Introduction
Lung cancer is a leading cause of tumor-related
morbidity and mortality worldwide (1,2).
Despite recent breakthroughs in the diagnosis and treatment of this
disease, the prognosis of patients remains poor, especially for
those with metastatic disease (2).
Standard treatment for lung cancer includes traditional
chemotherapeutic agents such as cisplatin and docetaxel; however,
their efficacy is somewhat hindered by the development of drug
resistance (3). In the case of
non-small cell lung cancer (NSCLC), newer targeted-molecular
therapies have shown great promise (4,5), yet
the majority of patients with advanced disease still develop drug
resistance within one year (6).
Therefore, there is a pressing need to develop new therapeutic
options in the fight against lung cancer, and to better understand
the mechanisms leading to drug resistance.
Drug resistance has been linked to changes in the
DNA repair capacity within tumor cells (7). Chemotherapeutic drugs such as
cisplatin induce DNA damage in tumor cells, and, in turn, activate
the DNA damage response. Activation of the DNA damage response by
chemotherapeutic drugs can have the desired effect of tumor cell
apoptosis, or the cells may repair the lesion, leading to
chemoresistance.
Components of the DNA damage response pathway have
not only been implicated in chemoresistance, but they themselves
may play oncogenic roles (8,9).
Indeed, the capacity to repair damaged DNA through homologous
recombination has previously been implicated in patient
susceptibility to lung cancer (10,11).
For example, NSCLC patients expressing high levels of RAD51, a
protein involved in homologous recombination and DNA repair, had
significantly poorer 5-year survival rates compared to those with
low RAD51 expression (12).
Similarly, the DNA repair protein RAD52 was implicated as an
oncogene in lung squamous cell carcinoma, with enhanced tumor cell
death observed in mouse cells ablated of RAD52 (9,13).
Therefore, it is worth exploring components of the DNA damage
response pathway, such as RAD1/2, as antitumor drug targets in lung
cancer.
In the present study, we examined the role of RAD52
motif-containing protein 1 (RDM1) in NSCLC. The RDM1 gene
was identified through a database search looking for proteins
similar to RAD52 (14). Like RAD52,
RDM1 is involved in DNA double-strand break repair and homologous
recombination (14,15). Previous research has shown that RDM1
binds to cisplatin-damaged DNA in vitro, suggesting its
possible role in the development of chemoresistance (14). As further evidence, RDM1 ablation in
a chicken B cell line DT40 resulted in a more than 3-fold increase
in cisplatin sensitivity (14).
However, RDM1-knockout cells were not hypersensitive to DNA damage
caused by ionizing radiation, ultraviolet irradiation, or
alkylation (14). Therefore, the
precise role of RDM1 in cisplatin resistance remains unclear.
Subsequent research has shown that RDM1 expression
is modulated by heat shock (16).
More recent studies revealed that the DNA-binding capability of
RDM1 is enhanced at low pH, indicating that it may function in
acidic conditions such as the tumor microenvironment (17). Furthermore, RDM1 has been shown to
be upregulated at the mRNA level in lung squamous cell carcinoma
(18). Nonetheless, despite the
growing evidence, the precise function of RDM1 in NSCLC remains
largely unknown. Therefore, this study aimed to clarify the role of
RDM1 in NSCLC.
Materials and methods
Patients and tissue specimens
Paraffin-embedded tumors and adjacent non-cancerous
tissue specimens were collected from a series of 103 patients with
NSCLC who had only undergone surgical resection, but had not
received chemotherapy between January 1, 2015 and 31 December 31,
2016 at the Inner Mongolia Autonomous Region People's Hospital,
Hohhot, Inner Mongolia, China. The study group consisted of 77 men
(75%) and 26 women (25%), and the mean age was 60.7 years (range,
32–81 years). The specimens were classified by their
tumor-node-metastasis (TNM) stage (19). Detailed patient information
including gender, age, tumor size, histological differentiation,
and lymph node metastasis is documented in Table I. The study was approved by the
Medical Ethics Committee of Inner Mongolia Autonomous Region
People's Hospital, and all participants provided written, informed
consent.
 | Table I.Association between RDM1 expression
and the clinicopathological characteristics of the NSCLC cases. |
Table I.
Association between RDM1 expression
and the clinicopathological characteristics of the NSCLC cases.
|
|
| RDM1
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Total | Negative
(n=45) | Positive
(n=58) | P-value |
|---|
| Age (mean ± SD,
years) | 103 | 61.8±10.2 | 59.6±11.7 | 0.368 |
| Gender, n (%) |
|
Male | 77 | 32 (41.5) | 45 (58.5) | 0.447 |
|
Female | 26 | 13 (50) | 13 (50) |
|
| Tumor size (cm), n
(%) |
| ≤3 | 38 | 27 (71.1) | 11 (28.9) | 0.007a |
|
>3 | 65 | 18 (27.7) | 47 (72.3) |
|
| Histological
differentiation, n (%) |
|
Well | 7 | 5 (71.4) | 2 (28.6) | 0.013a |
|
Moderate | 35 | 19 (54.3) | 16 (45.7) |
|
|
Poor | 61 | 21 (34.4) | 40 (65.6) |
|
| Histological
classification, n (%) |
|
Squamous carcinoma | 61 (59) | 24 (39.3) | 37 (61.7) | 0.653 |
|
Adenocarcinoma | 12 (12) | 7 (58.3) | 5 (41.7) |
|
|
Adenosquamous | 10 (10) | 4 (40) | 6 (60) |
|
|
Others | 16 (3) | 9 (56.2) | 7 (43.8) |
|
| Lymph node
metastasis, n (%) |
|
Absent | 42 | 24 (57.1) | 18 (42.9) | 0.034a |
|
Present | 61 | 21 (34.4) | 40 (65.6) |
|
| TNM stage, n
(%) |
|
I+II | 59 | 37 (62.7) | 22 (37.3) | 0.004a |
|
III+IV | 44 | 8 (18.2) | 36 (81.8) |
|
| 1-year survival
rate, n (%) | 48 (46.6) | 31 (68.9) | 16 (27.6) | 0.045a |
Immunohistochemistry
Paraffin-embedded 4-µm tissue sections were stained
with the anti-RDM1 antibody. In brief, deparaffinized tissue
sections were processed for antigen retrieval using target
retrieval solution (pH 9.0; Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA) at 120°C for 4 min. Non-specific immunoreactions
were blocked at room temperature for 30 min using the Dako
Protein-Block kit (Agilent Technologies, Inc.). Sections were
incubated at 4°C overnight with the Invitrogen RDM1 polyclonal
primary antibody (1:100; cat. no. PA5-50103; Thermo Fisher
Scientific, Inc., Waltham, MA, US)A, followed by incubation with
3,3′-diaminobenzidine tetrahydrochloride (Dako; Agilent
Technologies, Inc.) for 5 min. Nuclear counter staining was
conducted with hematoxylin. Images were captured using a light
microscope (Olympus BX50; Olympus Corp., Tokyo, Japan) and
processed using Photoshop software (version 7.0; Adobe, San Jose,
CA, USA).
