
Mann-Whitney U test was employed to calculate for statistical
significance. P<0.05 was considered to indicate statistically
significant differences.
Results
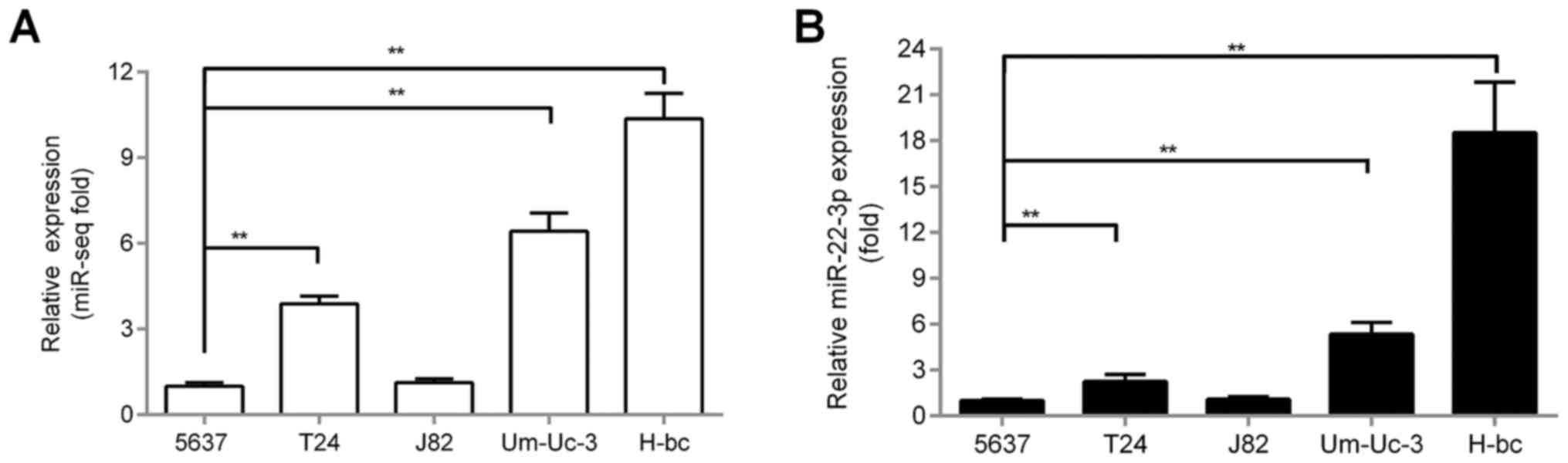
miR-22-3p levels were higher in
chemoresistant (H-bc) than in chemosensitive (5637) BCa cell
lines
Based on the fold differences of relative
chemoresistance indices (average of relative IC50 to
each drug) of BCa cell lines to seven chemotherapeutics (Pi, Pa,
Ad, EH, Ci, Hy and Ge), 5637 was the most multi-chemosensitive,
whereas H-bc was the most multi-chemoresistant cell line (Table I). 5637 and H-bc have been reported
as relatively multi-chemosensitive and multi-chemoresistant BCa
cell lines, respectively (10–13).
As revealed by both sequencing-based miR-omic analysis (miR-omic:
1.00:10.10; Fig. 1A) and qRT-PCR
validation (qRT-PCR: 1.00:18.52; Fig.
1B), miR-22-3p level was over 18-fold higher in H-bc than in
5637 cells (Fig. 1B). All these
observations suggest that miR-22-3p may promote BCa chemoresistance
similar to miR-193a-3p (10).
 | Table I.Chemo-resistance (IC50) to
each drug in the BCa cell lines. |
Table I.
Chemo-resistance (IC50) to
each drug in the BCa cell lines.
|
| Relative
IC50 values of the cell lines |
|---|
|
|
|
|---|
| Agents | 5637 | T24 | J82 | Um-Uc-3 | H-bc |
|---|
| Pi | 1.00 | 3.39 | 5.36 | 3.68 | 9.65 |
| Pa | 1.00 | 13.67 | 18.52 | 13.51 | 16.24 |
| Ad | 1.00 | 3.08 | 2.67 | 8.83 | 9.32 |
| EH | 1.00 | 1.74 | 25.22 | 22.05 | 37.96 |
| Hy | 1.00 | 1.18 | 2.85 | 1.85 | 120.82 |
| Ci | 1.00 | 1.18 | 1.77 | 1.02 | 1.71 |
| Ge | 3.88 | 1.00 | 3.19 | 15.94 | 3578 |
| Chemo-resistance
index | 1.41 | 3.61 | 8.51 | 9.55 | 539.10 |
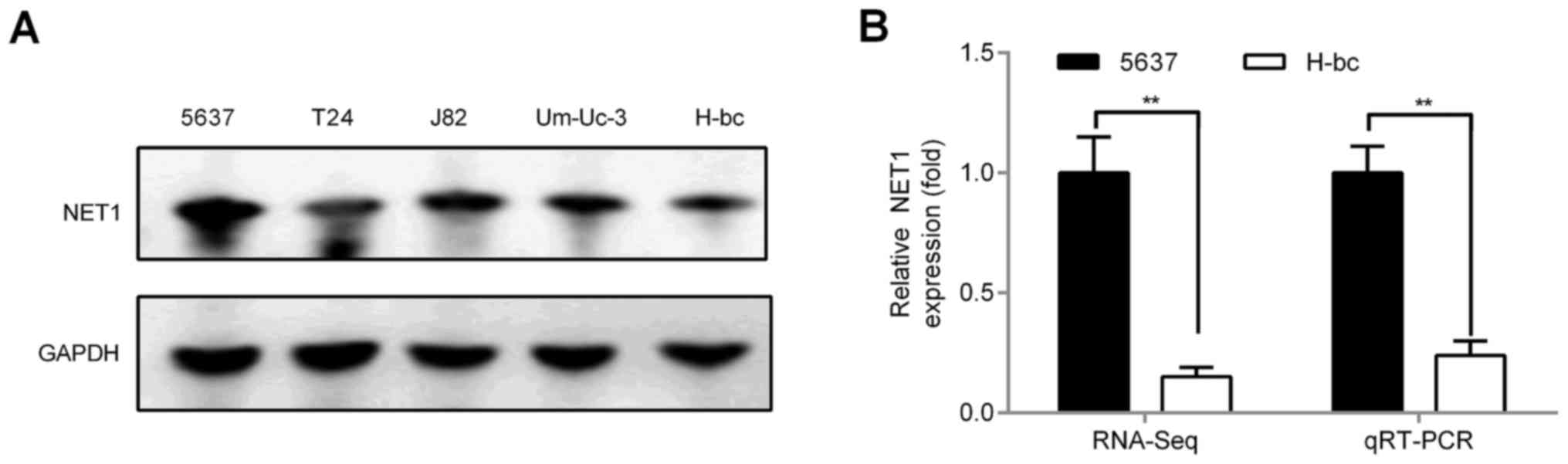
NET1 mRNA is a target of miR-22-3p in
BCa cells
To understand multiple chemoresistance mechanisms of
miR-22-3p-regulated BCa, we first used the bioinformatic software
miRDB and TargetScan to perform bioinformatic analysis. We studied
