Introduction
Ovarian cancer (OC) is a fatal gynecologic cancer
worldwide accounting for most cancer-associated mortalities in
women with gynecologic cancer (1).
Most patients diagnosed with OC are at advanced stages, leading to
unavailability of curative treatments. Therefore, the long-term
prognosis of OC patients is poor, with a low 5-year overall
survival rate of ~40% (2).
Therefore, investigating the molecular mechanisms underlying the
progression of OC may facilitate the discovery of novel biomarkers
and therapeutic targets of OC.
MicroRNAs (miRNAs), a group of small non-coding RNAs
with ~20 nucleotides (3), can
regulate the expression of target genes in a post-transcriptional
manner by interacting with the 3′-untranslated region (3′-UTR) of
mRNAs (4). miRNAs play versatile
roles in development and disease processes (5). miRNAs have been found to play critical
roles in various human cancers (6,7). In
OC, miRNAs can potentially serve as biomarkers and therapeutic
targets for OC patients (8).
However, the role of specific miRNAs in OC remain to be
investigated. Recently, miR-616 was identified to be a
cancer-associated miRNA. It promoted the migration, invasion and
epithelial-mesenchymal transition (EMT) of hepatocellular carcinoma
through inhibition of PTEN expression (9). In prostate cancer, miR-616 promoted
the growth of prostate cancer by inhibiting tissue factor pathway
inhibitor TFPI-2 (10). In lung
cancer, sulforaphane suppressed the EMT and metastasis of lung
cancer cells through the miR-616-mediated GSK3β/β-catenin signaling
pathways (11). However, the role
of miR-616 in OC and the underlying molecular mechanisms remain
unknown.
In the present study, we demonstrated that the level
of miR-616 in OC was elevated. Compared with patients without
metastasis, OC patients with metastasis had a significantly
increased miR-616 level. An increased level of miR-616 was
associated with poor tumor differentiation, advanced
tumor-node-metastasis (TNM) stages and poor prognosis of OC
patients. Functionally, miR-616 enhanced the ability of the
migration, invasion and EMT of OC cells. Furthermore, our data
revealed that miR-616 interacted with TIMP2 3′-UTR and inhibited
the expression of TIMP2. TIMP2 overexpression inhibited the
promoting effects of miR-616 overexpression on cell metastasis and
EMT while knockdown of TIMP2 reversed the inhibitory effects of
miR-616 knockdown on these cellular functions. Collectively, this
study demonstrated that miR-616 is an oncogenic miRNA in OC and
promotes the progression of OC by enhancing cell metastasis and
EMT.
Materials and methods
Clinical samples and cell culture
Sixty pairs of OC samples and non-tumor tissues were
collected from the Department of Gynecologic Oncology, Zhejiang
Cancer Hospital between January 2008 to December 2011. Only
patients with complete information of clinical features and
complete survival information were included. All clinical tissues
collected in this study were pathologically confirmed as OC.
Informed consent was obtained from every patient enrolled in this
study. The protocol of this study was approved by the Institutional
Research Ethics Committee of Zhejiang Cancer Hospital.
The FTE187 cell line, an immortalized human
fallopian tube epithelial cell line and five types of human OC
cells (A2780, CAOV3, HO-8910, SKOV-3 and ES-2) were purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal
bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), penicillin and streptomycin, were used for
all cell cultures of OC cells. Medium 199 and MCDB105 medium mixed
at a ratio of 1:1 supplemented with FBS and EGF (all from
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), were used for the
culture of FTE187 cells. Cell cultures were maintained in a
humidified incubator with 5% CO2 at 37°C.
Cell transfection
miR-616 mimics, miR-616 inhibitors, control mimics
and negative control inhibitors were obtained from Guangzhou
GeneCopoeia (Guangzhou, China). TIMP2 and the control vector, as
well as TIMP2 siRNA and negative control siRNA were purchased from
Addgene (Cambridge, MA, USA). All vectors or siRNAs were
transfected into OC cells using Lipofectamine® 2000
(Invitrogen, Carlsbad, CA, USA).
Real-time quantitative reverse
transcription-PCR (qRT-PCR)
TRIzol was used to extract the RNA from clinical
samples and OC cells. qRT-PCR assays in the present study were
performed using a miRNA Reverse Transcription kit and a Human miRNA
assay kit (both from Applied Biosystems; Thermo Fisher Scientific,
Inc.). Primers for miR-616, TIMP2, GAPDH and U6 were purchased from
GeneCopoeia. U6 and GAPDH were used as internal controls for
miR-616 and TIMP2, respectively. The primers sequences were as
follows: miR-616 forward, 5′-CTGTGTGCCACACAGTTTG-3′ and reverse,
5′-CGGCCCTTAACTCATTCTTT-3′; U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′
and reverse, 5′-CAGGGGCCATGCTAATCTT-3′; TIMP2 forward,
5′-CTCGGCAGTGTGTGGGGTC-3′ and reverse, 5′-CGAGAAACTCCTGCTTGGGG-3′;
and GAPDH forward, 5′-ATTCCATGGCACCGTCAAGGCTGA-3′ and reverse,
5′-TTCTCCATGGTGGTGAAGACGCCA-3′.
