Introduction
Breast cancer is the most common cancer among women
worldwide. Breast cancer represents a heterogeneous collection of
distinct diseases, and can be classified into distinct subgroups,
including luminal, HER2 positive and basal subtypes, based on
molecular markers ER/PR and Her2 status (1). Triple-negative breast cancers (TNBCs),
which are characterized by the lack of expression of estrogen and
progesterone receptors and an absence of HER2 amplification,
account for 15–20% of all diagnosed breast cancers (2). TNBC is the most invasive and
aggressive subtype of breast cancer, with a clinically observed
higher rate of distant metastasis and poor overall survival.
Unfortunately, there is no clinical therapy specific for TNBC
patients (3). Thus, understanding
the molecular mechanisms and identifying biological markers of TNBC
progression is urgently required to help provide more effective
treatments against this disease.
MicroRNAs (miRNAs) are a large set of small
endogenous non-coding RNAs of 20–22 nucleotides, which are involved
in gene expression by targeting the 3′-UTR (untranslated regions)
of mRNA (4). Emerging evidence has
suggested that miRNAs play fundamental roles in diverse biological
and pathological processes, including cell differentiation,
proliferation, apoptosis, tumorigenesis and metastasis (5). Recently, miRNA expression studies,
especially large-scale profiling, suggested that a dysfunction of
miRNA may be associated with breast cancer progression (6–9). Among
the differentially expressed miRNAs in breast cancer, miR-30 was
found to be dysregulated, and associated with lymph node metastasis
and poor prognosis, as well as drug resistance in breast cancer
(10–12). However, the mechanisms by which
miR-30 exerts its effects and its significance in breast cancer
remain largely unexplored.
In the present study, we demonstrated that miR-30a
was frequently downregulated in TNBC, and TNBC patients with low
expression of miR-30a demonstrated high histological grade and more
lymph node metastasis. Moreover, we found that overexpression of
miR-30a suppressed TNBC cell migration and invasion in
vitro, as well as inhibited tumor growth and metastasis in
vivo. We further discovered that miR-30a specifically targeted
ROR1, which in turn suppressed the epithelial-mesenchymal
transition (EMT). Our findings provided new evidence revealing
miR-30a as a tumor-metastasis suppressor miRNA and highlighted the
role of miR-30a as a novel target for TNBC treatment.
Materials and methods
Cell culture
MCF-7, T47D, BT474, MDA-MB-231 and BT549 breast
cancer cell lines as well as MCF-10A breast epithelial cell line
were obtained from the American Type Culture Collection (ATCC;
Manassas, VI, USA). Cells were maintained in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific), 100 U/ml penicillin sodium and 100 mg/ml streptomycin
sulfate at 37°C in a humidified incubator with 5%
CO2.
Cell transfection and virus
infection
miR-30a mimics, anti-miR-30a and negative control
were obtained from Guangzho Ruibo Bio Co., Ltd. (Guangzhou, China).
The expression plasmid for pCMV6-ROR1 was purchased from OriGene
Technologies Inc. (Rockville, MD, USA). Cell transfections were
conducted using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific) according to the manufacturer's protocols.
Recombinant lentiviruses containing miR-30a or
scrambled sequences were obtained from GeneCopeia Inc. (Guangzhou,
China). For selection of breast cancer stable cell lines,
miR-30a-expressing retroviruses were transduced into MDA-MB-231 and
BT549 cells in the presence of Polybrene (6 µg/ml; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany). Cells were selected with 2 µg/ml
puromycin for 14 days.
Clinical sample
All tumor tissues and adjacent normal tissues were
collected from breast cancer patients who underwent complete
resection at the Sixth Affiliated Hospital of Guangzhou Medical
University (Qingyuan, China). Follow-up information was obtained
from review of the medical records of the patients. The present
study was approved by the Ethics Committee of Southern Medical
University Authority.
Cell invasion assay
A cell invasion assay was conducted using 24-well
Boyden chambers with 8 µm inserts with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). Cells (1×105) were placed on
the inserts in the upper chambers with 200 µl serum-free RPMI-1640
medium at 37°C. A volume of 600 µl of RPMI-1640 containing 10% FBS
was placed in the lower chamber. After 48 h, the non-invading cells
and matrix were gently removed using cotton swabs. The invasive
cells that crossed the inserts to the lower surface, were fixed
with 4% paraformaldehyde, stained with 0.1% crystal violet solution
(Sigma-Aldrich, St. Louis, MO, USA) and 5 microscopic fields (at a
magnification of ×200) were used to count and photograph the
cells.
Wound-healing assay
Cells (5×105) were seeded in 6-well
plates to grow into a monolayer and cultured until 80–90%
confluence at 37°C. After starvation in serum-free medium for 24 h,
a linear wound was created using a P200 sterile pipette and washed
with PBS. Images were captured using an Olympus IX70 microscope
microscope (a magnification of ×200; Olympus Corp., Tokyo, Japan)
and the distance migrated was observed at time-points 0, 24 and 48
h.
Western blot analysis
Cells were lysed with radioimmunoprecipitation assay
(RIPA) buffer (Pierce, Rockford, IL, USA) containing mM
phenylmethylsulfonyl fluoride (PMSF) and quantified using the BCA
method (Pierce). Then proteins were centrifuged at 12,000 × g at
4°C for 15 min to remove insoluble fragments. The proteins (30 µg)
were separated by a 10% SDS-polyacrylamide gel and transferred onto
polyvinylidene difluoride membranes (PVDF; Millipore, Billerica,
MA, USA). The membrane was blocked with 5%-skim milk powder in TBST
for 1 h. Primary antibodies against vimentin (cat. no. 5741),
N-cadherin (cat. no. 13116), E-cadherin (cat. no. 3195), ZO-1 (cat.
no. 13663) and Snail (cat. no. 3879; all diluted at 1:1,000 and
were from Cell Signaling Technology, Danvers, MA, USA) were used.
ROR1 (cat. no. ab135669; Abcam, Cambridge, MA, USA) and β-actin
(cat. no. sc-1615; Santa Cruz Biotechnology, Santa Cruz, CA, USA)
were diluted at 1:1,000 and then incubated with the membranes
overnight at 4°C. After incubation with HPR-conjugated secondary
antibodies (anti-rabbit-IgG; 1:5,000; cat. no. 7074; Cell Signaling
Technology, Danvers, MA, USA) for 1 h at room temperature, the
blots were visualized with ECL reagent (Millipore).