240 target genes that appeared simultaneously in the two predictive
softwares and some target genes of miR-22-3p that have been
studied; these target genes of miR-22-3p include TET2 (18), NET1 (25), CD147 (20), Tcf7 (26), VE-cadherin (27), TGFβR (28), KDM3A (21), CD151 (16), MTHFR (15), HMGB1 (22), Sp1 (29) and SRF (30) (only some of the genes appeared among
the 240 target genes predicted by the website). These genes were
detected by RNA-seq in 5637 and H-bc cell lines to identify the
target of miR-22-3p in BCa. Among the two cell lines, NET1 was the
more pronounced target gene, and its expression was opposite to
that of miR-22-3p. qRT-PCR and western blot analysis indicated that
NET1 levels, including mRNA (RNA-seq analysis: 1.00:0.15 and
qRT-PCR analysis: 1.00:0.25; Fig.
2B) and protein levels (western blot analysis: 1.00:0.50;
Fig. 2A), were significantly higher
in 5637 than in H-bc cells.
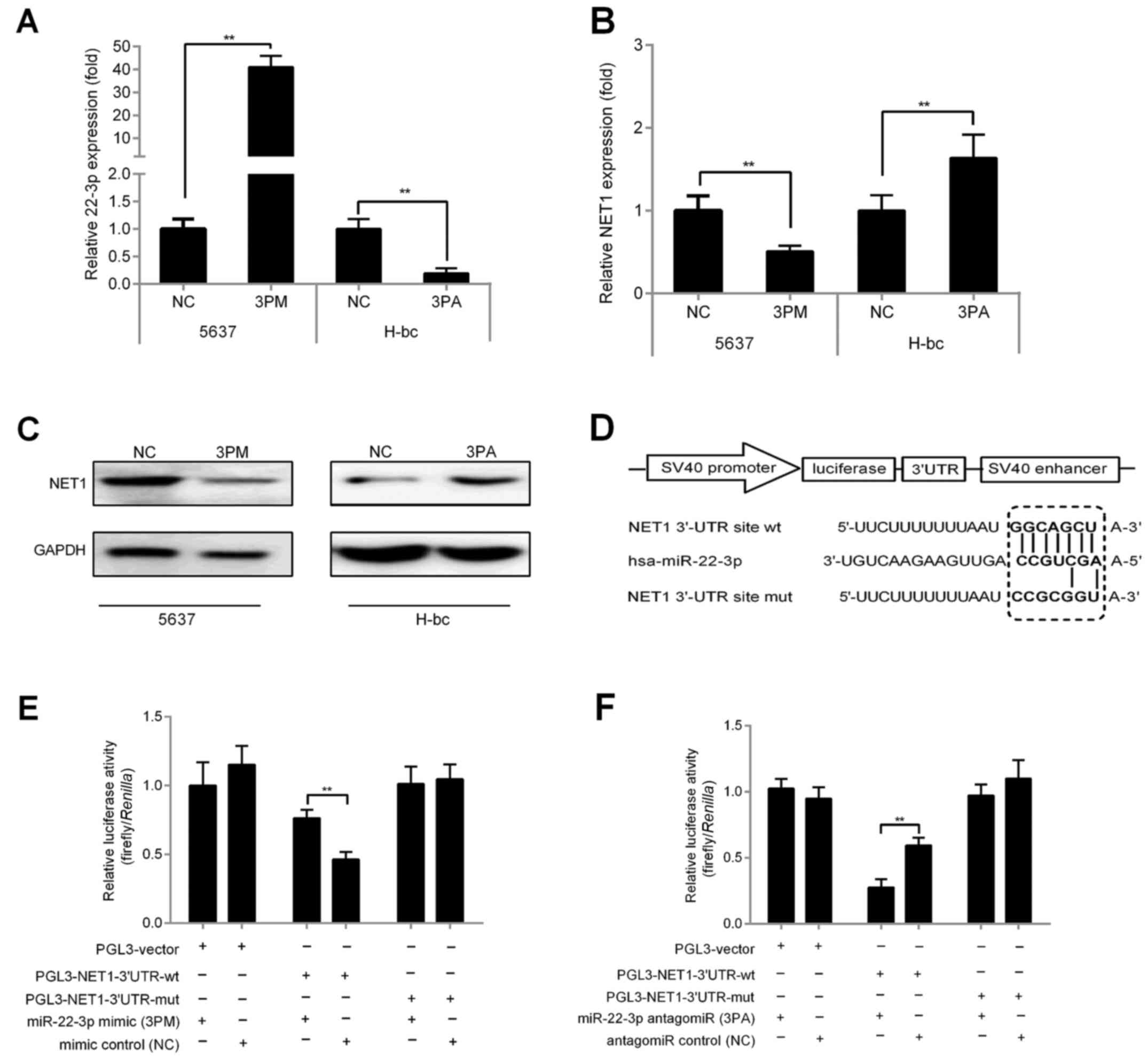
Given our above-mentioned studies, we observed that
NET1 and miR-22-3p levels are negatively correlated. Hence,
miR-22-3p may function by regulating NET1 expression to influence
BCa chemoresistance. To further detect whether NET1 is a genuine
target gene of miR-22-3p in BCa, we examined mRNA expression of
NET1 in 5637 and H-bc cells transfected with miR-22-3p mimic (3PM)
and miR-22-3p antagomir (3PA), respectively, vs. NC. qRT-PCR
results revealed that the expression of miR-22-3p-mimic-transfected
5637 cells increased by >37-fold vs. that of NC (Fig. 3A). By contrast, miR-22-3p expression
was significantly reduced by 78.0% in the
miR-22-3p-antagomiR-transfected H-bc cells vs. NC (Fig. 3A). As expected, miR-22-3p mimic
transfection decreased NET1 mRNA levels by 50.8% and protein levels
by 47.74% with respect to those of NC in 5637 cells (Fig. 3B and C). Similarly, miR-22-3p
antagomiR transfection increased NET1 mRNA levels by 64.0% and
protein levels by 2.29 times that of NC in H-bc cells (Fig. 3B and C).