Western blotting
Cellular protein was extracted using RIPA lysis
buffer, and a BCA kit (Pierce; Thermo Fisher Scientific, Inc.) was
employed for the measurement of protein concentration. After being
loaded and separated on 4–20% SDS gels, cellular proteins were
transferred to polyvinylidene fluoride (PVDF) membranes. The
membranes were incubated with the primary antibodies. Primary
antibodies used in this study included TIMP2 (1:1,000; cat. no.
5738; Cell Signaling Technology, Inc., Danvers, MA, USA),
E-cadherin (1:1,000; cat. no. sc-71009; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA), N-cadherin (1:1,000; cat. no. 4061;
Cell Signaling Technology, Inc.) and GAPDH (1:1,500; cat. no.
sc-51631; Santa Cruz Biotechnology, Inc.) overnight at 4°C. Then,
secondary antibodies (1:3,000; cat. no. sc-3744 and cat. no.
sc-2089; Santa Cruz Biotechnology, Inc.) were incubated with
membranes at room temperature for 2 h. The expression level of
detected proteins was visualized using ECL reagents (Amersham
Biosciences Corp., Piscataway, NJ, USA).
Transwell assays
The migratory and invasive abilities of OC were
evaluated by Transwell assay. The day prior to the Transwell assay,
OC cells were starved in serum-free DMEM media overnight. After
trypsinization, OC cells (5×104) were re-suspended in
serum-free DMEM and seeded in the upper chamber of the Transwell
inserts (Millipore, Billerica, MA, USA). The lower chamber of the
Transwell inserts were filled with 700 µl serum-containing DMEM
(20% FBS) as a chemoattractant. For the invasion assays, the upper
Transwell chamber was coated with 100 µl Matrigel (diluted in DMEM
at the ratio of 1:6). Twenty-four hours later, the cells which did
not migrate or invade through the membrane were removed with cotton
swab while the cells that had migrated or invaded through the
membrane were stained with crystal violet. The number of migrated
or invaded OC cells was counted under light microscope.
Immunohistochemical (IHC)
staining
To investigate the relationship between the
expression of miR-616 and E-cadherin, N-cadherin and TIMP2, 10
randomly selected OC tissues with a low miR-616 level and 10 OC
tissues with a high miR-616 level were used to perform IHC
staining. The paraformaldehyde-fixed clinical tissues were
subjected to IHC staining. Paraffin sections (4-µm thickness) were
subjected to deparaffinization and re-hydration through xylene and
graded ethanol. These slides were incubated with 3% hydrogen
peroxide for 10 min to quench the endogenous peroxidase activity.
Then, the slides were blocked with goat serum at room temperature
for 1 h and incubated with the E-cadherin (1:100; cat. no.
sc-71009), N-cadherin (1:100; cat. no. 4061) or TIMP2 antibodies
(1:50; cat. no. 5738) at 4°C overnight. Secondary antibodies (cat.
no. SAP-9101; ZSGB-Bio, Beijing, China) were then incubated with
these slides at room temperature for 1 h. Finally, these sections
were stained with diaminobenzidine, and then hematoxylin. The
intensity of IHC staining was classified into 4 grades: 0, none; 1,
weak; 2, moderate; and 3, strong. The percentage of positive
staining was divided into the 5 grades: 0 (<10%), 1 (10–30%), 2
(30–50%), 3 (51–70%) and 4 (>70%). IHC scores were calculated by
multiplying the percentage of positive cells (P) by the
intensity.
Luciferase reporter assay
A luciferase reporter assay was performed to
investigate whether miR-616 interacted with the 3′-UTR of TIMP2.
The binding sites of miR-616 within the TIMP2 3′-UTR construct in
antisense orientation was further mutated using Q5 Site Directed
Mutagenesis kit (New England Biolabs, Beverly, MA, USA). OC cells
were seeded in 24-well plates at the density of 1–3×105
cells/well. Then, the cells cultured in 24-well plates were
co-transfected with miR-616 mimics or miR-616 inhibitors along with
the wild-type 3′-UTR of TIMP2 or the mutated 3′-UTR of TIMP2, and
pRL-SV40 Renilla plasmid (Promega Corp., Madison, WI, USA).
Cell transfection was performed using Lipofectamine®
2000. Forty-eight hours later, the luciferase activities were
evaluated using the Dual-Luciferase Reporter Assay system (Promega,
Shanghai, China).
In vivo experiments
Tail vein injection experiments were performed in
nude mice to evaluate the in vivo metastatic capacity of OC
cells. OC cells (1×105) transfected with control vector
or miR-616 mimics, or, those transfected with negative control
vector or miR-616 inhibitors, were injected into nude mice through
tail veins. Eight weeks after tail vein injection, the mice were
sacrificed and the lungs were isolated for hematoxylin and eosin
(H&E) staining. The protocols regarding the in vivo
manipulations were approved by the Animal Care Committee of
Zhejiang Cancer Hospital.