Immunofluorescence staining
Cells grown on coverslips were fixed in 4%
paraformaldehyde for 10 min at room temperature, and then blocked
with phosphate-buffered saline (PBS) containing 1% bovine serum
albumin (BSA) for 30 min. The cells were incubated with E-cadherin,
N-cadherin or vimentin antibodies overnight at 4°C, followed by
incubation with CFTM555 goat anti-rabbit IgG (H+L) (1:1,000; cat.
no. 4413; Cell Signaling Technology) for 1 h at room temperature.
Cell nuclei were counterstained with DAPI for 10 min, and then
analyzed using an Olympus LX70 fluorescence microscope (Olympus
Corp.).
RNA extraction and real-time
RT-PCR
Total cellular RNA was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific). For mRNA detection, ROR1
and GAPDH mRNA expression was analyzed using SYBR-Green qRT-PCR
according to the manufacturer's instructions (Applied Biosystems;
Themo Fisher Scientific). The following primers were used: GAPDH
forward, 5′-GACTCATGACCACAGTCCATGC-3′ and reverse,
5′-AGAGGCAGGGATGATGTTCTG-3′; ROR1 forward,
5′-TGCCAGCCCAGTGAGTAATCT-3′ and reverse,
5′-GCCAATGAAACCAGCAATCTG-3′.
For miRNA detection, the reverse-transcribed cDNA
was synthesized with the All-in-One™ miRNA First-Strand cDNA
Synthesis kit (GeneCopoeia, Rockville, MD, USA). miR-30a expression
was determined with the All-in-One™ miRNA qRT-PCR Detection kit
(GeneCopoeia) and U6 snRNA was used as the internal control.
Dual-Luciferase reporter assay
The PmiRGLO Vector was used to construct the
wild-type and mutated type of ROR1 3′-UTR. While the wild-type
3′-UTR of ROR1 is complementary to the seed region of miR-30a
(UGUUUAC), the mutated type 3′-UTR is poorly complementary to the
seed region of miR-30a. miR-30a mimics or miR-30a NC (50 nmol/l)
and wild-type or mutated type of ROR1 3′-UTR vectors (500 ng) were
co-transfected into 293T cells or TNBC cells (ATCC) using
Lipotectamine™ 2000 (Invitrogen; Themo Fisher Scientific) following
the manufacturer's instructions. The luciferase activities were
quantified by Dual-Luciferase reporter assay (Promega, Madison, WI,
USA). Relative luciferase activities were calculated by firefly
luciferase activities/Renilla luciferase activities.
Animal studies
BT549/miR-30a or BT549/miR-SCR cells
(1×106) were subcutaneously injected into 4- to
6-week-old BALB/c nude mice (N=6 per group). Tumors were assessed
every 4 days and tumor volumes were calculated using the formula:
Volume=length × (width/2)2. The mice were sacrificed
after 32 days of implantation and the tumors were excised and
weighed. To assay the effect of miR-30a on tumor metastasis,
BT549/miR-30a or BT549/miR-SCR cells (1×106) were
injected into the tail vein of nude mice (N=6 per group). After 45
days, necropsies were performed. The number of micrometastases in
the lungs per tissue section in individual mice were determined
from morphological observation of H&E-stained sections. All
animal studies were approved by the Animal Experimentation Ethics
Committee of Southern Medical University.
Bioinformatics analysis
The prognostic value of miR-30a was analyzed by a
Web-based Kaplan-Meier plotter (http://www.kmplot.com/), which is a meta-analysis tool
of gene expression and survival data of 5,143 breast cancer
patients (2015 version) using multiple microarray data.
Statistical analysis
Statistical analyses were conducted using the SPSS
16.0 software (SPSS, Inc., Chicago, IL, USA). Comparisons between
the groups were analyzed using the t-test and the Chi-square test
(χ2). The differences were considered to be
statistically significant when P<0.05.
Results
miR-30a is frequently downregulated in
TNBC
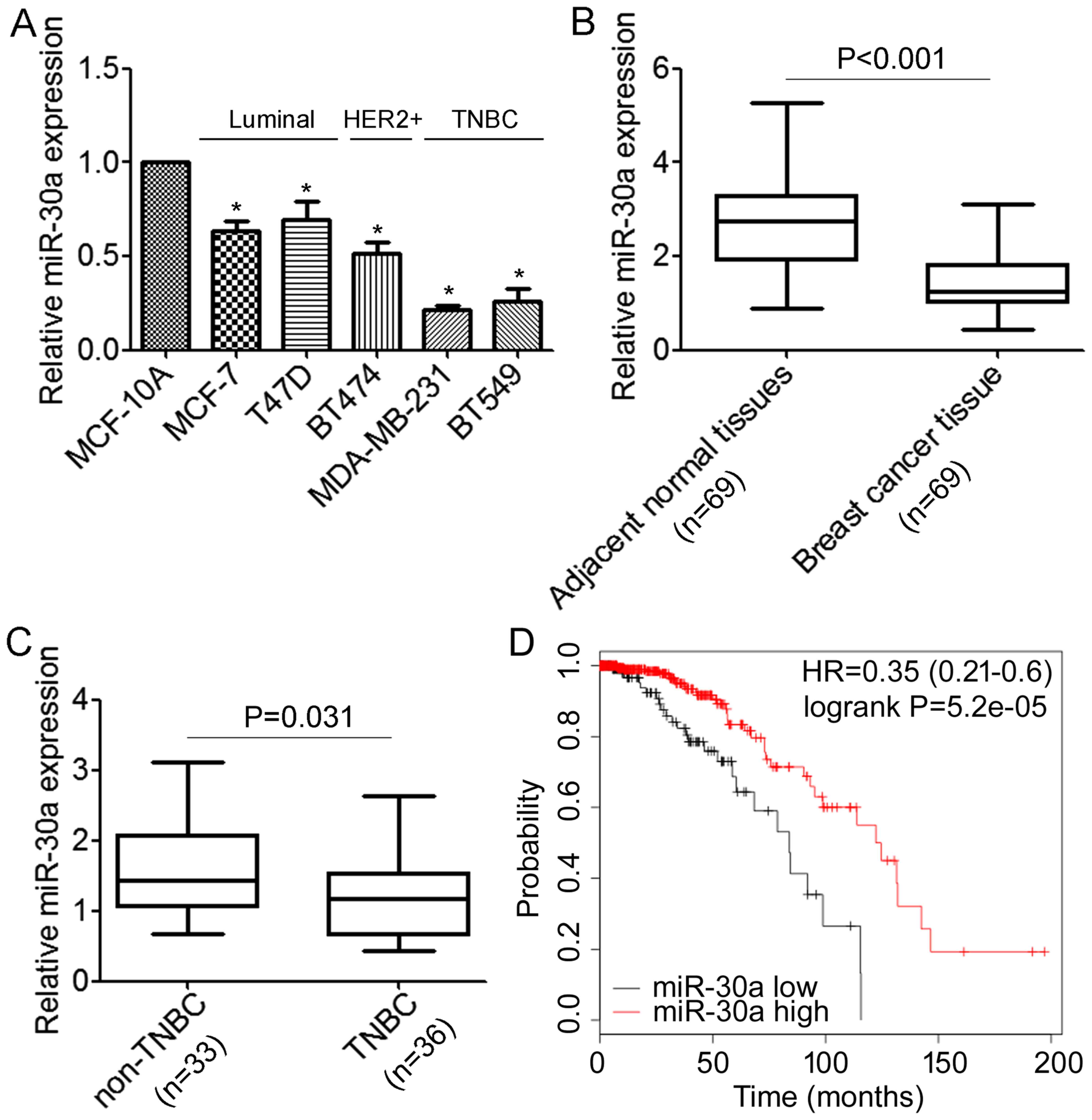
We first performed qRT-PCR analyses to determine the
expression levels of miR-30a in normal mammary cell lines MCF-10A
and a panel of breast cancer cells, including MCF-7, T47D, BT474,
MDA-MB-231 and BT549. We revealed that miR-30a was expressed at
lower levels in breast cancer cells when compared with the MCF-10A
cells (Fig. 1A). Notably, the
lowest expression of miR-30a was associated with the highly
migratory/invasive TNBC cells (Fig.