To confirm whether NET1 is a target gene of
miR-22-3p in BCa, we demonstrated that the 3′UTR region of NET1
contains a potential binding region of miR-22-3p (150–157 bp) by
sequence analysis (Fig. 3D). We
placed wild-type (wt) and mutant (mut) NET1 genes downstream the
luciferase gene of the control vector (pGL3-vector; Promega) to
produce PGL3-NET1-UTR-wt/mut (Fig.
3D). These constructs and control vector (PGL3-vector) were
transfected into 5637 and H-bc cells, respectively, for comparison
of luciferase activities. As shown in Fig. 3E and F, in mimic-transfected 5637
cells, PGL3-NET1-UTR-wt exhibited lower luciferase activity than
the other two reported constructs with respect to that of the NC.
By contrast, activities in antagomiR-transfected H-bc cells were
higher. In conclusion, our findings strongly indicated that NET1 is
a target of miR-22-3p in BCa.
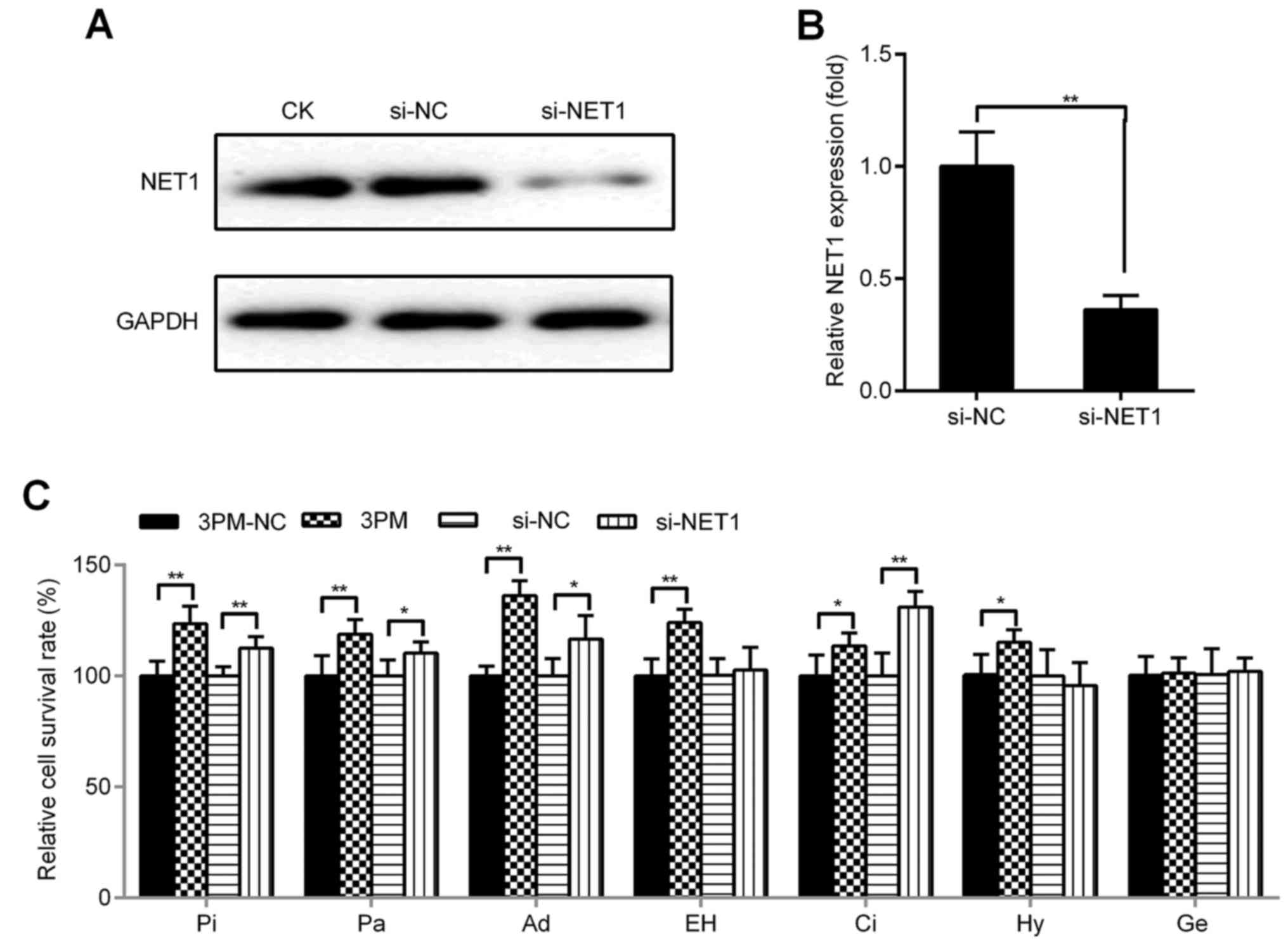
NET1 negatively correlates with the
promotive effect of miR-22-3p on BCa multi-chemoresistance
To investigate whether inhibition and overexpression
of NET1 produces steady effects with miR-22-3p mimic/antagomiR, we
designed, synthesized and transfected NET1 siRNA into 5637 cells.