Statistical analysis
All data were expressed as the mean ± standard error
of the mean (SEM) and GraphPad Prism 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA) was used for statistical analysis in this study.
After dividing OC patients into two groups based on the cut-off
value defined as the median level of miR-616, differences of the
Kaplan-Meier curves between the miR-616-high group and miR-616-low
group were detected using the log-rank test. P<0.05 was
considered to indicate a statistically significant result.
Results
miR-616 is upregulated in OC tissues
and cell lines
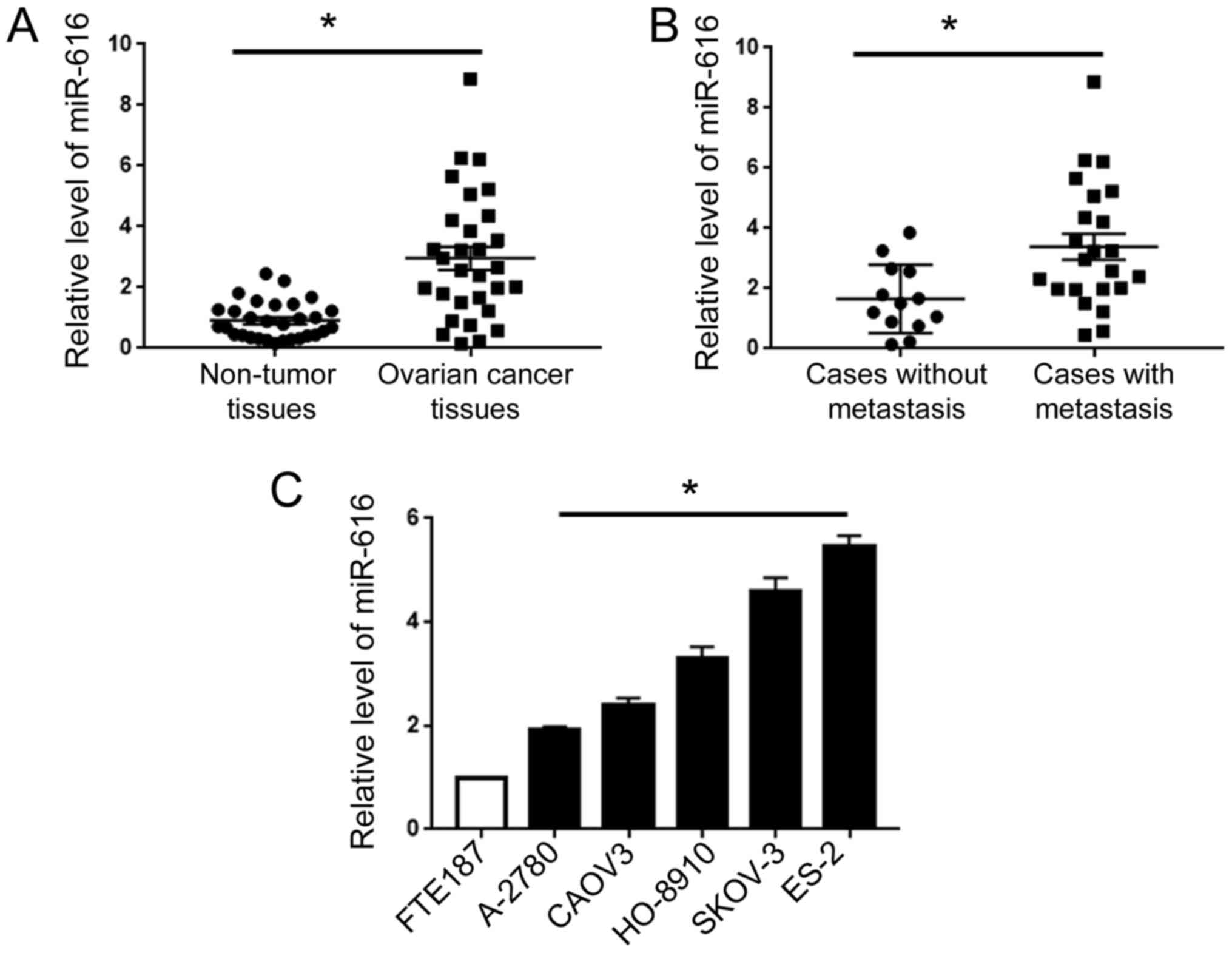
qRT-PCR for clinical tissues revealed that compared
with adjacent non-tumor tissues, OC tissues exhibited an increased
level of miR-616 (P<0.05; Fig.
1A). Then, we compared the level of miR-616 in patients with or
without metastasis. Compared with the patients without metastasis,
patients with metastasis exhibited an elevated level of miR-616
(P<0.05; Fig. 1B). Additionally,
we assessed the expression level of miR-616 in human fallopian tube
epithelial cells (FTE187 cells) and five types of human OC cells
(A2780, CAOV3, HO-8910, SKOV-3 and ES-2). Compared with the FTE187
cells, all five OC cell lines exhibited an increased level of
miR-616, with the highest level in ES-2 cells and the lowest level
in A-2780 cells (P<0.05; Fig.