Cell culture
Human lung cancer cell lines, including NCI-H1299,
A549, H1688 and H1975, were obtained from the Cell Bank of the
Chinese Academy of Science (Shanghai, China). Human normal lung
epithelial BEAS-2B cells were used as control. Cells were cultured
in Gibco™ RPMI-1640 medium (Thermo Fisher Scientific, Inc.),
containing 10% HyClone™ fetal bovine serum (FBS; GE Healthcare
Bio-Sciences, Pittsburgh, PA, USA) and 100 U/ml
penicillin/streptomycin. All cells were incubated at 37°C in a
humidified atmosphere containing 5% CO2.
Knockdown of RDM1 by lentivirus
The target sequence for RDM1-specific shRNA (shRDM1:
5′-GGCCCATCCTGGTTTCTAT-3′) and the negative control shRNA (shCtrl:
5′-TTCTCCGAACGTGTCACGT-3′) were obtained from GeneChem Biotech
(Shanghai, China). The NCI-H1299 cell line was transfected with
shRDM1 or shCtrl using transfection reagents (c1507; Applygen
Technologies, Inc., Beijing, China) according to the manufacturer's
instructions.
Western blot analysis. Lung tissue samples and human
lung cancer cell lines were homogenized in RIPA buffer (50 mmol/l
Tris-HCl, 150 mmol/l NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1%
SDS, 2 mmol/l Na3VO4, 500 mmol/l NaF, 58 µg/ml aprotinin, 10 µg/ml
leupeptin, and 2 mmol/l phenylmethylsulfonyl fluoride) using the
Multi Beads Shocker system (Yasui-Kikai, Osaka, Japan) at 4°C.
Protein samples (15 µg) were separated by SDS-PAGE using a gradient
gel (5–20%; ATTO, Tokyo, Japan), followed by electroblotting onto
polyvinylidene difluoride membranes (ATTO) and incubation in a
blocking buffer of 5% non-fat milk for 1 h at room temperature.
Membranes were incubated with the following primary
antibodies: rabbit anti-RDM1 (1:1,000; cat. no. ab201293; Abcam)
and mouse anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH;
1:5,000; Chemicon, Temecula, CA, USA) at 4°C overnight. Membranes
were visualized by incubation with appropriate horseradish
peroxidase-linked secondary antibodies, followed by enhanced
chemiluminescence (ECL) detection using ECL Plus western blotting
detection reagents (Amersham Biosciences, Little Chalfont, UK). The
immunoreactive bands of targeted proteins were quantified by VH
analyzer software (VH-H1A5; Keyence Corp., Osaka, Japan),
normalized by GAPDH levels, and determined relative to the sham
values. All experiments were repeated three times.
Quantitative polymerase chain reaction
(qPCR)
Total RNA was extracted from lung tissue samples and
human lung cancer cell lines using the RNeasy Mini Kit (Qiagen
Sciences, Inc., Gaithersburg, MD, USA), purified with the
RNase-Free DNase Set (Qiagen), and used as a template for cDNA
synthesis by the PrimeScript RT Reagent Kit (Takara, Shiga, Japan).
PCR reactions were performed in a Thermal Cycler Dice Real Time
System Tp800 (Takara) using SYBR Premix ex Taq (Takara)
according to the manufacturer's instructions as follows: 30 sec at
95°C, then 40 cycles of 5 sec at 95°C and 30 sec at 60°C; then for
the dissociation stage, 15 sec at 95°C, 30 sec at 60°C, and 15 sec
at 95°C. The sequences of primers were: RDM1 forward primer,
5′-GCCCATCCTGGTTTCTATGCC-3′ and reverse primer,
5′-AGACGAACCTTGACTGGAGAT-3′; GAPDH forward primer,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse primer,
5′-CACCCTGTTGCTGTAGCCAAA-3′. Relative gene expression data were
determined by the standard curve method, and fold-changes were
normalized by GAPDH, and relative to control values. Each sample
was tested in triplicate.
Detection of cell proliferation
The NCI-H1299 cell line was seeded into 96-well
plates at a density of 2000 cells per well, and infected with the
control or RDM1-siRNA lentivirus. Cell numbers were then counted
via Celigo S Image Cytometer (Nexcelom Bioscience, Ltd.,
Manchester, UK) every day for 5 days to determine the growth rate.
All samples were assayed in triplicate.
