Introduction
Degeneration of the intervertebral disc (IVD) is a
pathological process characterized by changes in architecture and
biochemical composition, which alters the ability of the IVD to
bear load and can lead to disk herniation (1–5). IVDs
have a gelatinous center, known as the nucleus pulposus (NP),
encompassed by coaxial lamellae that form the inner and outer
annulus fibrosus. Degeneration of the IVD can cause metabolic
changes in the extracellular matrix (ECM) leading to reduced water
content and a loss of the boundary between the outer annulus
fibrosus and the NP (6–8). The ECM is predominantly composed of
collagen type II and proteoglycans, mainly in the form of aggrecan
(9,10). Aggrecan occurs in the NP and in the
inner annulus, where it is associated with cartilage
differentiation (10). Studies
using tissue isolated from surgical patients have revealed that ECM
degeneration is associated with loss of proteoglycan content
(11). Spinal degenerative diseases
are characterized by bone marrow and endplate lesions that are
visible on magnetic resonance imaging (12). These degenerative alterations in the
vertebral end plates are known as modic changes and are considered
to contribute to pain development (13,14).
Modic changes are not only related to traumatic injury, bacterial
infection and genetics but are also associated with localized
inflammation, since proinflammatory crosstalk between the bone
marrow and IVDs has recently been identified (14–16).
Proinflammatory cytokines are reported to play an
important role in ECM breakdown in relation to IVD degeneration
(17,18). Levels of interleukin (IL)-1β, IL-6,
IL-10 and tumor necrosis factor (TNF)-α have been found to be
significantly elevated in patients with IVD degeneration (19) and played an important role in IVD
degeneration (20). IL-1β is a
predominate cytokine that is upregulated in degenerating IVDs and
is considered to be a principal mediator in the breakdown of ECM
(21). TNF-α can enhance
matrix-degrading enzyme activity and increase NP cell apoptosis
(17,22,23).
Li et al also reported that TNF-α induced NP cell
inflammation and apoptosis by activating NF-κB signalling (24). Heme oxygenase-1 (HO-1) has also been
reported to participate in the cellular anti-inflammatory processes
relating to IVD degeneration (25).
When HO-1 is induced, the effects of IL-1β on the expression of the
catabolic markers matrix metalloproteinases (MMP)-1, 3, 9 and 13
are reversed (25).
Toll-like receptor (TLR) signaling pathways were
also reported to increase the level of proinflammatory cytokines,
TNF-a, IL-1, IL-6 and IL-8, leading to inflammation and pain
(26,27). The functions of the toll-like
receptor 4 (TLR4) proteins included the activation of the innate
immune system (28). In addition,
TLR4 was highly expressed in cartilage with advanced OA and played
a key role in cartilage degradation (29,30).
IVD cells have been found to express TLR4 and respond to TLR4
activation induced by lipopolysaccharide (LPS) treatment by
upregulating a coordinated set of inflammatory cytokines and
inhibiting ECM expression in IVD in vitro and in vivo
(31). Ligand binding to TLRs
initiated a signaling cascade that led to the activation of nuclear
factor (NF)-κB and MAPKs, which promoted the production of
inflammatory cytokines, chemokines and degradative enzymes
(32). NF-κB is an heterodimer
consisting of p65 and p50 subunits and is sequestered in the
cytoplasm by inhibitor proteins, the IκBs (33). TLR4 is known to recognize LPS, a
component found in bacterial cells (34). As a TLR ligand, LPS can decrease
proteoglycan (PG) synthesis, aggrecan production, and the
expression of collagen II by TLR4 signaling in both murine and
human articular chondrocytes (30).
IL-1 and LPS-mediated NP cell inflammation and PG and
matrix-degrading enzyme production were antagonized by LfcinB
treatment (35). LPS can induce TLR
signalling in intervertebral disc cells, leading to increased
expression of proinflammatory cytokines (36). LPS-induced aggrecan and collagen II
downregulation was inhibited by carthamin yellow through the
suppression of the MAPK pathway activation (37). LPS stimulates NF-κB binding in the
TLR4 gene promoter, whereas TLR4 expression is blocked by NF-κB
inhibitors (31,38). Furthermore, LPS was reported to
induce inflammation in acute kidney injury (AKI) by activating the
TLR4/NF-κB signaling pathway (39).
MicroRNAs (miRNAs) are small (18–24 nucleotides
long) non-protein-coding RNAs that regulate gene expression at the
post-transcriptional level (40).
Microarray expression analyses have revealed a significant miRNA
dysregulation in osteoarthritis which indicated that miRNAs may be
involved in the pathology of degenerative joint diseases (41). miR-140 is known to be a
cartilage-specific miRNA, with a major role in pathogenesis
(42,43). In a recent study, miR-140 was found
to protect chondrocytes against the anti-proliferation and
cell-matrix signaling changes induced by cytokine IL-1β in an
osteoarthritis model (44). Karlsen
et al (45) reported that
overexpressing miR-140 can rescue IL-1β-suppressed aggrecan and the
expression of SOX9 and inhibit inflammation in OA. In a recent
study, miR-140 was found to bind to the 3′-UTR of SIRT1, a member
of the sirtuin family of proteins, in a dual-luciferase reporter
assay performed in 293 cell lines (46). SIRT1 has been shown to deacetylate
and thereby, deactivate the p53 protein (47), which is an essential metabolic
regulatory transcription factor (48). Li et al (49) reported that miR-140-5p inhibited
cell proliferation and inflammatory cytokine secretion by
downregulating TLR4 in synovial fibroblasts. In a previous study
using microarray analysis, we revealed that miR-140 was more
upregulated in articular chondrocytes than in mesenchymal stem
cells and modulated IL-1 response (50). We have also discovered that miR-140
targeted the TLR4 3′-UTR using TargetScan. In the present study, we
assessed the miRNA-140 3′-UTR binding site of TLR4 by a
dual-luciferase reporter assay performed in NP cells. TLR4,
inflammation cytokines, aggrecan and collagen type II expression
were detected by qRT-PCR induced by LPS stimulation in
vitro. TLR4 is a major receptor of LPS. We also assessed the
impact of miR-140 on proinflammatory cytokine levels, aggrecan and
collagen type II expression in relation to TLR4 expression induced
by LPS stimulation. Therefore, we assessed whether miR-140 could
prevent the progression of inflammation and degeneration in
LPS-induced NP cells by inhibiting the expression of TLR4.
Materials and methods
Human tissue samples
The Research Ethics Committee of Changzhou No. 2
People's Hospital (Changzhou, China) approved the resection of all
specimens. Written informed consent was obtained by the patients or
their relatives to obtain human intervertebral tissue at surgery.
NP samples (n=22) were obtained from patients (33–78 years old;
mean age, 53 years) who underwent disc resection surgery or spinal
fusion to relieve lower back pain. In addition the NP specimens
were grouped according to a grading system for IVD degeneration,
which was based on preoperative magnetic resonance images (MRI)
(51). No medications or
anti-inflammatory drugs were used before surgery. In the present
study, samples graded II–III (n=12) were designated the low
degeneration group (used as a relatively normal control); samples
graded IV–V (n=10) were designated the high degeneration group. All
tissues were snap-frozen in liquid nitrogen at the time of surgical
removal and stored at 80°C or were fixed in 10% neutral buffered
formalin until use.
Isolation of human NP cells
NP tissues were identified by their macroscopic
morphology and were carefully separated from any obvious
granulation tissue, cartilaginous endplates, or annulus fibrosus as
previously described (52).
Isolation of NP cells was performed as previously described
(25). Briefly, cells were
extracted from minced NP tissue specimens from 12 low graded
II–III, who underwent posterior discectomy surgery of lumbar
degenerative disease, by digesting in 2 U/ml protease in Dulbecco's
modified Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher
Scientific, Waltham, MA, USA) for 30 min at 37°C followed by 0.25
mg/ml type II collagenase (Gibco; Thermo Fisher Scientific) for 4 h
at 37°C. The cell suspension was centrifuged at 800 × g for 5 min
and NP cells were resuspended in DMEM/F12 (10% FBS, 100 U/ml
penicillin, 100 µg/ml streptomycin and 1% L-glutamine).
