Introduction
Colorectal cancer (CRC) remains the fourth most
common cause of cancer-related deaths worldwide (1), with more than 1.2 million new cases
being identified annually (2).
Although most cases of CRC are sporadic, 20–30% of individuals with
CRC carry inherited mutations in key tumor suppressors, such as APC
and TP53 (3,4). Compared with other solid malignancies,
CRC is characterized by slow development, which renders the tumor
curable and preventable. Additionally, the survival rate of
patients with CRC is critically dependent on the tumor stage at
diagnosis. Thus, early diagnosis of CRC has become a central
subject in the field of CRC studies. The identification of robust
molecular indicators associated with the proliferation and
tumor-node-metastasis (TNM) potential of CRC can lead to avoiding
understaging of the tumor and help to pinpoint patients with
early-stage CRC. In recent years, studies regarding microRNA (miR)
have indicated the potential of this non-coding RNA type in
categorizing the subtype and prognosis of CRC (5–7).
miR can suppress mRNA translation of targeted genes
by binding to the 3′ untranslated region (UTR) of target messenger
RNA (mRNA) (8). Dysregulation of
miRs has been demonstrated to be associated with tumorigenesis of
various human organs, including the colorectum (9,10). Liu
et al (6) revealed that
miR-124 upregulation reduced cell viability and proliferation of
CRC cells in vitro. In another clinical study, Sarlinova
et al (11) reported
significantly upregulated expression of miR-21 and
miR-221 and downregulation of miR-150 in blood
samples of CRC patients. As one of the most studied miRs in various
types of cancer (12–14), the positive correlation between the
expression of miR-150 and the survival of CRC patients has
been long revealed (15,16). Furthermore, the dysregulation of
miR-150 in CRC was subsequently studied by Wang et al
(7) and Feng et al (17), who revealed the antagonistic effect
of miR-150 on the oncogenesis and progression of CRC via
targeting MUC4 and c-Myb. Given that the above-mentioned studies
markedly indicated that miR-150 is a tumor suppressor gene
in CRC, it is reasonable to further explore the mechanism driving
the antitumor function of miR-150 in CRC.
Inhibitor of apoptosis stimulating protein of p53
(iASPP) belongs to the ASPP family (18). This factor can inhibit the normal
function of p53, which leads to oncogenesis in human organs
(19,20). Furthermore, iASPP can also
negatively regulate the p65 subunit of nuclear factor-κB (NF-κB),
which plays a vital function in inflammation and apoptosis
(21). Therefore, suppressing the
function of iASPP may serve as a promising therapeutic strategy for
the prevention and treatment of CRC. Based on bioinformatic
analysis, iASPP is a potential target of miR-150 and
regulation of iASPP by miR-150 may influence the biological
features of CRC cells. To verify our hypothesis in the present
study, we challenged the expression of miR-150 in clinical
samples and then detected the effect of miR-150
induction/iASPP inhibition on the viability, apoptosis and
mobility of CRC cells. The findings outlined in the present study
confirmed the direct regulation of iASPP by miR-150, which
would impair the growth and metastasis potential of CRC.
Materials and methods
Reagents and chemicals
Antibodies against iASPP (cat. no. ab115605) and
GAPDH (cat. no. KC-5G5) were purchased from Abcam (Cambridge, UK)
and Kangcheng Bio (Beijing, China), respectively. Hoechst staining
kit (cat. no. H1399) was purchased from Thermo Fisher Scientific
(Waltham, MA, USA). IgG-HRP antibody (cat. no. BA1054) was
purchased from Wuhan Boster Biological Technology Ltd. (Wuhan,
China).
Patient and CRC specimen
collection
CRC specimens were collected from 30 patients (from
July 2015 to October 2016) in The Chinese People's Liberation Army
General Hospital. The specimens were fixed and prepared in paraffin
sections. The patients enrolled in the present study were diagnosed
with primary CRC and had detailed clinicopathological and
prognostic information. Screening, inspection and data collection
were approved by the Ethics Committee of The Chinese People's
Liberation Army General Hospital, and a written informed consent
form was signed by all subjects. The procedures performed adhered
to the Declaration of Helsinki.
Immunohistochemistry
Immunohistochemistry was used to detect the
expression of iASPP protein in tissues, according to the operation
guide of immunohistochemistry. Paraformaldehyde (4%) fixed tissues
and paraffin embedded sections for antigen retrieval were used.
Samples were then incubated overnight at 4°C with iASPP antibodies
(1:400). After being washed with PBS, samples were incubated for 50
min at RT with IgG-HRP antibodies (1:500). Then the samples were
observed and images were captured by an optical microscope
(CX41-23C02; Olympus Corp., Tokyo, Japan).
Cell culture
Human CRC cell lines FHC, HCT116, HCT8, HT29, H1299
and SW480 were obtained from Shanghai Bioleaf Biotech, Co., Ltd.,
(Shanghai, China). The cells were cultured in minimum essential
medium (MEM, M2279; Sigma-Aldrich, St. Louis, MO, USA) with 15%
fetal bovine serum (FBS; 10099-141; Gibco, Carlsbad, CA, USA) and
1% (v/v) antibiotics mix and maintained in an atmosphere of 95% air
and 5% CO2 at 37°C. The expression level of
miR-150 was determined using reverse transcription real-time
PCR (qPCR) as described in the following sections. The cell line
with the lowest expression level of miR-150 was selected for
subsequent assays and, based on the results of qPCR, SW480 cell
line had the lowest expression level of miR-150 (Fig. 1D) and was employed as an in
vitro model for CRC.
Construction of vector, sequences of
siRNA and transfection
Specific siRNA targeting iASPP
(5′-AGTTCATGTCCAGAAAGTCCC-3′) and non-targeting siRNA
(5′-ACGUGACACGUUCGGAGAATT-3′) were used to knockdown the expression
of iASPP. Coding sequences were cloned through amplification
reaction using primers (iASPP forward,
5′-GGGGTACCATGGACAGCGAGGCATTCC-3′ and iASPP reverse,
5′-CCGCTCGAGCTAGACTTTACTCCTTTGAGGCTTCAC-3′). Subsequently, the PCR
product (2487 bp) was ligated to the pcDNA3.0 plasmid, and
recombinant plasmid was confirmed by sequencing after digestion
with KpnI/XhoI. SW480 cells were transfected with
different vectors using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific). The coding sequence of iASPP was ligated
into the pcDNA plasmid to form the pcDNA-iASPP vector for
overexpression of the gene.
Experimental design and grouping
To detect the function of miR-150 in the
oncogenesis of CRC, SW480 cells were divided into two groups: i) NC
group, SW480 cells transfected with NC mimics; and ii) mimics
group, SW480 cells transfected with miR-150 mimics. Each
group was represented by at least five replicates.
To elucidate the key role of iASPP in the
progression of CRC, SW480 cells were divided into three groups: i)
Blank group, SW480 cells; ii) NC group, SW480 cells transfected
with pcDNA-NC plasmid; and iii) siRNA group, SW480 cells
transfected with pcDNA-siiASPP plasmid. Each group was represented
by at least five replicates.
The interaction between miR-150 and iASPP was
further assessed with four groups: i) blank group, SW480 cells; ii)
NC group, SW480 cells transfected with NC mimics; iii) mimics
group, SW480 cells transfected with miR-150 mimics; and iv)
mimics+pcDNA group, miR-150 stably overexpressed in SW480
cells transfected with pcDNA-siiASPP plasmid.
