Introduction
Survivin, also known as baculovirus IAP
repeat-containing protein 5 (BIRC5) and apoptosis inhibitor 4
(API4), is a member of the inhibitor of apoptosis protein (IAP)
family (1), which all contain at
least one copy of a baculovirus IAP repeat (BIR) domain, and
suppress apoptosis when overexpressed in cells (2,3).
Previous studies have shown that survivin participates in the
suppression of apoptosis, as well as the regulation of cell
division (4–6).
Survivin is a homodimer of a 16.5-kDa protein
(7). Located at the tip of
chromosome 17 in humans (17q25), the survivin gene has four
dominant (1, 2, 3, and 4) and two hidden (2B and 3B) exons.
Alternative splicing of its pre-mRNA produces splice variants, five
of which are known as survivin wild-type (wt), survivin-2B,
survivin-DEx3 (8), survivin-3B
(9) and survivin 2α (10).
It has been demonstrated that the vast majority of
tumors express Survivin mRNA and protein at high levels, whereas
most normal adult tissues do not, suggesting that survivin
expression is commonly associated with cancer (1,11–13).
Survivin may be localized inside or outside the cell (14); inside the cell, survivin has been
observed in the cytoplasm, the nucleus and the mitochondria
(15–17), but it may also be released into the
extracellular space through vesicles (14–18).
In previous studies, it was demonstrated that active
caspase-3 and −7 co-immunoprecipitated with survivin, whereas their
inactive pro-forms did not (19,20).
This interaction disrupts the caspase cascade and cleavage mediated
by caspases, thereby resulting in decreased apoptosis (21). In a similar manner, survivin
inhibits cytochrome c- and caspase-8-induced DEVD
(Asp-Glu-Val-Asp)-cleavage activity (21). Previous studies also revealed that
survivin antisense oligonucleotides target and downregulate
survivin mRNA and induce apoptosis (22,23).
Survivin contains a CDE/CHR element, which is involved in cell
cycle-specific regulation, implying that survivin may be involved
in the cell cycle process (24).
During mitosis, survivin can interact with CDK1 (24). Survivin can also interact with the
cell cycle regulator CDK4, leading to CDK2/cyclin E activation and
Rb phosphorylation. In a previous study, forced overexpression of
survivin resulted in an accelerated S phase and resistance to G1
arrest (25). Survivin, Borealin,
INCENP and Aurora B kinase are components of the chromosomal
passenger complex (CPC), which is a key regulator of chromosome
segregation and cytokinesis during cell division (26,27).
Knockdown of survivin expression was found to inhibit cell
proliferation, arrest the cell cycle at the G2/M checkpoint and
induce cellular apoptosis (28).
Previous studies also showed that survivin participates in cell
autophagy. The survivin inhibitor YM155 induced cell death through
autophagy (26,29,30);
when mRNA and protein expression levels of survivin and BCL-2
decreased, the expression levels of caspase-3, poly(ADP-ribose)
polymerase (PARP), Beclin 1 and LC-3 increased (31). Survivin may also enhance DNA repair
capability by upregulating Ku70 and homologous recombination
(32,33).
Urinary bladder cancer (BCa) and kidney cancer are
among the most frequently diagnosed cancers and are the leading
causes of cancer-related death, ranking sixth and ninth,
respectively, in terms of estimated new cases worldwide (34). There have been a number of reports
concerning survivin as a tumor marker in the diagnosis of
urothelial carcinoma, although further research and confirmation
are required. Studies have shown that the serum levels of survivin
protein are close to the detection limits of commercial
enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems,
Inc., Minneapolis, MN, USA; and Abnova, Taipei, Taiwan) (35). In the present study, using
survivin-specific monoclonal antibodies (mAbs) made previously by
our laboratory, we aimed to establish methods for detecting the
expression of survivin in cancer cell lines, serum samples, urine
samples and cancer tissues from BCa and renal cell carcinoma (RCC)
patients, and to further evaluate the efficacy of survivin as a
tumor marker in the surveillance of BCa and RCC.
Materials and methods
Chemical reagents
Protein-A/G Sepharose (HiTrap Protein G HP, 1 ml)
was purchased from GE Healthcare Life Sciences (Little Chalfont,
UK). The enhanced chemiluminescence western blotting system and
bicinchoninic acid protein assay kit were obtained from Thermo
Fisher Scientific, Inc. (Waltham, MA, USA). Horseradish peroxidase
(HRP) (H1759) and the IgG Subclass kit were purchased from
Sigma-Aldrich (Merck KGaA, Darmstadt, German).
3,3′,5,5′-Tetramethylbenzidine (TMB) and ELISA stop buffer were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Phosphate-buffered saline (PBS), HRP-conjugated goat anti-mouse IgG
and the immunohistochemistry detection system were purchased from
ZSGB-BIO (Beijing, China). PBST (0.05% Tween-20 in PBS) was used as
ELISA washing buffer, Tris-buffered saline (TBS) (20 mM Tris-HCl,
pH 7.5, 150 mM NaCl) and TBST (0.05% Tween-20 in TBS) were used as
western blotting washing buffer.
Cell lines
The lung cancer cell line A549, esophageal carcinoma
cell line EC109 and human hepatoblastoma cell line HepG2 were
maintained in our laboratory. The BCa cell line 5637 was purchased
from the Cell Bank of the Chinese Academy of Sciences (Beijing,
China). A549, EC109 and HepG2 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum. The 5637 cells were cultured in RPMI-1640 supplemented with
10% fetal bovine serum.
Animals
The animal experiments were approved by the Animal
Care Committee of Peking University and conformed to the guidelines
of the National Institutes of Health. All efforts were made to
minimize animal suffering. Balb/c mice weighing 18–22 g were
purchased from the Laboratory Animal Centre of the Chinese Academy
of Medical Sciences.
Human specimen collection
All human specimens were obtained from Peking
University Cancer Hospital and Institute, diagnosed
histopathologically, and staged according to the
tumor-node-metastasis (TNM) classification released by the American
Joint Committee on Cancer (AJCC, 7th edition, 2010). A total of 105
and 125 urine samples, and 122 and 208 corresponding serum samples
from BC and RCC patients, respectively, were collected between
March 2015 and December 2015. A total of 10 cases of formalin-fixed
paraffin-embedded BCa tissue sections corresponding to the urine
samples were also obtained. The healthy control (HC) groups
included 131 urine samples and 198 serum samples from individuals
who were health-check examinees and showed no abnormalities on
laboratory examinations. On the day of collection, all urine
samples were centrifuged at 3,000 rpm for 5 min, and the
supernatant was acquired, aliquoted and frozen at −20°C until
detection. Each patient and healthy examinee signed an informed
consent form. All study procedures were in accordance with the
Helsinki Declaration and the study was approved by the Ethics
Committee of Peking University Cancer Hospital and Institute.
Antibodies and standard protein
Hybridomas (A6, D8, C6, A9 and E6) were prepared
previously. Culture supernatants of hybridomas were assessed for
survivin expression, immunoglobulin subclass and specificity by
ELISA as described below. Hybridoma cells with high signals on
ELISA were injected into the abdominal cavity of Balb/c mice. mAbs
from the ascites fluids of Balb/c mice were purified by protein G
affinity chromatography. The titer of the purified mAb was measured
using the ELISA method. Antibody concentrations were determined by
measuring the absorbance at 280 nm using BSA as a protein standard.
A recombinant human sequence survivin protein,
MS2-survivin, produced by our laboratory was used as a
protein standard (36,37).
ELISA for the expression in hybridoma supernatants
and titer of purified mAbs. Microplates (Costar; Corning Inc.,
Corning, NY, USA) were coated with 100 µl MS2-survivin
proteins (2.5 µg/ml) per well overnight at 4°C, and then washed 3
times and blocked with 200 µl 5% skimmed milk for 1 h at 37°C.
After three washes, 100 µl serially diluted hybridoma supernatants
(from 1:100, for the expression of mAbs) or 100 µl serially diluted
purified mAbs (from 1:1,000, for the titer of mAbs) were incubated
for 1 h at 37°C. Following three washes, 100 µl HRP-conjugated goat
anti-mouse IgG (1:4,000 dilution) was used as the secondary
antibody. Plates were incubated for another 1 h at 37°C, washed 3
times, and 100 µl substrate solution TMB was added. The reaction
was stopped with 50 µl stop solution for 20 min at 37°C, and the
absorbance was then measured at 450 nm using a microplate reader
(model 680; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
ELISA for the specificity and subclass
of mAbs
As described for the ELISA above, microplates were
coated with 100 µl 2.5 µg/ml MS2-survivin, GST-survivin,
GST-uPA, MS2-PAI, MS2-NSE, MS2-MK
or BSA overnight at 4°C. Following blocking, hybridoma supernatants
diluted 10-fold were added, and HRP-conjugated anti-mouse IgG was
used as the secondary antibody. The subclass of the mAbs was
identified in the hybridoma supernatants with the mouse mAb
isotyping kit (Sigma-Aldrich; Merck KGaA).