Subsequently, the NP cells were treated with 10 µg/ml of LPS
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Cell transfection
For miR-140 overexpression and knockdown, miR-140
mimics, miR-140 inhibitor and two scrambled miRNAs used as negative
controls (mimics NC for miR-140 mimics and inhibitor-NC for miR-140
inhibitor, respectively), were purchased from Shanghai GeneChem,
Co., Ltd. (Shanghai, China). The 20 nM miRNAs were transfected into
NP cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific) following the manufacturer's instructions.
qRT-PCR
Total RNA was extracted from cultured NP cells and
degenerative IVD tissues using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific) following the manufacturer's instructions. RNA
molecules smaller than 200 nucleotides in size were purified using
a mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according
to the manufacturer's instructions, as previously described
(53). Complementary DNA (cDNA) was
synthesized using an M-MLV kit (Life Technologies; Thermo Fisher
Scientific). qRT-PCR was carried out using the Platinum SYBR-Green
qPCR SuperMix-UDG kit (Life Technologies; Thermo Fisher Scientific)
on an ABI Prism 7500 PCR system (Applied Biosystems; Thermo Fisher
Scientific). Data were normalized to β-actin. qRT-PCR analysis of
mature miR-140 was performed using a TaqMan MicroRNA Assay kit
(Applied Biosystems; Thermo Fisher Scientific). Briefly,
TaqMan-based real-time quantification of miR-140 included two
steps: stem-loop RT and real-time PCR. Stem-loop RT primers bound
at the 3′ portion of miR-140 molecules and were reverse transcribed
with reverse transcriptase. Subsequenlty, the RT product was
quantified using conventional TaqMan PCR that included
miR-140-specific forward primer, reverse primer and a dye-labeled
TaqMan probe. The purpose of the tailed forward primer at the 5′
end was to increase the melting temperature (Tm) of the miR-140
molecules. Data were normalized to U6 snRNA. All primers are listed
in Table I. Each test was performed
in triplicate. Relative expression was calculated using the
2−∆∆Ct method (54).
 | Table I.The primer sequences. |
Table I.
The primer sequences.
| Gene | Primer
sequence |
|---|
| TLR4 | F:
5′-TACAGAAGCTGGTGGCTGTG-3′ |
|
| R:
5′-ACCCGCAAGTCTGTGCAATA-3′ |
| IL-6 | F:
5′-AGTGAGGAACAAGCCAGAGC-3′ |
|
| R:
5′-GTGCCCATGCTACATTTGCC-3′ |
| IL-1β | F:
5′-CTGAGCTCGCCAGTGAAATG-3′ |
|
| R:
5′-TCCATGGCCACAACAACTGA-3′ |
| TNF-α | F:
5′-TCTTCTCGAACCCCGAGTGA-3′ |
|
| R:
5′-CTTGGTCTGGTAGGAGACGG-3′ |
| Aggrecan | F:
5′-TCAGTGGTGACTTCACAGGC-3′ |
|
| R:
5′-TTCACCAACCGTAGGAGTGC-3′ |
| Collagen | F:
5′-TGGCAAGCAAGGAGACAGAG-3′ |
| type II | R:
5′-GACCTCTAGGGCCAGAAGGA-3′ |
| β-actin | F:
5′-CGTGACATTAAGGAGAAGCTG-3′ |
|
| R:
5′-CTAGAAGCATTTGCGGTGGAC-3′ |
| miR-140 | RT:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCA-3′ |
|
| F:
5′-GGCTAGTCAGTGGTTTTACCCT-3′ |
|
| R:
5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | F:
5′-CGCTTCGGCAGCACATATAC-3′ |
|
| R:
5′-AAATATGGAACGCTTCACGA-3 |
Western blot analysis
Total protein was extracted from NP cells using a
protein extraction kit and quantified using a BCA assay kit
(Sigma-Aldrich; Merck KGaA). Following separation by SDS-PAGE, 60
µg of protein was transferred to a PVDF membrane and blocked for 1
h with TBS buffer (10 mM Tris-HCl, pH 7.5 and 150 mM NaCl)
containing 5% skimmed milk. The membrane was then probed with
primary antibody at 4°C overnight. The primary antibodies were TLR4
(1:500; cat. no. 13556; Abcam) and IκΒα (1:500; cat. no. 4814),
p-IκΒα (1:500; cat. no. 9246), p65 (1:500; cat. no. 8242), and
p-p65 (1:500; cat. no. 3039) (all were from Cell Signaling
Technology, Beverly, MA, USA). Following three 5-min washes in TBS
containing Tween-20 (TBST), the membrane was probed with secondary
antibody, goat anti-rabbit IgG-HRP (1:10,000; cat. no. ab6721) and
rabbit anti-mouse IgG H&L (HRP) (1:10,000; cat. no. ab6728)
(all from Abcam) at room temperature for 2 h. After further washing
in TBST, immunolabeling was detected using Pierce ECL Western
Blotting Substrate (Pierce Biotechnology; Thermo Fisher Scientifc).
β-actin was included as a control and densitometric analysis was
performed to determine the relative expression of the target
proteins.
Luciferase reporter assay
The luciferase reporter assay was performed as
previously described (39). The
putative miR-140 binding sites in the 3′-UTR of TLR4 was predicted
using TargetScan (http://www.targetscan.org); 3′-UTR cDNA fragments of
TLR4 containing the putative wild-type or mutant miR-140 binding
sites were amplified using the following primers: wild-type 3′-UTR
of TLR4, forward, 5′-GTTTAAGACGTGCTTCAAATATCCA-3′ and reverse,
5′-TGATAAGACCAGGAAGCGGA-3′; mutant 3′-UTR of TLR4, forward,
5′-TCAAATACCATATTATGGTGTA-3′ (the bold text indicates the
mutation in the 3′-UTR of TLR4) and reverse
5′-AAACTTCTGCTGCAACTCTCATT-3′. The amplified cDNA fragments were
subcloned into a psiCHECK-2 vector (Promega, Madison, WI, USA) at
the XhoI and NotI sites downstream of the luciferase
gene. A total of 293 cell lines were co-transfected with the
luciferase reporter systems and miR-140 mimics or mimics NC as
indicated in the figure legends. Luciferase activity was detected
24 h after transfection using the Dual-Luciferase reporter assay
system (Promega) following the manufacturer's instructions. Data
were normalized to Renilla luciferase activity.
Immunohistochemistry (IHC)
Tissues were fixed in 10% neutral buffered formalin,
embedded in paraffin, dewaxed in xylene and dehydrated in alcohol.
Endogenous peroxidase activity was blocked by hydrogen peroxide
pretreatment for 15 min using avidin/biotin blocking kit (ab64212;
Abcam). Paraffin-embedded, formalin-fixed tissues were
immunostained with rabbit polyclonal primary antibody against human
TLR4 (1:500; cat. no. ab13556), IL-1β (1:300; cat. no. ab9722),
IL-6 (1:300; cat. no. ab6672), TNF-α (1:400; cat. no. ab6671),
aggrecan (1:500; cat. no. ab36861) and collagen type II (1:500;
cat. no. ab34712) (all from Abcam) overnight at 4°C. Rabbit IgG
(cat. no. ab109489; Abcam) replaced the primary antibody as
negative control (at an equal protein concentration) after washing
the samples in phosphate-buffered saline (PBS). The second goat
anti-rabbit HRP antibody (cat. no. ab80437; Abcam) was incubated
for 30 min at room temperature. Then, the signal was amplified and
visualized with diaminobenzidine-chromogen, followed by
counterstaining with hematoxylin. TLR4, inflammation cytokines and
ECM protein immunostaining were analyzed using intensity and
distribution measurements as previously described (55,56)
and the staining intensity was determined as follows: 0, no
staining; 1, weak; 2, moderate and 3, strong. The staining
distribution was determined as the percentage of positive cells (0,
none; 1, <25%; 2, 25–75%; and 3, 75–100%). The expression of
TLR4, IL-1β, IL-6, aggrecan and collagen II was evaluated by
multiplying the intensity and distribution. Cells were further
divided according to TLR4 expression into ‘low’ (TLR4 low) and
‘high’ (TLR4 high) groups, according to a cut-off point. The
cut-off point for TLR4 expression was calculated using the X-tile
software program (The Rimm Lab at Yale University (New Haven, CT,
USA); http://www.tissuearray.org/rimmlab) as previously
described (57).