1A). Next, we detected miR-30a in 69 primary tumor samples for
which estrogen receptor and HER2 expression were available and
their matched adjacent normal tissues, and we found that the
expression level of miR-30a was significantly lower in tumor
tissues compared with the matched adjacent tissues (Fig. 1B). Moreover, the expression level of
miR-30a was significantly decreased in TNBC compared with non-TNBC
(Fig. 1C). We further investigated
possible correlations between the expression of miR-30a and
clinicopathological factors. As shown in Table I, there was no association observed
between miR-30a and the age of patients (P=0.267). However, we
found that patients with low expression of miR-30a demonstrated
high histological grade and more lymph node metastasis (P=0.032 and
P<0.01, respectively). Notably, we found that decreased miR-30a
expression was significantly correlated with triple-negative breast
cancer (P<0.01). Furthermore, the association between the
expression of miR-30a and the prognosis of breast cancer patients
was analyzed using the online Kaplan-Meier survival analysis of the
expression data from 579 breast cancer patients (http://www.kmplot.com/). The results indicated that
patients with low miR-30a expression had a shorter overall survival
time compared to the patients with high miR-30a expression
(P<0.05, log-rank test) (Fig.
1D). Collectively, these data demonstrated that miR-30a is
downregulated in TNBC cells and tissues, and correlated with TNBC
metastasis.
 | Table I.Correlation between
clinicopathological features and the expression of miR-30a. |
Table I.
Correlation between
clinicopathological features and the expression of miR-30a.
|
|
| miR-30a
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Number of patients
(n=69) | Low (n=29) | High (n=40) |
P-valuea |
|---|