Labeling of mAbs with HRP
Anti-survivin mAbs that produced high signals on
ELISA (D8, C6, A9 and E6) were selected for labeling with HRP. mAbs
were dialyzed against several changes of carbonate buffer [0.1 M
sodium carbonate buffer
(NaHCO3/Na2CO3) pH 9.5] overnight
at 4°C. HRP protein was dissolved in deionized water immediately
prior to use (protecting solution from light, stirring for 20 min
at room temperature) at a concentration of 5 mg/ml, and dialyzed
against CH3COONa (1 mmol/l sodium acetate buffer, pH
4.4)overnight at 4°C. mAb and HRP solutions were combined in equal
quantities by gentle stirring, and incubated at room temperature
for 2 h. Next, 0.1 ml NaH4B (sodium borohydride) was
added and incubated at 4°C for 2 h. The reaction solution was
dialyzed against several changes of PBS buffer (0.01 M sodium
phosphate, 0.15 M sodium chloride, pH 7.4) overnight at 4°C. After
dialyzing, the reaction mixture was applied to a Sephacryl S-200
column to remove uncoupled HRP (38). The mAbs coupled with HRP were used
in the subsequent experiments.
Development of a sandwich ELISA using
a pair of mAbs
D8, C6, A9 and E6 (100 µl, 2.5 µg/ml) were coated on
96-well microplates overnight at 4°C. After blocking with 200 µl 5%
skimmed milk in PBS for 1 h at 37°C and three washes with PBST, 100
µl 0.5 µg/ml MS2-survivin was added to the corresponding
wells. After washing, 1,000- and 5,000-fold diluted HRP-labeled
mAbs (D8, C6, A9 and E6) were added. The plates were incubated for
1 h at 37°C, washed 3 times and substrate solution was added. The
absorbance was measured at 450 nm after the addition of stop
solution. A pair of mAbs was selected to develop a sandwich ELISA
system, which was evaluated according to intra-assay precision,
inter-assay precision and minimum detectable dose (MDD). By
replicating assays in 20 wells with 10 ng/ml survivin protein as a
standard substance, the intra-assay coefficient of variation (CV)
was obtained. The inter-assay CV was obtained by detecting the same
concentration of survivin protein 10 times.
Detection of the survivin protein with
the sandwich ELISA
Using the developed sandwich ELISA system, serum and
urine samples from patients and HCs were assessed for survivin
expression. Serially diluted MS2-survivin (2,000-0.24
ng/ml) was detected as a standard, with 0 ng/ml as blank, and 500
ng/ml BSA as a negative contrast.
Western blotting
A549, EC109, HepG2 and 5637 cells were harvested,
washed twice in ice-cold PBS and lysed using TPEB extraction
reagent (Tiangen Biotech Co., Ltd., Beijing, China) for 30 min on
ice with sonication every 10 min, after which the lysed mixture was
separated by centrifugation at 14,000 × g (4°C). The supernatants
were used as cell lysates. Protein concentration was determined
with a bicinchoninic acid protein assay kit. Cell lysates were
boiled in lysis buffer containing 2% SDS for 10 min.
MS2-survivin fusion proteins (10 ng) or cell lysates (30
µg) were concentrated by 5% SDS-PAGE (pH 6.8) at 60 V for 30 min,
fractionated by 12% SDS-PAGE (pH 8.8) at 100 V for ~2 h and
transferred to nitrocellulose membranes at 200 mA for 1.5 h.
Western markers (Beijing Transgen Biotech Co., Ltd., Beijing,
China) were run in parallel. The blotted membranes were blocked
with 5% non-fat milk in PBST and incubated overnight at 4°C with
enzyme-linked mAbs; anti-survivin mAb D8 (sc-17779; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) was used as a positive
control. After washing, the HRP-conjugated goat anti-mouse IgG was
used as the secondary antibody for D8 (Santa Cruz Biotechnology,
Inc.) and incubated for 1 h at room temperature. Following three
washes with PBST, bound antibodies were visualized using enhanced
chemiluminescence. For normalization of the target gene, β-actin
was used as an internal reference.
Immunohistochemistry
Paraffin sections of 4-µm thickness were baked for 2
h at 65°C. Deparaffinization was performed using xylene (15 min,
twice) and hydration was conducted using a series of graded ethanol
(100, 95, 85 and 75%; 5 min each) to distilled water. The antigens
were retrieved with pH 6.0 citrate buffer for 5 min at 125°C in a
pressure boiler. Following cooling and washing with PBST, blocking
for endogenous peroxidase was performed for 10 min in 0.3%
H2O2. After three further washes in PBST,
non-specific binding was blocked with PBST containing 5% skimmed
milk for 30 min at room temperature. The sections were then rinsed
in PBST 3 times and incubated at 4°C with mAbs, anti-survivin mAb
D8 (Santa Cruz Biotechnology, Inc.) as a positive control, or 5%
skimmed milk in PBST as negative control. Following three washes,
the sections were incubated with Polymer Helper for 20 min, and
then washed again 3 times prior to incubation for 30 min with
polyperoxidase-anti-mouse/rabbit IgG. After a further three washes,
the sections were sequentially developed in DAB solution for 5 min,
counterstained in hematoxylin for 1 min, washed in tap water,
rinsed in ethanol containing 1% hydrochloric acid, washed in tap
water for 30 min, and dehydrated in graded ethanol (75, 85, 95 and
100%) and xylene. Coverslips were applied to the samples, which
were then evaluated under light microscopy independently by two
pathologists from the Department of Pathology, Peking University
Cancer Hospital and Institute, without prior knowledge of the
patient clinical data. The intensity of the staining was scored on
a scale of no staining/negative, weak staining/(+), moderate
staining/(++) and strong staining/(+++).
Statistical analysis
Statistical analysis was carried out using SPSS for
Windows (version 16.0; SPSS, Inc., Chicago, IL, USA). The survivin
concentrations in patients and healthy individuals were compared by
Student's t-test and also assessed using the area under the
receiver operating characteristic (ROC) curve (AUC). The cut-off
value was determined by the optimal Youden's index (sensitivity +
specificity - 1). All tests were two-sided and P<0.05 was set as
the significance level.
Results
Expression, specificity, titer and
subclass of mAbs
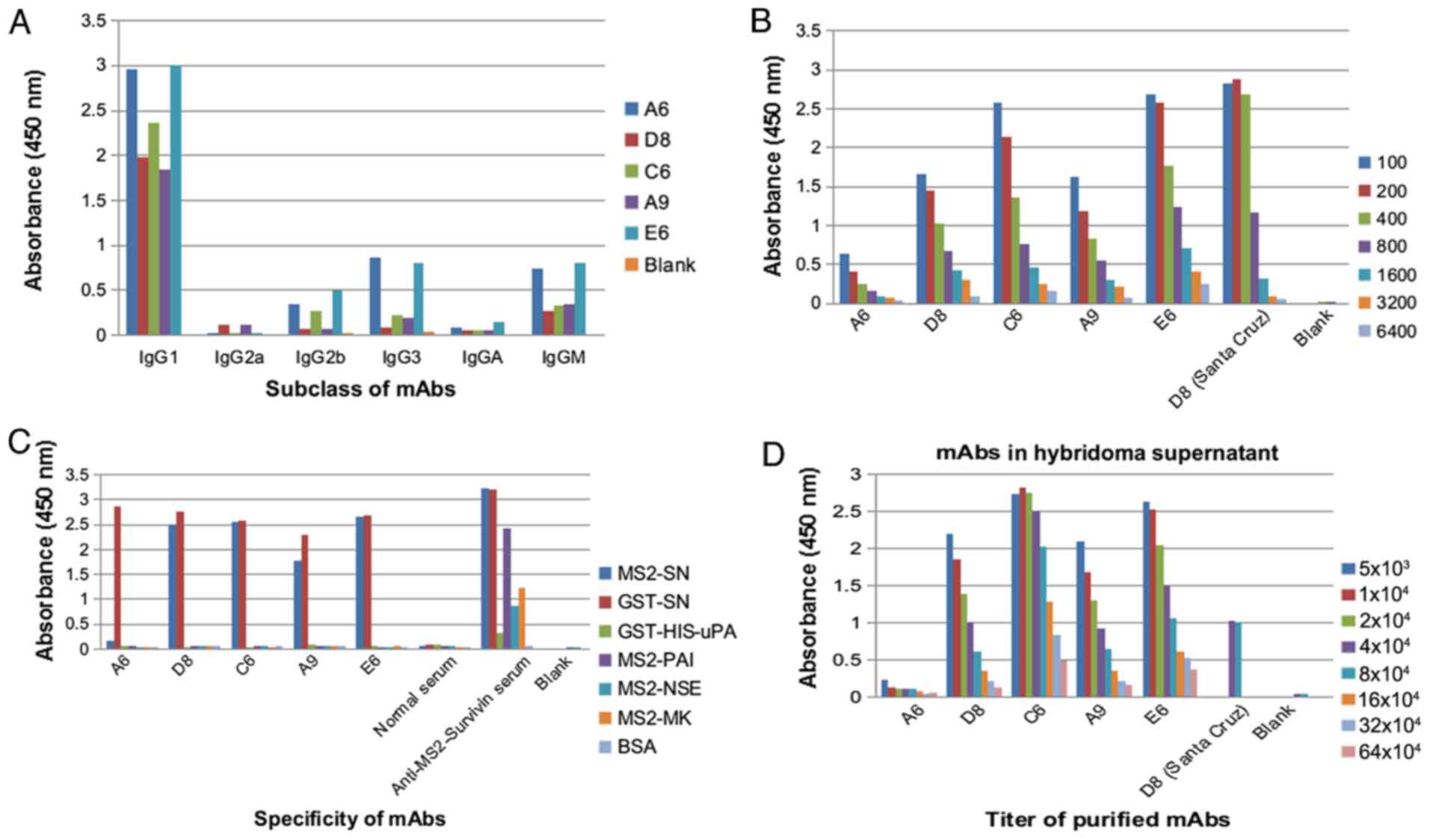
Hybridomas were tested for survivin subclass by
indirect ELISA. Hybridomas (A6, D8, C6, A9 and E6) with high
expression, specificity and antibody titer were selected for
further mAb pairing. The results showed that the subclass of these
mAbs was IgG1 (Fig. 1A). D8, C6, A9
and E6, which exhibited strong signals on ELISA, were chosen for
subsequent mAb pairing (Fig.