Cell immunofluorescence
For immunofluorescent staining, cells were first
fixed in 4% PFA for 10 min at room temperature, and then incubated
overnight in anti-aggrecan (1:100; cat. no. ab36861) or
anti-collagen II (1:100; cat. no. ab34712) antibody (all from
Abcam), respectively. The following day, cells were labeled with
the corresponding goat anti-rabbit IgG H&L (Cy3®)
preadsorbed ab6939 (Abcam) for 2 h at 37°C, counterstained with
DAPI and observed under an Olympus FluoView 2000 laser scanning
confocal microscope (Olympus Corp., Tokyo, Japan).
Statistical analysis
Results are presented as the mean ± standard
deviation (SD). Data analysis was performed using SPSS version 13.0
(SPSS, Inc., Chicago, IL, USA). Student's t-test or one-way ANOVA
was applied to test differences between the groups. Pearson's
Chi-square test was used to analyze the relationship between the
TLR4 expression and the clinicopathological features. P<0.05
were considered to indicate a statistically significant result.
Results
Higher expression of TLR4 and
inflammatory cytokines and lower expression of miR-140 and ECM in
high-grade IVD degeneration tissues compared with low-grade
tissue
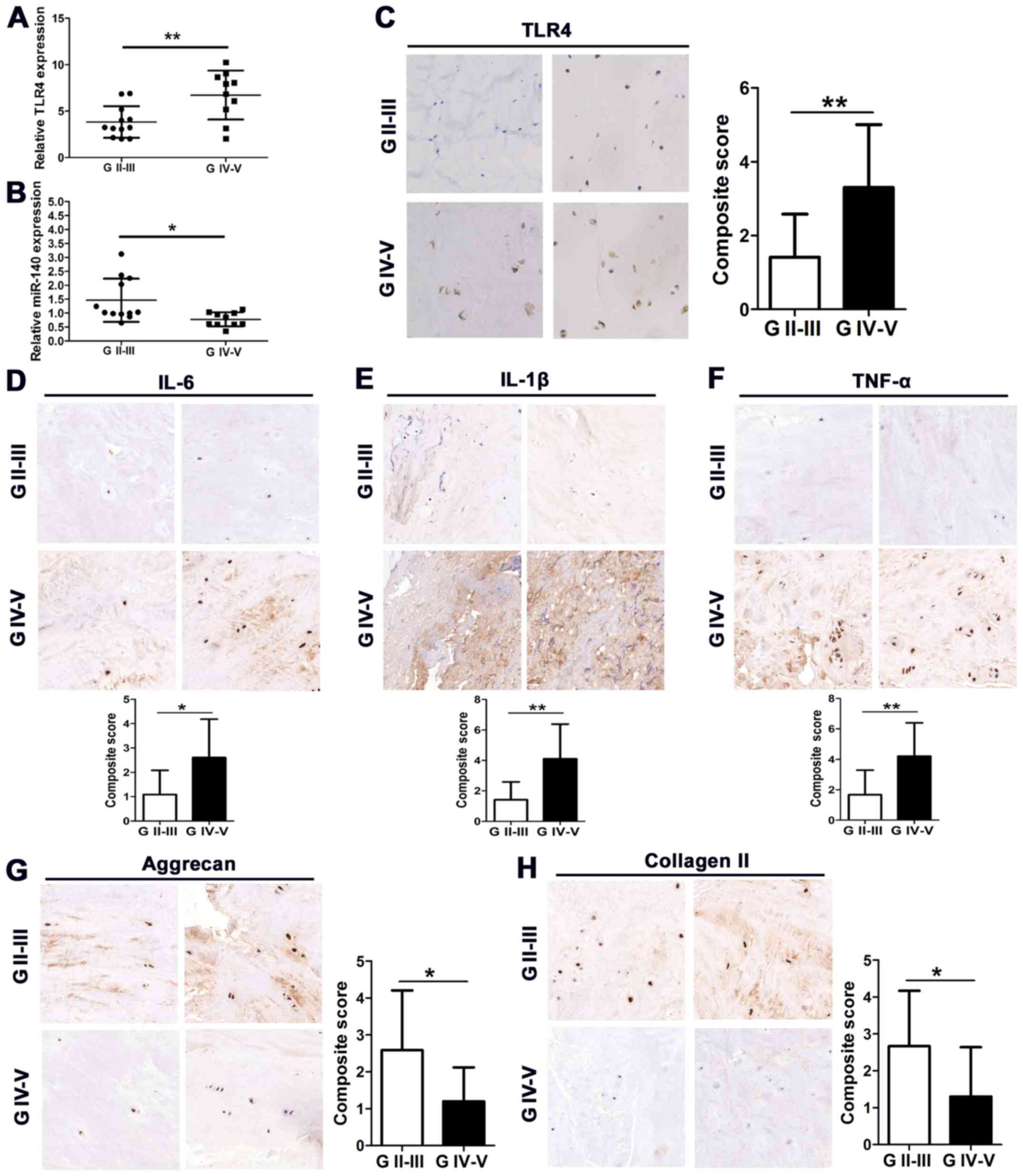
The expression of TLR4 in the clinical
characteristics of patients with high and low-grade IVD
degeneration are detailed in Table
II. There was no significant difference between the level of
TLR4 expression and age, sex, body mass index and disc level.
However, TLR4 was expressed at a significantly higher level in the
tissue of patients with high-grade IVD degeneration and at a
significantly lower level in the tissue of patients with low-grade
IVD degeneration. Relative expression of TLR4 and miR-140 was
assessed by quantitative real-time PCR (qRT-PCR) in high- and
low-grade (used as a relatively normal control) degenerative IVD
tissue (Fig. 1A and B). TLR4 was
found to be expressed at a higher level in grade IV–V tissues
compared to grade II–III (P<0.01). In contrast, miR-140 was
downregulated in grade IV–V tissues compared to grade II–III
tissues (P<0.05). TLR4 was also detected by immunohistochemistry
(IHC) in IVD degenerative tissues. Staining of 12 grade II–III
tissues was less intense than that of 10 grade IV–V tissues
indicating a high level of TLR4 in high-grade tissue (Fig. 1C). In addition, a higher level of
the inflammatory cytokines IL-6, IL-1β and TNF-α was detected in
high-grade tissues than in low-grade tissues (Fig. 1D-F). However, a lower level of
aggrecan and collagen II expression was detected in high-grade
tissues than in low-grade tissues (Fig.
1G and H). These results demonstrated that TLR4 and
inflammatory cytokines were upregulated in the process of IVD
tissue degeneration whereas miR-140 and ECM proteins were
downregulated.
 | Table II.TLR4 staining and clinicopathological
characteristics of 22 patients with IVD degeneration. |
Table II.