Simultaneously, we constructed a NET1 overexpression vector
(pINDUCER21-EGFP-NET1) by using the plasmid of tetracycline-induced
expression system with green fluorescent protein and transfected
this plasmid into H-bc cells. Protein and mRNA levels of NET1 were
downregulated by transfection of si-NET1 (western blot analysis:
0.12:1.00; qRT-PCR: 0.36:1.00; Fig. 4A
and B). This finding was consistent with that of miR-22-3p
mimic transfection (western blot analysis: 0.42:1.00; qRT-PCR:
0.51:1.00; Fig. 3B and C) in 5637
cells with respect to that of NC. Cell death triggered by all six
drugs (Pi, Pa, Ad, EH, Ci and Hy, but not Ge) was significantly
attenuated after transfection with miR-22-3p mimic (Fig. 4C), whereas cell death caused by Pi,
Pa, Ad and Ci was significantly attenuated after transfection with
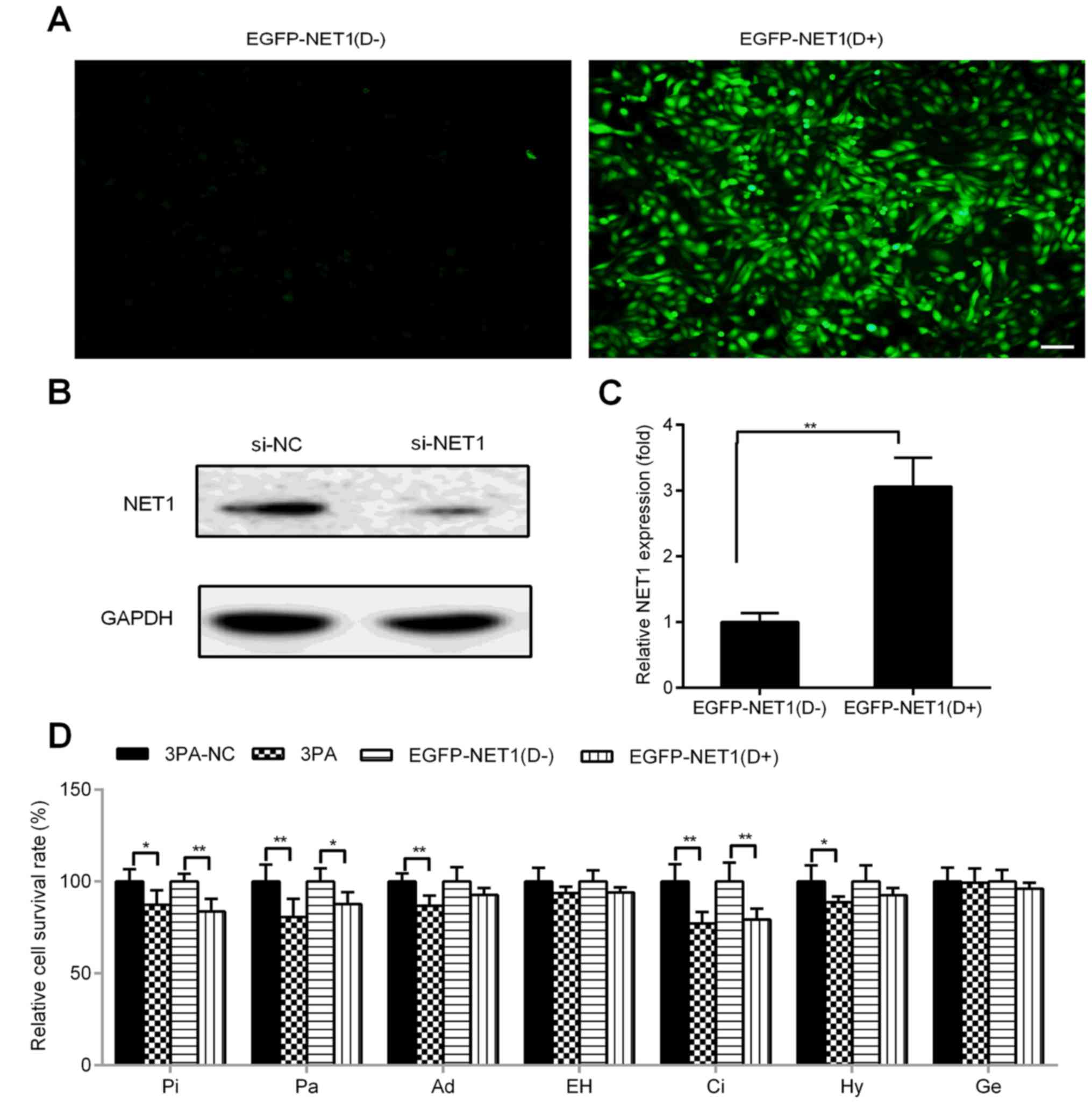
NET1 siRNA with respect to those of NC (Fig. 4C). Conversely, EGFP-NET1 was used
for transfection and successfully induced with docetaxel in H-bc
cells (Fig. 5A), where NET1 protein
and mRNA levels were upregulated (western blot analysis: 2.14:1.00;
qRT-PCR: 3.06:1.00; Fig. 5B and C,
respectively). H-bc became further amenable to cell death triggered
by Pi, Pa and Ci (Fig. 5D). Similar
to EGFP-NET1 transfection, miR-22-3p antagomiR transfection
sensitized H-bc cells to Pi, Pa, Ad, Ci and Hy but not to EH and Ge
(Fig. 5D).