| Age (years) |
|
|
| 0.267 |
|
<40 | 25 | 7 | 18 |
|
|
>40 | 44 | 12 | 32 |
|
| Histological
grade |
|
|
| 0.032 |
| I | 21 | 6 | 15 |
|
| II | 30 | 12 | 18 |
|
|
III | 18 | 11 | 7 |
|
| Lymph node
metastasis |
|
|
| <0.01 |
|
Negative | 32 | 9 | 23 |
|
|
Positive | 37 | 20 | 17 |
|
| Triple-negative
breast cancer |
|
|
| <0.01 |
|
Yes | 36 | 12 | 24 |
|
| No | 33 | 17 | 16 |
|
miR-30a inhibits the migration and
invasion of TNBC cells in vitro
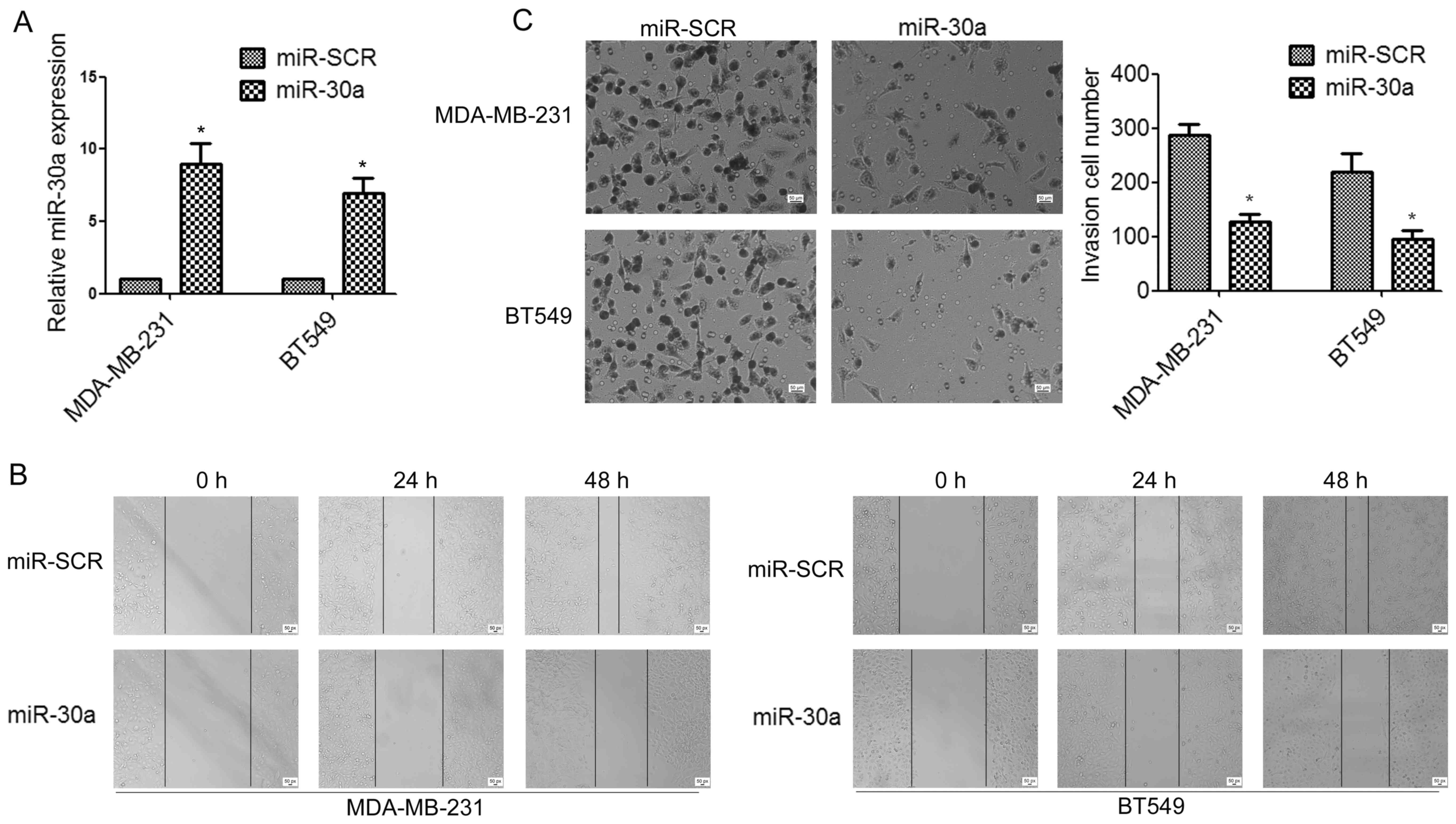
To determine whether miR-30a can affect TNBC cell
migration and invasion, we established stable miR-30a-precursor
expressing and negative control (miR-SCR) cell lines by lentiviral
transfections (Fig. 2A). Wound
healing assay indicated that the overexpression of miR-30a in
MDA-MB-231 and BT549 cells significantly suppressed cell migration,
compared to the control group (Fig.
2B). Moreover, Transwell assays demonstrated that ectopic
expression of miR-30a significantly impaired invasion of TNBC cells
(Fig. 2C).
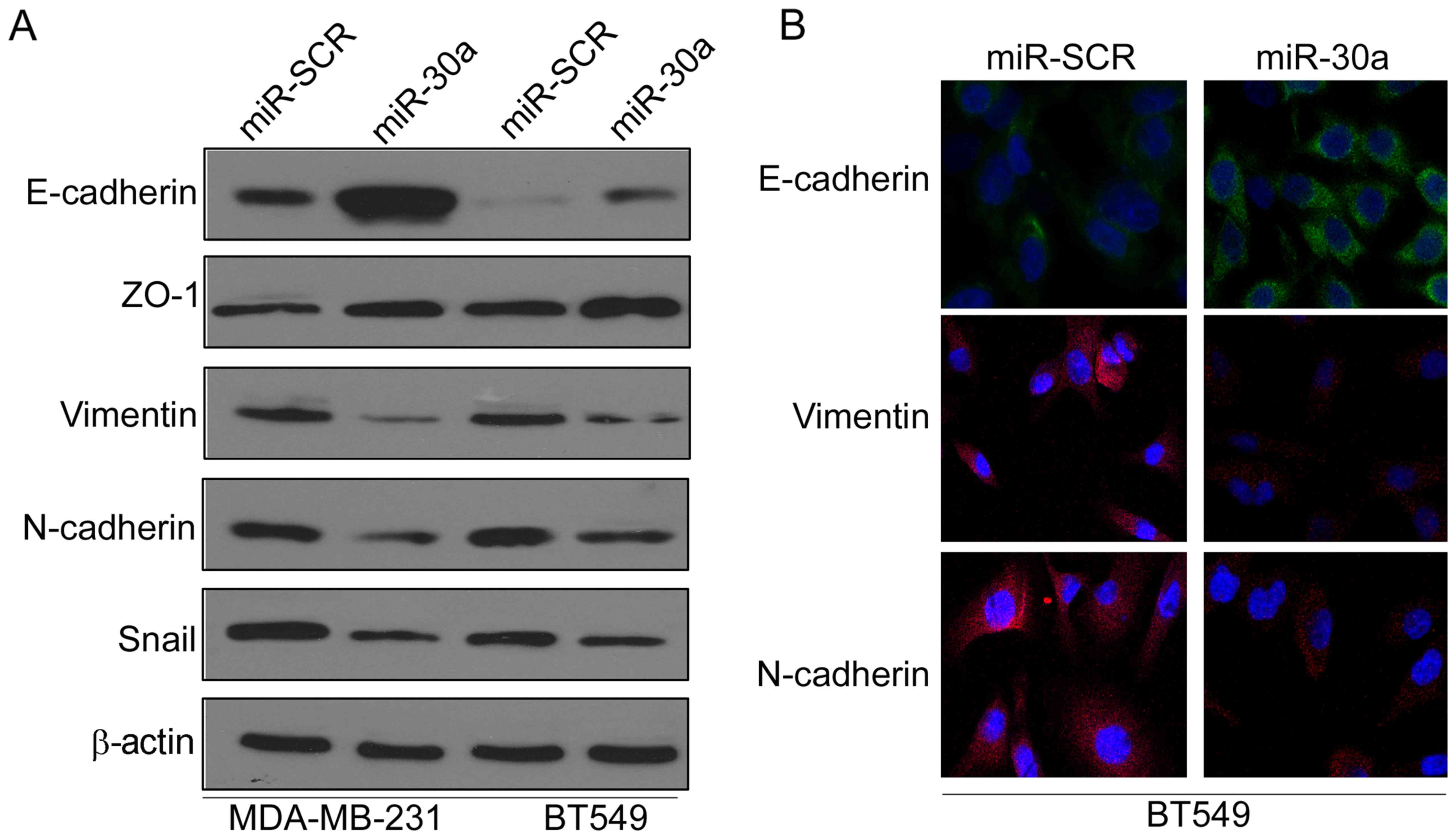
miR-30a inhibits
epithelial-mesenchymal transition of TNBC cells
Epithelial-mesenchymal transition (EMT) has been
revealed to be crucial in promoting cancer progression and
metastasis (13). Thus, we
investigated whether miR-30a inhibited EMT of TNBC cells by
investigating the expression levels of EMT-related markers. Western
blot analysis indicated that ectopic expression of miR-30a
significantly increased the expression of epithelial marker
E-cadherin, but reduced the expression of mesenchymal markers
vimentin and N-cadherin in MAD-MB-231 and BT549 cells (Fig. 3A). Moreover, immunofluorescence
analysis further confirmed that the expression of E-cadherin was
increased, whereas the expression of vimentin and N-cadherin was
decreased in miR-30a-expressing BT549 cells (Fig. 3B).