Dual-Luciferase assay
The direct regulating function of miR-150 on
the 3′UTR of iASPP was determined with a Dual-Luciferase
assay. Luciferase activity was detected by Dual-Luciferase assay
kit (E1960; Promega, Madison, WI, USA) after 24 h of transfection
and co-transfection of Renilla luciferase plasmid, used as
the internal control for transfection efficiency.
Real-time PCR
Total RNA in the cells was extracted by RNA
purification using RNA Extraction kit (9109; Takara Bio, Inc.,
Otsu, Japan) accordingly. β-actin and U6 were selected as the
internal reference genes. cDNA templates were achieved by using
Super MMLV Reverse Transcriptase (DBI-2342; DBI Bioscience,
Shanghai, China), and the final RT-qPCR reaction mix contained 10
µl Bestar® SYBR Green qPCR Master Mix, 0.5 µl of each
primer (miR-150, forward
5′-ACACTCCAGCTGGGTCTCCCAACCCTTGTACC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; iASPP, forward,
5′-GAAAGCCTGGAACGAGTCTGA-3′ and reverse, 5′-GCGCTAGTGAGGTTGTCCT-3′;
U6, forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH, forward,
5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse,
5′-ATGGCATGGACTGTGGTCAT-3′), 1 µl cDNA template and 8 µl RNase-free
H2O. Amplification was performed as follows: a
denaturation step at 94°C for 2 min, followed by 40 cycles of
amplification at 94°C for 20 sec, 58°C for 20 sec and 72°C for 20
sec. The reaction was stopped at 25°C for 5 min. The relative
expression levels were detected and analyzed by Exicycler™ 96
(Bioneer Corp., Daejeon, Korea) based on the formula of
2−∆∆ct.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was performed to detect cell viability.
Briefly, SW480 cells (1×105 cells/ml) that underwent
different treatments were seeded into one well of a 96-well plate
and incubated for 72 h. Every 24 h, 10 µl of CCK-8 solution was
added to every well and incubated at 37°C for a minimum of another
4 h. The OD values were detected at 450 nm and employed as the
representative of cell viability.
Flow cytometry
Cell cycle distribution was determined using flow
cytometry. Cells were stained with propidium iodide (PI) in the
dark for 20 min at room temperature. The results were analyzed
using a FACS flow cytometer (BD Accuri C6; BD Biosciences, San
Jose, CA, USA).
Hoechst staining
DNA damage in cell nuclei was detected using a
Hoechst staining method after cells were transfected for 24 h.
Cells were stained with Hoechst 33258 (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
instructions.
Scratch assay
To evaluate cell mobility, a scratch assay was
performed on the transfected cells. Cells at a density of
2×104 cells/well were incubated in one well of a 24-well
plate. After marking reference points, cells were cultured to
confluence at 37°C for 48 h. Then, a cell-free line was made
(regarded to as a scratch), and debris at the edges was removed.
Twenty-four hours after the scratch was made, gap distances between
the midline were assessed using an optical microscope (Olympus
Corp.) in reference to the reference points.
Transwell assays
Transwell assays were performed to detect the
mobility of SW480 cells. In brief, cells at a density of
2×104 cells/well were incubated in the upper chamber
(Corning Costar, Cambridge, MA, USA) after a 24-h serum-free
incubation, and the chamber was pre-coated with 40 µl Matrigel (0.8
µg/µl) at 37°C for 2 h. Then, the system was placed at 37°C for 24
h and, subsequently, cells in the upper surface were completely
removed. The lower surfaces of the chamber were stained for 5 min
using 1% (w/v) crystal violet and the cell number was recorded
using Image-Pro Plus 6.0 software (Media Cybernetics, Inc.,
Rockville, MD, USA).
Western blotting
Total cellular protein was extracted using the
protein lysate. Western blot analysis was conducted according to
Lin et al (22). The
membranes were incubated with primary antibodies against iASPP
(1:4,000) and GAPDH (1:10,000) for 1 h at room temperature.
Secondary HRP-conjugated IgG antibodies (1:20,000) were then added
and incubated for 45 min at room temperature. The blots were
developed and the results were recorded in the Gel Imaging System
(ScanWizard Bio; Microtek International, Inc., Taipei, Taiwan).
Then, data were analyzed using Image-Pro Plus 6.0 (Media
Cybernetics, Inc.).
Statistical analysis
Data were expressed as the mean ± standard deviation
(SD). Student's t-test was performed with a significance level of
0.05. Statistical analyses and graphing were performed using
GraphPad Prism 6.01 (GraphPad Software, Inc., Chicago, IL,
USA).
Results
The expression of miR-150 is induced
and the expression of iASPP is upregulated in clinical CRC
samples
The expression status of miR-150 was
determined with qPCR validation in clinical CRC samples and
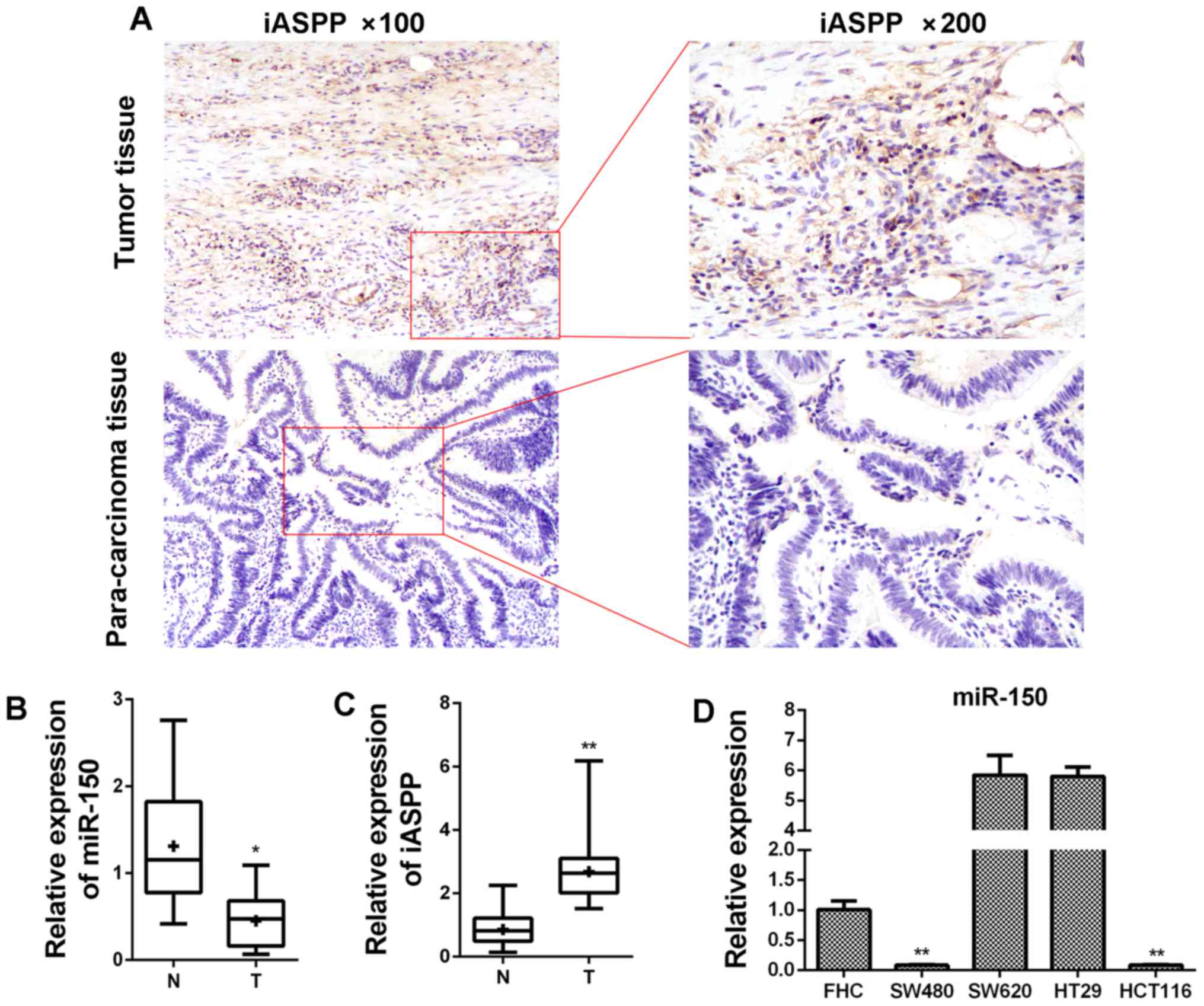
corresponding para-carcinoma tissues from patients (Table I). As displayed in Fig. 1, the level of miR-150 in CRC
samples was lower than that in para-carcinoma samples and the
difference was statistically significant (Fig. 1B; P<0.05). On the contrary, the
expression of iASPP at the mRNA level was induced in CRC
samples (Fig. 1C). Based on the
detection in clinical samples, it was inferred that miR-150
had an anti-CRC effect during the progression of the tumor.