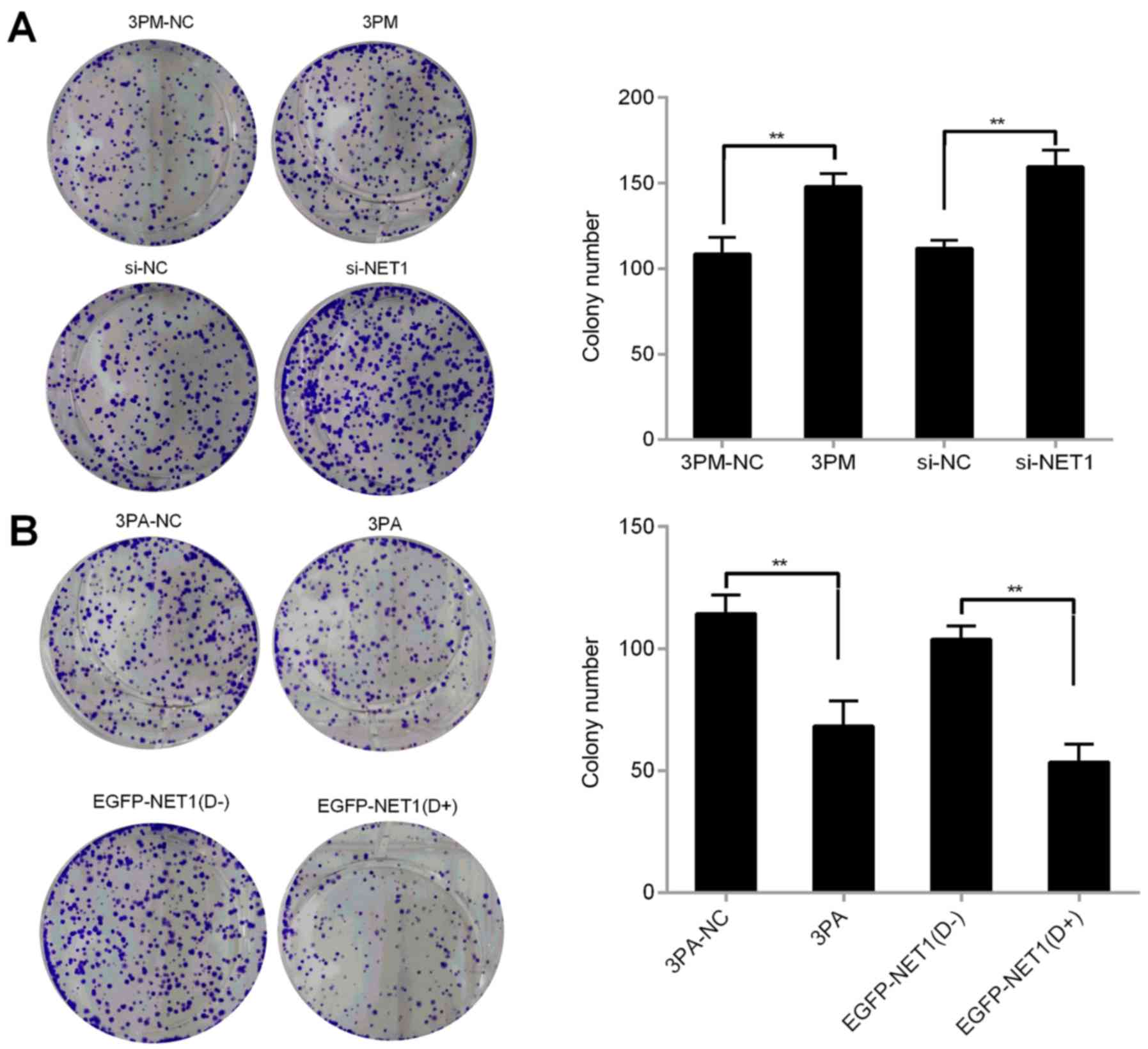
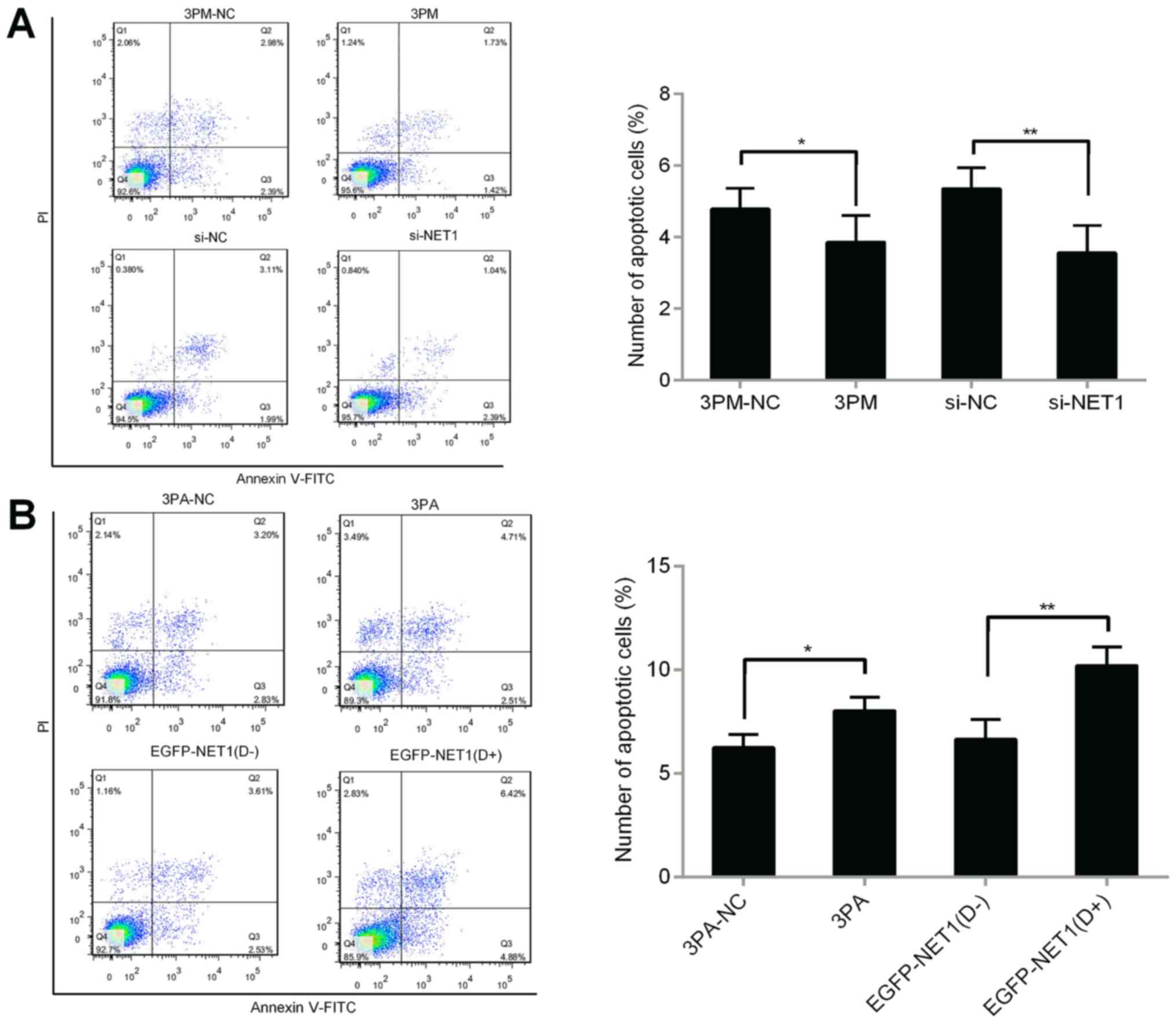
Forced reversal of miR-22-3p and NET1
on colony formation and apoptosis in chemoresistant BCa cells
We then investigated whether miR-22-3p is involved
in colony formation and apoptosis of BCa cells in H-bc and 5637
cells. miR-22-3p mimic or si-NET1 was transfected into 5637 cells
to detect colony formation and apoptosis. Compared with the control
cells, miR-22-3p-mimic- or si-NET1-transfected cells significantly
increased the number of cell colonies and decreased the number of
apoptotic cells (Figs. 6A and
7A). We then transfected miR-22-3p
antagomiR or overexpressed EGFP-NET1 into H-bc cells and performed
the same experiment. The results revealed a decreased number of
colonies and increased apoptotic rate in the H-bc cells (Figs. 6B and 7B). miR-22-3p possibly enhanced colonies
formation of BCa cells and inhibited cell apoptosis by modulating
the NET1 gene. Overall, the NET1 gene contributed significantly in
promoting miR-22-3p-mediated tolerance to BCa chemotherapy.
Discussion
Tumor resistance to chemotherapy is a key reason
underlying the poor prognosis of patients with clinical cancer.
With the increase in the research of the association of miRNAs and
tumor behavior, an increasing number of studies have revealed
information in regards to the abnormal expression of miRNAs and
clinical chemotherapy drug tolerance. miRNA mutations, abnormal
expression or abnormal processing affect the normal functions of
miRNAs and lead to ectopic expression of target proteins. When
target genes in tumor cells are regulated by miRNAs, the
sensitivity of tumor cells to drug changes. Studies have shown that
miR-22 affects chemotherapeutic tolerance of osteosarcoma cells by
regulating its target gene HMGB1 (22). However, understanding the role of
miR-22-3p in cancer chemoresistance, especially in BCa, remains
elusive.
In BCa, a new panel of miR-22-3p-targeting genes may
play pivotal roles in multi-drug chemoresistance of BCa. We
identified bioinformatic-predicted target genes at the RNA-seq omic
level in multi-chemosensitive 5637 and multi-chemoresistant H-bc
cells which exhibited differential expression of miR-22-3p. Results
were then verified by qRT-PCR of the target gene (Figs. 1 and 2). Findings indicated that NET1 levels