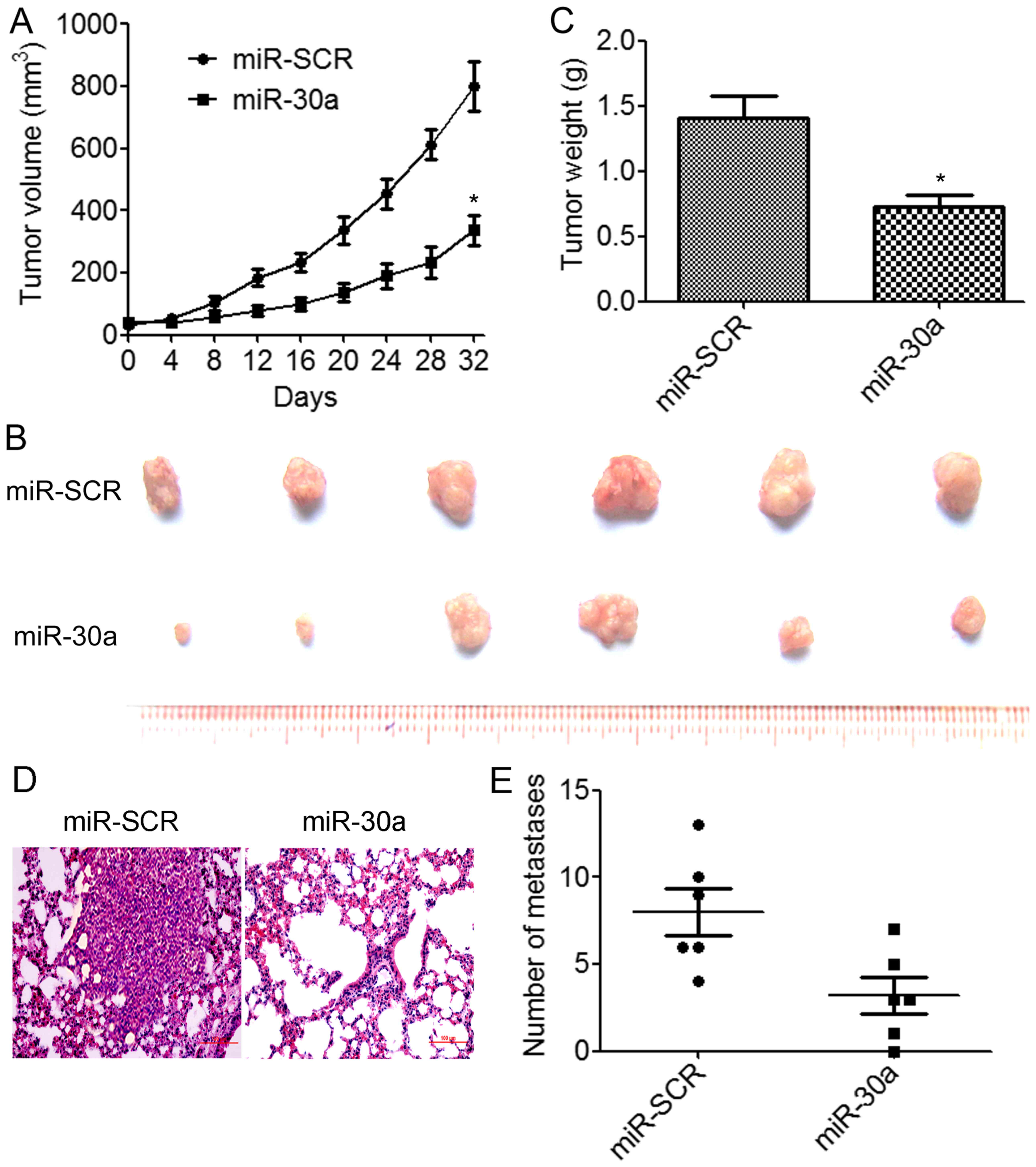
miR-30a inhibits TNBC tumor growth and
metastasis in vivo
We further assessed the effects of miR-30a
overexpression on tumor growth and metastasis in vivo.
BT549/miR-30a cells that stably expressed miR-30a or BT549/miR-SCR
control cells were implanted into the right flanks of nude mice by
subcutaneous injections (N=6 animals/group). We determined that the
overexpression of miR-30a resulted in a significant decrease in
tumor volumes compared to the control group (Fig. 4A). The weights and size of excised
tumors from the miR-30a-overexpressing group were significantly
decreased than those from the control group (Fig. 4B and C). We further investigated
whether miR-30a suppressed TNBC metastasis in vivo.
BT549/miR-30a cells or BT549/miR-SCR cells were injected into the
lateral tail vein of nude mice. The results indicated that lung
tumor nodules were significantly reduced in the mice injected with
the BT549/miR-30a cells compared to the BT549/miR-SCR cells
(Fig. 4D and E). These results
demonstrated that miR-30a suppressed TNBC growth and metastasis
in vivo.