TLR4 staining and clinicopathological
characteristics of 22 patients with IVD degeneration.
|
| TLR4 |
|
|
|---|
|
|
|
|
|
|---|
| Parameters | Low (%) | High (%) | Total | P-value |
|---|
| Age (years) |
|
|
| 0.096 |
|
≤45 | 6 (27.3) | 3 (13.6) | 9 |
|
|
>45 | 4 (18.2) | 9 (40.9) | 13 |
|
| Sex |
|
|
| 0.145 |
|
Female | 2 (9.1) | 6 (27.3) | 8 |
|
|
Male | 8 (36.4) | 6 (27.3) | 14 |
|
| Body mass
index |
|
|
| 0.937 |
| ≤24
kg/m2 | 4 (18.2) | 5 (22.7) | 9 |
|
| >24
kg/m2 | 6 (27.3) | 7 (31.8) | 13 |
|
| Disc level |
|
|
| 0.190 |
|
L3/4 | 5 (22.7) | 3 (13.6) | 8 |
|
|
L4/5 | 2 (9.1) | 7 (31.8) | 9 |
|
|
L5/S1 | 3 (13.6) | 2 (9.1) | 5 |
|
| MRI grade |
|
|
| 0.029a |
| G
(II–III) | 8 (36.4) | 4 (18.2) | 12 |
|
| G
(IV–V) | 2 (9.1) | 8 (36.4) | 10 |
|
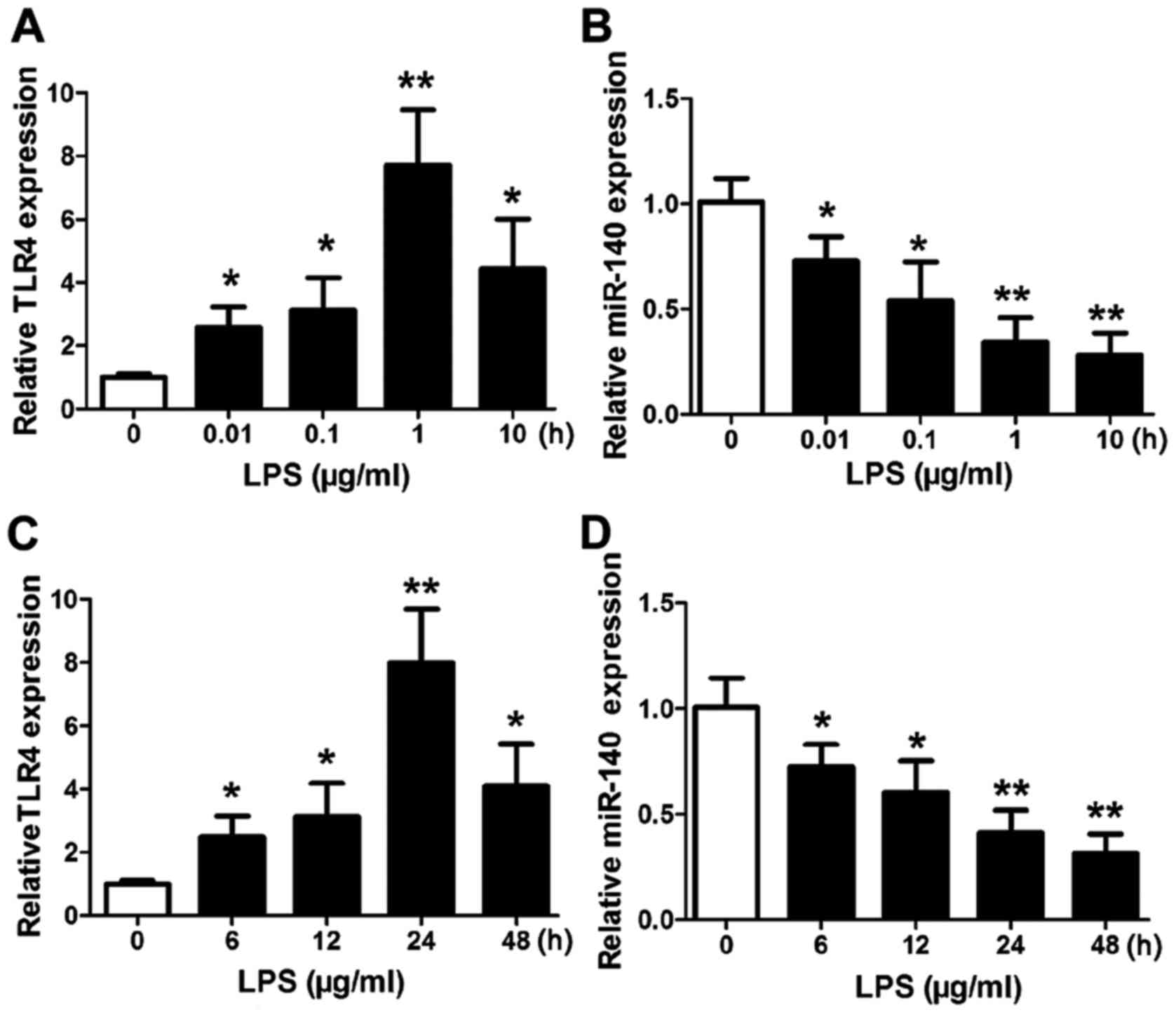
TLR4 is induced and miR-140 is
downregulated in response to LPS stimulation
Subsequently, we assessed the expression of
TLR4 and miR-140 in human NP cells stimulated with LPS. As
expected, qRT-PCR analysis revealed that TLR4 expression was
significantly increased in cells treated with a series of LPS
concentrations (0.01–10 µg/ml) (Fig.
2A). The highest level of TLR4 expression was found in
response to 1 µg/ml LPS (P<0.01 vs. the control). In contrast,
the expression levels of miR-140 decreased significantly with
increasing concentrations of LPS (Fig.
2B) with the lowest expression level in response to 10 µg/ml
LPS (P<0.01 vs. the control). A time series of LPS (1 µg/ml)
stimulation over 48 h revealed the highest levels of TLR4
expression were at 24 h (P<0.01 vs. the control), after which
the levels began to decrease (Fig.
2C). Whereas, the expression of miR-140 was decreased (Fig. 2D) and continued to decrease with the
lowest level at 48 h (P<0.01 vs. the control). These results
indicated that the presence of LPS induced TLR4 activation which
reached a peak at a specific level. In contrast, the continued
presence of LPS inhibited the expression of miR-140.