1B-D).
Sandwich ELISA development and
evaluation
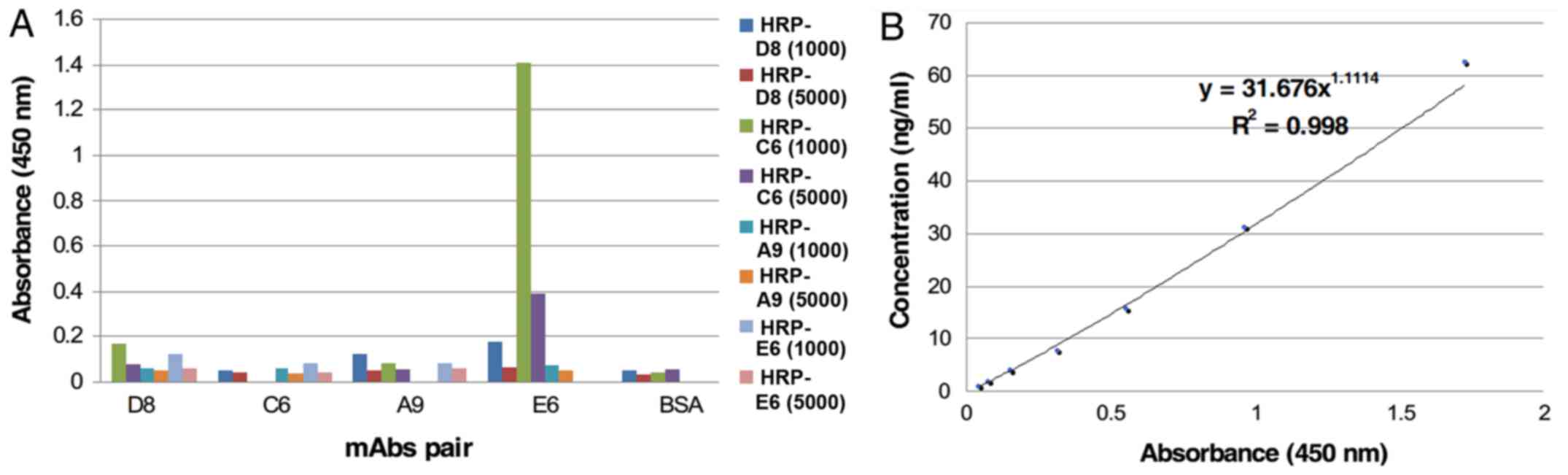
E6 was selected as the capture mAb and HRP-C6 was
selected as the detecting mAb to develop the sandwich ELISA
(Fig. 2A). The intra-assay CV was
7.28% and the inter-assay CV was 9.58%, indicating that the
sandwich ELISA had good reproducibility. According to the standard
protein curve (Fig. 2B), the MDD of
the assay was 0.98 ng/ml.
Expression levels of survivin in urine
and serum samples from patients
Urine samples from 105 cases of BCa and 125 cases of
RCC, as well as 122 and 208 corresponding serum samples, were
assessed. The HC groups included 131 urine samples and 198 serum
samples from health-check examinees who showed no abnormalities on
laboratory examination results. The basic characteristics,
including the age and sex of the patients and HCs, are summarized
in Table I.
 | Table I.Basic characteristics (age and sex)
of the BCa and RCC patients and HCs. |
Table I.
Basic characteristics (age and sex)
of the BCa and RCC patients and HCs.
|
|
|
| Sex |
|---|
|
|
|
|
|
|---|
| Samples | N | Age in years [mean
(range)] | Male | Female |
|---|
| Healthy urine | 131 | 48.1679
(24–66) | 108 | 23 |
| Healthy serum | 198 | 36.9141
(22–66) | 69 | 129 |
| BCa urine | 105 | 61.8544
(29–84) | 71 | 34 |
| BCa serum | 122 | 62.1721
(29–81) | 96 | 26 |
| RCC urine | 124 | 57.0000
(24–85) | 83 | 31 |
| RCC serum | 208 | 57.1394
(27–84) | 132 | 76 |
In BCa and RCC patients, survivin concentrations
were significantly higher compared with those in HCs in both the
urinary and serum samples (P<0.05) (Table II).
 | Table II.Survivin level in BCa and RCC
patients and HCs in both urinary and serum samples. |
Table II.
Survivin level in BCa and RCC
patients and HCs in both urinary and serum samples.
| Samples (n) | Survivin level
(mean±SD) | P-value | AUC | Cut-off value | Sensitivity | Specificity |
|---|
| Urine samples |
|
|
|
|
|
|
| HC (131) |
28.7327±75.56408 |
|
|
|
|
|
| BCa (105) |
131.1819±150.13326 |
<0.001 | 0.800 | 8.2765 | 0.762 | 0.886 |
| RCC (124) |
173.4632±161.66956 |
<0.001 | 0.812 | 9.4985 | 0.71 | 0.84 |
| Serum samples |
|
|
|
|
|
|
| HC (198) | 1.6221±3.45691 |
|
|
|
|
|
| BCa (122) | 3.4660±8.78510 | 0.009 | 0.691 | 1.2385 | 0.713 | 0.561 |
| RCC (208) | 2.8443±7.12991 | 0.028 | 0.600 | 1.1625 | 0.620 | 0.475 |
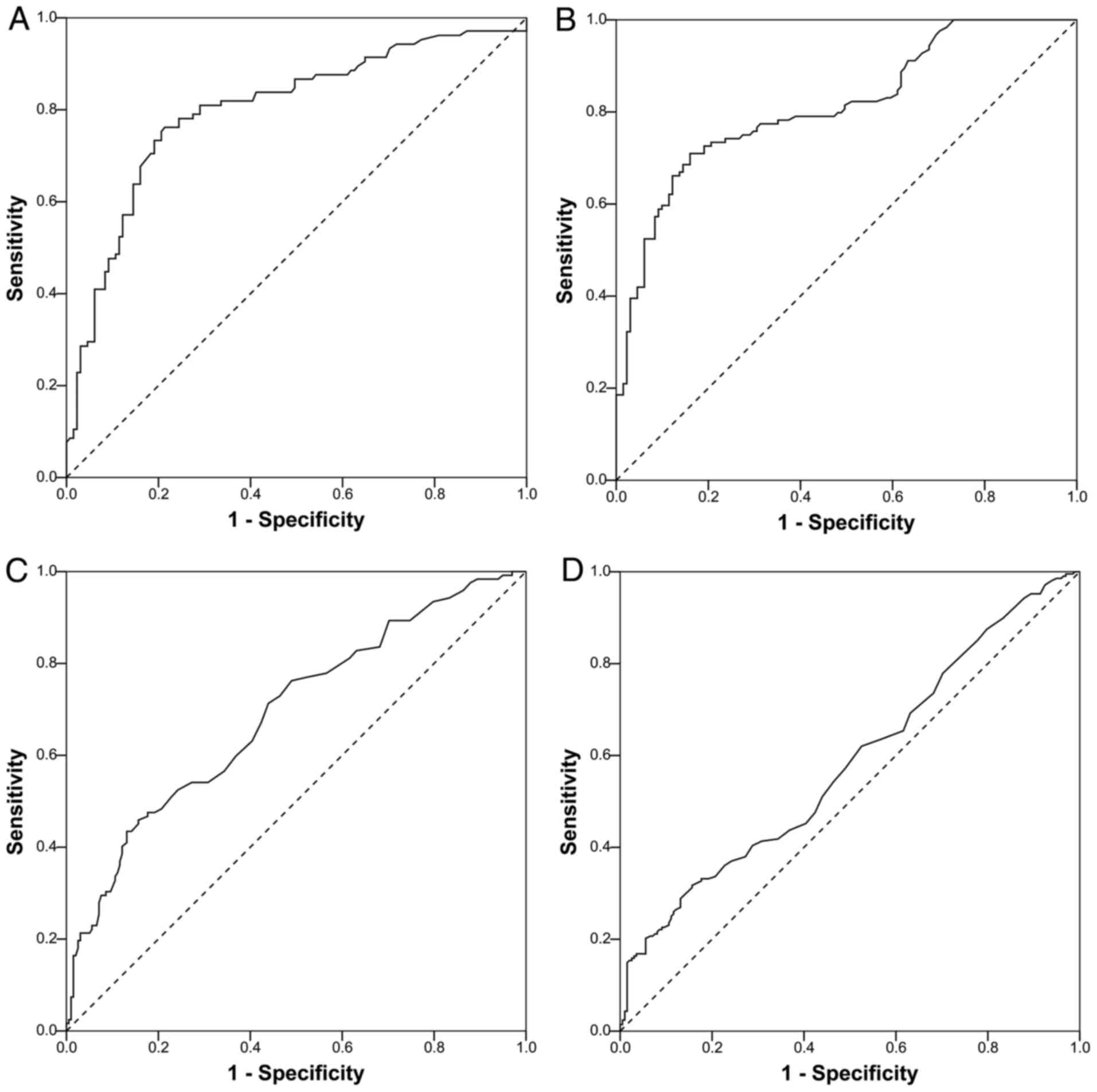
ROC curves based on the detection of survivin in
urine and serum samples from cancer patients and HCs are shown in
Fig. 3. The AUCs were 0.800, 0.812,
0.691 and 0.600, respectively, in BCa urine, RCC urine, BCa serum
and RCC serum samples. According to the optimal Youden's index,
cut-off values of 8.2765 and 9.4985 ng/ml in urine samples were
proposed for BCa and RCC, respectively, corresponding to
sensitivity values of 76.20 and 71.00%, and specificity values of
88.60 and 84.00%. In BCa and RCC serum samples, cut-off values of
1.2385 and 1.1625 ng/ml, respectively, resulted in sensitivity
values of 71.3 and 62.00%, and specificity values of 56.10 and
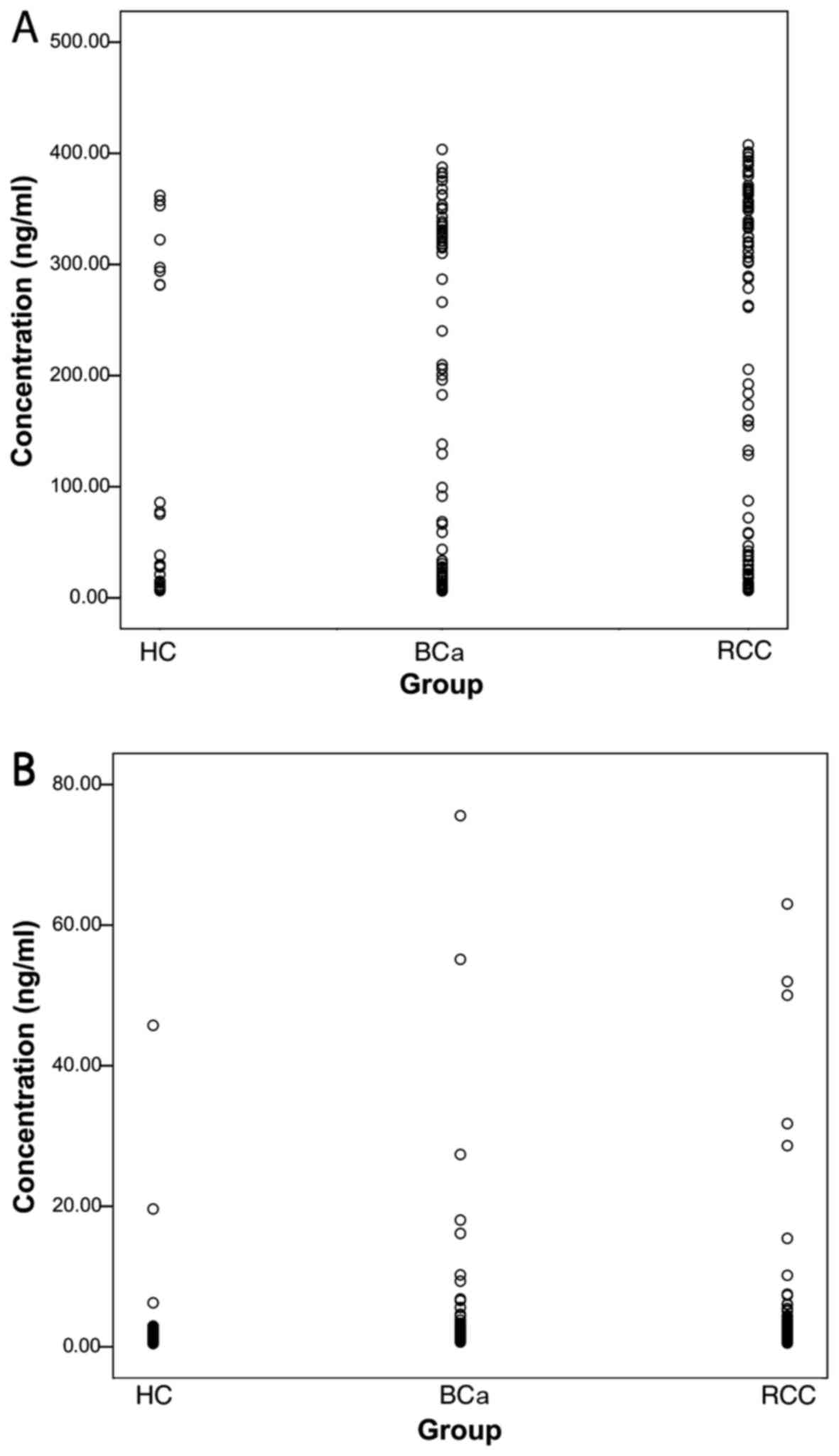
47.50% (Table II). The scatter
plot showing the survivin concentrations in samples from BCa and
RCC patients and HCs is shown in Fig.
4.
Survivin concentrations higher than the cut-off
value were defined as positive. Among the 50 positive urine samples
from patients with BCa, 39 (78%) of their corresponding serum
samples were also positive, while in RCC, 27 serum samples (41.54%)
were positive out of the 65 patients with positive urine samples.
This indicated that survivin concentration in urine was fairly
consistent with that in serum. No significant differences in the
expression of survivin were observed between patients with primary
and recurrent BCa (Tables III and
V). Before and after surgery,