miR-30a directly inhibits the
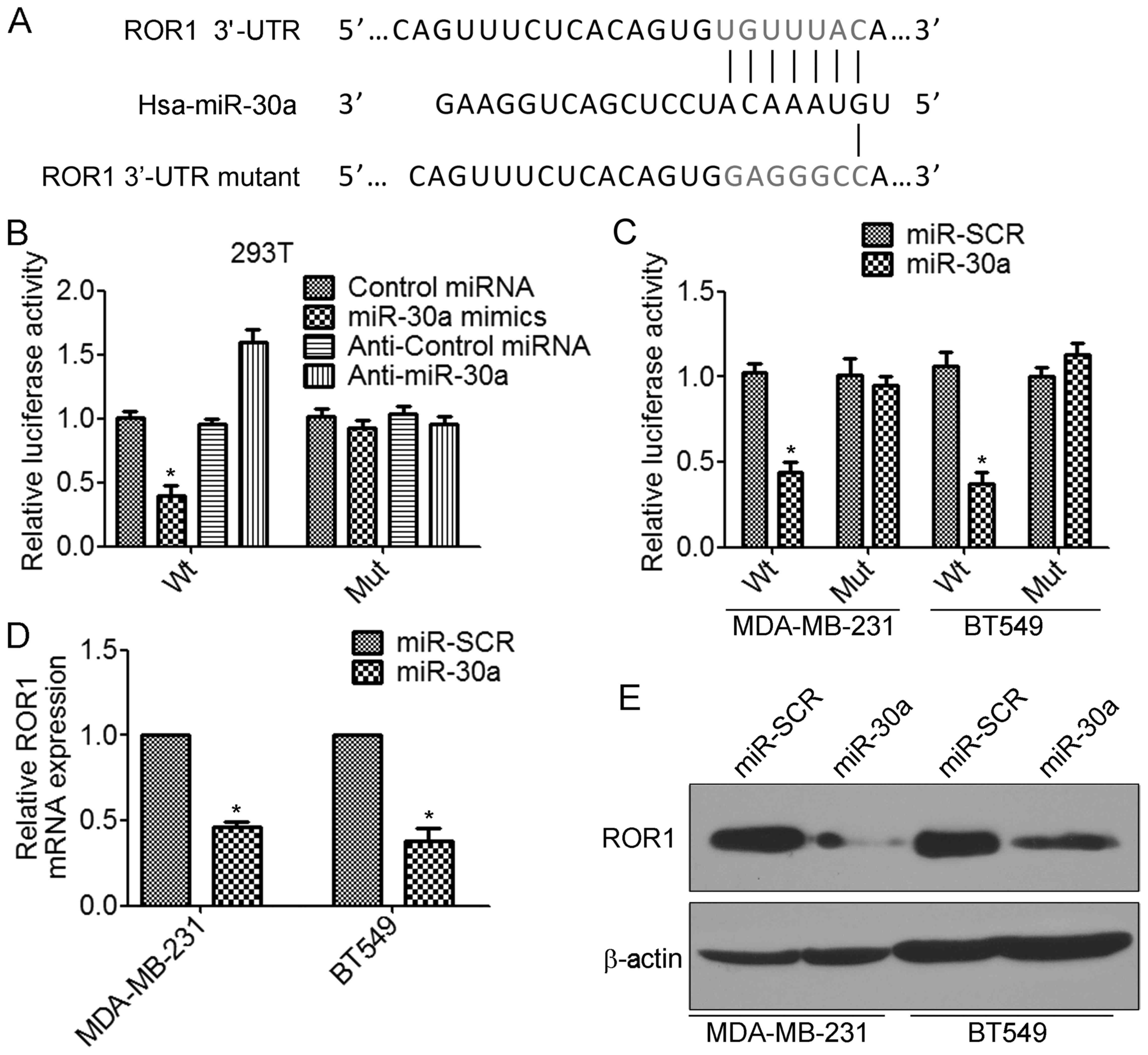
expression of ROR1 through its 3′-UTR
We further investigated the molecular mechanism by
which miR-30a inhibits the invasion and metastasis of TNBC cells.
According to the prediction by TargetScan database, the 3′-UTRs of
receptor tyrosine kinase-like orphan receptor 1 (ROR1) mRNA contain
putative miR-30a binding sites (Fig.
5A). To determine whether miR-30a regulates ROR1 by binding to
the corresponding 3′-UTRs, we cloned the 3′-UTR from ROR1 into the
pmirGLO luciferase reporter vector. We also generated the mutant
ROR1-3′-UTR vector, in which the sequence for miR-30a binding on
the 3′-UTR was mutated (Fig. 5A).
The luciferase assay revealed that transfection of miR-30a mimics
significantly decreased the ROR1-3′-UTR wild-type but not the
ROR1-3′-UTR mutant luciferase levels in 293T cells (Fig. 5B). In contrast, transfection of
anti-miR-30a increased the ROR1-3′-UTR wild-type luciferase
activity (Fig. 5B). Similar results
were found in TNBC cells. Overexpression of miR-30a significantly
inhibited ROR1-3′-UTR wild-type luciferase activity (Fig. 5C). Furthermore, we found that
overexpression of miR-30a significantly decreased the levels of
ROR1 mRNA in MDA-MB-231 and BT549 cells (Fig. 5D). There results were confirmed by
western blot analysis (Fig. 5E).
Collectively, these results indicated that miR-30a inhibited the
expression of ROR1 by directly targeting the 3′-UTR of ROR1.
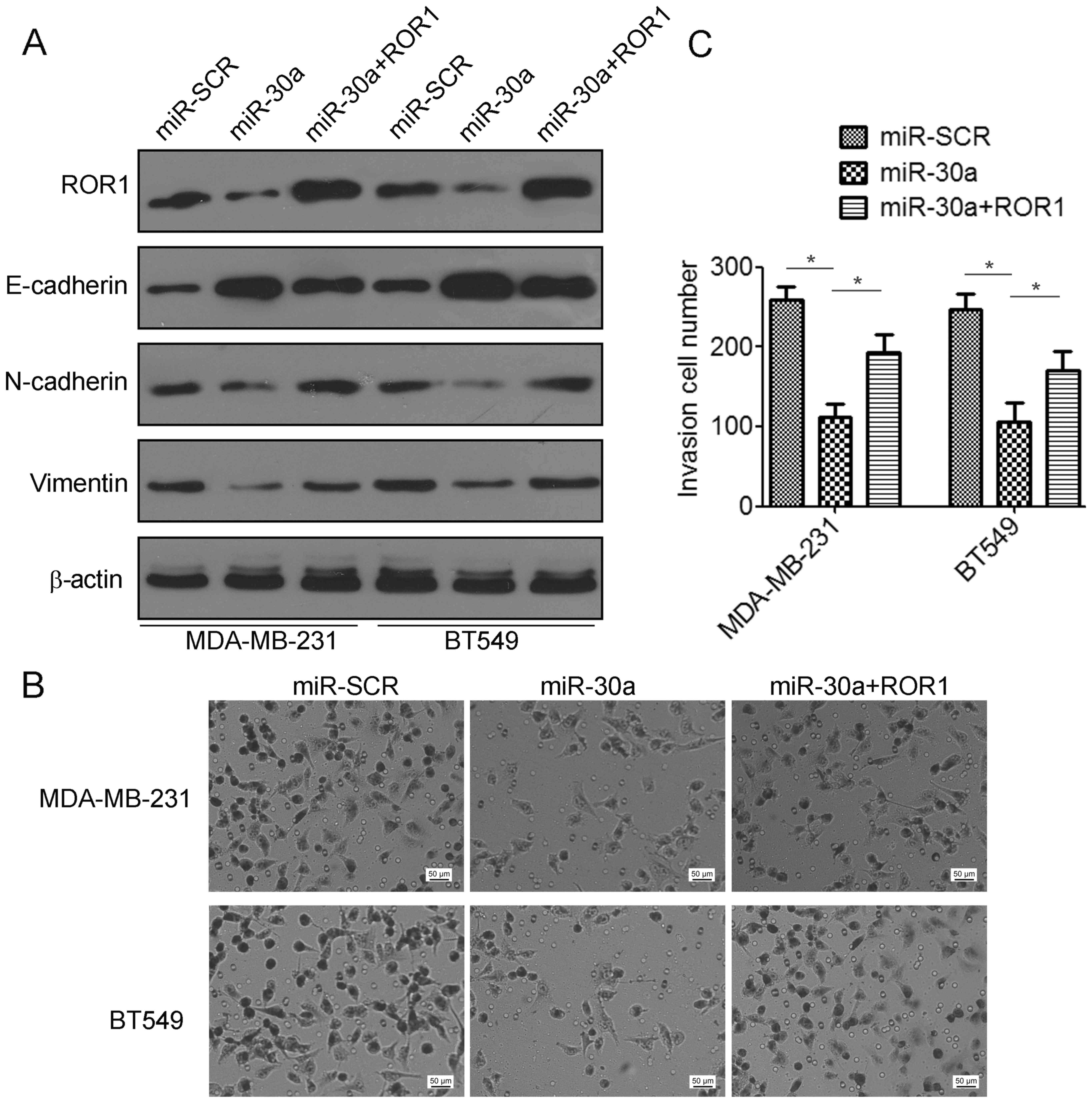
ROR1 contributes to mir-30a-mediated
suppression of TNBC cell invasion and migration
To elucidate whether the migration and invasion
suppressive effects of miR-30a were mediated by inhibition of ROR1
in TNBC cells, gain-of-function studies were performed. We found
that co-transfection with ROR1-expressing plasmids and miR-30a
indicated a significant decrease in the expression of E-cadherin
but an increase in the expression of N-cadherin and vimentin
compared to the miR-30a-transfected cells (Fig. 6A). Using Matrigel invasion assays,
we found that overexpression of ROR1 partially restored cell
invasion suppression by miR-30a (Fig.
6B and C). Collectively, these data revealed that miR-30a
suppressed TNBC migration and invasion by targeting ROR1.
Discussion
TNBC is an aggressive breast cancer subtype that
metastasizes early and is associated with poor overall survival
(14). Thus, understanding the
molecular mechanisms and identifying biological markers of TNBC
progression is urgently required to help provide more effective
treatments against the disease. miRNAs have been shown to play
important roles in cancer development and progression (15). Recently, several studies have
demonstrated that dysregulated miRNA expression was associated with
the clinical outcome in TNBC patients, which suggested that miRNAs
play a key role in TNBC development and progression (16–18).
Recently, the miR-30 family has been frequently found to be
downregulated in several types of tumors (19–22).
The prognostic features of miR-30a were investigated in 221