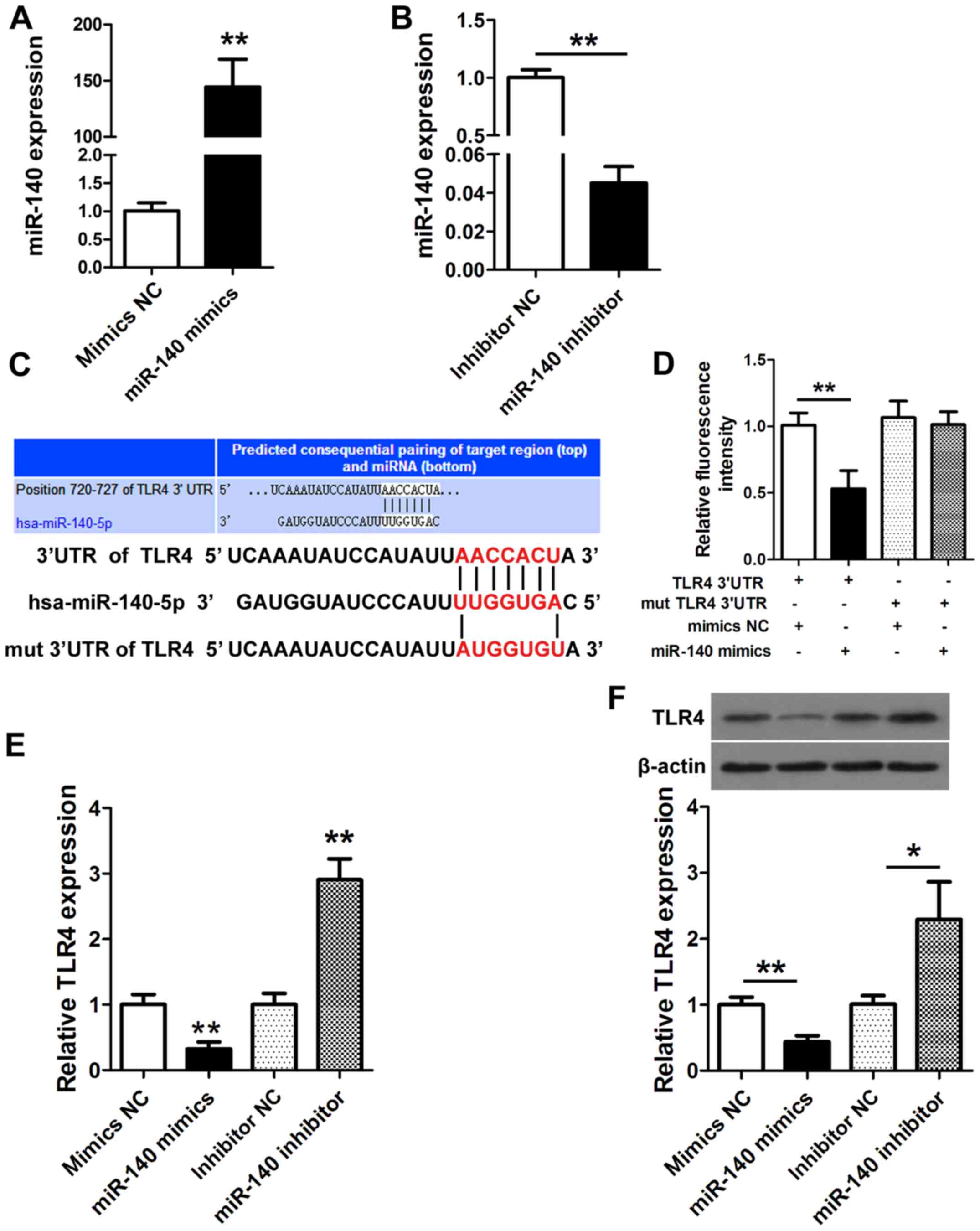
miR-140 directly regulates TLR4 by
targeting its 3′-UTR
NP cells were transfected with miR-140 mimics,
miR-140 mimics-negative control (mimics NC), miR-140 inhibitor or
miR-140 inhibitor-negative control (inhibitor NC) for 24 h. The
expression of miR-140 was detected by qRT-PCR (Fig. 3A and B) and confirmed the activity
of the miR-140 mimic and miR-140 inhibitor. Fig. 3C displays the putative miR-140
binding site in the TLR4 3′-UTR. A luciferase reporter
plasmid containing wild-type or mutant TLR4 was
co-transfected into NP cells with miR-140 mimics or mimics NC.
Luciferase activity was determined at 24 h after transfection using
a dual-luciferase assay and demonstrated as the relative luciferase
activity normalized to Renilla activity. A significantly
lower luciferase activity was found with the miR-140 mimic then
with the wild-type TLR4 3′-UTR (P<0.01; Fig. 3D). We then assessed the expression
and protein levels of TLR4 in NP cells transfected with the miR-140
mimic and miR-140 inhibitor. TLR4 was significantly downregulated
with the miR-140 mimic (P<0.01) while it was upregulated with
the miR-140 inhibitor (P<0.01; Fig.
3E). Similar results were obtained with western blotting
(P<0.01; Fig. 3F). The
upregulation of TLR4 in the absence of miR-140 or when the
putative 3′-UTR binding site of miR-140 was mutated indicated that
miR-140 may regulate TLR4.
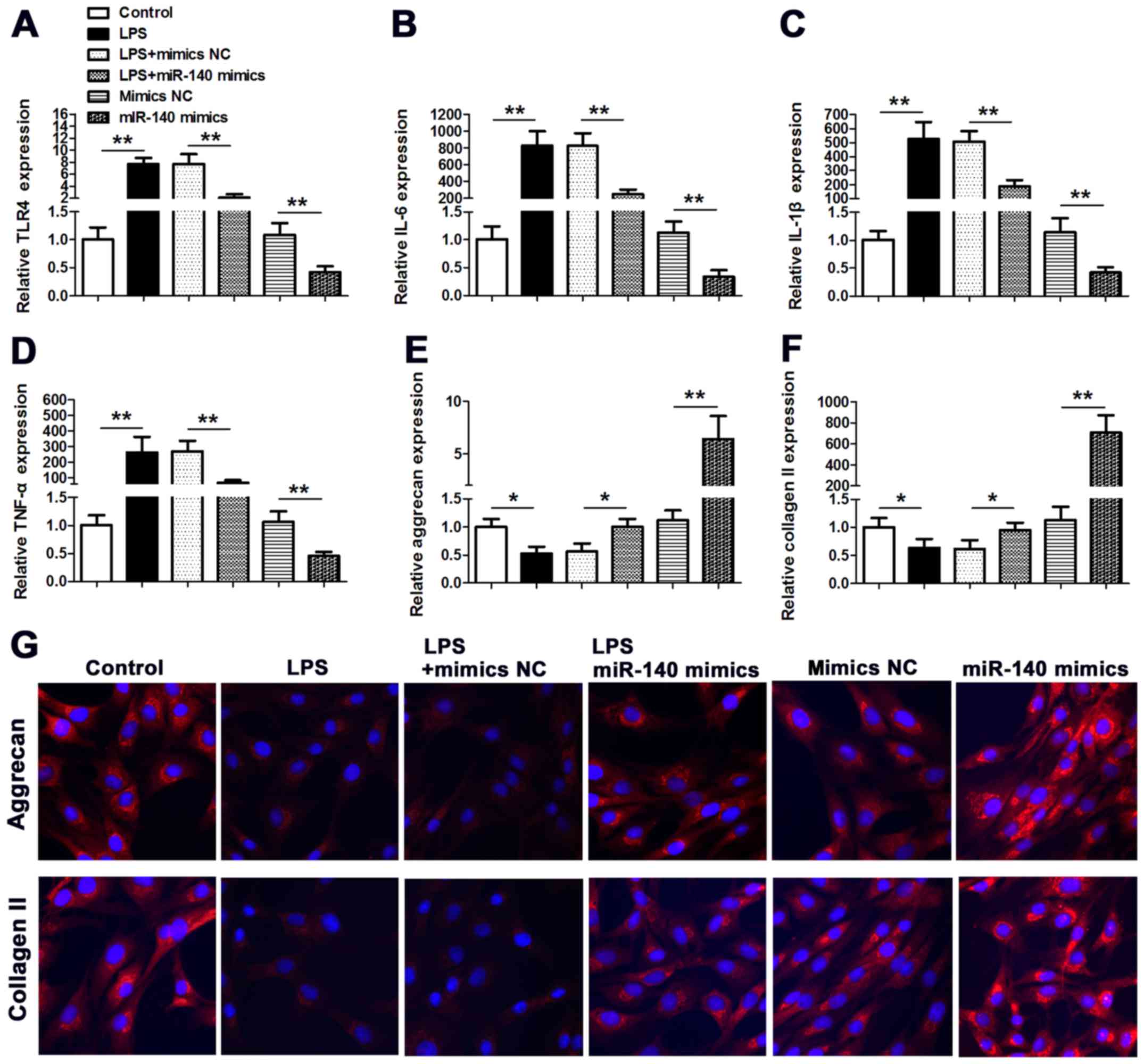
Overexpression of miR-140 inhibits the
effect of TLR4 on IVD inflammation and degeneration induced by LPS
stimulation
The influence of miR-140 on the expression of
inflammation cytokines was assessed in NP cells stimulated with
LPS. The expression of TLR4, interleukin (IL)-1β, IL-6 and tumor
necrosis factor (TNF)-α was significantly increased by LPS
(P<0.01 vs. the control), however, these increased levels of
expression were lower in cells co-transfected with miR-140 mimic.