survivin concentration also showed no significant differences in
BCa or RCC patients (Tables
III–VI).
 | Table III.Correlation between the level of
survivin in urine and clinicopathological characteristics of the
BCa patients. |
Table III.
Correlation between the level of
survivin in urine and clinicopathological characteristics of the
BCa patients.
| Clinicopathological
characteristics | n | Survivin level
[mean ± SD (ng/ml)] |
P-valuea |
|---|
| Sex |
|
|
|
|
Male | 68 |
147.533±150.6453 | 0.2308 |
|
Female | 34 |
109.4359±150.0653 |
|
| Age (years) |
|
|
|
|
≤60 | 44 |
131.6626±158.9245 | 0.8543 |
|
>60 | 58 |
137.2398±145.6814 |
|
| Tumor number |
|
|
|
| 1 | 36 |
134.1215±151.2537 | 0.8094 |
| ≥2 | 29 |
143.405±156.5502 |
|
| Tumor size
(mm) |
|
|
|
|
≤30 | 48 |
152.9562±156.843 | 0.5104 |
|
>30 | 25 |
127.7777±149.2811 |
|
| Primary or not |
|
|
|
|
Primary | 70 |
139.0455±154.1364 | 0.775 |
|
Recurrent | 27 |
129.2186±143.5481 |
|
| Primary |
|
|
|
|
Preoperation | 29 |
131.741±160.0746 | 0.7414 |
|
Postoperation | 41 |
144.212±151.591 |
|
| Recurrent |
|
|
|
|
Preoperation | 8 |
181.713±144.7057 | 0.3015 |
|
Postoperation | 17 |
115.344±147.2315 |
|
| Tumor grade |
|
|
|
|
G1-G2 | 20 |
151.8126±156.8784 | 0.5698 |
| G3 | 68 |
129.7054±151.0077 |
|
| Tumor thrombus |
|
|
|
|
Visible | 12 |
116.5475±126.4072 | 0.4722 |
|
Invisible | 23 |
155.526±161.2349 |
|
| Nodal status |
|
|
|
|
Positive | 14 |
102.4099±141.4549 | 0.4438 |
|
Negative | 91 |
135.6084±151.6818 |
|
| Tumor stage |
|
|
|
|
<pT2 | 25 |
170.8168±167.305 | 0.1397 |
|
≥pT2 | 50 |
116.1421±139.9187 |
|
| TNM stage |
|
|
|
|
I–II | 27 |
155.4603±161.8041 | 0.2152 |
|
III–IV | 32 |
107.7036±130.888 |
|
| NPM22 |
|
|
|
|
Positive | 14 |
154.2549±166.2502 | 0.7092 |
|
Negative | 29 |
174.1876±161.573 |
|
| Smoking status |
|
|
|
|
Yes | 33 |
164.6349±159.3603 | 0.2442 |
| No | 56 |
125.2063±149.5392 |
|
| Hypertension |
|
|
|
|
Yes | 37 |
151.438±160.473 | 0.5581 |
| No | 52 |
131.9741±149.1533 |
|
 | Table V.Corrrelation between the level of
survivin in serum and the clinicopathological characteristics of
the BCa patients. |
Table V.
Corrrelation between the level of
survivin in serum and the clinicopathological characteristics of
the BCa patients.
| Clinicopathological
characteristics | n | Survivin level
[mean ± SD (ng/ml)] |
P-valuea |
|---|
| Sex |
|
|
|
|
Male | 96 |
3.696406±9.550458 | 0.5799 |
|
Female | 26 |
2.615346±5.097275 |
|
| Age (years) |
|
|
|
|
≤60 | 51 |
2.582745±2.824331 | 0.2792 |
|
>60 | 71 |
4.100479±11.25758 |
|
| Tumor number |
|
|
|
| 1 | 46 |
4.352109±11.68199 | 0.2803 |
| ≥2 | 44 |
2.414568±2.853097 |
|
| Tumor size
(mm) |
|
|
|
|
≤30 | 65 |
3.578169±9.72505 | 0.3181 |
|
>30 | 37 |
2.277784±2.857489 |
|
| Primary or not |
|
|
|
|
Primary | 95 |
3.238726±9.413477 | 0.42 |
|
Recurrent | 24 |
4.5705±6.482018 |
|
| Primary |
|
|
|
|
Preoperation | 38 |
5.188211±14.65777 | 0.1816 |
|
Postoperation | 57 |
1.93907±1.578393 |
|
| Tumor grade |
|
|
|
|
G1-G2 | 20 |
6.2087±16.74663 | 0.3343 |
| G3 | 91 |
2.483747±3.440776 |
|
| Tumor thrombus |
|
|
|
|
Visible | 19 |
3.250842±3.698517 | 0.1065 |
|
Invisible | 26 |
1.769385±1.140501 |
|
| Nodal status |
|
|
|
|
Positive | 21 |
2.638762±3.288208 | 0.223 |
|
Negative | 18 |
1.716389±0.6870672 |
|
| Tumor grade |
|
|
|
|
<pT2 | 24 |
2.504208±5.329035 | 0.5181 |
|
≥pT2 | 70 |
3.5015±9.065315 |
|
| TNM stage |
|
|
|
|
I–II | 61 |
3.486115±10.05631 | 0.2152 |
|
III–IV | 35 |
2.697457±2.812385 |
|
| Smoking status |
|
|
|
|
Yes | 42 |
2.236857±3.004294 | 0.2349 |
| No | 71 |
3.682845±9.411087 |
|
| Hypertension |
|
|
|
|
Yes | 42 |
2.595571±4.7085 | 0.5244 |
| No | 73 |
3.406726±8.902606 |
|
 | Table VI.Correlation between the level of
survivin in serum and the clinicopathological characteristics of
the RCC patients. |
Table VI.