Furthermore, miR-150 expression was assessed, and lower
expression was detected in SW480 and HCT116 cells, and relatively
higher expression presented in SW620 and HT29 cells compared with
the FHC cell line (Fig. 1D). The
expression of iASPP was also investigated using
immunohistochemistry in clinical tissues from CRC patients. As
displayed in Fig. 1A, the
expression of iASPP was relatively higher in tumor tissue than
para-carcinoma tissue.
 | Table I.Characteristics of patients with
CRC. |
Table I.
Characteristics of patients with
CRC.
|
| Expression of
miR-150 |
|---|
|
|
|
|---|
|
Characteristics | Total | Low | High |
|---|
| Age (years) |
|
<60 | 18 | 10 | 8 |
|
≥60 | 12 | 5 | 7 |
| Sex |
|
Male | 17 | 9 | 8 |
|
Female | 13 | 6 | 7 |
| Tumor size
(cm) |
|
<5 | 11 | 9 | 2 |
| ≥5 | 19 | 6 | 13 |
| TNM stage |
|
I+II | 13 | 10 | 3 |
|
III+IV | 17 | 5 | 10 |
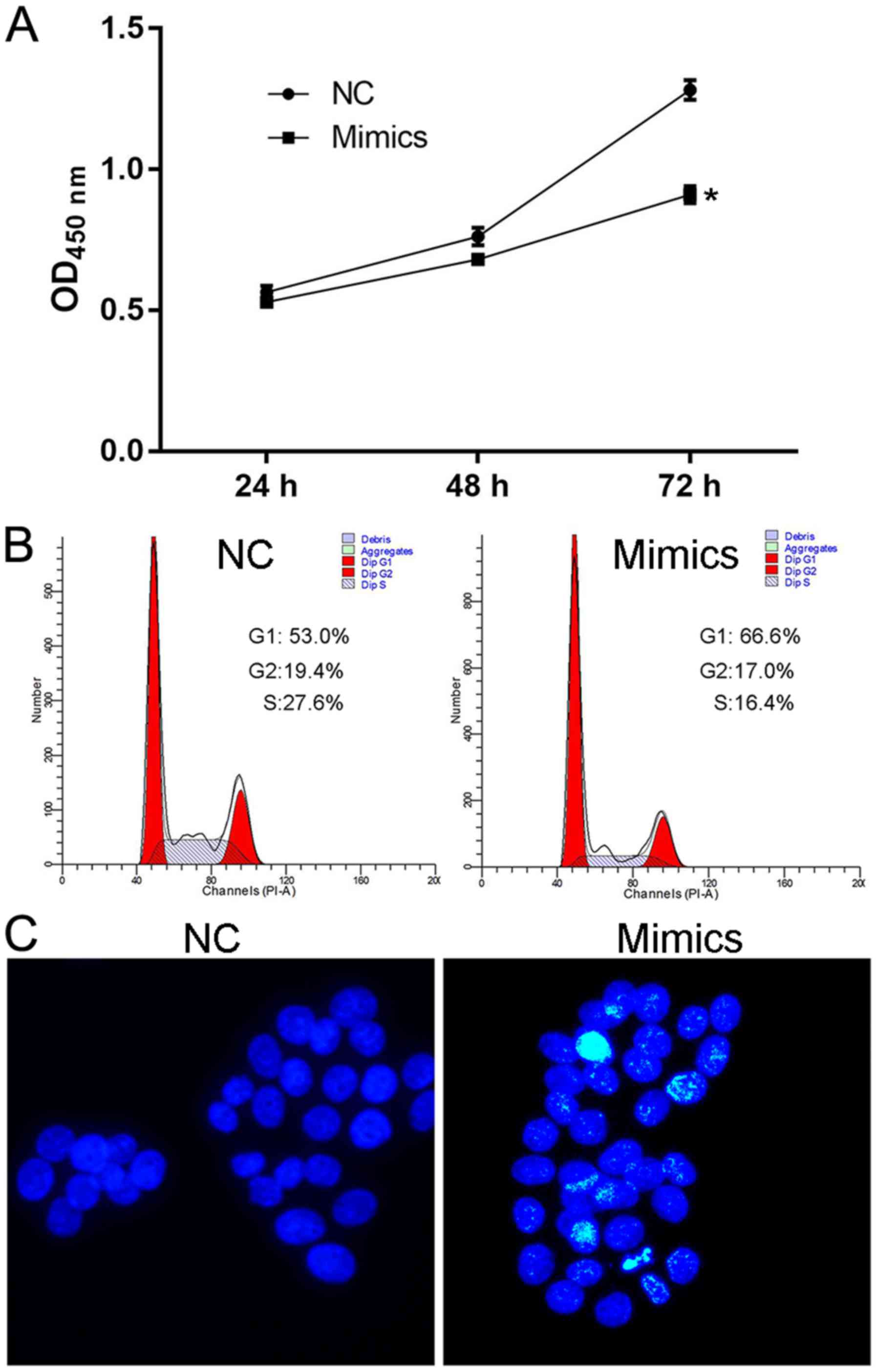
Augmented expression of miR-150
suppresses cell viability and induces cell apoptosis and G1
cell-cycle arrest in SW480 cells
The effect of miR-150 overexpression on
normal cell features was determined to validate the antitumor
effect of miR. Being induced by transfection of specific mimics,
the upregulated expression of miR-150 significantly
decreased the OD450 value in the mimics group at 72 h (Fig. 2A), representing impaired viability
of SW480 cells. Furthermore, the overexpression of miR-150
induced G1 cell cycle arrest and cell apoptosis in the mimics
group, as a larger proportion of cells were distributed in the G1
phase (Fig. 2B) and more
Hoechst-positive cells (Fig. 2C)
were detected in cells transfected with miR-150. Detection
focusing on the proliferation potential of SW480 cells confirmed
the conclusion that miR-150 is an anti-CRC molecule that
influences CRC cells both by decreasing cell proliferation and
inducing cell apoptosis.