Furthermore, without LPS treatment, the expression of TLR4, IL-1β,
IL-6 and TNF-α was also significantly lower in cells transfected
with miR-140 mimic (P<0.01 vs. mimic NC) (Fig. 4A-D). The expression levels of the
ECM components aggrecan and collagen II were also assessed in
LPS-stimulated NP cells (Fig. 4E and
F). Aggrecan and collagen II were both downregulated in
response to LPS stimulation (P<0.05 vs. control for both),
whereas the expression of aggrecan and collagen II was reversed by
co-transfecting cells with miR-140 mimic. In addition, without LPS
stimulation, the aggrecan and collagen II expression were also
significantly increased when the cells were transfected with
miR-140 mimics (P<0.01 vs. mimic NC). Aggrecan and collagen II
expression was also visualized in NP cells by immunofluorescence
staining (Fig. 4G). Nuclei were
stained with DAPI and untransfected NP cells were used as a
control. LPS stimulation clearly reduced the expression of aggrecan
and collagen II in NP cells. However, in cells co-transfected with
miR-140 mimic, aggrecan and collagen II levels were similar to
levels in control cells under LPS treatment; however, without LPS
stimulation, Collectively, these results imply that overexpression
of miR-140 can inhibit the effect of TLR4 on IVD inflammation and
degeneration induced by LPS stimulation.
Overexpression miR-140 modulates
LPS-induced TLR4 expression and NF-κB activation
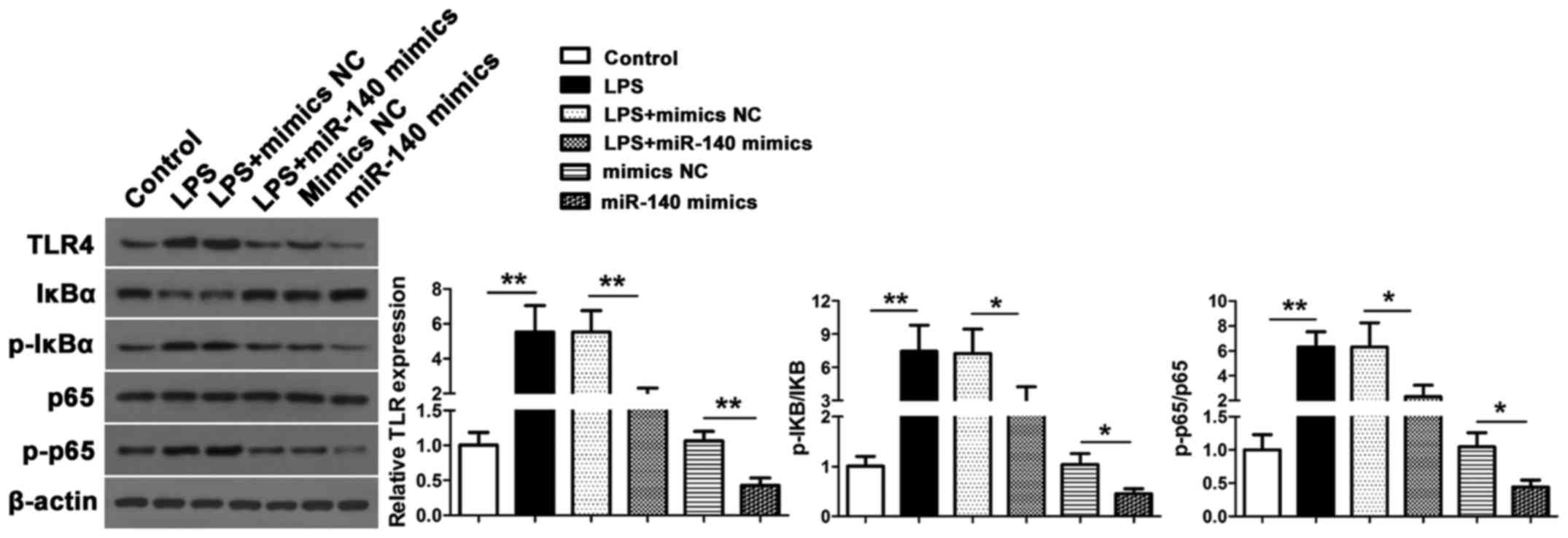
Finally, we assessed the TLR4 expression and NF-κB
activation in response to LPS stimulation in NP cells by measuring
levels of IκBα, p-IκBα, p65 and p-p65 protein expression by western
blotting (Fig. 5). Increased levels
of TLR4, p-IκBα and p-p65 were found in NP cells after LPS
stimulation (P<0.01 vs. the control) but these levels were lower
in miR-140 mimic co-transfected cells for TLR4 (P<0.01 vs. mimic
NC), p-IκBα/IκBα (P<0.05 vs. mimic NC) and p-p65/p65 (P<0.05
vs. mimic NC). These results indicated that miR-140 inhibited
LPS-induced TLR4 expression and NF-κB activation. Furthermore,
without LPS stimulation, overexpression of miR-140 can also inhibit
TLR4 expression and inhibit NF-κB activation.
Discussion
Multiple factors are reported to give rise to IVD
degeneration and at present, many of these are irreversible, such
as genetic disposition, aging or traumatic damage. Therefore, a
method to alleviate the burden of this condition would be a
suitable compromise. Collagen fibers are constantly challenged by
direct radial pressure from the NP and cranial-caudal stretch from
the separation of the two endplates (5,9).
Therefore, degenerated IVDs have a fibrous dehydrated nucleus with
a bulging annulus fibrosus. In advanced stages of disc
degeneration, inflammatory cytokines contribute to neurovascular
in-growth and there is a progressive decrease in the expression of
proteoglycans and collagen type II genes, which are important
structural elements of the NP (9,19).
Furthermore, traumatic damage to the matrix leads to the
accumulation of cytokines and several studies have implicated the
role of TLRs in IVD degeneration (31,35,36,58).
The expression levels of TLR1/2/4/6 were found to be dependent on
the degree of IVD degeneration (58).
In the present study, we have investigated the
relationship between TLR4 and miR-140-5p in IVD degeneration. TLR4
expression was higher in high grades of IVD degeneration tissues
than in low-grade IVD degeneration tissues whereas the opposite
occurred with miR-140. Furthermore, the expression of TLR4 was
associated with MRI grade in clinical characteristics of patients,
but there was no significant difference in age, sex, body mass
index and disc level. The expression of TNF-α, IL-1β and IL-6 was
also higher in high-grade than in low-grade IVD tissues. miR-140
was expressed at a lower level in high-grade IVD degeneration
tissue compared to lower grade IVD degeneration. The expression of
aggrecan and collagen II was also lower in high-grade than
low-grade IVD degeneration tissues. TLR4 is a major receptor of
LPS. LPS stimulation resulted in a significant increase in the
expression of TLR4 and decrease in the expression of miR-140 in NP
cells. Furthermore, LPS induced an increasing inflammation cytokine
expression and decreasing aggrecan and collagen II expression.
These results are consistent with previous studies that revealed
that LPS inhibited aggrecan and collagen II synthesis in IVD and
murine chondrocytes (30,31). We mutated a putative binding site of
miR-140 in 3′-UTR TLR4 and confirmed that the mutant gave a higher
luciferase activity than the wild-type 3′-UTR TLR4 when the cells
were transfected with miR-140, and indicated that miR-140 targeted
TLR4; this result was consistent with the previous study by Li
et al (49).
Overexpression of miR-140 inhibited the upregulation
of TNF-α, IL-1β, IL-6, TLR4 expression and NF-κB activation and
rescued the downregulation of aggrecan and collagen II induced by
LPS stimulation. In a similar way, Li et al (44) revealed that miR-29a and miR-140
significantly reversed the effect of IL-1β pretreatment on
chondrocytes, by influencing MMP13 and TIMP1 expression in both
mRNA and protein levels, and subsequently affected the content of
collagen II and aggrecan in chondrocytes. Karlsen et al
(45) reported that overexpressing
miR-140 increased the levels of proteins involved in the synthesis
of hyaline ECM and reduced the levels of aggrecanases and other
proteins that degraded the ECM, which could explain the extremely
elevated levels of aggrecan and collagen II in response to miR-140
overexpression and the observation that the overexpression of
miR-140 rescued the reduction of ECM expression induced by LPS
stimulation in the present study.