Correlation between the level of
survivin in serum and the clinicopathological characteristics of
the RCC patients.
| Clinicopathological
characteristics | n | Survivin level
[mean ± SD (ng/ml)] |
P-valuea |
|---|
| Sex |
|
|
|
|
Male | 132 |
3.238803±8.425232 | 0.2117 |
|
Female | 76 |
2.159039±3.945889 |
|
| Age (years) |
|
|
|
|
≤50 | 53 |
3.308321±7.91286 | 0.5843 |
|
>50 | 155 |
2.6856±6.862163 |
|
| TNM stage |
|
|
|
|
I–II | 63 | 3.338±9.992344 | 0.6743 |
|
III–IV | 87 |
2.740908±6.052562 |
|
| Fuhrman grade |
|
|
|
|
I–II | 109 |
3.31945±8.594069 | 0.08494 |
|
III–IV | 56 |
1.84625±1.528992 |
|
| Histologic
category |
|
|
|
| Clear
cell | 24 |
2.77989±6.906926 | 0.7456 |
|
Other | 172 |
3.211257±8.32561 |
|
| Tumor size
(mm) |
|
|
|
|
≤50 | 98 |
3.723214±9.906343 | 0.1146 |
|
>50 | 80 |
2.036437±3.158853 |
|
| Tumor thrombus |
|
|
|
|
Visible | 27 |
3.507778±9.377017 | 0.5324 |
|
Invisible | 139 |
2.951094±7.665209 |
|
| Smoking status |
|
|
|
|
Yes | 54 |
2.6185±3.955068 | 0.6389 |
| No | 136 |
3.042706±8.449693 |
|
| Hypertension |
|
|
|
|
Yes | 68 |
1.542221±0.9502749 | 0.01248 |
| No | 128 |
3.605889±9.051269 |
|
The associations between the expression of survivin
and the clinicopathological characteristics of BCa and RCC patients
were analyzed by Student's t-test. No associations were identified,
except association between hypertension and the presence of
survivin in the serum of RCC patients was found (P=0.012) (Tables III–VI).
In addition, previous studies have reported on the
use of nuclear matrix protein 22 (NMP22) in the diagnosis of BCa
(39,40). In the present study, no association
between NMP22 and survivin level was found (Table III).
Expression of survivin in cancer cell
lines
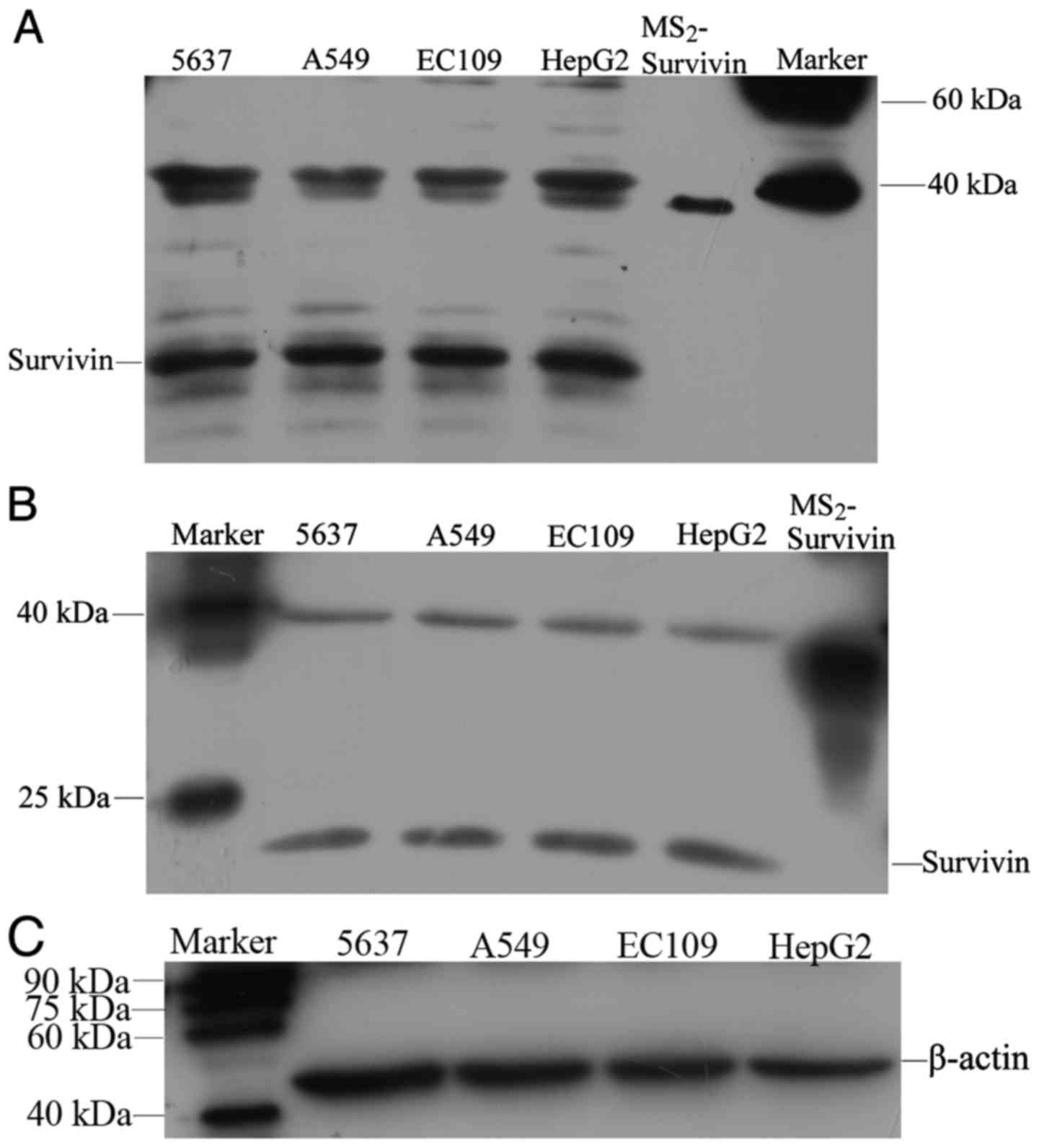
Western blotting was applied to determine whether
survivin was expressed in cancer cell lines and whether the
HRP-conjugated mAbs produced in the present study could be used to
detect survivin. The western blotting results indicated that
standard MS2-survivin was detectable as a 30-kDa band,
while survivin in the cell lines was observed as a 16.5-kDa band
and β-actin as a 42-kDa band (Fig.
5C). 5637, A549, EC109 and HepG2 cells all expressed survivin,
and the positive signals detected by the HRP-conjugated mAb
(Fig. 5A) were consistent with
those detected by the commercial D8 antibody (Santa Cruz
Biotechnology, Inc.) (Fig. 5B). In
addition, bands at 30–40 kDa were present in all of the cell lines
with both mAbs, which may represent heterodimers or aggregates of
survivin (Fig. 5A and B).