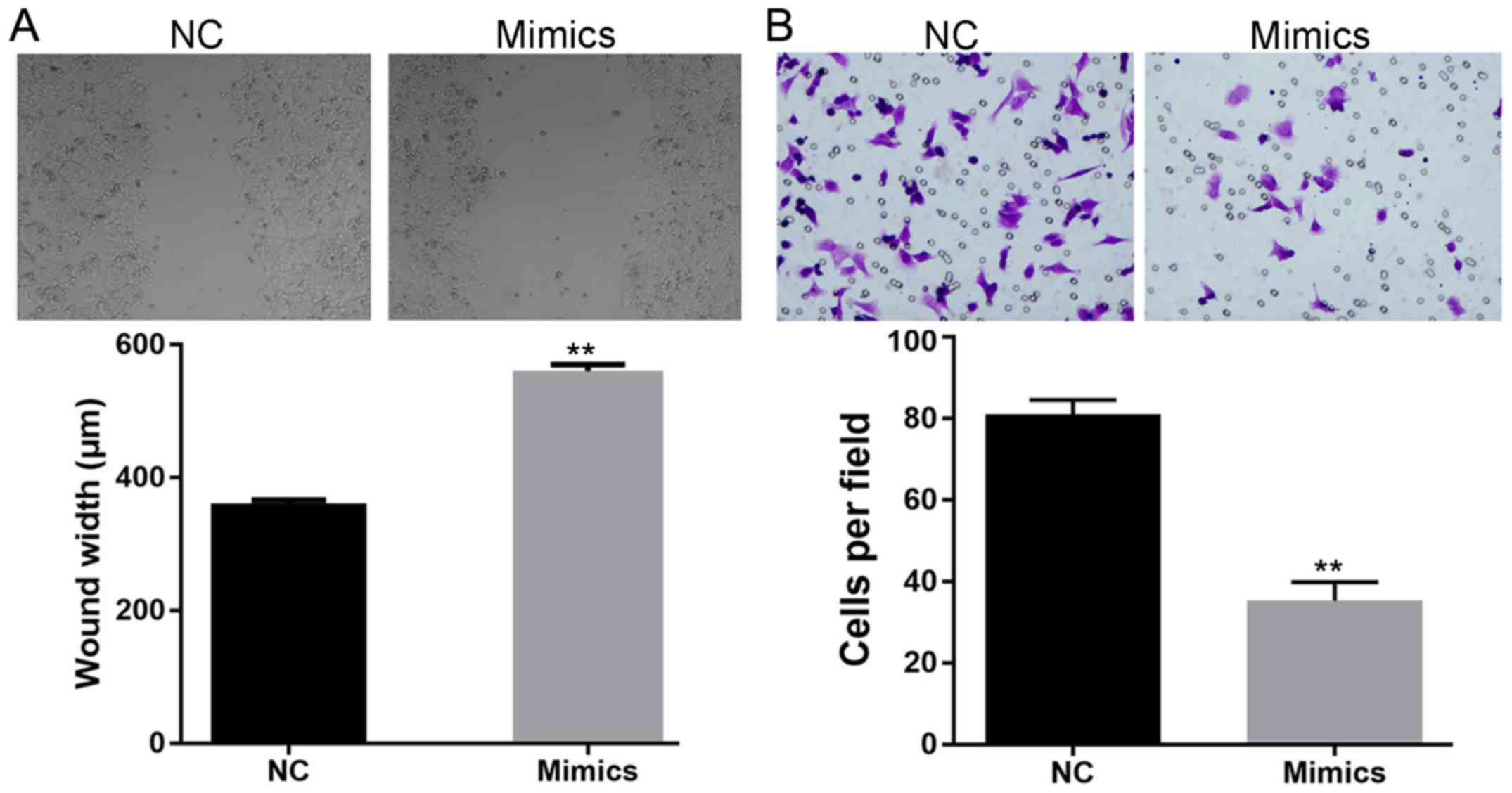
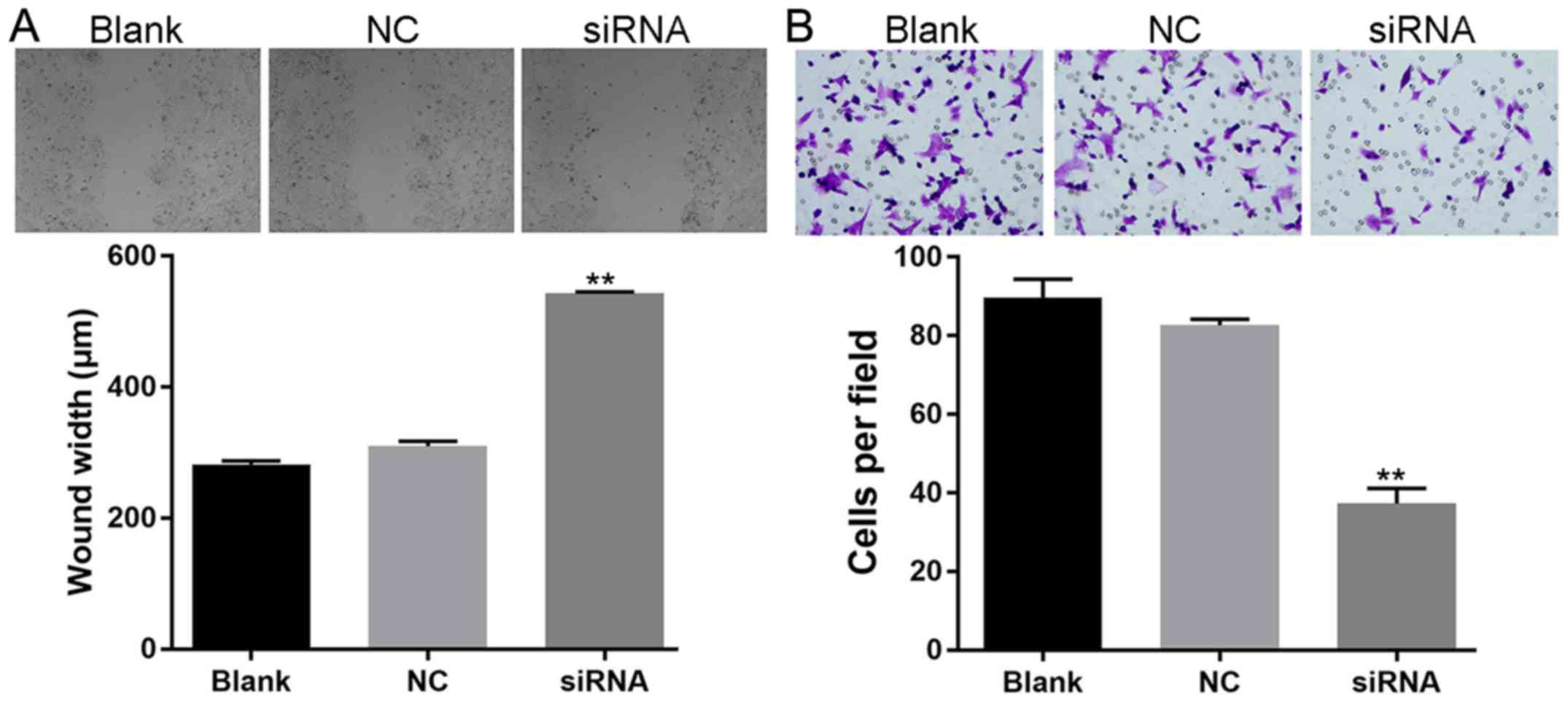
Augmented expression of miR-150
impairs the migration and invasion ability of SW480 cells
The effect of miR-150 overexpression was
further assessed by detecting its effect on the metastasis
potential of SW480 cells. For cells subjected to the scratch assay,
induced expression of miR-150 resulted in a delayed closure
rate of the gap (wider gap width) (Fig.