Fu et al (39) assessed the expression of TLR4 in an
LPS-induced kidney model. Compared with the control group, the
phosphorylation levels of NF-κB and IκBα were obviously increased
in the LPS group. However, they revealed that the drug Tenuigenin
dose-dependently inhibited LPS-induced TLR4 expression and NF-κB
activation. NF-κB is usually located in the cytoplasm with IκBα,
but when it is stimulated by LPS it becomes detached from IκB and
regulates cytokine production from the nucleus (59). TLR4 is a major receptor of LPS and
regulates the NF-κB pathway (60).
Recently, cordycepin, an extract from Cordyceps militaris,
was found to suppress the LPS-induced activation of the NF-κB
pathway in NP cells (61). TLR4
activated the NF-κB signaling pathway and induced the release of
inflammatory cytokines production and NF-κB was reported to be
responsible for the regulation of inflammatory cytokines production
(39,60,62).
We have found that TLR4 expression is increased in
IVD degeneration and that the levels of TLR4 were negatively
regulated by miR-140 with or without LPS stimulation. Li et
al (49) reported in 2017 that
miR-140 inhibited the expression of inflammation by downregulating
the expression of TLR4. However, in the present study, we used an
LPS-induced NP cell inflammation and degeneration model to activate
TLR4 expression in vitro. In a previous study LPS was
reported to increase inflammation expression and inhibit ECM
expression in IVD in an in vitro and in vivo model
(31). More functional in
vivo model experiments are definitely needed in a further study
to detect whether miR-140 can inhibit inflammation expression and
ECM reduction through the downregulation of the expression of TLR4
which was induced by LPS stimulation. Furthermore, with or without
LPS stimulation, miR-140 alleviated inflammation and degeneration
by decreasing the level of cytokines and increasing the expression
of aggrecan and collagen II by inhibiting TLR4 expression in NP
cells. The present study may lead to a greater understanding of IVD
degeneration and miRNAs may prove to be useful as markers or
treatment agents in disease management.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Health and
Family Planning Commission of Changzhou Major Science and
Technology Projects (ZD201504), the H-level Medical Talents
Training Project (2016CZBJ033) and The Nineteenth Batch of
Changzhou Science and Technology Plan (applied basic research)
(CJ20160047).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
DZ and NX conceived and designed the study. QZ, YW,
YJ and SZ performed the experiments. QZ and YW wrote the paper. DZ
and NX reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of Changzhou No. 2 People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang F, Zhao X, Shen H and Zhang C:
Molecular mechanisms of cell death in intervertebral disc
degeneration (Review). Int J Mol Med. 37:1439–1448. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rannou F, Lee TS, Zhou RH, Chin J, Lotz
JC, Mayoux-Benhamou MA, Barbet JP, Chevrot A and Shyy JY:
Intervertebral disc degeneration: The role of the mitochondrial
pathway in annulus fibrosus cell apoptosis induced by overload. Am
J Pathol. 164:915–924. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kadow T, Sowa G, Vo N and Kang JD:
Molecular basis of intervertebral disc degeneration and
herniations: What are the important translational questions? Clin
Orthop Relat Res. 473:1903–1912. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feng C, Liu H, Yang M, Zhang Y, Huang B
and Zhou Y: Disc cell senescence in intervertebral disc
degeneration: Causes and molecular pathways. Cell Cycle.
15:1674–1684. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi H, Johnson ZI and Risbud MV:
Understanding nucleus pulposus cell phenotype: A prerequisite for
stem cell based therapies to treat intervertebral disc
degeneration. Curr Stem Cell Res Ther. 10:307–316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walter BA, Purmessur D, Moon A,
Occhiogrosso J, Laudier DM, Hecht AC and Iatridis JC: Reduced
tissue osmolarity increases TRPV4 expression and pro-inflammatory
cytokines in intervertebral disc cells. Eur Cell Mater. 32:123–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Freemont AJ: The cellular pathobiology of
the degenerate intervertebral disc and discogenic back pain.
Rheumatology (Oxford). 48:5–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vergroesen PP, Kingma I, Emanuel KS,
Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieën JH and Smit TH:
Mechanics and biology in intervertebral disc degeneration: A
vicious circle. Osteoarthritis Cartilage. 23:1057–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayes AJ, Benjamin M and Ralphs JR:
Extracellular matrix in development of the intervertebral disc.
Matrix Biol. 20:107–121. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roughley PJ, Alini M and Antoniou J: The
role of proteoglycans in aging, degeneration and repair of the
intervertebral disc. Biochem Soc Trans. 30:869–874. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Franceschi C and Bonafè M: Centenarians as
a model for healthy aging. Biochem Soc Trans. 31:457–461. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Walker BF: The prevalence of low back pain
in Australian adults. A systematic review of the literature from
1966–1998. Asia Pac J Public Health. 11:45–51. 1999.
|
|
14
|
Crockett MT, Kelly BS, van Baarsel S and
Kavanagh EC: Modic type 1 vertebral endplate changes: Injury,
inflammation, or infection? AJR Am J Roentgenol. 209:167–170. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dudli S, Haschtmann D and Ferguson SJ:
Fracture of the vertebral endplates, but not equienergetic impact
load, promotes disc degeneration in vitro. J Orthop Res.
30:809–816. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanna RM, Shanmuganathan R, Rajagopalan
VR, Natesan S, Muthuraja R, Cheung KMC, Chan D, Kao PYP, Yee A and
Shetty AP: Prevalence, patterns, and genetic association analysis
of modic vertebral endplate changes. Asian Spine J. 11:594–600.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Johnson ZI, Schoepflin ZR, Choi H, Shapiro
IM and Risbud MV: Disc in flames: Roles of TNF-α and IL-1β in
intervertebral disc degeneration. Eur Cell Mater. 30:104–116;
discussion 116–117. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Le Maitre CL, Hoyland JA and Freemont AJ:
Catabolic cytokine expression in degenerate and herniated human
intervertebral discs: IL-1beta and TNFalpha expression profile.
Arthritis Res Ther. 9:R772007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altun I: Cytokine profile in degenerated
painful intervertebral disc: Variability with respect to duration
of symptoms and type of disease. Spine J. 16:857–861. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Markova D, Anderson DG, Zheng Z,
Shapiro IM and Risbud MV: TNF-α and IL-1β promote a
disintegrin-like and metalloprotease with thrombospondin type I
motif-5-mediated aggrecan degradation through syndecan-4 in
intervertebral disc. J Biol Chem. 286:39738–39749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Le Maitre CL, Freemont AJ and Hoyland JA:
The role of interleukin-1 in the pathogenesis of human
intervertebral disc degeneration. Arthritis Res Ther. 7:R732–R745.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu F, Gao F, Liu Y, Wang Z, Zhuang X, Qu
Z, Ma H, Liu Y, Fu C, Zhang Q, et al: Bioinformatics analysis of
molecular mechanisms involved in intervertebral disc degeneration
induced by TNF-α and IL-1β. Mol Med Rep. 13:2925–2931. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang C, Yu X, Yan Y, Yang W, Zhang S,
Xiang Y, Zhang J and Wang W: Tumor necrosis factor-α: A key
contributor to intervertebral disc degeneration. Acta Biochim
Biophys Sin (Shanghai). 49:1–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li X, Cheng S, Wu Y, Ying J, Wang C, Wen
T, Bai X, Ji W, Wang D and Ruan D: Functional self-assembled
peptide scaffold inhibits tumor necrosis factor-alpha-induced
inflammation and apoptosis in nucleus pulposus cells by suppressing
nuclear factor-κB signaling. J Biomed Mater Res A. 106:1082–1091.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu B, Shi C, Xu C, Cao P, Tian Y, Zhang Y,
Deng L, Chen H and Yuan W: Heme oxygenase-1 attenuates IL-1β
induced alteration of anabolic and catabolic activities in
intervertebral disc degeneration. Sci Rep. 6:211902016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
O'Neill LA and Dinarello CA: The IL-1
receptor/toll-like receptor superfamily: Crucial receptors for
inflammation and host defense. Immunol Today. 21:206–209. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Janeway CA Jr and Medzhitov R: Innate
immune recognition. Annu Rev Immunol. 20:197–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takeda K, Kaisho T and Akira S: Toll-like
receptors. Annu Rev Immunol. 21:335–376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim HA, Cho ML, Choi HY, Yoon CS, Jhun JY,
Oh HJ and Kim HY: The catabolic pathway mediated by Toll-like
receptors in human osteoarthritic chondrocytes. Arthritis Rheum.