1C).
An increased miR-616 level confers
poor clinical features and prognosis of OC patients
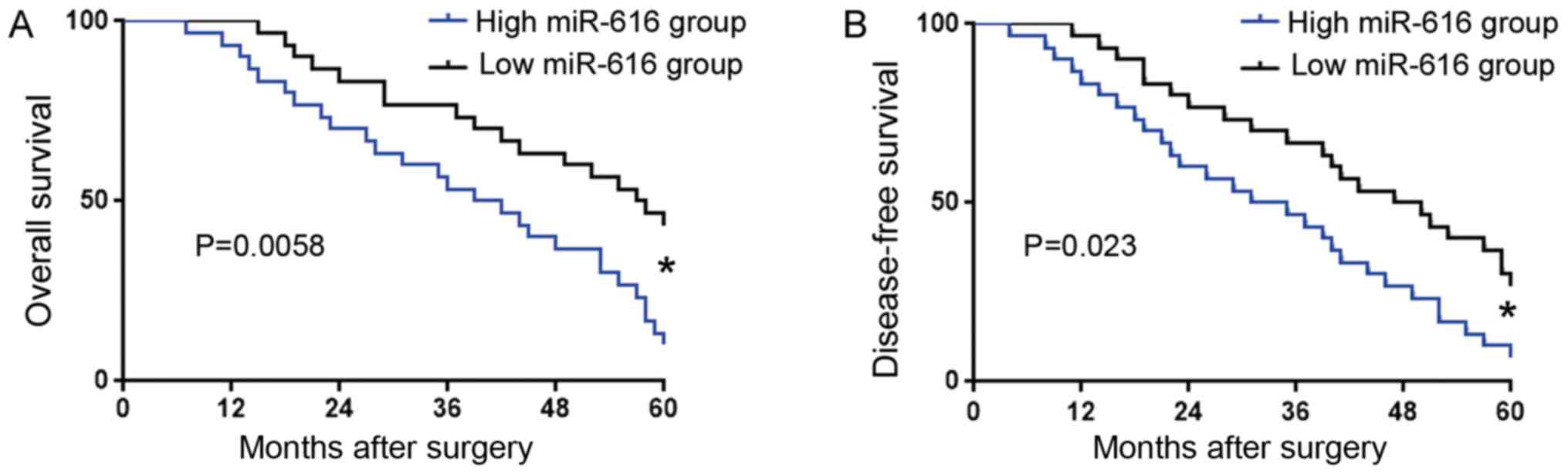
We further investigated the prognostic significance
of miR-616 in OC. OC patients were divided into two groups based on
the cut-off value defined as the median level of miR-616: low
miR-616 group (n=30) and high miR-616 group (n=30). Association
analysis revealed that a high miR-616 level was associated with
poor tumor differentiation (P=0.048) and advanced TNM stage
(P=0.017) (Table I). Kaplan-Meier
analysis (Fig. 2) revealed that
compared with those in the low miR-616 group, patients with a high
miR-616 level had significantly decreased overall survival (OS) and
disease-free survival (DFS). These results revealed that the
expression level of miR-616 could indicate the unfavorable clinical
features and poor prognosis of OC patients.
 | Table I.Association analysis between the
expression level of miR-616 and the clinical features of OC
patients. |
Table I.
Association analysis between the
expression level of miR-616 and the clinical features of OC
patients.