3A). Furthermore, impaired invasion ability was also detected
in SW480 cells with overexpression of miR-150 (Fig. 3B), as less cells penetrating the
membranes were recorded in the mimics group. The results of the
scratch and Transwell assays together indicated the inhibitory
effect of miR-150 on the metastasis potential of CRC
cells.
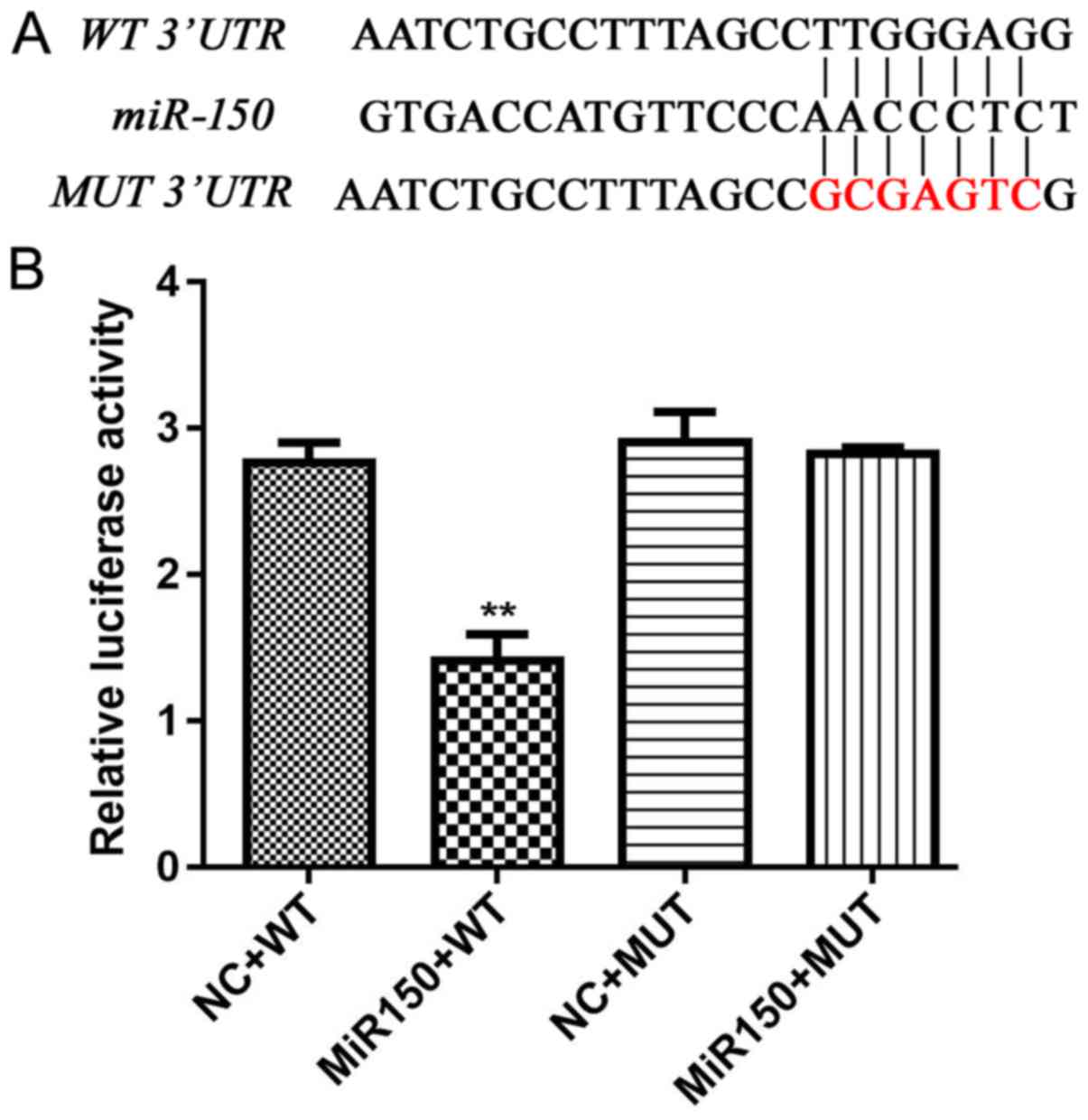
iASPP is directly regulated by miR-150
and plays a promoting role in the oncogenesis of CRC
Based on bioinformatic analysis, iASPP was a
potential target of miR-150. In the present study, a
possible interaction between the two indicators was determined
using a dual-luciferase assay. The results indicated that only
cells transfected with miR-150 mimics and wild-type iASPP
3′UTR demonstrated a decreased relative luciferase activity
(Fig. 4), which indicated a direct
and specific modulating effect of miR-150 on the
iASPP gene.
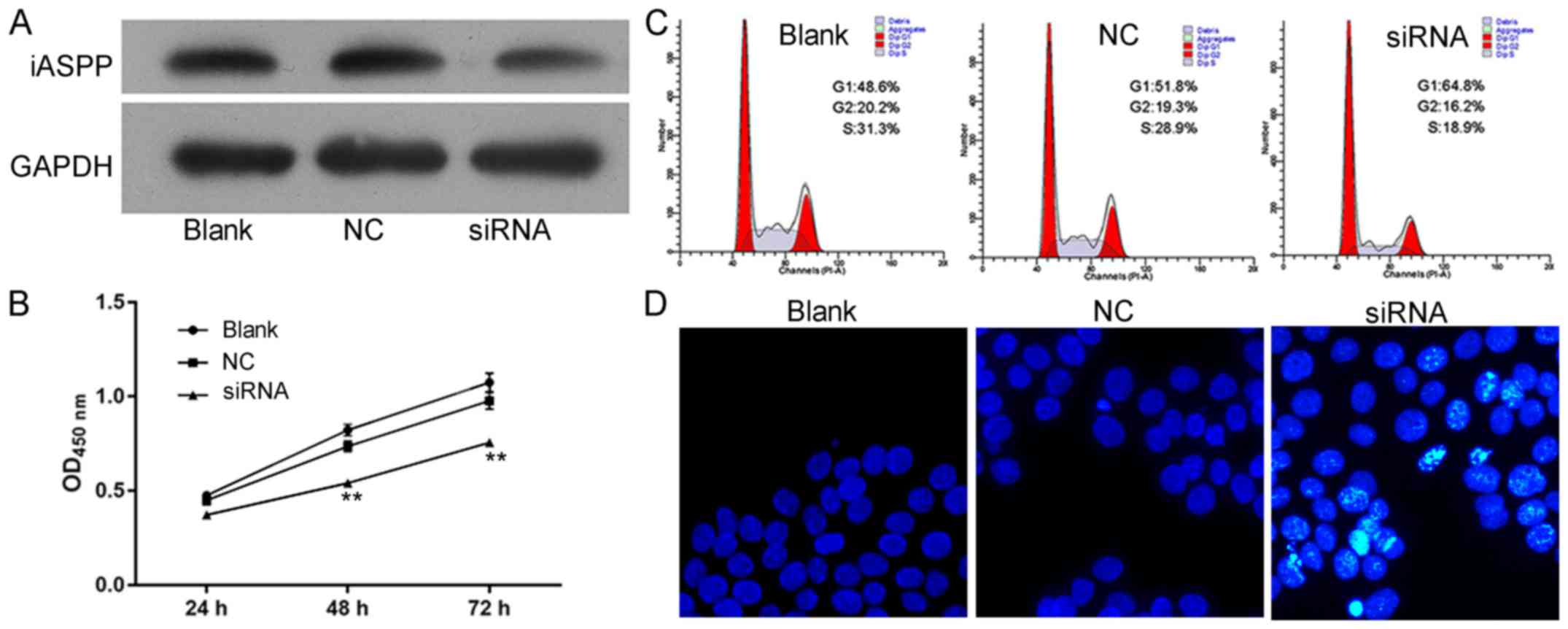
To confirm the results that iASPP promoted the
progression of CRC, the effects of iASPP knockdown on cell
viability, cell apoptosis, cell cycle distribution and cell
migration and invasion were also assessed. The expression of iASPP
was confirmed by western blot analysis after transfection with
siRNA (Fig. 5A). The results
indicated that iASPP knockdown exhibited a similar effect on
SW480 cells to that of miR-150 overexpression: in the siRNA
group, cell viability was decreased (Fig. 5B), G1 cell cycle arrest and
apoptosis were induced (Fig. 5C and
D) and cell migration and invasion were impaired (Fig. 6).