were significantly higher in 5637 cells than in H-bc cells and were
negatively correlated with miR-22-3p expression. Subsequently, we
revealed that NET1 is a direct target gene of miR-22-3p and plays a
catalytic role in BCa chemoresistance (Figs. 3–7).
In colon cancer/gastric cancer, NET1 was also found to be a target
gene of miR-638/miR-573 (31,32).
The above-mentioned study indicated that miR-22-3p plays a similar
role to that of miR-193a-3p in BCa chemoresistance (10).
NET1 is among the seven members of the transmembrane
4 superfamily (TM4SF), which was discovered by Serru et al
in the EST database (33). NET1 is
located on chromosome 1p34.1 and features a total length of 1297
bp, a coding sequence of 128–853 bp, a 241-amino-acid ORF (GenBank
accession no. NM005727.3), which are typical structural
characteristics of TM4SF. The amino acid sequence of NET1 is
homologous to those of other TM4SF members by 36% (CD82), 32%
(CO-029) and 29% (Talla-1). NET1 mediates extracellular signal
transduction through cysteine residues in its transmembrane domain
(34). Previous studies have shown
that NET1 is a novel tumor-related gene associated with many
malignancies, including hepatocellular carcinoma (35), skin squamous cell carcinoma (SSCC)
(36), breast (37), colorectal (31), lung (38) and gastric cancer (32). NET1 knockout was found to reduce the
proliferation, invasion and tumor growth of SSCC cells (36). NET1 gene overexpression in breast
adenocarcinoma cell lines also led to excessive cell migration
(37). NET1 is related to
chemoresistance in BCa through a highly complex mechanism. Hence,
we investigated the drug resistance mechanism of the NET1 gene in
BCa cells. Reversal of NET1/miR-22-3p expression in 5637 cells
inhibited cell death triggered by each of the six drugs used in the
present study except for gemcitabine (Fig. 4C). Similarly, reversing expression
of NET1/miR-22-3p in H-bc cells promotes drug-triggered cell death
except for epirubicin hydrochloride and gemcitabine (Fig. 5D). This result indicated that the
influence of miR-22-3p/NET1 on the drug resistance of different
drugs may not be consistent among different types of tumor cells
due to heterogeneity. Therefore, understanding the role of
miR-22-3p/NET1 in the sensitivity of different drugs and different
types of tumors is crucial for its future clinical application.