Detection of caspase-3/-7
activity
After lentivirus infection, caspase-3/-7 activity
was detected using the Caspase-Glo® 3/7 Assay (G8091;
Promega, Madison, WI, USA) according to the manufacturer's
instructions. In brief, the NCI-H1299 cell line was seeded into
96-well plates at a density of 2,000 cells per well, and harvested
when the confluence reached 80%. Caspase-Glo® 3/7
Reagent (100 µl) was added to each well of a white-walled 96-well
plate containing 100 µl of blank, negative control cells, or
treated cells in culture medium, and incubated in the dark at room
temperature for 30 min to 3 h. The reaction product was detected at
405 nm using a plate-reading luminometer (M2009PR, Tecan Infinite,
Switzerland). All samples were assayed in triplicate.
Determination of apoptosis by flow
cytometry
The Invitrogen™ eBioscience™ Annexin V Apoptosis
Detection Kit APC (88–8007; Thermo Fisher Scientific, Inc.) was
used for labeling of apoptotic cells according to the
manufacturer's protocol. The cells from each group were collected
after lentivirus infection, washed twice with ice-cold
phosphate-buffered saline (PBS) and resuspended in 200 µl binding
buffer containing 10 µl Annexin V-APC. Fluorescence intensity was
measured by flow cytometry (BD Biosciences). All samples were
assayed in triplicate.
Cell cycle analysis by flow
cytometry
After lentivirus infection, the NCI-H1299 cell line
was seeded into 6-well plates at a density of 2×105
cells per well, and harvested when the confluence reached 80%. The
collected cells were fixed in 75% cold ethanol (10009269; Sinopharm
Chemical Reagent, Co., Ltd., Shanghai, China) for 1 h at 4°C. The
cells were then washed with cold PBS and stained with 500 µl
propidium iodide (PI) buffer (P4170 Sigma; Merck KGaA, Darmstadt,
Germany) for 1 h at 37°C in the dark. The cell cycle stages were
analyzed by flow cytometry. All samples were assayed in
triplicate.
Statistical analysis
All experiments were repeated three times. SPSS
software v. 17.0 (SPSS, Inc., Chicago, IL, USA) was used to analyze
the results using either Student's t-test or one-way analysis of
variance. A P-value <0.05 was considered statistically
significant.
Results
RDM1 is highly expressed in lung
cancer tissues and cell lines
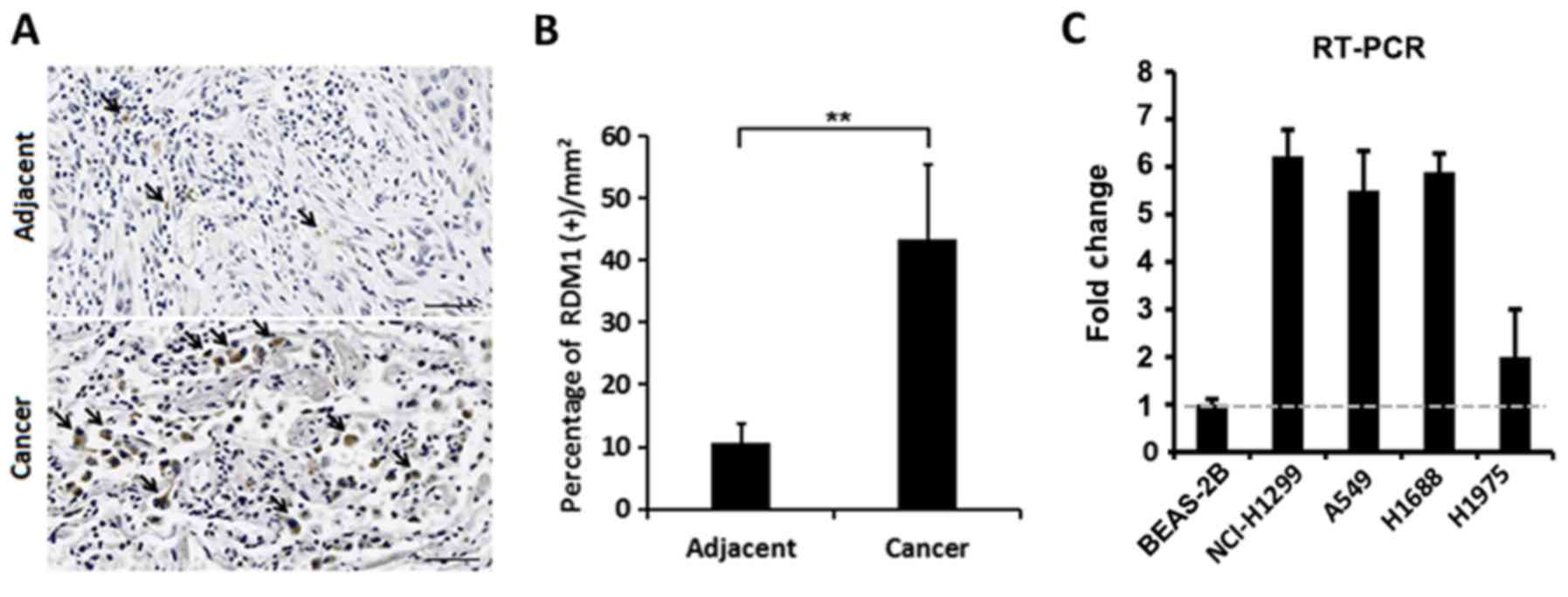
Using immunohistochemistry, we found that RDM1
protein expression was significantly elevated in the cell nuclei of
lung cancer tissues compared to that in adjacent non-cancerous
tissue (Fig. 1A and B). We also
found that expression of RDM1 mRNA was elevated in lung cancer cell
lines (NCI-H1299, A549, H1688 and H1975) when compared with that
noted in the human normal lung epithelial BEAS-2B cells (Fig. 1C). Therefore, the expression of RDM1
appears to be upregulated in lung cancer.
RDM1 expression correlates with tumor
size, histological differentiation, lymph node metastasis and TNM
staging in lung cancer
We then assessed the correlation between RDM1
expression and the clinicopathological parameters of lung cancer
patients. Among the 103 tissue samples obtained from NSCLC
patients, 58 showed positive RDM1 protein expression and 45 were
negative. As shown in Table I, RDM1
expression in NSCLC tissues was significantly correlated with tumor
size, histological differentiation, the degree of lymph node
metastasis, TNM stage and 1-year survival rate (all P<0.05);
however, it was not directly associated with age, gender or
histological classification.