|
| miR-616 expression
level |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
features | No. of patients | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
|
<50 | 25 | 15 | 10 | 0.295 |
| ≥50 | 35 | 15 | 20 |
|
| Histological
type |
|
|
|
|
|
High-grade serous | 36 | 20 | 16 | 0.516 |
| Clear
cell | 12 | 5 | 7 |
|
|
Endometrioid | 7 | 4 | 3 |
|
|
Mucinous | 3 | 1 | 2 |
|
| Low-grade
serous | 2 | 0 | 2 |
|
| Tumor
differentiation |
|
|
|
|
| Low | 33 | 12 | 21 | 0.048 |
|
Moderate | 15 | 9 | 6 |
|
| High | 12 | 9 | 3 |
|
| TNM stage |
|
|
|
|
| I–II | 24 | 17 | 7 | 0.017 |
|
III–IV | 36 | 13 | 23 |
|
miR-616 potentiates the metastatic
ability of OC cells
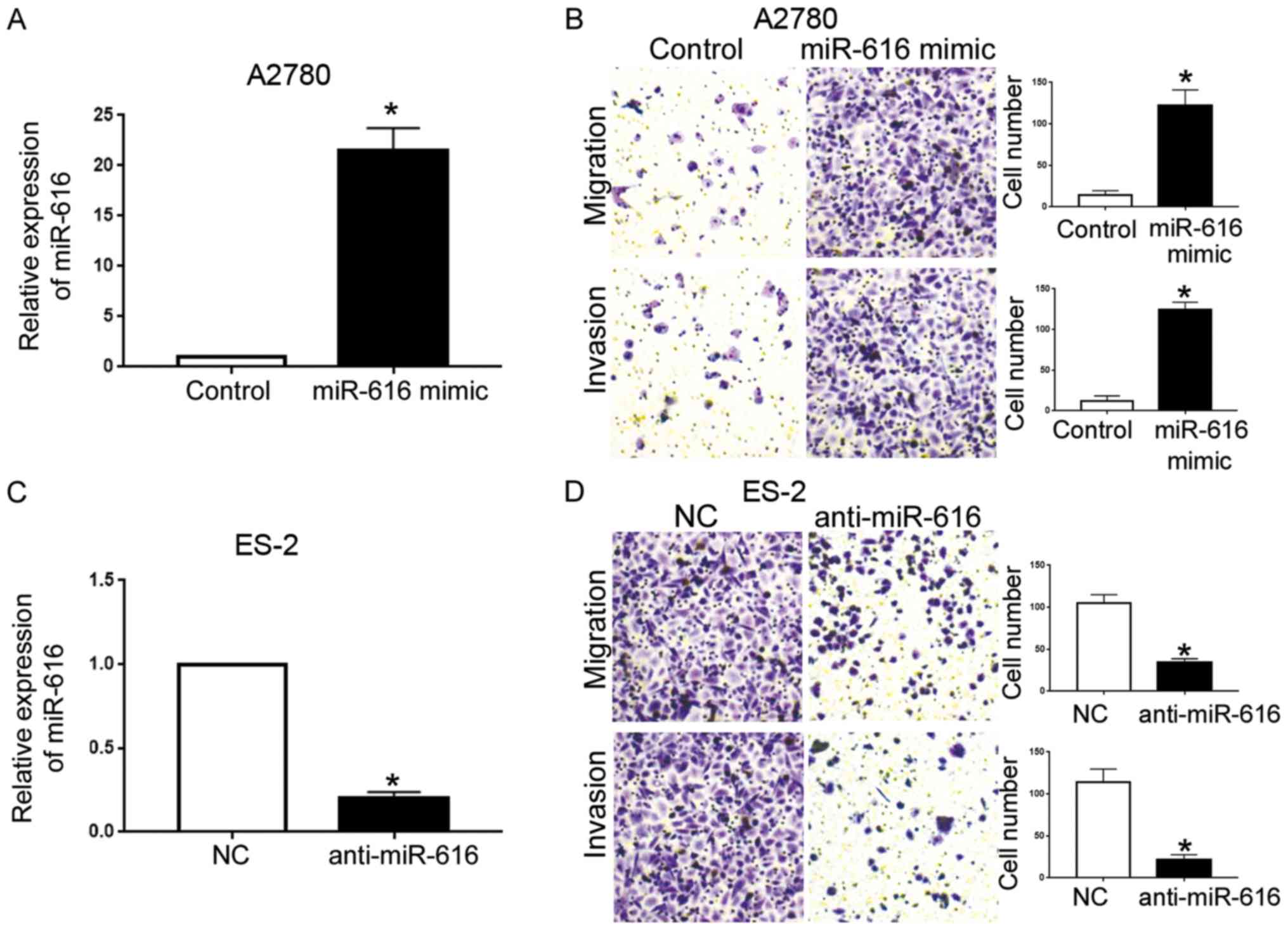
Next, we transfected A2780 cells with miR-616 mimics
to overexpress miR-616, and, transfected ES-2 cells with miR-616
inhibitors to knockdown miR-616. Compared with the control vector,
transfection of miR-616 mimics led to a significantly increased
miR-616 level in A2780 cells (P<0.05; Fig. 3A). Transwell assays further revealed
that forced miR-616 expression in A2780 cells led to increased
migration and invasion of A2780 cells (P<0.05; Fig. 3B). Conversely, transfection of
miR-616 inhibitors effectively knocked down miR-616 in ES-2 cells
(P<0.05; Fig. 3C), and
subsequently resulted in reduced migration and invasion of ES-2
cells (P<0.05; Fig. 3D).
miR-616 promotes the EMT of OC
cells
Since EMT has been widely accepted as an important
mechanism of cancer metastasis (12–14),
we further investigated whether miR-616 regulated EMT of OC cells.
Overexpression of miR-616 in A2780 cells decreased the level of
E-cadherin and increased the level of N-cadherin (P<0.05;
Fig. 4A). Conversely, knockdown of
miR-616 in ES-2 cells led to increased E-cadherin expression and
decreased N-cadherin expression (P<0.05; Fig. 4B). Furthermore, we randomly selected
10 OC tissues with low miR-616 levels and 10 OC tissues with high
miR-616 levels to perform IHC staining for E-cadherin and
N-cadherin, and compared their expression level in OC tissues with
low and high miR-616 levels. Compared with the tissues expressing
low miR-616, tissues expressing high miR-616 exhibited decreased
E-cadherin level and increased N-cadherin level (P<0.05;
Fig. 4C). Collectively, these data
indicated that miR-616 promoted the EMT of OC cells.
miR-616 promotes the lung metastasis
of OC cells in nude mice
To further elucidate the influence of miR-616 on the
in vivo metastatic ability of OC cells, we performed a tail
vein injection assay using A2780 cells overexpressing miR-616 and
ES-2 cells with miR-616 knockdown. Compared with the A2780 cells in
the control group, the tail vein injection of A2780 cells
overexpressing miR-616 resulted in an increased number of lung
metastatic nodules (P<0.05; Fig.