patients with invasive ductal carcinoma of the breast, revealing
that reduced miR-30a expression was associated with unfavorable
outcome (decreased RFS and DFS) and suppressed breast tumor growth
and metastasis (12,23). Undoubtedly, our results demonstrated
that miR-30a was significantly decreased in breast cancer, and
statistically significant correlations between low levels of
miR-30a expression with histological grade and lymph node
metastasis were revealed. Notably, the expression levels of miR-30a
in TNBC were markedly lower than those in non-TNBC, and forced
expression of miR-30a suppressed cell invasion and metastasis in
TNBC cells both in vitro and in vivo, which suggested
that miR-30a is a potential tumor metastasis suppressor miRNA in
TNBC.
Epithelial-mesenchymal transition (EMT),
characterized by the loss of epithelial characteristics and the
acquisition of mesenchymal phenotype, plays an important role in
cancer invasion and metastasis (13). Recently, studies have shown that
miR-30a negatively regulated the expression of Snail, a
transcriptional regulator that suppresses E-cadherin expression
during EMT, which suggested that miR-30a plays critical roles in
EMT (24,25). In fact, we found that forced
expression of miR-30a significantly suppressed the EMT of TNBC
cells, increasing the expression of epithelial marker E-cadherin,
but decreasing the expression of mesenchymal markers (vimentin and
N-cadherin) in miR-30a-overexpressing TNBC cells, thereby
decreasing cell motility and invasion. These results are consistent
with previous studies, which revealed that the suppressive effect
of miR-30a on vimentin expression led to decreased migration and
invasiveness of breast cancer cells (23).
Subsequently, we identified receptor tyrosine
kinase-like orphan receptor 1 (ROR1) as a potential functional
target of miR-30a. ROR1 is an important oncofetal protein in normal
embryonal development (26).
Recently studies revealed that ROR1 is upregulated in several types
of human cancers (27–33). In breast cancer, high expression of
ROR1 has been demonstrated to be associated with aggressive tumor
phenotypes such as TNBC (34).
Similarly, Chien et al further suggested that the expression
of ROR1 was significantly increased in TNBC and correlated with
poor survival of TNBC patients, and that targeting ROR1 may serve
as a potential therapy for the treatment of TNBC patients (35). Moreover, high levels of ROR1
expression were revealed to be associated with genes involved in
EMT, and silencing of ROR1 in TNBC cells reduced the expression of
EMT-related markers such as Snail, Slug, Zeb and vimentin, which in
turn suppressed cell invasion and metastasis both in vitro
and in vivo (34). In the
present study, we observed that overexpression of ROR1 in
miR-30a-expressing BT549 and MDA-MB-231 cells reversed the
inhibitory effects of miR-30a in EMT and abrogated the
miR-30a-mediated suppression of TNBC cell invasion and migration,
which suggested that miR-30a suppressed EMT, as well as suppressed
TNBC cell migration and invasion by targeting ROR1.