Survivin expression in human BCa
tissue
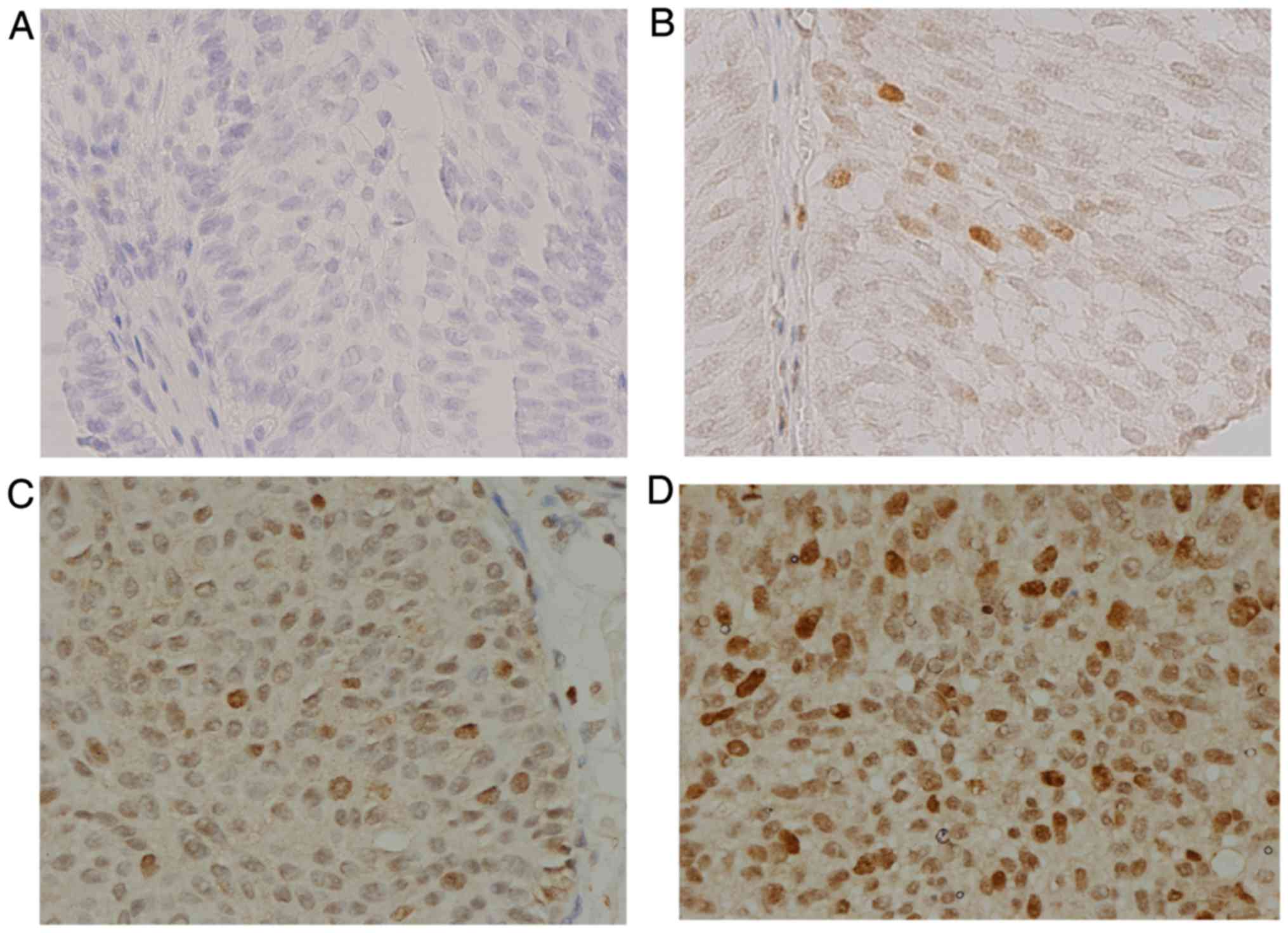
In order to verify that survivin mAbs could identify
survivin expression in human tissues, immunohistochemistry was used
to detect survivin expression in BCa tissue, with the D8 antibody
(Santa Cruz Biotechnology, Inc.) used as a control. Among 10 BCa
samples, all samples displayed positive staining of survivin
protein in the cancer cells at different expression levels using
both the survivin mAbs and the D8 antibody (Santa Cruz
Biotechnology, Inc.). The results revealed that survivin was
distributed in the nuclei and cytoplasm of BCa cells, although
predominantly in the cell nuclei. The intensity of immunostaining
with the survivin mAbs was weak/(+) in 1 case (10%), moderate/(++)
in 4 cases (40%), and strong/(+++) in 5 cases (50%) (Fig. 6), whereas 7 (70%) and 3 (30%) cases
showed moderate/(++) and strong/(+++) staining, respectively, with
the D8 antibody (Santa Cruz Biotechnology, Inc.). The corresponding
urine and serum samples of the 1 weak/(+) positive BCa tissue were
both negative on ELISA. In the 9 patients with cancer tissues
expressing moderate or strong survivin levels, the following
results were observed: the serum samples of 4 patients were not
collected, while their urine samples were all positive for survivin
on ELISA; in 3 of the patients, both urine and serum samples were
positive on ELISA; and in the remaining 2 patients, urine samples
were positive and serum samples were negative on ELISA.
Further findings suggested a positive correlation
between the intensity of immunostaining and tumor grade (G1, G2,
G3). Among 4 patients with tumor grade G2, the intensity of
immunostaining was weak/(+) in 1 and moderate/(++) in 3; whereas,
among 6 patients with tumor grade G3, 2 exhibited moderate/(++) and
4 exhibited strong/(+++) immunostaining.
Discussion
Survivin has been shown to have significance in
clinical applications. Recent studies have demonstrated the
diagnostic role of survivin in urogenital and urinary bladder
cancer (41–43), and survivin overexpression may be an
important prognostic factor for recurrence in certain cancers
(44–46). Serum survivin levels before and
during chemotherapy may serve as a predictive biomarker for the
treatment response in malignant mesothelioma (47). Furthermore, studies have also shown
that survivin mediates multidrug resistance and reduces apoptosis
(48,49). In recent years, a number of studies
have focused on targeting survivin as a therapeutic strategy, which
has included the use of small-molecule inhibitors and
peptidomimetics (YM155, shepherdin) (50,51),
transcriptional inhibitors such as survivin antisense
oligonucleotides (LY2181308, EZN-3042) (52,53),
gene therapy and immunotherapy (54). Many studies have also investigated
the mechanism of action of survivin. The BIR domain of survivin
interferes with caspase-3 and −7 and induces inhibition of
apoptosis (21). Survivin can
interact with the cell cycle regulator Cdk4, leading to Cdk2/cyclin
E activation and Rb phosphorylation (55). Survivin overexpression also
activates NF-κB p65, which is important for the acquisition and
maintenance of the oncogenic characteristics of cancer (56). In addition, the HER2-STAT3-survivin
axis could serve as a predictive marker and therapeutic target to
overcome radiotherapy resistance in HER2-positive breast cancer
(57). However, further
investigations are still required to fully elucidate the role of
survivin in different types of cancers.
Previous studies have demonstrated that a
detrimental feature of bladder cancer is its high recurrence rate,
which necessitates frequent surveillance imaging and repetitive
transurethral resections (58). In
the present study, using a sandwich ELISA method developed with E6
and HRP-C6 antibodies, survivin expression in both urine and serum
samples was demonstrated to be significantly higher in patients
with bladder cancer or renal cell carcinoma than that noted in
healthy controls, and this difference was more pronounced in urine
samples. In both bladder cancer and renal cell carcinoma patients,
survivin expression showed no significant differences in primary
vs. recurrent cancer or before vs. after surgery. These results
implicate survivin as a potential tumor marker for the diagnosis
and prognosis of bladder cancer or renal cell carcinoma. In
addition, hypertension is a significant risk factor for renal cell
carcinoma. Several studies have shown a dose-dependent increase in
renal cell carcinoma with increasing blood pressure level (59,60),
and the present study demonstrated that the expression of survivin
in the serum of renal cell carcinoma patients was associated with
hypertension.
It has been shown that different splice variants of
survivin give rise to distinct protein isoforms: survivin-2B and
survivin-ΔEx3 retain anti-apoptotic activity (8); survivin-3B exerts cytoprotective
functions (9); and survivin-2α is
not assumed to exert any anti-apoptotic activity (10). The expression levels of the five
survivin splice variants were all significantly higher in cancer
tissues compared with these levels in normal tissues in previous
studies (61,62). In the present study, western
blotting was used to assess survivin expression in the cancer cell
lines 5637, A549, EC109 and HepG2. A band at 30–40 kDa was detected
using both the HRP-conjugated mAbs generated in our laboratory and
the commercial antibody purchased from Santa Cruz; this band was
assumed to represent heterodimers or aggregates of survivin.
Previous studies have shown that, in the case of wt survivin, ~94%
of wt survivin consisted of dimers containing some monomers, and
the remaining 6% of wt survivin consisted of large aggregates
(63). Monomers in mammalian cells
can form heterodimers by binding to other proteins, such as CRM1
(63), and survivin splice variants
may also heterodimerize with survivin to regulate its functions
(64,65).
Previous studies have shown that survivin
localization in cells is consistent with its function in cell
division (nucleus) and cell viability (cytoplasm), as well as
confirming the presence of different isoforms which had distinct
cellular localizations (66).
Immunohistochemical analysis in the present study illustrated that
survivin was distributed in the nucleus and cytoplasm of bladder
cancer cells, although predominantly in the cell nucleus. The
expression of survivin in tissues may be consistent with that in
urine and serum. Previous studies have found that the presence of
nuclear survivin may be an independent biomarker for disease
recurrence and overall survival in cancer patients (67,68).
In post-chemoradiotherapy tissues, nuclear survivin expression
disappeared completely and cytoplasmic expression increased,
particularly in treatment-responsive patients (69). A positive correlation between the
intensity of immunostaining and tumor grade (G1, G2, G3) was found
in the present study, which further confirmed the role of survivin
in tumors.
In conclusion, the sandwich ELISA established in the
present study had high sensitivity and specificity for the
detection of survivin expression. Survivin expression in urine and
serum samples from bladder cancer and renal cell carcinoma patients
was significantly higher than that in healthy controls. Western
blotting of cancer cell lines with HRP-conjugated mAbs and
immunohistochemistry of cancer tissues confirmed survivin
expression in bladder cancer. Our study further suggests that
survivin is a potential tumor marker for the surveillance of
bladder cancer and renal cell carcinoma. The availability of these
survivin mAbs would be of use in a wide range of studies on
survivin.
Acknowledgements
The authors thank Spandidos Publications for their
assistance with language editing of our manuscript.