RDM1 knockdown significantly reduces
the cellular proliferation rate and increases apoptosis in a lung
cancer cell line
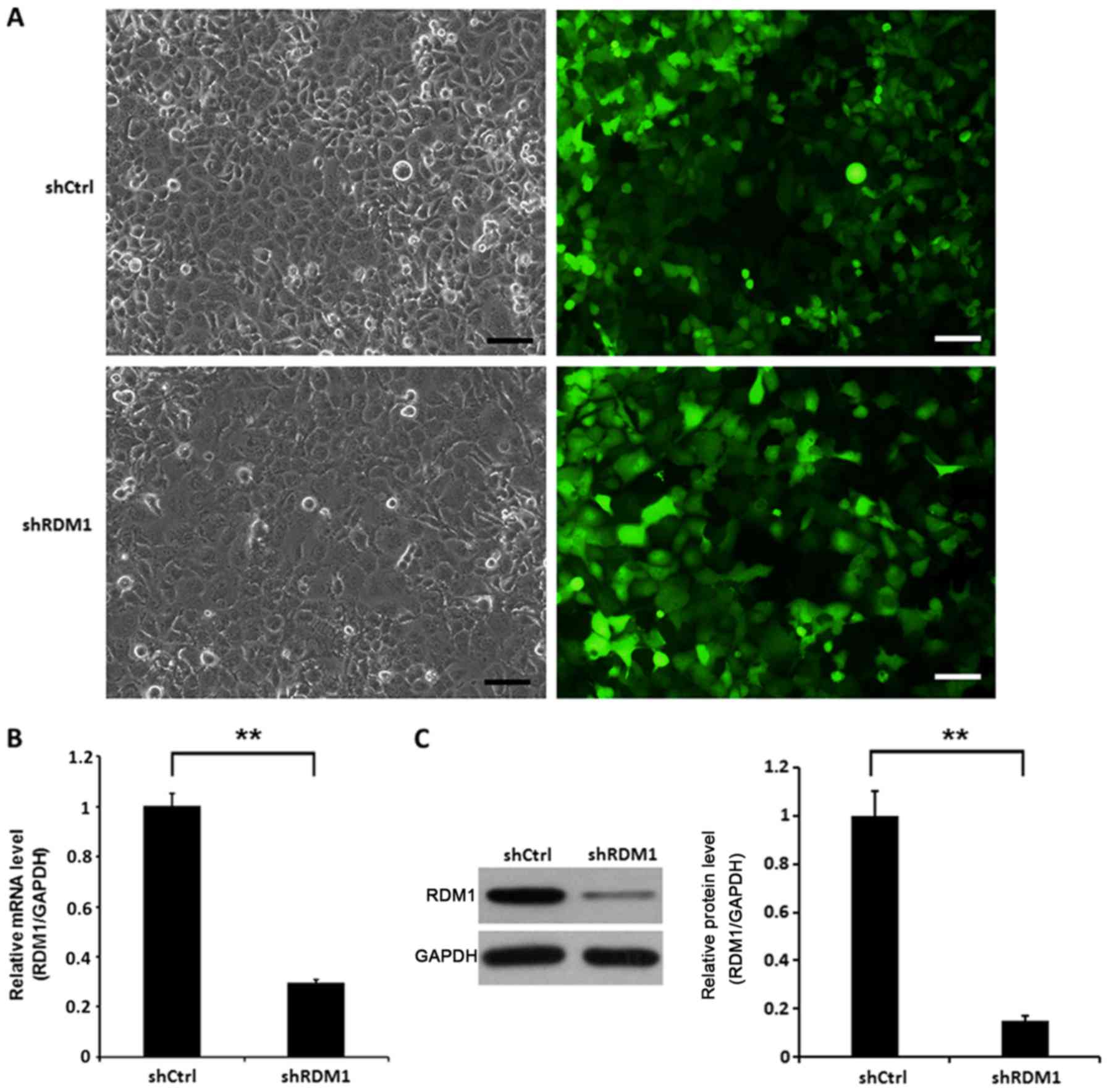
To investigate the role of RDM1 in vitro, we
knocked down RDM1 in a human NSCLC cell line (NCI-H1299)
using lentiviral vector-expressed siRNA. NCI-H1299 cells infected
with RDM1-siRNA showed significantly reduced RDM1 mRNA and protein
levels at day 3 post-infection compared to the controls (P<0.01;
Fig. 2).