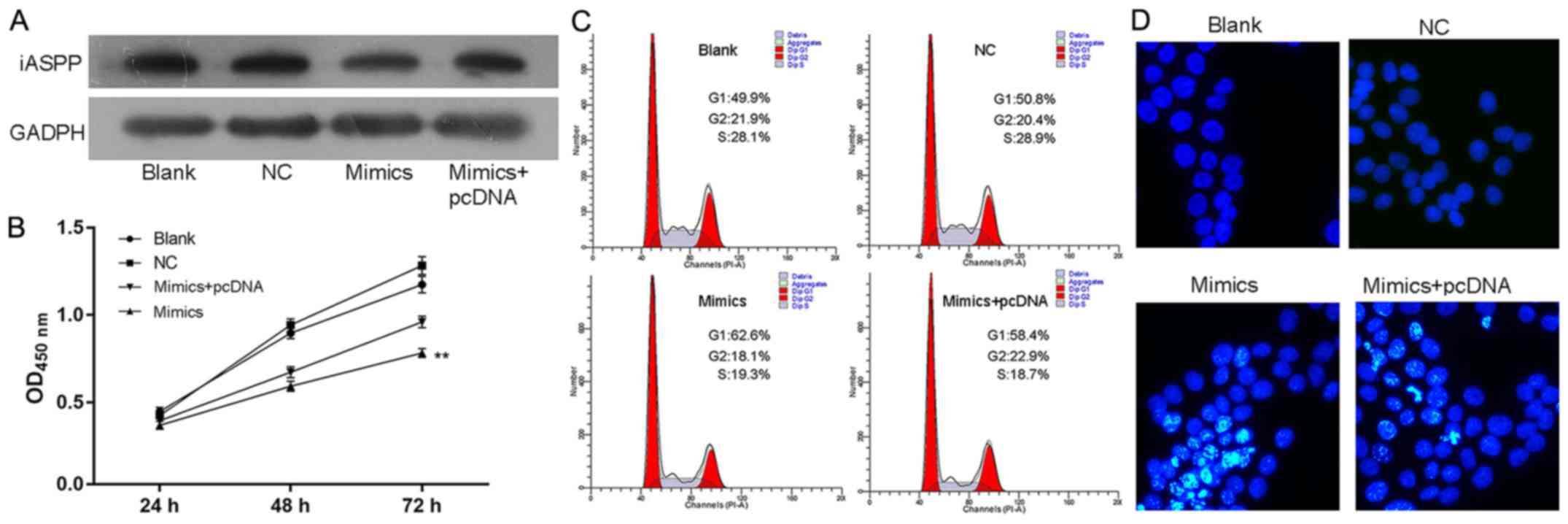
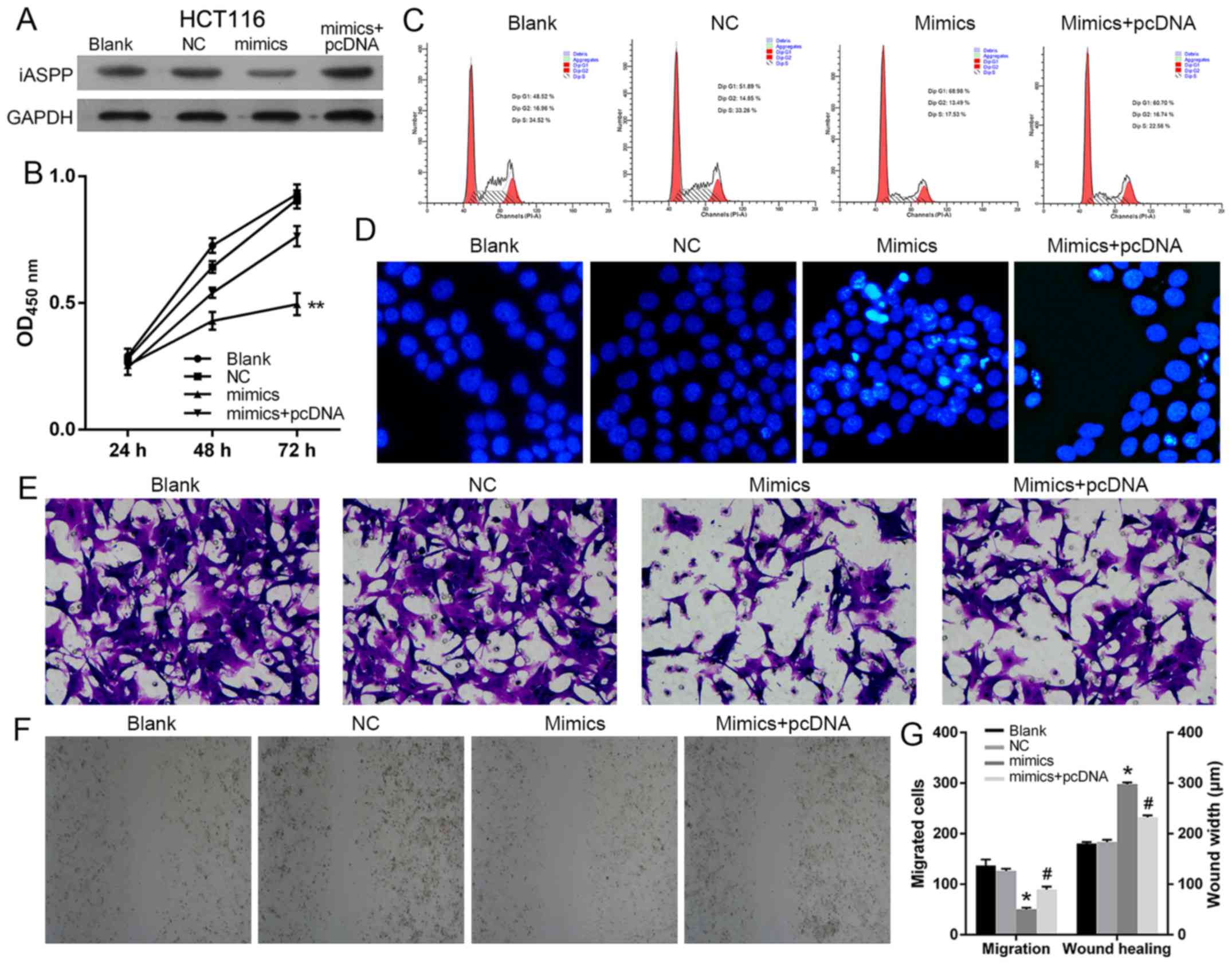
miR-150 exerts its suppressing effect
on CRC cells by targeting iASPP
Given the direct regulating function of
miR-150 in the transcription of iASPP, it was
hypothesized that the antitumor effect of miR-150 on CRC was
dependent on the suppressed function of iASPP. Therefore,
the expression of iASPP was induced in miR-150 overexpressed
SW480 and HCT116 cells (Figs. 7A
and 9A, respectively).