Funding
This study was supported by the Capital Laboratory
Medicine Clinical Characteristic Fund (no. Z121107005112004).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
QZ conceived and designed the study. DC and JX
performed the experiments. DC wrote the paper. QZ and DC reviewed
and edited the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All study procedures were in accordance with the
Helsinki Declaration and the study was approved by the Ethics
Committee of Peking University Cancer Hospital and Institute. The
animal experiments were approved by the Animal Care Committee of
Peking University and conformed to the guidelines of the National
Institutes of Health.
Consent for publication
Each patient and healthy examinee provided written
informed consent for the publication of any associated data and
accompanying images.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar
|
|
2
|
Deveraux QL and Reed JC: IAP family
proteins - suppressors of apoptosis. Genes Dev. 13:239–252. 1999.
View Article : Google Scholar
|
|
3
|
Miller LK: An exegesis of IAPs: Salvation
and surprises from BIR motifs. Trends Cell Biol. 9:323–328. 1999.
View Article : Google Scholar
|
|
4
|
Li F, Ambrosini G, Chu EY, Plescia J,
Tognin S, Marchisio PC and Altieri DC: Control of apoptosis and
mitotic spindle checkpoint by survivin. Nature. 396:580–584. 1998.
View Article : Google Scholar
|
|
5
|
Altieri DC and Marchisio PC: Survivin
apoptosis: An interloper between cell death and cell proliferation
in cancer. Lab Invest. 79:1327–1333. 1999.
|
|
6
|
Li F, Ackermann EJ, Bennett CF, Rothermel
AL, Plescia J, Tognin S, Villa A, Marchisio PC and Altieri DC:
Pleiotropic cell-division defects and apoptosis induced by
interference with survivin function. Nat Cell Biol. 1:461–466.
1999. View Article : Google Scholar
|
|
7
|
Chantalat L, Skoufias DA, Kleman JP, Jung
B, Dideberg O and Margolis RL: Crystal structure of human survivin
reveals a bow tie-shaped dimer with two unusual alpha-helical
extensions. Mol Cell. 6:183–189. 2000. View Article : Google Scholar
|
|
8
|
Mahotka C, Wenzel M, Springer E, Gabbert
HE and Gerharz CD: Survivin-deltaEx3 and survivin-2B: Two novel
splice variants of the apoptosis inhibitor survivin with different
antiapoptotic properties. Cancer Res. 59:6097–6102. 1999.
|
|
9
|
Badran A, Yoshida A, Ishikawa K, Goi T,
Yamaguchi A, Ueda T and Inuzuka M: Identification of a novel splice
variant of the human anti-apoptopsis gene survivin. Biochem Biophys
Res Commun. 314:902–907. 2004. View Article : Google Scholar
|
|
10
|
Caldas H, Honsey LE and Altura RA:
Survivin 2alpha: A novel Survivin splice variant expressed in human
malignancies. Mol Cancer. 4:112005. View Article : Google Scholar
|
|
11
|
Reed JC: The Survivin saga goes in vivo. J
Clin Invest. 108:965–969. 2001. View
Article : Google Scholar
|
|
12
|
Satoh K, Kaneko K, Hirota M, Masamune A,
Satoh A and Shimosegawa T: Expression of survivin is correlated
with cancer cell apoptosis and is involved in the development of
human pancreatic duct cell tumors. Cancer. 92:271–278. 2001.
View Article : Google Scholar
|
|
13
|
Tanaka C, Uzawa K, Shibahara T, Yokoe H,
Noma H and Tanzawa H: Expression of an inhibitor of apoptosis,
survivin, in oral carcinogenesis. J Dent Res. 82:607–611. 2003.
View Article : Google Scholar
|
|
14
|
Dallaglio K, Marconi A and Pincelli C:
Survivin: A dual player in healthy and diseased skin. J Invest
Dermatol. 132:18–27. 2012. View Article : Google Scholar
|
|
15
|
Dohi T, Beltrami E, Wall NR, Plescia J and
Altieri DC: Mitochondrial survivin inhibits apoptosis and promotes
tumorigenesis. J Clin Invest. 114:1117–1127. 2004. View Article : Google Scholar
|
|
16
|
Fortugno P, Wall NR, Giodini A, O'Connor
DS, Plescia J, Padgett KM, Tognin S, Marchisio PC and Altieri DC:
Survivin exists in immunochemically distinct subcellular pools and
is involved in spindle microtubule function. J Cell Sci.
115:575–585. 2002.
|
|
17
|
Dohi T, Okada K, Xia F, Wilford CE, Samuel
T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, et al: An
IAP-IAP complex inhibits apoptosis. J Biol Chem. 279:34087–34090.
2004. View Article : Google Scholar
|
|
18
|
Khan S, Jutzy JM, Aspe JR, McGregor DW,
Neidigh JW and Wall NR: Survivin is released from cancer cells via
exosomes. Apoptosis. 16:1–12. 2011. View Article : Google Scholar
|
|
19
|
Wright ME, Han DK and Hockenbery DM:
Caspase-3 and inhibitor of apoptosis protein(s) interactions in
Saccharomyces cerevisiae and mammalian cells. FEBS Lett.
481:13–18. 2000. View Article : Google Scholar
|
|
20
|
Song Z, Yao X and Wu M: Direct interaction
between survivin and Smac/DIABLO is essential for the
anti-apoptotic activity of survivin during taxol-induced apoptosis.
J Biol Chem. 278:23130–23140. 2003. View Article : Google Scholar
|
|
21
|
Tamm I, Wang Y, Sausville E, Scudiero DA,
Vigna N, Oltersdorf T and Reed JC: IAP-family protein survivin
inhibits caspase activity and apoptosis induced by Fas (CD95), Bax,
caspases, and anticancer drugs. Cancer Res. 58:5315–5320. 1998.
|
|
22
|
Chen J, Wu W, Tahir SK, Kroeger PE,
Rosenberg SH, Cowsert LM, Bennett F, Krajewski S, Krajewska M,
Welsh K, et al: Down-regulation of survivin by antisense
oligonucleotides increases apoptosis, inhibits cytokinesis and
anchorage-independent growth. Neoplasia. 2:235–241. 2000.
View Article : Google Scholar
|
|
23
|
Olie RA, Simões-Wüst AP, Baumann B, Leech
SH, Fabbro D, Stahel RA and Zangemeister-Wittke U: A novel
antisense oligonucleotide targeting survivin expression induces
apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer
Res. 60:2805–2809. 2000.
|
|
24
|
Chandele A, Prasad V, Jagtap JC, Shukla R
and Shastry PR: Upregulation of survivin in G2/M cells and
inhibition of caspase 9 activity enhances resistance in
staurosporine-induced apoptosis. Neoplasia. 6:29–40. 2004.
View Article : Google Scholar
|
|
25
|
Suzuki A, Hayashida M, Ito T, Kawano H,
Nakano T, Miura M, Akahane K and Shiraki K: Survivin initiates cell
cycle entry by the competitive interaction with Cdk4/p16(INK4a) and
Cdk2/cyclin E complex activation. Oncogene. 19:3225–3234. 2000.
View Article : Google Scholar
|
|
26
|
Jeyaprakash AA, Klein UR, Lindner D, Ebert
J, Nigg EA and Conti E: Structure of a Survivin-Borealin-INCENP
core complex reveals how chromosomal passengers travel together.
Cell. 131:271–285. 2007. View Article : Google Scholar
|
|
27
|
D'Avino PP and Capalbo L: New Auroras on
the roles of the chromosomal passenger complex in cytokinesis:
Implications for cancer therapies. Front Oncol. 5:2212015.
|
|
28
|
Li Y, Liu D, Zhou Y, Li Y, Xie J, Lee RJ,
Cai Y and Teng L: Silencing of survivin expression leads to reduced
proliferation and cell cycle arrest in cancer cells. J Cancer.
6:1187–1194. 2015. View Article : Google Scholar
|
|
29
|
Hagenbuchner J, Kiechl-Kohlendorfer U,
Obexer P and Ausserlechner MJ: BIRC5/Survivin as a target for
glycolysis inhibition in high-stage neuroblastoma. Oncogene.
35:2052–2061. 2016. View Article : Google Scholar
|
|
30
|
Véquaud E, Séveno C, Loussouarn D,
Engelhart L, Campone M, Juin P and Barillé-Nion S: YM155 potently
triggers cell death in breast cancer cells through an
autophagy-NF-kB network. Oncotarget. 6:13476–13486. 2015.
View Article : Google Scholar
|
|
31
|
Ding YH, Fan XD, Wu JJ, Deng ZK, Wei B and
Li YF: Effect of YM155 on Apoptosis and Autophagy of K562 Cells.
Zhongguo Shi Yan Xue Ye Xue Za Zhi. 23:375–380. 2015.(In
Chinese).
|
|
32
|
Jiang G, Ren B, Xu L, Song S, Zhu C and Ye
F: Survivin may enhance DNA double-strand break repair capability
by up-regulating Ku70 in human KB cells. Anticancer Res.
29:223–228. 2009.
|
|
33
|
Véquaud E, Desplanques G, Jézéquel P, Juin
P and Barillé-Nion S: Survivin contributes to DNA repair by
homologous recombination in breast cancer cells. Breast Cancer Res
Treat. 155:53–63. 2016. View Article : Google Scholar
|
|
34
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
35
|
Jia X, Gao Y, Zhai D, Liu J, Wang Y, Jing
LI and Du Z: Survivin is not a promising serological maker for the
diagnosis of hepatocellular carcinoma. Oncol Lett. 9:2347–2352.