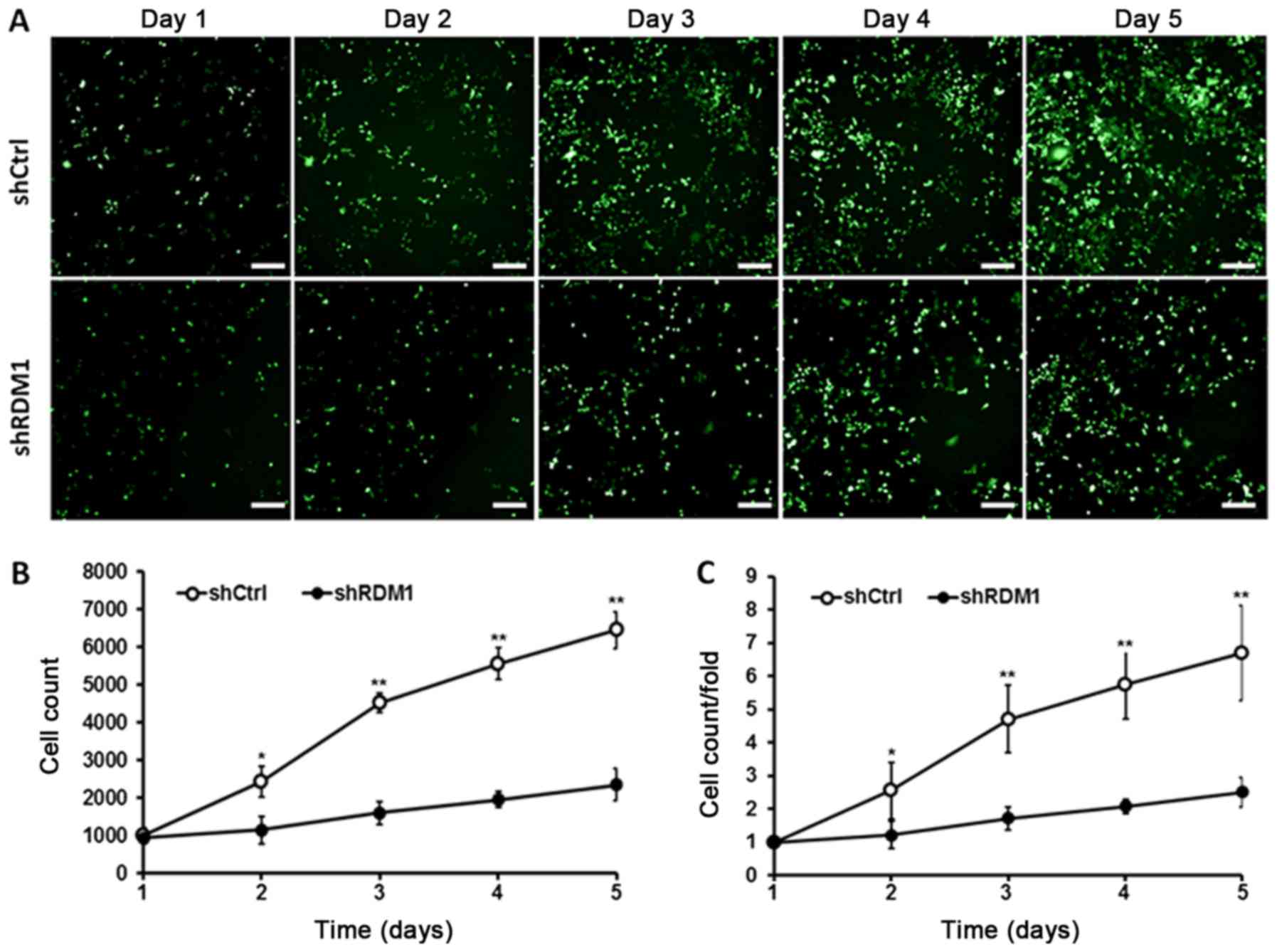
We then monitored the effect of RDM1
knockdown on lung cancer cell growth and apoptosis. We found that
NCI-H1299 cells transfected with RDM1-siRNA showed a significant
reduction in the cell growth rate over 5 days compared to the
controls (P<0.01; Fig. 3). In
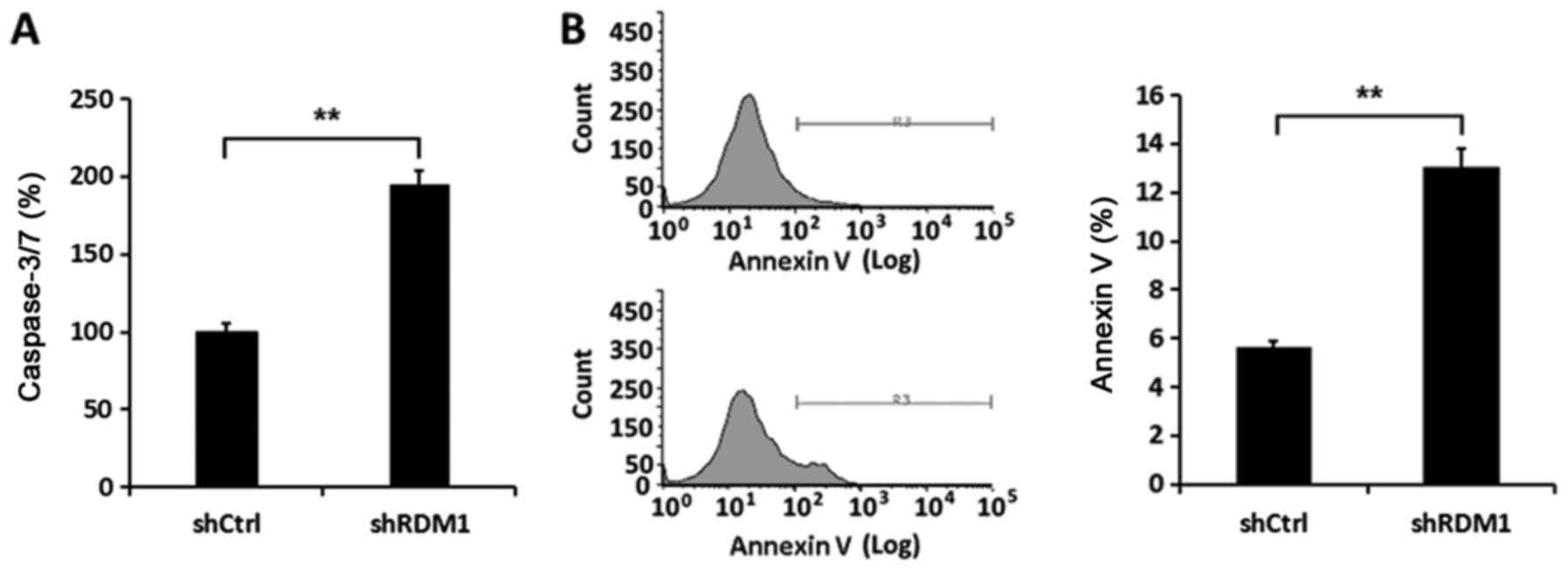
addition, the activity of caspase-3/-7 in NCI-H1299 cells infected
with RDM1-siRNA was enhanced compared to that noted in the controls
(P<0.01; Fig. 4A). Flow
cytometric analysis confirmed there was a significant increase in
the apoptosis rate in the NCI-H1299 cells in which RDM1 had
been knocked down (P<0.01; Fig.