5A). Knockdown of miR-616 in ES-2 cells led to a decreased
number of lung metastasis nodules (P<0.05; Fig. 5B).
TIMP2 is a direct target of miR-616 in
OC
After elucidating the expression and function of
miR-616 in OC, we further investigated the mechanisms underlying
the functions of miR-616 in OC. We searched the databases in the
websites TargetScan 6.2 and miRanda to identify the potential
target of miR-616. The data on these two websites revealed that
miR-616 contained the complementary sequences mediating its
interaction with TIMP2 3′-UTR (Fig.
6A). A luciferase activity assay revealed that miR-616
overexpression decreased the luciferase activity of the wild-type
(wt) TIMP2 3′-UTR (P<0.05; Fig.
6B) without affecting that of the mutated (mt) TIMP2 3′-UTR
(Fig. 6B). Conversely, miR-616
knockdown resulted in increased luciferase activity of the wt TIMP2
3′-UTR (P<0.05; Fig. 6C) and did
not affect that of the mt TIMP2 3′-UTR (Fig. 6C). These results indicated that
miR-616 interacts with TIMP2 3′-UTR through complementary
sequences. The results of qRT-PCR and western blotting revealed
that forced expression of miR-616 led to decreased mRNA and protein
levels of TIMP2 in A2780 cells (P<0.05; Fig. 6D and F) while miR-616 knockdown led
to increased mRNA and protein levels of TIMP2 in ES-2 cells
(P<0.05; Fig. 6E and G).
Furthermore, IHC in OC tissues revealed that compared with tissues
expressing a low miR-616 level, tissues with a high miR-616 level
exhibited a significantly decreased level of TIMP2 (P<0.05;
Fig. 6H). Collectively, these data
indicated that TIMP2 is a downstream target of miR-616 in OC.
miR-616 exerts the promoting effects
on OC metastasis by inhibiting TIMP2 expression
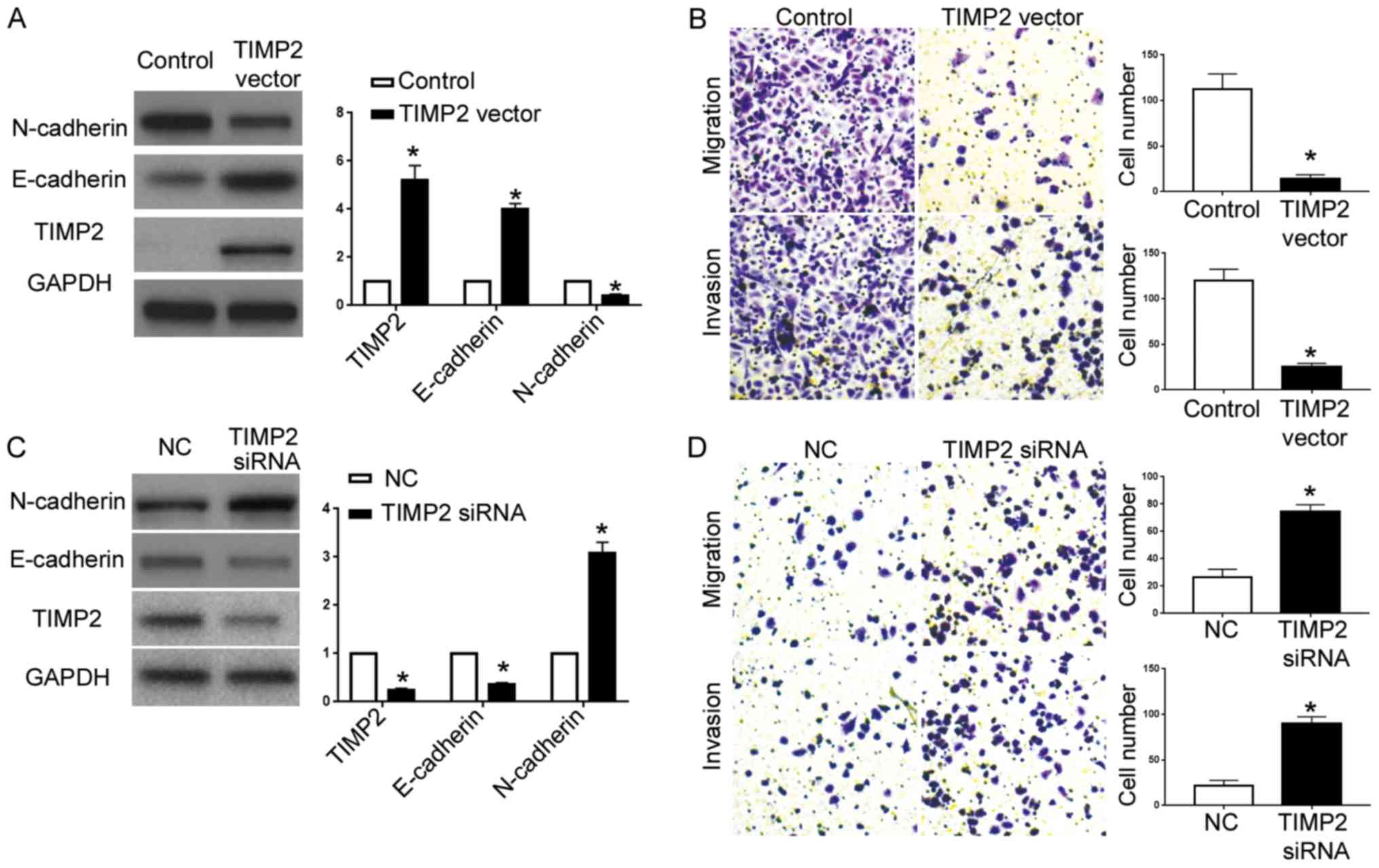
Lastly, we overexpressed TIMP2 in A2780 cells with
miR-616 overexpression. In the A2780 cells overexpressing miR-616,
transfection of the TIMP2 vector significantly increased TIMP2
expression (P<0.05; Fig. 7A),
and resulted in increased E-cadherin expression and decreased
N-cadherin expression (P<0.05; Fig.
7A). Transwell assays revealed that overexpression of TIMP2
abrogated the effects of miR-616 overexpression on cell migration
and invasion (P<0.05; Fig. 7B).
On the other hand, TIMP2-siRNA significantly decreased TIMP2
expression in ES-2 cells with miR-616 knockdown (P<0.05;
Fig. 7C) and resulted in decreased
E-cadherin and increased N-cadherin (P<0.05; Fig. 7C). Functionally, TIMP2 knockdown
abrogated the inhibiting effects of miR-616 knockdown on cell
metastatic ability (P<0.05; Fig.
7D).
Discussion
Cancer metastasis is an important cause for the poor
survival of cancer patients. The mechanisms underlying the
metastatic processes of cancer cells are complex and remain largely
unknown. Accumulating evidence has demonstrated that miRNAs play