As indicated above, NET1 exerts carcinogenic effects
and may play a fundamental role in cancer development. However, the
role of NET1 in BCa has been only slightly studied. In the present
study, our data found that si-NET1 restrained expression of NET1
and consequently inhibited BCa apoptosis and promoted colony
formation. miR-22-3p mimic exerted the same effect in 5637 cells
(Figs. 6A and 7A). Successful induction of the NET1
overexpression vector with green fluorescent protein and a
tetracycline-inducing system resulted in NET1 overexpression, which
then promoted BCa apoptosis and inhibited colony formation.
miR-22-3p antagomiR exerted the same effect on H-bc cells (Figs. 6B and 7B). These results demonstrated that
miR-22-3p-mediated NET1 regulation may promote BCa chemoresistance.
In the past few years, the research on molecular mechanism of
apoptosis made breakthrough progress. It has been shown that many
apoptosis-related genes participate in apoptotic processes,
including caspases, death receptors and Bcl-2. The role of these
genes on the miR-22-3p signal axis in BCa chemoresistance awaits
further research.
In conclusion, miR-22-3p expression induces
chemoresistance in BCa cell lines. miR-22-3p may partially promote
chemotherapeutic tolerance by targeting NET1. Thus, miR-22-3p and
NET1 may serve as predictive chemopathic markers for BCa patients.
The detailed mechanism of BCa chemoresistance through
miR-22-3p-regulated NET1 targeting must be explored, and further
studies on cell signaling and in vitro nude
mouse-transplanted tumor systems will be conducted.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81502191 and 81702540)
and the Anhui Provincial Natural Science Foundation (nos.
1608085MH166, 1508085QH178, 1508085QH183, 1708085QH202 and
1708085QH179).
Availability of data and materials
The datasets are available from Dr Tao Tao upon
reasonable request.
Authors' contributions
TT and JX conceived and designed the study. SN, JX,
JZ, DP, DS, CX and HD performed the experiments. SN and LL wrote
the manuscript. TT, ZS and JX reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study did not involve human participants and
animals.
Consent for publication
This study did not involve human participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang JS, Lara PN Jr and Pan CX: Progress
in personalizing chemotherapy for bladder cancer. Adv Urol.
2012:3649192012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bambury RM, Power DG and O'Reilly S:
Intratumor heterogeneity and branched evolution. N Engl J Med.
366:2132–2133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Setoyama T, Ling H, Natsugoe S and Calin
GA: Non-coding RNAs for medical practice in oncology. Keio J Med.
60:106–113. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan B, Manley J, Lee J and Singh SR: The
emerging roles of microRNAs in cancer metabolism. Cancer Lett.
356:301–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Allen KE and Weiss GJ: Resistance may not
be futile: microRNA biomarkers for chemoresistance and potential
therapeutics. Mol Cancer Ther. 9:3126–3136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vinall RL, Ripoll AZ, Wang S, Pan CX and
deVere White RW: MiR-34a chemosensitizes bladder cancer cells to
cisplatin treatment regardless of p53-Rb pathway status. Int J
Cancer. 130:2526–2538. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tao J, Lu Q, Wu D, Li P, Xu B, Qing W,
Wang M, Zhang Z and Zhang W: microRNA-21 modulates cell
proliferation and sensitivity to doxorubicin in bladder cancer
cells. Oncol Rep. 25:1721–1729. 2011.PubMed/NCBI
|
|
9
|
Su SF, Chang YW, Andreu-Vieyra C, Fang JY,
Yang Z, Han B, Lee AS and Liang G: miR-30d, miR-181a and
miR-199a-5p cooperatively suppress the endoplasmic reticulum
chaperone and signaling regulator GRP78 in cancer. Oncogene.
32:4694–4701. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv L, Li Y, Deng H, Zhang C, Pu Y, Qian L,
Xiao J, Zhao W, Liu Q, Zhang D, et al: MiR-193a-3p promotes the
multi-chemoresistance of bladder cancer by targeting the HOXC9
gene. Cancer Lett. 357:105–113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y,
Xiao J, Qian L, Zhao W, Liu Q, et al: The miR-193a-3p regulated
PSEN1 gene suppresses the multi-chemoresistance of bladder
cancer. Biochim Biophys Acta. 1852:520–528. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y,
Xiao J, Qian L, Zhao W, Liu Q, et al: miR-193a-3p regulates the
multi-drug resistance of bladder cancer by targeting the LOXL4 gene
and the oxidative stress pathway. Mol Cancer. 13:2342014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Deng H, Lv L, Zhang C, Qian L, Xiao
J, Zhao W, Liu Q, Zhang D, Wang Y, et al: The miR-193a-3p-regulated
ING5 gene activates the DNA damage response pathway and inhibits
multi-chemoresistance in bladder cancer. Oncotarget. 6:10195–10206.
2015.PubMed/NCBI
|
|
14
|
Gurha P, Abreu-Goodger C, Wang T, RamiRez
MO, Drumond AL, van Dongen S, Chen Y, Bartonicek N, Enright AJ, Lee
B, et al: Targeted deletion of microRNA-22 promotes stress-induced
cardiac dilation and contractile dysfunction. Circulation.