In summary, the present study demonstrated that
miR-30a is frequently downregulated in TNBC, and decreased miR-30a
expression was associated with histological grade and lymph node
metastasis. Moreover, we discovered that miR-30a specifically
targeted ROR1, which in turn suppressed epithelial-mesenchymal
transition, as well as cell invasion and metastasis both in
vitro and in vivo. Our findings provide an experimental
basis for investigating miR-30a as a potential therapeutic target
for TNBC.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the
National Natural Science Foundation of China (grant no.
81672992).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LC and XW conceived and designed the study. XW, HQ,
RT performed the experiments. HS and HP were involved in patient
data acquisition, analyzed and interpreted data. ZF participated in
the design of the study, and assisted with drafting the manuscript.
LC and XW wrote the paper. LC, XW, HQ, RT, HS, HP and ZF reviewed
and edited the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by Southern
Medical University (Guangzhou, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch A, Eroles P, Zaragoza R, Viña JR and
Lluch A: Triple-negative breast cancer: Molecular features,
pathogenesis, treatment and current lines of research. Cancer Treat
Rev. 36:206–215. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Teoh SL and Das S: The role of MicroRNAs
in diagnosis, prognosis, metastasis and resistant cases in breast
cancer. Curr Pharm Des. 23:1845–1859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li P, Xu T, Zhou X, Liao L, Pang G, Luo W,
Han L, Zhang J, Luo X, Xie X and Zhu K: Downregulation of miRNA-141
in breast cancer cells is associated with cell migration and
invasion: Involvement of ANP32E targeting. Cancer Med. 6:662–672.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bockhorn J, Dalton R, Nwachukwu C, Huang
S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al:
MicroRNA-30c inhibits human breast tumour chemotherapy resistance
by regulating TWF1 and IL-11. Nat Commun. 4:13932013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu
Z, Hu G and Yang Q: MicroRNA-30a suppresses breast tumor growth and
metastasis by targeting metadherin. Oncogene. 33:3119–3128. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Radojicic J, Zaravinos A, Vrekoussis T,
Kafousi M, Spandidos DA and Stathopoulos EN: MicroRNA expression
analysis in triple-negative (ER, PR and Her2/neu) breast cancer.
Cell Cycle. 10:507–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mathe A, Scott RJ and Avery-Kiejda KA:
MiRNAs and other epigenetic changes as biomarkers in triple
negative breast cancer. Int J Mol Sci. 16:28347–28376. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cascione L, Gasparini P, Lovat F, Carasi
S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, et
al: Integrated microRNA and mRNA signatures associated with
survival in triple negative breast cancer. PLoS One. 8:e559102013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agrawal R, Tran U and Wessely O: The
miR-30 miRNA family regulates Xenopus pronephros development and
targets the transcription factor Xlim1/Lhx1. Development.
136:3927–3936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baffa R, Fassan M, Volinia S, O'Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM and
Rosenberg A: MicroRNA expression profiling of human metastatic
cancers identifies cancer gene targets. J Pathol. 219:214–221.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kao CJ, Martiniez A, Shi XB, Yang J, Evans
CP, Dobi A, deVere White RW and Kung HJ: miR-30 as a tumor
suppressor connects EGF/Src signal to ERG and EMT. Oncogene.
33:2495–2503. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Zhang H, Liu J, Tu X, Zang Y, Zhu
J, Chen J, Dong L and Zhang J: miR-30 inhibits TGF-β1-induced
epithelial-to-mesenchymal transition in hepatocyte by targeting
Snail1. Biochem Biophys Res Commun. 417:1100–1105. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumarswamy R, Mudduluru G, Ceppi P,
Muppala S, Kozlowski M, Niklinski J, Papotti M and Allgayer H:
MicroRNA-30a inhibits epithelial-to-mesenchymal transition by
targeting Snai1 and is downregulated in non-small cell lung cancer.
Int J Cancer. 130:2044–2053. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Borcherding N, Kusner D, Liu GH and Zhang
W: ROR1, an embryonic protein with an emerging role in cancer
biology. Protein Cell. 5:496–502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
O'Connell MP, Marchbank K, Webster MR,
Valiga AA, Kaur A, Vultur A, Li L, Herlyn M, Villanueva J, Liu Q,
et al: Hypoxia induces phenotypic plasticity and therapy resistance
in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer
Discov. 3:1378–1393. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou JK, Zheng YZ, Liu XS, Gou Q, Ma R,
Guo CL, Croce CM, Liu L and Peng Y: ROR1 expression as a biomarker
for predicting prognosis in patients with colorectal cancer.
Oncotarget. 8:32864–32872. 2017.PubMed/NCBI
|
|
29
|
Balakrishnan A, Goodpaster T,
Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK, Berger C,
Kosasih PL, Rajan A, Sommermeyer D, et al: Analysis of ROR1 protein
expression in human cancer and normal tissues. Clin Cancer Res.
23:3061–3071. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang S, Cui B, Lai H, Liu G, Ghia EM,
Widhopf GF II, Zhang Z, Wu CC, Chen L, Wu R, et al: Ovarian cancer
stem cells express ROR1, which can be targeted for
anti-cancer-stem-cell therapy. Proc Natl Acad Sci USA.
111:17266–17271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hojjat-Farsangi M, Moshfegh A,
Daneshmanesh AH, Khan AS, Mikaelsson E, Osterborg A and Mellstedt
H: The receptor tyrosine kinase ROR1-an oncofetal antigen for
targeted cancer therapy. Semin Cancer Biol. 29:21–31. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gentile A, Lazzari L, Benvenuti S,
Trusolino L and Comoglio PM: The ROR1 pseudokinase diversifies
signaling outputs in MET-addicted cancer cells. Int J Cancer.
135:2305–2316. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang S, Chen L, Wang-Rodriguez J, Zhang
L, Cui B, Frankel W, Wu R and Kipps TJ: The onco-embryonic antigen
ROR1 is expressed by a variety of human cancers. Am J Pathol.
181:1903–1910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui B, Zhang S, Chen L, Yu J, Widhopf GF
II, Fecteau JF, Rassenti LZ and Kipps TJ: Targeting ROR1 inhibits
epithelial-mesenchymal transition and metastasis. Cancer Res.
73:3649–3660. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chien HP, Ueng SH, Chen SC, Chang YS, Lin
YC, Lo YF, Chang HK, Chuang WY, Huang YT, Cheung YC, et al:
Expression of ROR1 has prognostic significance in triple negative
breast cancer. Virchows Arch. 468:589–595. 2016. View Article : Google Scholar : PubMed/NCBI
|