4B). Therefore, RDM1 promoted cell proliferation and inhibited
apoptosis in a lung cancer cell line.
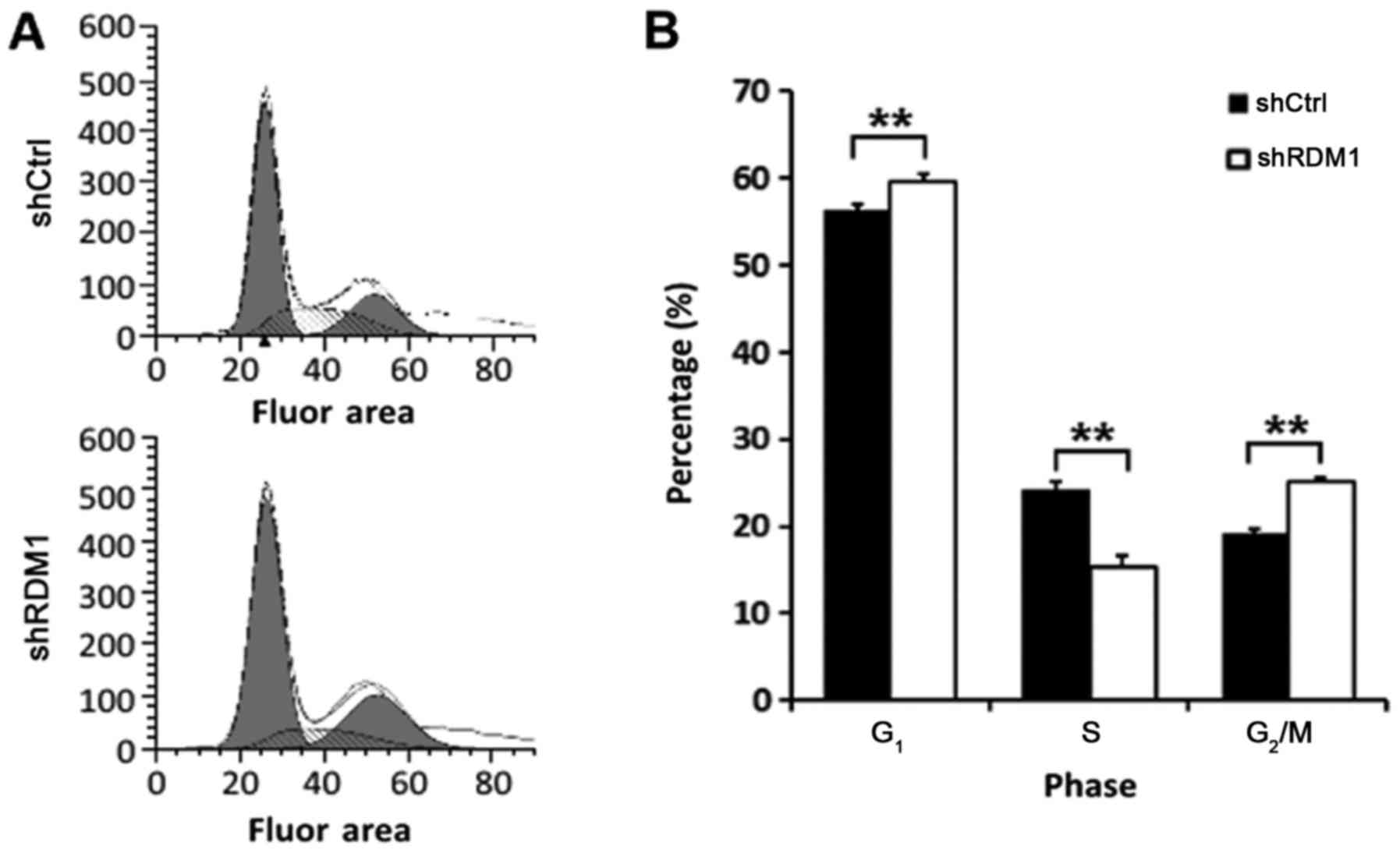
RDM1 regulates the cell cycle
Next, we examined the effect of RDM1
knockdown on the cell cycle using flow cytometry. RDM1
knockdown resulted in a significant increase in the proportion of
lung cancer cells in the G1 and G2/M phases
and a decreased proportion in the S phase (P<0.01; Fig. 5). These results suggest that RDM1
may be involved in DNA repair in lung cancer cells, thereby
promoting progression through the DNA damage checkpoint
(G2/M) of the cell cycle and avoiding apoptosis.
Discussion
The present study aimed to clarify the role of RAD52
motif-containing protein 1 (RDM1) in non-small cell lung cancer
(NSCLC). We found elevated expression of RDM1 at both the mRNA and
protein levels in NSCLC tissues and cell lines. To the best of our
knowledge, this study first reported that RDM1 protein expression
in NSCLC tissues correlates with tumor size, histological
differentiation, lymph node metastasis, TNM stage and 1-year
survival rate. Meanwhile, RDM1 knockdown in a human NSCLC
cell line significantly reduced the cellular proliferation rate and
increased apoptosis. In addition, RDM1 knockdown enhanced
the activity of caspase-3/-7. Finally, we provide evidence that
RDM1 may function as a DNA repair protein that promotes cell cycle
progression of lung cancer cells. Taken together, our results
indicate that RDM1 could be a molecular marker of NSCLC and may
represent a novel drug target.