Subsequently, the growth and metastasis potential of SW480 and
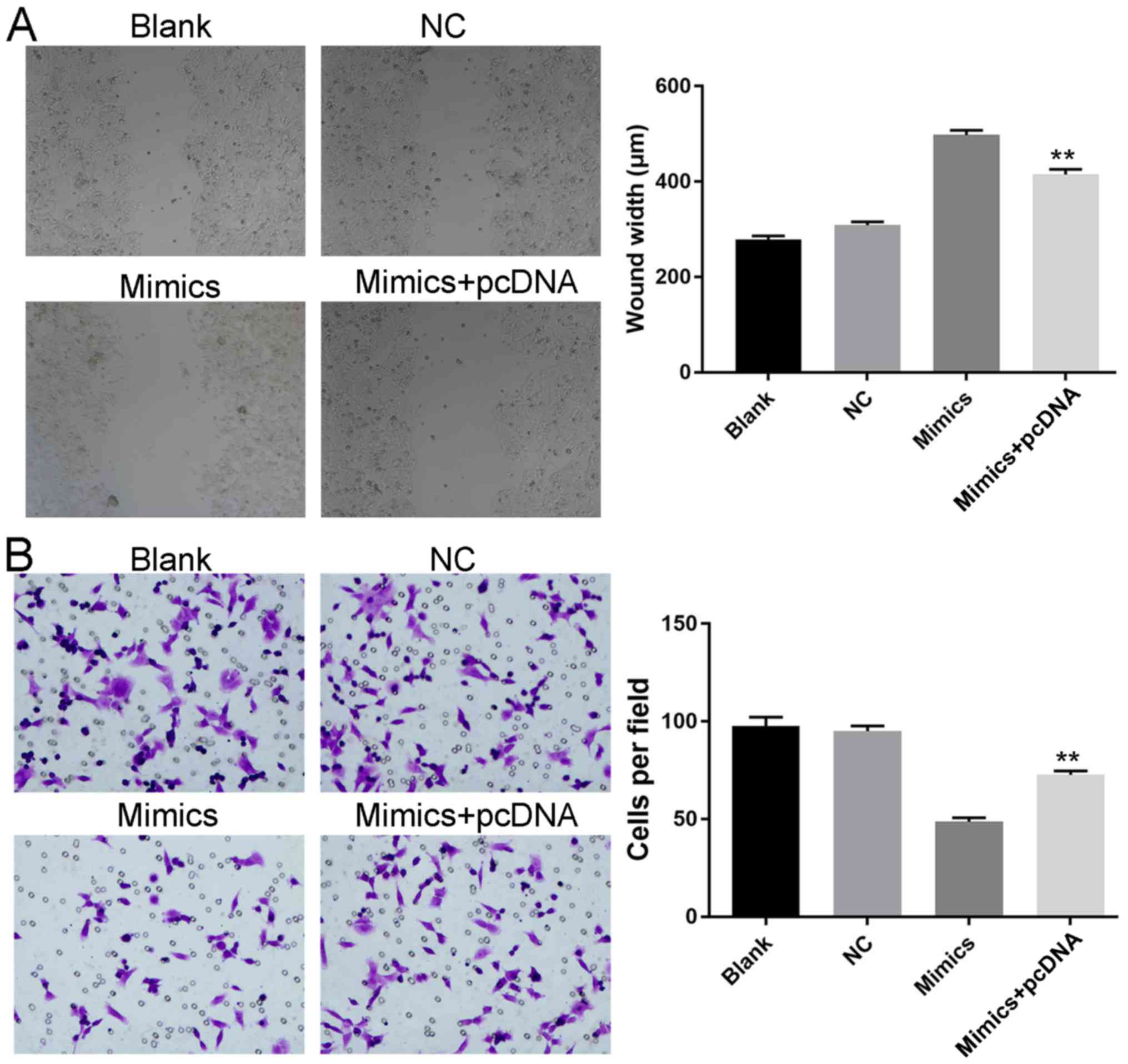
HCT116 cells in different groups were assessed. Based on the
results, induced expression of iASPP counteracted the effect
of miR-150 overexpression on SW480 and HCT116 cells.
Concerning growth potential, induced iASPP expression increased
cell viability, relieved cells from G1 cell cycle arrest and
inhibited cell apoptosis in the miRNA+pcDNA group when compared
with the miRNA group (Figs. 7B-D
and 9B-D). Additionally,
upregulated iASPP levels also improved the metastatic potential of
SW480 and HCT116 cells, as a faster closure rate and more cells
penetrating the membrane were detected in the miRNA+pcDNA group as
demonstrated by scratch and Transwell assays (Figs. 8 and 9E-G). It was hypothesized that, without
the low level of iASPP, the suppressing function of miR-150
on CRC cells was confounded, which indicated that the anti-CRC
function of miR-150 was exerted via the inhibition of
iASPP.
Discussion
The suppressed expression of miRs in colorectal
cancer (CRC) may represent a novel therapeutic avenue for the
treatment of CRC (5,23). Among all reported downregulated miRs
in CRC, the potential of miR-150 as a biomarker for
diagnosing and predicting CRC has been reported in several studies
(7,17). In the present study, the data
confirmed the decreased level of miR-150 in clinical CRC
samples. Furthermore, reintroduction of miR-150 markedly
supressed viability, induced apoptosis and inhibited migration and
invasion of CRC cells in vitro. The effect of miR-150
on SW480 cells depended on the function of iASPP, the
overexpression of which blocked the impairments of miR-150
mimics on SW480 cells. The present study clearly indicated that
miR-150 was capable of predicting and suppressing CRC, a
finding that deserves further investigation.
Dysregulation of miR-150 has been reported in
diverse tumor types. However, the exact function of miR-150
varies by tumor type. For gastric, breast and lung cancer,
miR-150 plays a role in promoting the progression of cancer
(13,24,25).