critical roles in cancer metastasis (15). miRNAs have been revealed to regulate
the migration, invasion, extracellular matrix degradation and EMT
of cancer cells, to facilitate the occurrence of cancer metastasis
(16). Therefore, miRNAs have been
demonstrated to be promising biomarkers and therapeutic targets of
OC.
miR-616 was recently identified to be a
cancer-associated miRNA. miR-616 was found to promote metastasis
and EMT of HCC cells by targeting PTEN expression (9). miR-616 was demonstated to promote the
growth of prostate cancer cells by inhibiting the expression of
TFPI-2 (10). The latest research
in non-small cell lung cancer revealed that miR-616 promoted the
growth and metastasis by targeting SOX7. This present study
revealed that miR-616 expression was increased in OC cells and
tissues. An elevated miR-616 level in OC was associated with poor
prognosis of OC patients. Forced expression of miR-616 increased
the migration and invasion of A2780 cells while miR-616 knockdown
inhibited the metastasis of ES-2 cells. In vivo experiments
in nude mice revealed that miR-616 overexpression increased the
lung metastasis of A2780 cells while miR-616 knockdown decreased
the lung metastasis of ES-2 cells. These results indicated that
miR-616 plays oncogenic roles in OC by enhancing cell metastasis
both in vitro and in vivo.
EMT, characterized as a reduction of epithelial
marker (E-cadherin) expression and an increase of mesenchymal
marker (N-cadherin) expression (17), is an important cause for the
enhanced ability of cancer metastasis (18). In this study, miR-616 overexpression
was found to promote EMT of A2780 cells as suggested by decreased
E-cadherin expression and increased N-cadherin expression. In
contrast, miR-616 knockdown inhibited EMT of ES-2 cells. Data in OS
tissues revealed that compared with tissues with low miR-616
expression, tissues with high miR-616 levels exhibited decreased
E-cadherin expression and increased N-cadherin expression. These
data demonstrated that miR-616 enhanced the metastasis of OC cells
by promoting EMT.
Tissue inhibitor of metalloproteinase (TIMPs) are a
group of proteins inhibiting the activity of matrix
metalloproteinases (MMPs) (19,20).
TIMP-2 belongs to the TIMP family and inhibits MMP-2 activity
(21). It has been revealed to
inhibit the angiogenesis and growth of cancer cells (22). A study on pancreatic cancer revealed
that TIMP2 was involved in the metastasis and EMT of cancer cells
regulated by miR-106a (23). In the
present study, we found that miR-616 interacted with TIMP2 3′-UTR
and inhibited the expression of TIMP2 in OC cells. Notably, tissues
expressing a high miR-616 level exhibited decreased expression of
TIMP2. Furthermore, we demonstrated that TIMP2 was involved in the
promoting effects of miR-616 on cell migration, invasion and EMT.