DNA repair proteins, such as RAD52 and RDM1, have
previously been implicated in the development of resistance to
cancer therapy, including resistance to platinum-based chemotherapy
(14,15,20)
and radiotherapy (21). In addition
to drug resistance, proteins involved in DNA repair pathways have
been implicated in tumor progression (22). For example, RAD52 has been linked to
both the progression and risk of small cell lung cancer (SCLC) and
NSCLC (8,9,23). In
addition, RDM1 was previously shown to undergo stress-induced
nucleolar accumulation, indicating it may function in the
heat-shock response that is implicated in tumorigenesis (16).
As further evidence of the role of RDM1 in lung
cancer progression, here we showed that RDM1 expression was
correlated with tumor size and metastasis in NSCLC patients. In
addition, we found that increased RDM1 expression promoted cell
growth and reduced apoptosis in an NSCLC cell line. This indicates
that RDM1 plays a similar role to RAD52 in promoting the
progression of lung cancer, through increasing cell survival and
proliferation. However, the mechanism by which RDM1 promotes cell
proliferation requires further investigation.
A previous study found an abundance of RDM1
transcripts in the testis, which implies that RDM1 may play a
possible role in spermatogenesis (14). During spermatogenesis, immature
sperm cells undergo rounds of mitotic and meiotic division and the
control of genetic stability is paramount. We hypothesize that RDM1
may have a similar function in controlling genomic stability in
NSCLC cells, which allows the cells to proliferate. Indeed, we
found that knockdown of RDM1 resulted in a decreased proportion of
NSCLC cells in the S phase, with an increased proportion in the
G1 and G2/M phases. This indicates that the
cells were arrested at the G2/M DNA damage checkpoint,
which serves to protect genomic integrity and prevent cells with
damaged DNA from entering the S phase. In turn, cells with
unrepairable DNA lesions undergo permanent arrest or apoptosis,
whereas if the damage is repaired, for example by RDM1,
checkpoint-arrested cells may resume cell cycle progression
(24). Therefore, RDM1 appears to
promote cell cycle progression, likely through repairing DNA
double-strand breaks.
Our results are similar to those previously observed
for the related DNA repair protein, RAD52, in lung cancer (8,9). RAD52
is predominantly recruited for DNA repair during the S phase of the
cell cycle and plays a crucial role in the regulation of homologous
recombination-related genomic instability in humans (25). Indeed, RAD52 has been shown to
promote mitotic DNA synthesis following replication stress, such as
that occurring during tumor proliferation (26). In particular, RAD52 appears to
repair collapsed DNA replication forks in cancer cells (27). In addition, the recruitment of Rad52
to stalled DNA replication forks in yeast has been shown to be
controlled by various checkpoint and cell cycle signals (28). Therefore, the recruitment of RDM1 in
proliferating lung cancer cells may be under similar control by
cell cycle regulators or signals.
Similar findings have been made for RAD51.
Inhibition of RAD51 transcription in a chicken DT40 cell line
resulted in a high G2/M phase arrest, with high levels
of chromosome-type breaks (29).
Conversely, overexpression of RAD51 protein in tumor cells was
associated with high DNA repair capacity and elevated recombination
rates (30). Therefore, together,
these results imply that lung cancer cells may specifically
overexpress and recruit DNA repair proteins such as RDM1, RAD51 and
RAD52 in order to generate genetically stable clones suitable for
sustained proliferation.
While inhibiting RDM1 seems a promising therapeutic
option for NSCLC, we must remember that DNA repair proteins are
multifunctional. Therefore, any RDM1 inhibitor that is developed
may have unwanted side effects or toxicities. Furthermore, multiple
isoforms of RDM1 have been identified, each with a different
subcellular location, and potentially possessing different
functions (16). Thus, unraveling
the specific functions of each of these isoforms will be key for
reducing any off-target effects. Indeed, specific inhibitors of
proteins involved in homologous recombination for the treatment of
cancer remain in their infancy (22). Nonetheless, better characterization
of DNA repair proteins, such as that performed here for RDM1, will
aid in the discovery of not only more effective DNA repair
inhibitors, but also more efficient anticancer combination
therapies that overcome the issues of drug resistance.
In summary, our data revealed that RDM1 plays a
pivotal role in the survival and proliferation of NSCLC cells by
regulating the cell cycle, likely through its DNA repair
capabilities. Further studies are warranted to clarify the role of
RDM1 in various subtypes of NSCLC, as well as to determine its
prognostic and therapeutic value.
Acknowledgements
We thank Haiyan Zhao, Zhiqiang Wang for the
technical assistance (Inner Mongolia Medical University).
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81560013) and the
Hospital Fund from Inner Mongolia Autonomous Region People's
Hospital (grant no. 201712), Inner Mongolia Autonomous Region,
China.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
GX, DS and JS conceived and designed the study. GX,
JD, EW, FZ and RH performed the experiments. GX, DS and JS wrote
the paper. GX, JD, EW, FZ, RH, DS and JS reviewed and edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Inner Mongolia Autonomous Region People's Hospital,
and all participants provided written, informed consent.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GAPDH
|
glyceraldehyde 3-phosphate
dehydrogenase
|
|
NSCLC
|
non-small cell lung cancer
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
RDM1
|
RAD52 motif-containing protein 1
|
|
NSCLC
|
non-small cell lung cancer
|
|
TNM
|
tumor-node-metastasis
|
References
|
1
|
Ridge CA, McErlean AM and Ginsberg MS:
Epidemiology of lung cancer. Semin Intervent Radiol. 30:93–98.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Siegel RL and Jemal A: Lung
cancer statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calikusu Z, Yildirim Y, Akcali Z, Sakalli
H, Bal N, Unal I and Ozyilkan O: The effect of HER2 expression on
cisplatin-based chemotherapy in advanced non-small cell lung cancer
patients. J Exp Clin Cancer Res. 28:972009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan BA and Hughes BG: Targeted therapy
for non-small cell lung cancer: Current standards and the promise
of the future. Transl Lung Cancer Res. 4:36–54. 2015.PubMed/NCBI
|
|
5
|
Shtivelman E, Hensing T, Simon GR, Dennis
PA, Otterson GA, Bueno R and Salgia R: Molecular pathways and
therapeutic targets in lung cancer. Oncotarget. 5:1392–1433. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maione P, Sacco PC, Sgambato A, Casaluce
F, Rossi A and Gridelli C: Overcoming resistance to targeted
therapies in NSCLC: Current approaches and clinical application.