Conversely, for pancreatic cancer, miR-150 is able to
suppress the growth of the tumor by targeting MUC4 (26). The possible involvement of
miR-150 in CRC was first reported by Ogata-Kawata et
al (27) however, in their
study, serum exosomal levels of miR-150 were markedly higher
in primary CRC patients than in healthy controls. With emerging
attention being paid to the function of miR-150 in CRC,
several other researchers have consistently revealed a reduced
level of miR-150 in CRC tumors (5,15,17),
which indicated that miR-150 is a biomarker associated with
CRC prognosis (15). In the present
study, qPCR validation of clinical CRC and para-carcinoma samples
confirmed the conclusion that miR-150 was suppressed in CRC
tissues. Subsequently, induced miR-150 in human CRC cell
line SW480 further verified the inhibitory effect of miR-150
on the growth and metastatic potential of CRC cells. To further
uncover the downstream pathways involved in the antagonizing effect
of miR-150 on CRC, we also detected the effects of
interaction between miR-150 and iASPP on CRC cells.
iASPP acts as a negative regulator of the tumor
suppressor p53 and its overexpression has been associated to poor
prognosis and survival in some types of cancers (18,21).
The gene can suppress apoptosis by deactivating the function of p53
on the promoters of proapoptotic genes (21). In addition to regulating p53, iASPP
has been also proven to inhibit the transcription of RelA/p65 and
reduce inflammation (28). Induced
expression of miR-124 in CRC cells attenuated cell viability,
proliferation and colony formation via inhibition of iASPP protein
expression and forced overexpression of iASPP rescued CRC cells
from the inhibitory effect of miR-124 (6). Therefore, targeted suppression of
iASPP may serve as the mechanism by which its upstream miR prevents
oncogenesis of CRC. In the present study, we proved that
overexpression of iASPP contributed to the enhanced growth and
metastasis of CRC cells. By performing a dual-luciferase assay,
iASPP was validated to be a direct target of miR-150 in CRC
cells and induced expression of miR-150 restricted the
expression of iASPP both at the mRNA and protein levels. However,
the impairment of miR-150 in SW480 cells was partially
obscured by re-expression of iASPP. Collectively, these
results indicated that miR-150 can act as a suppressor on
the growth and metastasis of CRC and this effect was exerted by
direct inhibition of the expression of iASPP.
In conclusion, the present study provided more
evidence supporting the anti-CRC function of miR-150. The
low expression miR-150 in clinical samples highlighted the
possibility that the molecule may predict poor prognosis in CRC
patients. In addition, the suppressed viability, proliferation,
migration and invasion in SW480 cells due to enforced expression of
miR-150 supported the treatment potential of miR-150
in CRC. The present study also revealed a key role of iASPP in the
oncogenesis of CRC, which can be targeted by miR-150. The
findings of the present study provided a supplementary mechanism
underlying the suppressing effect of miR-150 on CRC and
offered a therapeutic target for future exploration.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (no. 61471397).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CL and XD conceived and designed the study. CL and
SX performed the experiments. CL and LC wrote the study. CL and XD
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures were approved by the Chinese People's
Liberation Army General Hospital from May 2015 to October 2016.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Reference
|
1
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DA Silva FC, Wernhoff P, Dominguez-Barrera
C and Dominguez-Valentin M: Update on Hereditary Colorectal Cancer.
Anticancer Res. 36:4399–4405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Strubberg AM and Madison BB: MicroRNAs in
the etiology of colorectal cancer: Pathways and clinical
implications. Dis Model Mech. 10:197–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aherne ST, Madden SF, Hughes DJ, Pardini
B, Naccarati A, Levy M, Vodicka P, Neary P, Dowling P and Clynes M:
Circulating miRNAs miR-34a and miR-150 associated with colorectal
cancer progression. BMC Cancer. 15:3292015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu K, Zhao H, Yao H, Lei S, Lei Z, Li T
and Qi H: MicroRNA-124 regulates the proliferation of colorectal
cancer cells by targeting iASPP. Biomed Res Int.
2013:8675372013.PubMed/NCBI
|
|
7
|
Wang WH, Chen J, Zhao F, Zhang BR, Yu HS,
Jin HY and Dai JH: MiR-150-5p suppresses colorectal cancer cell
migration and invasion through targeting MUC4. Asian Pac J Cancer
Prev. 15:6269–6273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sarlinova M, Halasa M, Mistuna D, Musak L,
Iliev R, Slaby O, Mazuchova J, Valentova V, Plank L and Halasova E:
miR-21, miR-221 and miR-150 Are Deregulated in Peripheral Blood of
Patients with Colorectal Cancer. Anticancer Res. 36:5449–5454.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin YC, Kuo MW, Yu J, Kuo HH, Lin RJ, Lo
WL and Yu AL: c-Myb is an evolutionary conserved miR-150 target and
miR-150/c-Myb interaction is important for embryonic development.
Mol Biol Evol. 25:2189–2198. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Watanabe A, Tagawa H, Yamashita J, Teshima
K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T,
et al: The role of microRNA-150 as a tumor suppressor in malignant
lymphoma. Leukemia. 25:1324–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pizzini S, Bisognin A, Mandruzzato S,
Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M,
Pilati P, et al: Impact of microRNAs on regulatory networks and
pathways in human colorectal carcinogenesis and development of
metastasis. BMC Genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng J, Yang Y, Zhang P, Wang F, Ma Y, Qin
H and Wang Y: miR-150 functions as a tumour suppressor in human
colorectal cancer by targeting c-Myb. J Cell Mol Med. 18:2125–2134.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gillotin S: iASPP, a potential drug target
in cancer therapy. Leuk Res. 33:1175–1177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Slee EA, Gillotin S, Bergamaschi D, Royer
C, Llanos S, Ali S, Jin B, Trigiante G and Lu X: The N-terminus of
a novel isoform of human iASPP is required for its cytoplasmic
localization. Oncogene. 23:9007–9016. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bell HS and Ryan KM: iASPP inhibition:
Increased options in targeting the p53 family for cancer therapy.
Cancer Res. 68:4959–4962. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai Y, Qiu S, Gao X, Gu SZ and Liu ZJ:
iASPP inhibits p53-independent apoptosis by inhibiting
transcriptional activity of p63/p73 on promoters of proapoptotic
genes. Apoptosis. 17:777–783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin W, Zhao Z, Ni Z, Zhao Y, Du W and Chen
S: IFI16 restoration in hepatocellular carcinoma induces tumour
inhibition via activation of p53 signals and inflammasome. Cell
Prolif. 50:e123922017. View Article : Google Scholar
|
|
23
|
Amirkhah R, Schmitz U, Linnebacher M,
Wolkenhauer O and Farazmand A: MicroRNA-mRNA interactions in
colorectal cancer and their role in tumor progression. Genes
Chromosomes Cancer. 54:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang S, Chen Y, Wu W, Ouyang N, Chen J,
Li H, Liu X, Su F, Lin L and Yao Y: miR-150 promotes human breast
cancer growth and malignant behavior by targeting the pro-apoptotic
purinergic P2X7 receptor. PLoS One. 8:e807072013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao M, Hou D, Liang H, Gong F, Wang Y, Yan
X, Jiang X, Wang C, Zhang J, Zen K, et al: miR-150 promotes the
proliferation and migration of lung cancer cells by targeting SRC
kinase signalling inhibitor 1. Eur J Cancer. 50:1013–1024. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu Y, Ge W, Wang X, Sutendra G, Zhao K,
Dedeić Z, Slee EA, Baer C and Lu X: Caspase cleavage of iASPP
potentiates its ability to inhibit p53 and NF-κB. Oncotarget.
6:42478–42490. 2015. View Article : Google Scholar : PubMed/NCBI
|