54:2152–2163. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bobacz K, Sunk IG, Hofstaetter JG, Amoyo
L, Toma CD, Akira S, Weichhart T, Saemann M and Smolen JS:
Toll-like receptors and chondrocytes: The
lipopolysaccharide-induced decrease in cartilage matrix synthesis
is dependent on the presence of toll-like receptor 4 and
antagonized by bone morphogenetic protein 7. Arthritis Rheum.
56:1880–1893. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rajan NE, Bloom O, Maidhof R, Stetson N,
Sherry B, Levine M and Chahine NO: Toll-Like Receptor 4 (TLR4)
expression and stimulation in a model of intervertebral disc
inflammation and degeneration. Spine. 38:1343–1351. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu YC, Yeh WC and Ohashi PS: LPS/TLR4
signal transduction pathway. Cytokine. 42:145–151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thompson JE, Phillips RJ,
Erdjument-Bromage H, Tempst P and Ghosh S: I kappa B-beta regulates
the persistent response in a biphasic activation of NF-kappa B.
Cell. 80:573–582. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lien E, Means TK, Heine H, Yoshimura A,
Kusumoto S, Fukase K, Fenton MJ, Oikawa M, Qureshi N, Monks B, et
al: Toll-like receptor 4 imparts ligand-specific recognition of
bacterial lipopolysaccharide. J Clin Invest. 105:497–504. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim JS, Ellman MB, Yan D, An HS, Kc R, Li
X, Chen D, Xiao G, Cs-Szabo G, Hoskin DW, et al: Lactoferricin
mediates anti-inflammatory and anti-catabolic effects via
inhibition of IL-1 and LPS activity in the intervertebral disc. J
Cell Physiol. 228:1884–1896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang F and Jiang D: IL-1β/HMGB1 signalling
promotes the inflammatory cytokines release via TLR signalling in
human intervertebral disc cells. Biosci Rep. 36:pii. e003792016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen B, Wang HT, Yu B, Zhang JD and Feng
Y: Carthamin yellow inhibits matrix degradation and inflammation
induced by LPS in the intervertebral disc via suppression of MAPK
pathway activation. Exp Ther Med. 14:1614–1620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wan J, Shan Y, Fan Y, Fan C, Chen S, Sun
J, Zhu L, Qin L, Yu M and Lin Z: NF-κB inhibition attenuates
LPS-induced TLR4 activation in monocyte cells. Mol Med Rep.
14:4505–4510. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fu H, Hu Z, Di X, Zhang Q, Zhou R and Du
H: Tenuigenin exhibits protective effects against LPS-induced acute
kidney injury via inhibiting TLR4/NF-κB signaling pathway. Eur J
Pharmacol. 791:229–234. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Díaz-Prado S, Cicione C, Muiños-López E,
Hermida-Gómez T, Oreiro N, Fernández-López C and Blanco FJ:
Characterization of microRNA expression profiles in normal and
osteoarthritic human chondrocytes. BMC Musculoskelet Disord.
13:1442012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Miyaki S and Asahara H: Macro view of
microRNA function in osteoarthritis. Nat Rev Rheumatol. 8:543–552.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang R, Ma J and Yao J: Molecular
mechanisms of the cartilage-specific microRNA-140 in
osteoarthritis. Inflamm Res. 62:871–877. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, Zhen Z, Tang G, Zheng C and Yang G:
MiR-29a and miR-140 protect chondrocytes against the
anti-proliferation and cell matrix signaling changes by IL-1β. Mol
Cells. 39:103–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Karlsen TA, de Souza GA, Ødegaard B,
Engebretsen L and Brinchmann JE: microRNA-140 inhibits inflammation
and stimulates chondrogenesis in a model of interleukin 1β-induced
osteoarthritis. Mol Ther Nucleic Acids. 5:e3732016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pando R, Even-Zohar N, Shtaif B, Edry L,
Shomron N, Phillip M and Gat-Yablonski G: MicroRNAs in the growth
plate are responsive to nutritional cues: Association between
miR-140 and SIRT1. J Nutr Biochem. 23:1474–1481. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Conrad E, Polonio-Vallon T, Meister M,
Matt S, Bitomsky N, Herbel C, Liebl M, Greiner V, Kriznik B,
Schumacher S, et al: HIPK2 restricts SIRT1 activity upon severe DNA
damage by a phosphorylation-controlled mechanism. Cell Death
Differ. 23:110–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rodgers JT, Lerin C, Haas W, Gygi SP,
Spiegelman BM and Puigserver P: Nutrient control of glucose
homeostasis through a complex of PGC-1alpha and SIRT1. Nature.
434:113–118. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li H, Guan SB, Lu Y and Wang F: MiR-140-5p
inhibits synovial fibroblasts proliferation and inflammatory
cytokines secretion through targeting TLR4. Biomed Pharmacother.
96:208–214. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Miyaki S, Nakasa T, Otsuki S, Grogan SP,
Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK and Asahara H:
MicroRNA-140 is expressed in differentiated human articular
chondrocytes and modulates interleukin-1 responses. Arthritis
Rheum. 60:2723–2730. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine. 26:1873–1878. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee JS, Jeong SW, Cho SW, Juhn JP and Kim
KW: Relationship between initial telomere length, initial
telomerase activity, age, and replicative capacity of nucleus
pulposus chondrocytes in human intervertebral discs: What is a
predictor of replicative potential? PLoS One. 10:e01441772015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
VanGuilder HD, Vrana KE and Freeman WM:
Twenty-five years of quantitative PCR for gene expression analysis.
Biotechniques. 44:619–626. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sive JI, Baird P, Jeziorsk M, Watkins A,
Hoyland JA and Freemont AJ: Expression of chondrocyte markers by
cells of normal and degenerate intervertebral discs. Mol Pathol.
55:91–97. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhu J, Ma J, Wang X, Ma T, Zhang S, Wang
W, Zhou X and Shi J: High expression of PHGDH predicts poor
prognosis in non-small cell lung cancer. Transl Oncol. 9:592–599.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun R, Wang X, Zhu H, Mei H, Wang W, Zhang
S and Huang J: Prognostic value of LAMP3 and TP53 overexpression in
benign and malignant gastrointestinal tissues. Oncotarget.
5:12398–12409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Klawitter M, Hakozaki M, Kobayashi H,
Krupkova O, Quero L, Ospelt C, Gay S, Hausmann O, Liebscher T,
Meier U, et al: Expression and regulation of toll-like receptors
(TLRs) in human intervertebral disc cells. Eur Spine J.
23:1878–1891. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang J, Guo C, Wei Z, He X, Kou J, Zhou E,
Yang Z and Fu Y: Morin suppresses inflammatory cytokine expression
by downregulation of nuclear factor-κB and mitogen-activated
protein kinase (MAPK) signaling pathways in
lipopolysaccharide-stimulated primary bovine mammary epithelial
cells. J Dairy Sci. 99:3016–3022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fu Y, Hu X, Cao Y, Zhang Z and Zhang N:
Saikosaponin a inhibits lipopolysaccharide-oxidative stress and
inflammation in Human umbilical vein endothelial cells via
preventing TLR4 translocation into lipid rafts. Free Radic Biol
Med. 89:777–785. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li Y, Li K, Mao L, Han X, Zhang K, Zhao C
and Zhao J: Cordycepin inhibits LPS-induced inflammatory and matrix
degradation in the intervertebral disc. PeerJ. 4:e19922016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
DiDonato JA, Mercurio F and Karin M: NF-κB
and the link between inflammation and cancer. Immunol Rev.
246:379–400. 2012. View Article : Google Scholar : PubMed/NCBI
|