Ther Adv Med Oncol. 7:263–273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Salehan MR and Morse HR: DNA damage repair
and tolerance: A role in chemotherapeutic drug resistance. Br J
Biomed Sci. 70:31–40. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lieberman R and You M: Corrupting the DNA
damage response: A critical role for Rad52 in tumor cell survival.
Aging. 9:1647–1659. 2017.PubMed/NCBI
|
|
9
|
Lieberman R, Pan J, Zhang Q and You M:
Rad52 deficiency decreases development of lung squamous cell
carcinomas by enhancing immuno-surveillance. Oncotarget.
8:34032–34044. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rajaee-Behbahani N, Schmezer P, Risch A,
Rittgen W, Kayser KW, Dienemann H, Schulz V, Drings P, Thiel S and
Bartsch H: Altered DNA repair capacity and bleomycin sensitivity as
risk markers for non-small cell lung cancer. Int J Cancer.
95:86–91. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spitz MR, Wei Q, Dong Q, Amos CI and Wu X:
Genetic susceptibility to lung cancer: The role of DNA damage and
repair. Cancer Epidemiol Biomarkers Prev. 12:689–698.
2003.PubMed/NCBI
|
|
12
|
Qiao GB, Wu YL, Yang XN, Zhong WZ, Xie D,
Guan XY, Fischer D, Kolberg HC, Kruger S and Stuerzbecher HW:
High-level expression of Rad51 is an independent prognostic marker
of survival in non-small-cell lung cancer patients. Br J Cancer.
93:137–143. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Benson FE, Baumann P and West SC:
Synergistic actions of Rad51 and Rad52 in recombination and DNA
repair. Nature. 391:401–404. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hamimes S, Arakawa H, Stasiak AZ, Kierzek
AM, Hirano S, Yang YG, Takata M, Stasiak A, Buerstedde JM and Van
Dyck E: RDM1, a novel RNA recognition motif (RRM)-containing
protein involved in the cell response to cisplatin in vertebrates.
J Biol Chem. 280:9225–9235. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hamimes S, Bourgeon D, Stasiak AZ, Stasiak
A and Van Dyck E: Nucleic acid-binding properties of the
RRM-containing protein RDM1. Biochem Biophys Res Commun. 344:87–94.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Messaoudi L, Yang YG, Kinomura A, Stavreva
DA, Yan G, Bortolin-Cavaillé ML, Arakawa H, Buerstedde JM, Hainaut
P, Cavaillé J, et al: Subcellular distribution of human RDM1
protein isoforms and their nucleolar accumulation in response to
heat shock and proteotoxic stress. Nucleic Acids Res. 35:6571–6587.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Stubbs M, McSheehy PM, Griffiths JR and
Bashford CL: Causes and consequences of tumour acidity and
implications for treatment. Mol Med Today. 6:15–19. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang F, Chen X, Wei K, Liu D, Xu X, Zhang
X and Shi H: Identification of key transcription factors associated
with lung squamous cell carcinoma. Med Sci Monit. 23:172–206. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (Eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li HM, Yuan P, Yu D, Ma F, Tan WW, Feng T,
Yang J, Huang Y, Lin DX, Xu BH and Tan W: Genetic variation in DNA
repair gene RAD52 is associated with the response to platinum-based
chemotherapy in SCLC patients. Zhonghua Zhong Liu Za Zhi.
38:504–509. 2016.(In Chinese). PubMed/NCBI
|
|
21
|
Ghosh S and Krishna M: Role of Rad52 in
fractionated irradiation induced signaling in A549 lung
adenocarcinoma cells. Mutat Res. 729:61–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kelley MR, Logsdon D and Fishel ML:
Targeting DNA repair pathways for cancer treatment: What's new?
Future Oncol. 10:1215–1237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han S, Gao F, Yang W, Ren Y, Liang X,
Xiong X, Pan W, Zhou L, Zhou C, Ma F and Yang M: Identification of
an SCLC susceptibility rs7963551 genetic polymorphism in a
previously GWAS-identified 12p13.33 RAD52 lung cancer risk locus in
the Chinese population. Int J Clin Exp Med. 8:16528–16535.
2015.PubMed/NCBI
|
|
24
|
Branzei D and Foiani M: Regulation of DNA
repair throughout the cell cycle. Nat Rev Mol Cell Biol. 9:297–308.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lok BH and Powell SN: Molecular pathways:
Understanding the role of Rad52 in homologous recombination for
therapeutic advancement. Clin Cancer Res. 18:6400–6406. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhowmick R, Minocherhomji S and Hickson
ID: RAD52 facilitates mitotic DNA synthesis following replication
stress. Mol Cell. 64:1117–1126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sotiriou SK, Kamileri I, Lugli N,
Evangelou K, Da-Ré C, Huber F, Padayachy L, Tardy S, Nicati NL,
Barriot S, et al: Mammalian RAD52 functions in break-induced
replication repair of collapsed DNA replication forks. Mol Cell.
64:1127–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barlow JH and Rothstein R: Timing is
everything: Cell cycle control of Rad52. Cell Div. 5:72010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sonoda E, Sasaki MS, Buerstedde JM,
Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y and
Takeda S: Rad51-deficient vertebrate cells accumulate chromosomal
breaks prior to cell death. EMBO J. 17:598–608. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Henning W and Sturzbecher HW: Homologous
recombination and cell cycle checkpoints: Rad51 in tumour
progression and therapy resistance. Toxicology. 193:91–109. 2003.
View Article : Google Scholar : PubMed/NCBI
|