125:2751–2761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li C, Ni J, Liu YX, Wang H, Liang ZQ and
Wang X: Response of MiRNA-22-3p and MiRNA-149-5p to folate
deficiency and the differential regulation of MTHFR
expression in normal and cancerous human hepatocytes. PLoS One.
12:e01680492017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Yu H, Lu X, Zhang P, Wang M and Hu
Y: MiR-22 suppresses the proliferation and invasion of gastric
cancer cells by inhibiting CD151. Biochem Biophys Res Commun.
445:175–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song SJ, Poliseno L, Song MS, Ala U,
Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al:
MicroRNA-antagonism regulates breast cancer stemness and metastasis
via TET-family-dependent chromatin remodeling. Cell. 154:311–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song SJ, Ito K, Ala U, Kats L, Webster K,
Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J,
Avigan DE, et al: The oncogenic microRNA miR-22 targets the TET2
tumor suppressor to promote hematopoietic stem cell self-renewal
and transformation. Cell Stem Cell. 13:87–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et
al: Identification of the miR-106b~25 microRNA cluster as a
proto-oncogenic PTEN-targeting intron that cooperates with
its host gene MCM7 in transformation. Sci Signal.
3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo LJ, Zhang LP, Duan CY, Wang B, He NN,
Abulimiti P and Lin Y: The inhibition role of miR-22 in
hepatocellular carcinoma cell migration and invasion via targeting
CD147. Cancer Cell Int. 17:172017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parrish JK, Sechler M, Winn RA and
Jedlicka P: The histone demethylase KDM3A is a
microRNA-22-regulated tumor promoter in Ewing Sarcoma. Oncogene.
34:257–262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li X, Wang S, Chen Y, Liu G and Yang X:
miR-22 targets the 3′UTR of HMGB1 and inhibits the HMGB1-associated
autophagy in osteosarcoma cells during chemotherapy. Tumour Biol.
35:6021–6028. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bennett G, Sadlier D, Doran PP, Macmathuna
P and Murray DW: A functional and transcriptomic analysis of NET1
bioactivity in gastric cancer. BMC Cancer. 11:502011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He S, Wei YZ, Wang GL, Xu YY, Zhou JM,
Zhang YX and Chen L: Study of RNA interference targeting NET-1
combination with sorafenib for hepatocellular carcinoma therapy in
vitro and in vivo. Gastroenterol Res Pract. 2013:6851502013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahmad HM, Muiwo P, Ramachandran SS, Pandey
P, Gupta YK, Kumar L, Kulshreshtha R and Bhattacharya A: miR-22
regulates expression of oncogenic neuro-epithelial transforming
gene 1, NET1. FEBS J. 281:3904–3919. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kaur K, Vig S, Srivastava R, Mishra A,
Singh VP, Srivastava AK and Datta M: Elevated hepatic miR-22-3p
expression impairs gluconeogenesis by silencing the wnt-responsive
transcription factor Tcf7. Diabetes. 64:3659–3669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu W, Zhan H, Zhou XY, Yao L, Yan M, Chen
A, Liu J, Ren X, Zhang X, Liu JX and Liu G: MicroRNA-22 regulates
inflammation and angiogenesis via targeting VE-cadherin. FEBS Lett.
591:513–526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hong Y, Cao H, Wang Q, Ye J, Sui L, Feng
J, Cai X, Song H, Zhang X and Chen X: MiR-22 may suppress
fibrogenesis by targeting TGFbetaR I in cardiac fibroblasts. Cell
Physiol Biochem. 40:1345–1353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai
T, Li LQ and Fan XH: Berberine upregulates miR-22-3p to suppress
hepatocellular carcinoma cell proliferation by targeting Sp1. Am J
Transl Res. 8:4932–4941. 2016.PubMed/NCBI
|
|
30
|
Xu D, Guo Y, Liu T, Li S and Sun Y: miR-22
contributes to endosulfan-induced endothelial dysfunction by
targeting SRF in HUVECs. Toxicol Lett. 269:33–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang J, Fei B, Wang Q, Song M, Yin Y,
Zhang B, Ni S, Guo W, Bian Z, Quan C, et al: MicroRNA-638 inhibits
cell proliferation, invasion and regulates cell cycle by targeting
tetraspanin 1 in human colorectal carcinoma. Oncotarget.
5:12083–12096. 2014.PubMed/NCBI
|
|
32
|
Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen
X, Ma L, Bi J, Wei G, Fang G and Xue X: TSPAN1 functions as an
oncogene in gastric cancer and is downregulated by miR-573. FEBS
Lett. 589:1988–1994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Serru V, Dessen P, Boucheix C and
Rubinstein E: Sequence and expression of seven new tetraspans.
Biochim Biophys Acta. 1478:159–163. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Todres E, Nardi JB and Robertson HM: The
tetraspanin superfamily in insects. Insect Mol Biol. 9:581–590.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ye K, Chang S, Li J, Li X, Zhou Y and Wang
Z: A functional and protein-protein interaction analysis of
neuroepithelial cell transforming gene 1 in hepatocellular
carcinoma. Tumour Biol. 35:11219–11227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
zhang J, Wang J, Chen L, Wang G, Qin J, Xu
Y and Li X: Expression and function of NET-1 in human skin squamous
cell carcinoma. Arch Dermatol Res. 306:385–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ecimovic P, Murray D, Doran P and Buggy
DJ: Propofol and bupivacaine in breast cancer cell function in
vitro-role of the NET1 gene. Anticancer Res. 34:1321–1331.
2014.PubMed/NCBI
|
|
38
|
Fang L, Zhu J, Ma Y, Hong C, Xiao S and
Jin L: Neuroepithelial transforming gene 1 functions as a potential
prognostic marker for patients with non-small cell lung cancer. Mol
Med Rep. 12:7439–7446. 2015. View Article : Google Scholar : PubMed/NCBI
|