2015. View Article : Google Scholar
|
|
36
|
You WANG, Qing-yun ZHANG, Ya-ming WANG and
Jian-jun XU: Cloning of survivin gene and preparation its
monoclonoal antibodies as well as checking survivin expression in
liver carcinoma cells. Clin J Lab Med. 29:258–262. 2006.
|
|
37
|
Li X, Wang Y, Xu J and Zhang Q: Sandwich
ELISA for detecting urinary Survivin in bladder cancer. Chin J
Cancer Res. 25:375–381. 2013.
|
|
38
|
Liu C, Guo J, Qu L, Bing D, Meng L, Wu J
and Shou C: Applications of novel monoclonal antibodies specific
for synuclein-gamma in evaluating its levels in sera and cancer
tissues from colorectal cancer patients. Cancer Lett. 269:148–158.
2008. View Article : Google Scholar
|
|
39
|
Jamshidian H, Kor K and Djalali M: Urine
concentration of nuclear matrix protein 22 for diagnosis of
transitional cell carcinoma of bladder. Urol J. 5:243–247.
2008.
|
|
40
|
Önal B, Han Ü, Yilmaz S, Köybasioglu F and
Altuğ U: The use of urinary nuclear matrix protein 22 (NMP22) as a
diagnostic adjunct to urine cytology for monitoring of recurrent
bladder cancer-institutional experience and review. Diagn
Cytopathol. 43:307–314. 2015. View
Article : Google Scholar
|
|
41
|
Sah NK and Seniya C: Survivin splice
variants and their diagnostic significance. Tumour Biol.
36:6623–6631. 2015. View Article : Google Scholar
|
|
42
|
Srivastava AK, Singh PK, Srivastava K,
Singh D, Dalela D, Rath SK, Goel MM and Bhatt Brahma ML: Diagnostic
role of survivin in urinary bladder cancer. Asian Pac J Cancer
Prev. 14:81–85. 2013. View Article : Google Scholar
|
|
43
|
Eissa S, Swellam M, Shehata H, El-Khouly
IM, El-Zayat T and El-Ahmady O: Expression of HYAL1 and survivin
RNA as diagnostic molecular markers for bladder cancer. J Urol.
183:493–498. 2010. View Article : Google Scholar
|
|
44
|
Plewka D, Jakubiec-Bartnik B, Morek M,
Bogunia E, Bienioszek M, Wolski H, Kotrych D, Dziekan K,
Seremak-Mrozikiewicz A and Plewka A: Survivin in ovary tumors.
Ginekol Pol. 86:525–530. 2015. View Article : Google Scholar
|
|
45
|
Zhang Y, Wang J, Sui X, Li Y, Lu K, Fang
X, Jiang Y and Wang X: Prognostic and clinicopathological value of
survivin in diffuse large B-cell lymphoma: A meta-analysis.
Medicine (Baltimore). 94:e14322015. View Article : Google Scholar
|
|
46
|
Akhtar M, Gallagher L and Rohan S:
Survivin: Role in diagnosis, prognosis, and treatment of bladder
cancer. Adv Anat Pathol. 13:122–126. 2006. View Article : Google Scholar
|
|
47
|
Goričar K, Kovač V, Franko A, Dodič-Fikfak
M and Dolžan V: Serum survivin levels and outcome of chemotherapy
in patients with malignant mesothelioma. Dis Markers.
2015:3167392015. View Article : Google Scholar
|
|
48
|
Yu CJ, Ou JH, Wang ML, Jialielihan N and
Liu YH: Elevated survivin mediated multidrug resistance and reduced
apoptosis in breast cancer stem cells. J BUON. 20:1287–1294.
2015.
|
|
49
|
Lee MR, Ji SY, Mia-Jan K and Cho MY:
Chemoresistance of CD133(+) colon cancer may be related with
increased survivin expression. Biochem Biophys Res Commun.
463:229–234. 2015. View Article : Google Scholar
|
|
50
|
Cheng Q, Ling X, Haller A, Nakahara T,
Yamanaka K, Kita A, Koutoku H, Takeuchi M, Brattain MG and Li F:
Suppression of survivin promoter activity by YM155 involves
disruption of Sp1-DNA interaction in the survivin core promoter.
Int J Biochem Mol Biol. 3:179–197. 2012.
|
|
51
|
Kudchadkar R, Ernst S, Chmielowski B,
Redman BG, Steinberg J, Keating A, Jie F, Chen C, Gonzalez R and
Weber J: A phase 2, multicenter, open-label study of sepantronium
bromide (YM155) plus docetaxel in patients with stage III
(unresectable) or stage IV melanoma. Cancer Med. 4:643–650. 2015.
View Article : Google Scholar
|
|
52
|
Carrasco RA, Stamm NB, Marcusson E,
Sandusky G, Iversen P and Patel BK: Antisense inhibition of
survivin expression as a cancer therapeutic. Mol Cancer Ther.
10:221–232. 2011. View Article : Google Scholar
|
|
53
|
Hansen JB, Fisker N, Westergaard M,
Kjaerulff LS, Hansen HF, Thrue CA, Rosenbohm C, Wissenbach M, Orum
H and Koch T: SPC3042: A proapoptotic survivin inhibitor. Mol
Cancer Ther. 7:2736–2745. 2008. View Article : Google Scholar
|
|
54
|
Coumar MS, Tsai FY, Kanwar JR, Sarvagalla
S and Cheung CH: Treat cancers by targeting survivin: Just a dream
or future reality? Cancer Treat Rev. 39:802–811. 2013. View Article : Google Scholar
|
|
55
|
Li F: Survivin study: What is the next
wave? J Cell Physiol. 197:8–29. 2003. View Article : Google Scholar
|
|
56
|
Zeng W, Li H, Chen Y, Lv H, Liu L, Ran J,
Sun X, Bieerkehazhi S, Liu Y, Li X, et al: Survivin activates NF-κB
p65 via the IKKβ promoter in esophageal squamous cell carcinoma.
Mol Med Rep. 13:1869–1880. 2016. View Article : Google Scholar
|
|
57
|
Kim JS, Kim HA, Seong MK, Seol H, Oh JS,
Kim EK, Chang JW, Hwang SG and Noh WC: STAT3-survivin signaling
mediates a poor response to radiotherapy in HER2-positive breast
cancers. Oncotarget. 7:7055–7065. 2016.
|
|
58
|
Johnson DC, Greene PS and Nielsen ME:
Surgical advances in bladder cancer: At what cost? Urol Clin North
Am. 42:235–252, ix. 2015.ix. View Article : Google Scholar
|
|
59
|
Macleod LC, Hotaling JM, Wright JL,
Davenport MT, Gore JL, Harper J and White E: Risk factors for renal
cell carcinoma in the VITAL study. J Urol. 190:1657–1661. 2013.
View Article : Google Scholar
|
|
60
|
Setiawan VW, Stram DO, Nomura AM, Kolonel
LN and Henderson BE: Risk factors for renal cell cancer: The
multiethnic cohort. Am J Epidemiol. 166:932–940. 2007. View Article : Google Scholar
|
|
61
|
Antonacopoulou AG, Floratou K, Bravou V,
Kottorou A, Dimitrakopoulos FI, Marousi S, Stavropoulos M, Koutras
AK, Scopa CD and Kalofonos HP: The survivin −31 snp in human
colorectal cancer correlates with survivin splice variant
expression and improved overall survival. Anal Cell Pathol (Amst).
33:177–189. 2010. View Article : Google Scholar
|
|
62
|
Ge QX, Li YY, Nie YQ, Zuo WG and Du YL:
Expression of survivin and its four splice variants in colorectal
cancer and its clinical significances. Med Oncol. 30:5352013.
View Article : Google Scholar
|
|
63
|
Muchmore SW, Chen J, Jakob C, Zakula D,
Matayoshi ED, Wu W, Zhang H, Li F, Ng SC and Altieri DC: Crystal
structure and mutagenic analysis of the inhibitor-of-apoptosis
protein survivin. Mol Cell. 6:173–182. 2000. View Article : Google Scholar
|
|
64
|
Caldas H, Jiang Y, Holloway MP, Fangusaro
J, Mahotka C, Conway EM and Altura RA: Survivin splice variants
regulate the balance between proliferation and cell death.
Oncogene. 24:1994–2007. 2005. View Article : Google Scholar
|
|
65
|
Necochea-Campion R, Chen CS, Mirshahidi S,
Howard FD and Wall NR: Clinico-pathologic relevance of Survivin
splice variant expression in cancer. Cancer Lett. 339:167–174.
2013. View Article : Google Scholar
|
|
66
|
Li F, Yang J, Ramnath N, Javle MM and Tan
D: Nuclear or cytoplasmic expression of survivin: What is the
significance? Int J Cancer. 114:509–512. 2005. View Article : Google Scholar
|
|
67
|
Piras F, Murtas D, Minerba L, Ugalde J,
Floris C, Maxia C, Colombari R, Perra MT and Sirigu P: Nuclear
survivin is associated with disease recurrence and poor survival in
patients with cutaneous malignant melanoma. Histopathology.
50:835–842. 2007. View Article : Google Scholar
|
|
68
|
Hou Y, Hu Q, Liu AG, Zhang LQ and Liu SY:
Expression of survivin and its location in bone marrow cells of
childhood acute leukemia: Relationship to therapeutic efficacy.
Zhongguo Dang Dai Er Ke Za Zhi. 8:101–104. 2006.(In Chinese).
|
|
69
|
Takasu C, Shimada M, Kurita N, Iwata T,
Sato H, Nishioka M, Morimoto S, Yoshikawa K, Miyatani T, Kashihara
H, et al: Survivin expression can predict the effect of
chemoradiotherapy for advanced lower rectal cancer. Int J Clin
Oncol. 18:869–876. 2013. View Article : Google Scholar
|