Restoring TIMP2 expression reduced the effects of miR-616
overexpression on the metastasis and EMT of A2780 cells.
Conversely, knockdown of TIMP2 abrogated the inhibiting effects of
miR-616 knockdown on the metastasis and EMT of ES-2 cells.
In conclusion, out results revealed that the
expression level of miR-616 is elevated in OC. A high miR-616 level
was associated with unfavorable clinical features and poor
prognosis of OC patients. miR-616 promoted the migration, invasion
and EMT of OC cells. Animal experiments demonstrated that miR-616
enhanced the occurrence of lung metastasis of OC cells in nude
mice. Furthermore, TIMP2 was identified to be the downstream target
of miR-616. Inhibition of TIMP2 expression was critical for the
promoting effects of miR-616 on cell migration, invasion and EMT.
This study demonstrated that miR-616 is an oncogenic miRNA in OC
and promotes the progression of OC by enhancing the metastatic
ability of OC cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Zhejiang
Medical and Health Research fund (no. 2017KY248) and the Zhejiang
Traditional Chinese Medicine Research fund (no. 2017ZA035).
Availability of data and materials
The datasets used duing the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZBC conceived and designed this study, ZBC and JQZ
performed the in vitro and in vivo experiments, YMZ
performed the data analysis, ZBC and JJW wrote and revised the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The protocol of this study was approved by the
Institutional Research Ethics Committee of Zhejiang Cancer Hospital
and informed consent was obtained from every patient enrolled in
this study. The protocols regarding the in vivo
manipulations were approved by the Animal Care Committee of
Zhejiang Cancer Hospital.
Consent for publication
Not applicable
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Bast RC Jr, Hennessy B and Mills GB: The
biology of ovarian cancer: New opportunities for translation. Nat
Rev Cancer. 9:415–428. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holschneider CH and Berek JS: Ovarian
cancer: Epidemiology, biology, and prognostic factors. Semin Surg
Oncol. 19:3–10. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cai Y, Yu X, Hu S and Yu J: A brief review
on the mechanisms of miRNA regulation. Genomics Proteomics
Bioinformatics. 7:147–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancerMicroRNA Cancer Regulation. 774.
Schmitz U, Wolkenhauer O and Vera J: Springer; Dordrecht: pp. 1–20.
2013, View Article : Google Scholar
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang D, Zhou P, Wang W, Wang X, Li J, Sun
X and Zhang L: MicroRNA-616 promotes the migration, invasion and
epithelial-mesenchymal transition of HCC by targeting PTEN. Oncol
Rep. 35:366–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma S, Chan YP, Kwan PS, Lee TK, Yan M,
Tang KH, Ling MT, Vielkind JR, Guan XY and Chan KW: MicroRNA-616
induces androgen-independent growth of prostate cancer cells by
suppressing expression of tissue factor pathway inhibitor TFPI-2.
Cancer Res. 71:583–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang DX, Zou YJ, Zhuang XB, Chen SX, Lin
Y, Li WL, Lin JJ and Lin ZQ: Sulforaphane suppresses EMT and
metastasis in human lung cancer through miR-616-5p-mediated
GSK3β/β-catenin signaling pathways. Acta Pharmacol Sin. 38:241–251.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moreno-Bueno G, Portillo F and Cano A:
Transcriptional regulation of cell polarity in EMT and cancer.
Oncogene. 27:6958–6969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sánchez-Tilló E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.PubMed/NCBI
|
|
16
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalluri R: EMT: When epithelial cells
decide to become mesenchymal-like cells. J Clin Invest.
119:1417–1419. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gotzmann J, Mikula M, Eger A,
Schulte-Hermann R, Foisner R, Beug H and Mikulits W: Molecular
aspects of epithelial cell plasticity: Implications for local tumor
invasion and metastasis. Mutat Res. 566:9–20. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vynios DH: TIMP2 (TIMP metallopeptidase
inhibitor 2). Atlas Genet Cytogenet Oncol Haematol. 13:229–231.
2009.
|
|
21
|
Nakopoulou L, Tsirmpa I, Alexandrou P,
Louvrou A, Ampela C, Markaki S and Davaris PS: MMP-2 protein in
invasive breast cancer and the impact of MMP-2/TIMP-2 phenotype on
overall survival. Breast Cancer Res Treat. 77:145–155. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duffy MJ, McGowan PM and Gallagher WM:
Cancer invasion and metastasis: Changing views. J Pathol.
214:283–293. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li P, Xu Q, Zhang D, Li X, Han L, Lei J,
Duan W, Ma Q, Wu Z and Wang Z: Upregulated miR-106a plays an
oncogenic role in pancreatic cancer. FEBS Lett. 588:705–712. 2014.
View Article : Google Scholar : PubMed/NCBI
|